Abstract
Hydrogen sulfide is a member of the growing family of gasotransmitters. Once regarded as a noxious molecule predominantly present in the atmosphere, H2S is now known to be synthesized endogenously in mammals. H2S participates in a myriad of physiological processes ranging from regulation of blood pressure to neuroprotection. Its chemical nature precludes H2S from being stored in vesicles and acting on receptor proteins in the fashion of other chemical messengers. Thus, novel cellular mechanisms have evolved to mediate its effects. This article focuses on sulfhydration (or persulfidation), which appears to be the principal post-translational modification elicited by H2S.
Keywords: Hydrogen sulfide, sulfhydration, gasotransmitter, cysteine
Hydrogen sulfide in biological systems
Mention of the word hydrogen sulfide (H2S) evokes images of rotten eggs with a foul odor, sewage drains, intestinal bacteria, swamps and volcanoes. Before the discovery that eukaryotes synthesize H2S, it was believed that the gas was present predominantly in the atmosphere and metabolized by microbes such as bacteria and archaea. H2S was considered a byproduct of metabolic processes and viewed as a toxic molecule until the discovery that it regulates vascular tone and blood pressure [1]. Like nitric oxide (NO) and carbon monoxide (CO), H2S functions as a gasotransmitter (see Glossary) that has cytoprotective roles at low concentrations. Also like NO, the direct precursor of H2S is an amino acid. While NO is produced from arginine, H2S is formed from cysteine. CO, by contrast, is generated from heme. Three enzymes generate H2S from L-cysteine [2] via independent reactions: cystathionine γ-lyase (CSE), cystathionine β-synthase (CBS) and 3-mercaptopyruvate sulfurtransferase (3-MST). More recently, D-cysteine was found to be a substrate for H2S production [3]. Although it was previously believed that CBS and 3-MST are predominantly present in the brain and CSE is largely associated with peripheral or non-nervous tissues, it is now known that all three enzymes are ubiquitous, and H2S impacts almost all cellular processes [2,4,5]. This review focuses on the biochemistry, detection and physiologic roles of sulfhydration.
Regulation of H2S production
Among the three enzymes that produce H2S, CSE seems to be the most inducible, similar to the inducible nitric oxide synthase (iNOS), which generates NO. Expression of CSE is modulated in diverse conditions such as inflammation mediated by tumor necrosis factor-α(TNF-α) and lipopolysaccharides (LPS), metabolites such as glucocorticoids and glucose, as well as dietary restriction and endoplasmic reticulum (ER) stress [6, 7, 8, 9, 10]. CBS, the principal enzyme producing H2S in the brain, is not subject to such induction and is regulated predominantly by metabolites binding to it. For instance, S-adenosylmethionine (SAM) allosterically activates the enzyme to increase H2S production [11, 12]. In addition, the other gasotransmitters (CO and NO) interact with the heme center of CBS and modulate its activity, affording a node for crosstalk between the gases [13, 14, 15].
Interplay between the gases regulates microvascular circulation in the brain during hypoxia. Under normoxic conditions, CO produced by the oxygen-dependent heme oxygenase 2 (HO-2) binds to the heme center of CBS to inhibit H2S production. During hypoxia, when the activity of HO-2 drops, tonic inhibition of CBS is relieved leading to elevated H2S production, which mediates arteriolar vasodilation [13]. Since the redox potential of the Fe3+/Fe2+ couple in CBS is very low (−350 mV), the feasibility of CBS entering the ferrous state has been a subject of controversy. Reversible inhibition of CBS by CO in the presence of methionine synthase reductase and NADPH, an endogenous reducing system, has been reported to mediate this reduction [16].
In addition to regulating H2S via the expression and activity of biosynthetic enzymes, these enaymes are expressed in distinct cell types. In the brain, CBS and CSE are almost exclusively expressed in astrocytes [13, 17] and neurons [13], respectively. 3-MST is found in both neurons and glia [18]. H2S can also be produced from cellular stores such as the sulfane sulfur and the acid labile pools [19]. Specifically, 3-MST generates H2S under alkaline conditions to produce sulfane sulfur, a store of H2S that can be mobilized by endogenous reductants such as thioredoxin and dihydrolipoic acid [20, 21]. Other sources of H2S include the microbes residing in the mammalian intestine [22, 23], which produce H2S and contribute to both pro and anti-inflammatory effects [24].
Protein sulfhydration
One of the modes by which H2S signals is by modification of cysteine residues on target proteins in a process designated as sulfhydration or persulfidation [25], which functions in a manner analogous to nitrosylation [26]. While NO targets cysteines to form –SNO groups, H2S mediates the conversion of the –SH group of reactive cysteines to form a persulfide or –SSH group. The nomenclature for this modification suggested by Mustafa et al. [25] has been a subject of debate. In a true chemical sense, persulfidation is the appropriate term, while sulfhydration can be regarded as the common or trivial name. Sulfhydration typically increases the reactivity of the cysteine residue being modified, whereas nitrosylation decreases its reactivity. Sulfhydration is a highly prevalent modification with about 25% of GAPDH being modified in hepatic lysates as assessed by the modified biotin switch assay [25].
A cysteine must be oxidized prior to modification with H2S, as H2S cannot directly react with reduced thiols. Although the cytosol is a predominantly reducing environment, sulfhydration can occur under certain conditions when the generation of reactive oxygen species occurs in response to physiological stimuli. The reactivity of cysteine residues to modifications depends on solvent exposure and the protonation status of its functional groups [27]. There is a negative correlation between the reactivity of a cysteine residue and its acid dissociation constant (pKa), which depends on the microenvironment of the protein and its local architecture. Sulfhydration typically occurs at cysteine residues with a low pKa, although recently the modification was also shown to occur on some cysteine residues with a high pKa [28]. Low pKa cysteines are more susceptible to sulfhydration, because they exist as thiolate anions (S−) at physiologic conditions. However, they are also more reactive with oxidants such as H2O2 that generate sulfenic acid (SOH), sulfinic acid (SO2H) or sulfonic acid derivatives [29, 30, 31]. Formation of SOH moieties is usually reversible, but higher oxidation states of the thiol group of cysteine are irreversible. Thus, an important function of sulfhydration could be to protect reactive cysteine residues on proteins from irreversible oxidation by generating a persulfide group [2]. The CySSO2H (perthiosulfinic) and CySSO3H (perthiosulfonic) oxidation products of persulfides can be recycled back by the reduction of their S-S moieties, which does not occur in the case of the CySO3H oxidation product of unmodified (unsulfhydrated) cysteine residues on proteins. We originally proposed this protective function of sulfhydration [2], which was tested in the case of phosphatase and tensin homolog deleted on chromosome 10, PTEN [32].
The biochemistry of sulfhydration
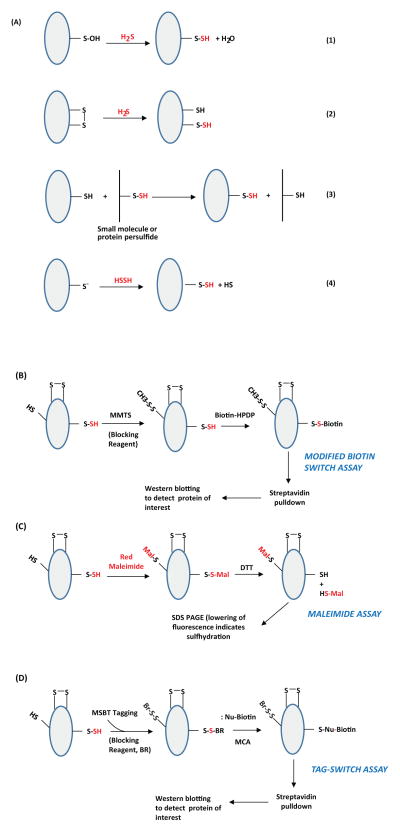
Although H2S mediates sulfhydration, H2S cannot directly modify reduced cysteine residues. H2S has been reported to generate persulfides when it reacts with oxidized thiols and disulfides [33]. Sulfhydration (persulfidation) can occur in several ways: i) by nucleophilic attack of sulfide on oxidized cysteine residues such as cysteine sulfenic acids (Cys-SOH) or cysteine disulfides (-S-S-) (Figure 1A, reactions 1–2) ii) through reaction between oxidized sulfide species such as polysulfides and cysteine thiols [33, 34](Figure 1A, reaction 3) iii) through reaction of H2S2 with a cysteine thiol [35](Figure 1A, reaction 4). An emerging concept in the field of H2S biology is that the gasotransmitter mediates its effects through the formation of polysulfides [36]. H2S can be readily oxidized to form polysulfides where the sulfur has zero valence charge. Solutions of H2S used to study the effects were found to contain polysulfides, which could be responsible for the observed effects [28].
Figure 1. Potential mechanisms of protein sulfhydration and its detection.(A) Mechanisms for sulfydration/persulfidation.
Sulfhydration can occur by the reaction of sulfide with oxidized cysteine residues such as cysteine sulfenic acid or disulfides (Reactions 1 and 2 respectively). Sulfhydration may also occur when an existing persulfide (on either a small molecule or a protein) reacts with a cysteine thiol (Reaction 3). Reaction of H2S2 with cysteine thiolates may also lead to sulfhydration (Reaction 4). (B) The modified biotin switch assay. The illustration depicts a protein with unmodified cysteines (-SH), sulfhydrated cysteines (-SSH) and disulfide bonded cysteines (S-S). Purified protein, or cell or tissue lysate, is incubated with methyl methanethiosulfonate (MMTS), to block unmodified cysteines. Unreacted MMTS is then removed by acetone precipitation or by gel filtration followed by treatment of the sulfhydrated protein with biotin-HPDP, which reacts with the protein at the site of sulfhydration. The biotinylated protein is enriched using streptavidin conjugates and analyzed by western blot analysis. (C) Maleimide assay. In this assay, the protein is first immunoprecipitated and treated with a fluorescent version of maleimide, which reacts with thiols under conditions that preserve the native conformation of the protein. After removing excess maleimide, the reaction mixture is treated with dithiothreitol (DTT), which reduces the disulfide bond resulting in the removal of the maleimide and a decrease in fluorescence that can be observed by SDS-PAGE. (D) The tag switch assay. The assay is a variation of the modified biotin switch assay. The reaction mixture is treated with the thiol blocking reagent (BR): methylsulfonyl benzothiazole (MSBT), followed by treatment with a methylcyanoacetate (MCA) derivative that comprises a nucleophilic component and a biotin moiety as a reporter. The biotinylated protein is then captured using streptavidin beads and analyzed by western blotting. The modifications caused by sulfide or its derivatives are shown in red.
Detection of sulfhydration
As cysteine residues undergo several different kinds of modifications such as nitrosylation, sulfhydration, glutathionylation and palmitoylation, it becomes critical to develop methods that distinguish the various posttranslational modifications. Because the specificity of several of the original assays utilized to detect sulfhydration was questioned, there have been multiple innovations in the methods utilized to detect sulfhydration or persulfidation.
Modified biotin switch assay
Several methods have been developed to detect sulfhydration. In the modified biotin switch assay, a variant of the assay developed to detect S-nitrosylation of proteins [37, 38], the sulfhydrated protein is treated with methyl methanethiosulfonate (MMTS) to block unmodified cysteine residues (Figure 1B). As a control, a reaction is treated with dithiothrietol (DTT) to reduce persulfide groups, and then blocked with MMTS. After removal of excess MMTS, the samples are labeled with biotin, which interacts with the thiol group. The biotinylated proteins are enriched by affinity capture using streptavidin conjugates, which specifically bind to biotinylated proteins, and then analyzed by western blotting [25,39]. The control samples treated with DTT, where the persulfide group is reduced back to the corresponding thiol, will not yield a signal on western blots. Using the modified biotin switch and densitometric analysis, 10–25% of GAPDH, actin and β-tubulin were found to be endogenously sulfhydrated in liver lysates [25]. The abundant expression of CSE in the liver could explain the high basal levels of protein sulfhydration.
When the modified biotin switch assay was developed, the reactivity of MMTS with persulfide groups was not considered, because mass spectrometry analysis did not reveal that sulfhydrated cysteines could be modified by MMTS (although the modification of –SSH groups by MMTS is chemically feasible). Modification of –SSH groups by MMTS was subsequently demonstrated by Caroll and associates [40]. Their study revealed that MMTS can not only react with persulfides on low molecular weight molecules such as GSSH, but also with persulfides on larger protein molecules (papain and Gpx3). In the modified biotin switch assay, where cell lysates harboring other proteins are utilized, the final readout relies on western blotting using antibodies directed against the protein under study. The increase in signal relative to the untreated lysates (samples exposed to H2S donors or in which H2S production is induced) is monitored.
Because MMTS can react with both cysteine thiols as well as cysteine persulfides of proteins, caution must be exercised while utilizing the modified biotin switch assay. The use of this assay in conjunction with mass spectrometry enhances reliability. In addition, the use of mice deleted for CSE can further validate sulfhydration. Mass spectrometry can confirm the type of modification occurring on cysteine residues. Conventional mass spectrometry on intact proteins is insufficient to distinguish sulfhydration from oxidation products such as sulfenic acid oxidative derivatives. High resolution mass spectrometry may distinguish between bona fide persulfide modification and oxidation products on side chains. In addition, one should include control reactions employing DTT to diminish modification of the persulfide back to the thiol state. The loss of mass can yield information regarding the type of S-thiolation on the cysteine residue. The sulfinic derivatives can be distinguished from the persulfides in this case, because sulfinic forms cannot be reduced by DTT. Reactions with iodoacetamide can also be included as the sulfinic forms cannot react with this molecule. However, iodoacetamide can react with cys-sulfenic acids - hence studies utilizing iodoacetamide should take this possible reaction into consideration [41]
Maleimide assay
Sulfhydration can also be detected using fluorescent thiol modifying reagents (Figure 1C). Fluorescent maleimide selectively labels sulfhydryl groups, both modified and unmodified. Treatment with DTT reduces only the modified cysteines, resulting in a decrease of the fluorescent signal that can be detected by separating and imaging the proteins on polyacrylamide gels under non-reducing conditions. The maleimide assay can be used to detect both sulfhydration and nitrosylation in the same sample [6, 39]. Recently however, the interaction of maleimides with sulfenic acids has been reported [41], which can pose a problem in the detection of sulfhydration.
Tag Switch assay
The tag switch assay utilizes methylsulfonylbenzothiazole (MSBT) to block thiols, after which a reagent comprising of a nucleophile and a biotin moiety is reacted with the protein solution [34, 42]. MSBT labels persulfide groups, which are then enriched using streptavidin conjugates and analyzed by western blotting (Figure 1D).
These detection methods have been utilized to study the physiologic functions of H2S where sulfhydration plays a role. H2S also signals via pathways that do not involve sulfhydration and are the subject of other reviews in the field [4, 43]. The study of sulfhydration, still in its infancy, would benefit from highly specific probes to uniquely identify this modification. Mass spectrometry is a valuable tool for confirming the type of modification.
Physiologic roles of sulfhydration
Sulfhydration is a highly prevalent modification and thus participates in a myriad of physiological processes ranging from regulation of blood pressure to neuronal function (Table 1).
Table 1.
Effects of sulfhydration
| Protein | Residues Sulfhydrated | Effect on Function | Reference |
|---|---|---|---|
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | C150 | Increases catalytic activity. In brain, increases association with the E3 ubiquitin ligase, Siah1 and targets PSD95 for degradation. | 25,73 |
| β-Actin | Not determined | Increased actin polymerization | 25 |
| Protein tyrosine phosphatase 1B (PTP1B) | C215 | Reduces phosphatase activity | 10 |
| Kir6.1 subunit of KATP | C43 | Activates channel activity by enhancing its interaction with PIP2 | 45 |
| IKCa | Not determined | Activates channel | 45 |
| p65 subunit of NF-κB | C38 | Promotes its interaction with the coactivator protein, ribosomal protein S3 (Rps3) | 6 |
| Parkin | C59, C95, C182 | Enhances its E3 ubiquitin ligase activity | 69 |
| Keap1 | C151 | Promotes dissociation from the transcription factor Nrf2 and releases it from sequestration | 64 |
| MEK1 | C341 | Facilitates PARP activation | 82 |
| Cu/Zn superoxide dismutase | C111 | Prevents oxidation induced aggregation | 83 |
| p66Shc | C59 | Prevents H2O2-induced phosphorylation and generation of ROS | 84 |
| Androgen receptor | C611, C614 | Prevents dimerization of the receptor | 85 |
| TRPV6 | C172, C329 | Maintains calcium homeostasis in bone marrow stem cells | 86 |
| PTEN | C71, C124 | Maintains enzyme activity. Prevents further oxidation by nitric oxide | 32 |
| Interferon regulatory factor 1 (IRF1) | C53 | Enhances the binding of the transcription factor (IS THAT WHAT YOU MEAN?) to the DNA methyl transferase 3A (DNMT3A) promoter to reduce methylation of target genes such as TFAM, a mitochondrial transcription factor, which is involved in mitochondrial biogenesis. | 55 |
Sulfhydration and the vascular system
Similar to NO, H2S regulates vasorelaxation, and inhibitors of CSE prevent vasorelaxation. Definitive evidence for the involvement of H2S in vasorelaxation was provided by studies using CSE knockout mice, which are impaired in their ability to produce H2S and develop age-dependent hypertension [1]. An independent study, however, reported no changes in blood pressure in CSE knockout mice [44], which could reflect differences in strain background and methodology. The aforementioned study utilized the tail cuff method to monitor blood pressure on C57BL/6J mice, whereas Yang et al. utilized the intra-carotid artery catheterization method on mice of a 129/SvEv-C57BL/6J mixed background.
H2S, like NO, has characteristics of an endothelium-derived relaxing factor (EDRF). H2S mediates vasorelaxation by sulfhydrating ATP-dependent potassium channels (KATP) present on vascular smooth muscle. Sulfhydration of Cys43 of the inwardly rectifying potassium channel subunit Kir6.1 prevents its association with ATP and promotes its binding to phosphatidylinositol-4,5-bisphosphate (PIP2), thereby keeping it in an open conformation to mediate vasorelaxation [45]. In addition to its effects on KATP channels, H2S also mediates vascular and other cytoprotective functions through HS− anions.
At physiological pH (7.4) and 37 °C, ~20% of sulfide exists as H2S gas, while at pH 7.4 and 25 °C ~40% of sulfide exists as H2S gas. At alkaline pH (e.g. 9.5), sulfide predominantly exists as HS− anion. H2S dissociates to HS− and H+ ions at physiologic pH [reviewed in 46]. HS- anions can react with various electrophilic compounds such as 8-nitro-cGMP, 15-deoxy-Δ12,14-prostaglandin J2, 4-hydroxy-2-nonenal, fatty acid nitroalkene derivatives and acrolein to modulate signaling or to prevent oxidative damage [47]. 8-nitro-cGMP can modify reactive cysteines in a process termed S-guanylation. Depletion of CSE and CBS increases the levels of S-guanylation in mammalian cells treated with 8-nitro-cGMP. Sulfhydration of 8-nitro-cGMP to 8-SH-cGMP not only decreases S-guanylation of proteins, but also generates a molecule which is resistant to phosphodiesterases and still retains cGMP activity. 8-SH-cGMP is the predominant cGMP-derivative in the brain, heart, and liver of mice [48]. Thus, besides influencing proteins, sulfhydration can modify small molecules to mediate cytoprotective effects.
H2S and sulfhydration in energy disposition and mitochondrial function
Although the H2S biosynthetic enzymes, with the exception of 3-MST, are predominantly cytosolic, H2S plays significant roles in mitochondrial metabolism and energy disposition. Some bacteria and archaea also use H2S as a fuel for energy production. In eukaryotes, mitochondria generate energy. One of the first known functions of H2S was its effect on the mitochondrial electron transport chain component cytochrome c oxidase. High concentrations of H2S inhibit the enzyme, leading to diminished ATP production and a state of suspended animation [49]. By contrast, lower concentrations of H2S stimulate mitochondrial energy metabolism [50, 51]. This stimulation can be ascribed to the inhibition of phosphodiesterase 2A (PDE2A), leading to elevation of cAMP and activation of protein kinase A [52]. Whether sulfhydration contributes to this effect remains to be elucidated. In mammalian vascular smooth muscle cells certain conditions, such as hypoxia and calcium influx, cause CSE to translocate to the mitochondria, which stimulates ATP production [53]. Similarly, in liver tissue and hepatocyte cell cultures, CBS translocates to the mitochondria during ischemia and prevents toxicity [54]. In a physiological context, localized production of H2S may avoid scavenging or sequestration of this molecule and allow for more efficient signaling.
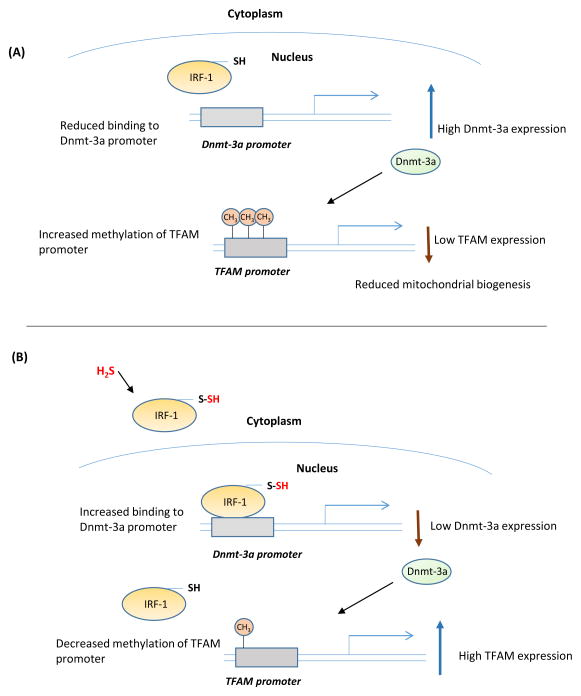
Recently, H2S was shown to be involved in the disposition of mitochondrial DNA (mtDNA) [55]. Mice with deletion of CSE have lower mtDNA content and copy number due to decreased expression of mitochondrial transcription factor A (TFAM). Under basal conditions, H2S sulfhydrates the transcriptional repressor interferon regulatory factor 1 (IRF-1) and enhances its binding to the promoter of DNA methyltransferase 3a (Dnmt 3a) to inhibit its expression. When H2S levels are low, the inhibition of Dnmt 3a transcription is relieved, resulting in its increased expression and methylation of target promoters such as TFAM, whose expression is thereby diminished (Figure 2).
Figure 2. Sulfhydration regulates expression of genes involved in mitochondrial metabolism.
(A) Interferon regulatory factor 1 (IRF-1) is a transcriptional repressor of the DNA methyltransferase 3a (Dnmt-3a). IRF-1 is regulated by sulfhydration. When H2S levels and consequently sulfhydration levels are low, IRF-1 is unable to bind its site on the Dnmt-3a promoter, leading to increased expression of Dnmt-3a, which methylates its target promoters, including the mitochondrial transcription factor A (TFAM), leading to reduced mitochondrial biogenesis. (B) When H2S production is increased, it sulfhydrates IRF-1 and enhances its interaction with the Dnmt-3a promoter to repress its expression. Consequently, methylation of the TFAM promoter is decreased, leading to a higher expression of TFAM and increased mitochondrial biogenesis.
Mitochondria are also sites of sulfide catabolism (56). The balance between biogenesis and degradation of H2S regulates its levels in cells. The main oxidation products of H2S breakdown are thiosulfate and sulfate [57]. Levels of sulfide oxidation products vary in different tissues. While thiosulfate is the major product in colonic mucosa [58], sulfate predominates in the liver and kidney [59]. The sulfide oxidation pathway includes the enzymes sulfide quinone oxidoreductase (SQR), sulfur dioxygenase (ETHE1 or persulfide dioxygenase) and sulfite oxidase. Sulfide is first acted on by SQR and converted to zero-valent sulfur with the involvement of an endogenous sulfur acceptor, which was shown to be GSH, leading to the formation of GSSH [60]. Sulfur dioxygenase or sulfur transferase can then act on GSSH to form sulfite and thiosulfate, respectively.
Role of sulfhydration in stress signaling
CSE is highly inducible, and its expression is regulated in response to a wide variety of stimuli. CSE is induced during ER stress [10,61], and the resulting H2S mediates sulfhydration of protein tyrosine phosphatase 1B (PTP1B) at Cys215, which inhibits its activity [10]. Consequently, the protein kinase RNA-like ER kinase (PERK), a target of the phosphatase activity of PTP1B, retains its active phosphorylation status and stimulates the unfolded protein response (UPR). PhosphoPERK catalyzes the phosphorylation of the eukaryotic translation initiation factor 2α (eIF2α) resulting in attenuation of global protein synthesis. Only a few select mRNAs are translated, such as that encoding activating transcription factor 4 (ATF4). ATF4 transcriptionally activates CSE, forming a feedforward loop that potentiates the ER stress response.
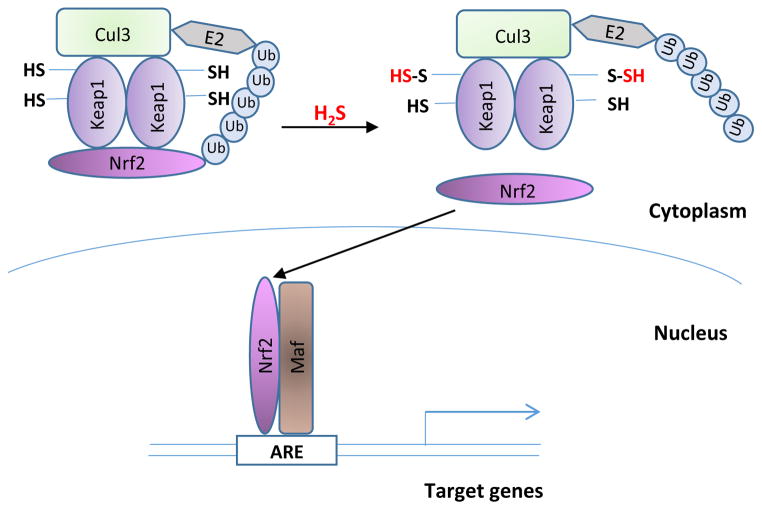
H2S mediates responses to ischemic stress in the cardiovascular system by promoting nuclear translocation of the nuclear factor erythroid 2-related factor 2 (Nrf2) [62]. Nrf2 is the central transcription factor that regulates the expression of phase II antioxidant response genes [63]. Under basal conditions, Nrf2 is sequestered in the cytosol by the kelch-like ECH-associated protein (Keap1), which targets it for proteasomal degradation (Figure 3). Keap1 has reactive cysteines that can be targeted by a wide variety of electrophiles. H2S sulfhydrates Cys151 of Keap1, resulting in dissociation of Keap1 from Nrf2 and translocation of Nrf2 to the nucleus to transcribe cytoprotective genes. Sulfhydration of Keap1 is diminished in CSE knockout mice, and mouse embryonic fibroblasts derived from the CSE knockout mice exhibit higher levels of oxidative stress and signs of senescence [64]. Thus, H2S regulates cellular senescence.
Figure 3. Sulfhydration regulates the expression of phase II cytoprotective genes.
Nuclear factor erythroid 2-related factor 2 (Nrf2) is the master regulator of a battery of genes, including the phase II genes, which respond to stressful conditions such as oxidative stress. Under basal conditions, Nrf2 is sequestered in the cytosol by kelch-like ECH-associated protein (Keap1), which targets it for proteasomal degradation involving Cul3 and E2 ubiquitin ligases. Keap1 has reactive cysteines, whose sulfhydration results in dissociation from Nrf2. Released Nrf2 translocates to the nucleus to regulate transcription of stress-responsive genes.
In addition to delaying senescence, H2S promotes longevity mediated by dietary restriction (DR), a phenomenon that appears to be evolutionarily conserved. In particular, depriving mice of sulfur amino acids protected against hepatic ischemia-reperfusion (IR) injury through elevation of CSE expression and H2S production [9]. In this study, elevated H2S production was necessary and sufficient in a mouse model of DR-mediated stress resistance. Mice injected with the H2S donor NaHS before ischemia had reduced liver damage. Injection of mice with propargylglycine (PAG), the inhibitor of CSE, reduced H2S production and abrogated the benefits of DR on IR injury. Injection of H2S donors prior to injection of PAG and induction of IR prevented liver damage [9]. Specifically, flux through the transsulfuration pathway, the biosynthetic pathway by which cysteine is synthesized, was found to be important for longevity in fruit flies and Caenorhabditis elegans [9,65]. It is quite likely that sulfhydration plays a role in the process.
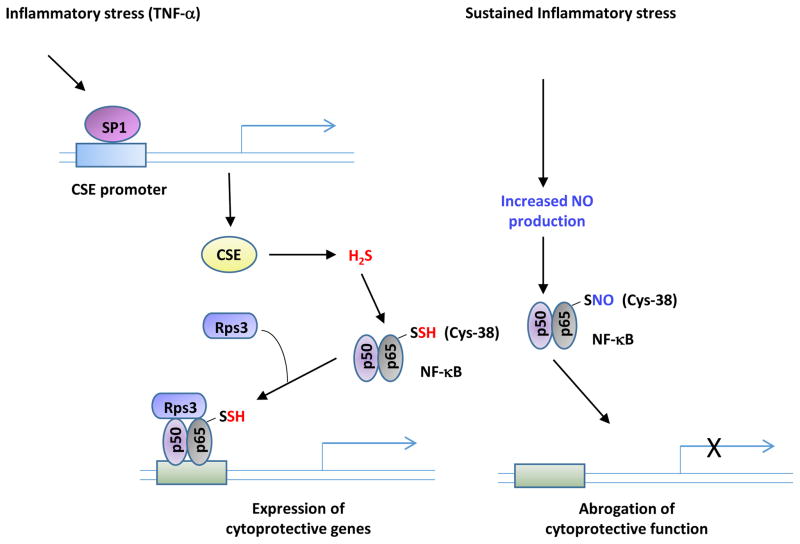
H2S regulates several cytoprotective functions. Apart from its role(s) in oxidative stress, H2S modulates responses to inflammation by sulfhydrating the p65 subunit of nuclear factor kappa B (NF-κB) [6]. When macrophages are stimulated with tumor necrosis factor α (TNF-α), the transcription factor SP1 induces expression of CSE. This leads to production of H2S, which sulfhydrates the p65 subunit of NF-κB. Sulfhydrated NF-κB binds to the ribosomal protein S3 (rps3) resulting in increased association with the promoters of cytoprotective target genes.
Sulfhydration in the nervous system
The three enzymes responsible for H2S production are present in the brain. While CSE is expressed in neurons, CBS is localized to astrocytes, and 3-MST is present both in neurons and astrocytes. One of the first known effects of H2S on the nervous system was its action on the N-methyl D-aspartate (NMDA) subtype of glutamate receptors to synergize with weak electrical stimulation and induce long-term potentiation (LTP) [66]. That H2S influences active (but not quiescent) synapses implicates a role for H2S in associative learning. Although the mechanism through which H2S stimulates NMDA receptors has not been shown, these receptors possess reactive cysteine residues that could be modified by sulfhydration.
H2S impacts diverse signaling processes in the brain, raising the possibility that dysregulation of its metabolism contributes to neurodegeneration. Aberrant H2S signaling has been observed in Alzheimer’s disease (AD) and Parkinson’s disease (PD). Moreover, H2S donors have neuroprotective effects in some mouse models of AD, acting by reducing inflammation and improving spatial memory [67]. The cellular mechanism of these effects may be through regulation of oxidative stress, which is a hallmark of age-dependent neurodegeneration. H2S donors have been found to alleviate oxidative stress in neurons by diminishing reactive oxygen species (ROS), stimulating the synthesis of glutathione, and increasing the uptake of cystine [68].
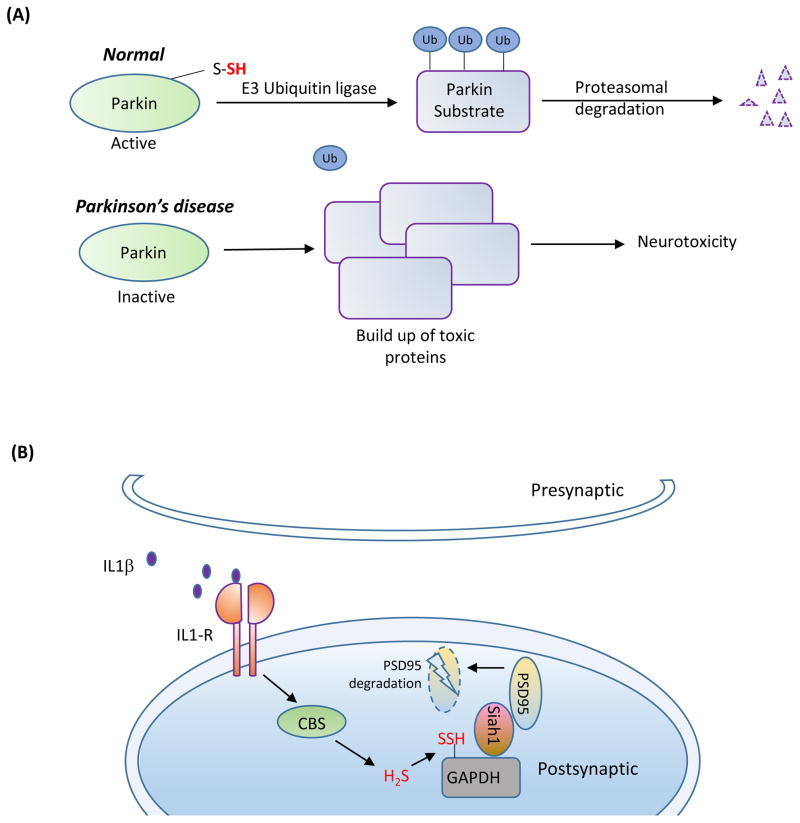
Sulfhydration might affect certain neurodegenerative diseases through its role in protein turnover, which is impaired in protein misfolding diseases such as PD. In PD, damage to the substantia nigra of the brainstem leads to motor dysfunction and tremor, which may reflect compromised ubiquitylation. Under basal conditions parkin, an E3 ubiquitin ligase, is activated by sulfhydration at Cys95, Cys59 and Cys182 [69]. PD can be associated with mutations in parkin that abolish parkin sulfhydration, leading to lowered catalytic activity. Supporting this idea, diminished sulfhydration of parkin is observed in post-mortem patient striata. As a result, there is accumulation of parkin targets such as α-synuclein, a constituent of the characteristic Lewy bodies, which mediates neurotoxicity (Figure 4A).
Figure 4. Sulfhydration in the brain.
(A) Sulfhydration is dysregulated in Parkinson’s disease (PD). In normal subjects, the E3 ubiquitin ligase, Parkin, is sulfhydrated under basal conditions, which enhances its catalytic activity. Parkin mediates ubiquitylation of substrates such as α-synuclein (a component of the Lewy bodies found in PD) and targets them for degradation. In sporadic forms of PD, sulfhydration of Parkin is diminished, leading to decreased catalytic activity, which results in accumulation of toxic proteins and neurotoxicity.(B) Sulfhydration regulates synaptic function. The proinflammtory cytokine interleukin-1β (IL-1β, purple circles) plays key roles in learning and memory and is involved in promoting long term potentiation (LTP). IL-1β activates the transcription factor specificity protein 1 (SP1), which stimulates the transcription of cystathionine β-synthase (CBS), the major H2S producing enzyme in the brain, leading to sulfhydration of the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Sulfhydrated GAPDH binds to seven in absentia homolog-1 (siah1), an E3 ubiquitin ligase, which targets post-synaptic density 95 protein (PSD95) for degradation. PSD95 is a scaffolding protein that participates in synaptic functions. Degradation of PSD95 leads to spine retraction and associated cognitive deficits.
Sulfhydration may also play a role in Huntington’s disease (HD), another protein misfolding neurodegenerative disease. It was recently shown that decreased expression of CSE and concomitant reductions in cysteine and H2S generation mediate neurodegeneration in HD, wherein selective atrophy of the corpus striatum occurs [70,71]. HD is caused by expansion of polyglutamine repeats in the huntingtin protein (Htt), which causes it to aggregate, resulting in widespread damage in neurons. Reduction in the expression of CSE occurs presumably by the influence of mutant huntingtin (mHtt) on specificity protein 1 (Sp1), the transcriptional factor for CSE under basal conditions [72]. Supplementing the diet of mouse models of HD with cysteine as well as providing N-acetylcysteine, a precursor of cysteine, in the drinking water of HD mice delays motor deficits and striatal atrophy. In addition to being a substrate for the production of H2S, cysteine is also a building block for the synthesis of the major cellular antioxidant glutathione. Additionally, cysteine is a precursor for the synthesis of other metabolites and macromolecules such as taurine, lanthionine and Coenzyme A. Cysteine by itself has substantial antioxidant efficacy. Thus, cysteine supplementation could mediate neuroprotection via several pathways.
H2S production is tightly regulated in the nervous system, because too much H2S can have deleterious consequences. For instance, inflammatory cytokines, such as interleukin 1β (IL-1β), induce memory loss by stimulating the expression of CBS, which produces H2S. H2S sulfhydrates GAPDH at Cys150, promoting its association with the E3 ubiquitin ligase Siah1. This enhances Siah1 binding to the synaptic scaffolding molecule postsynaptic density 95 (PSD95), causing PSD95 degradation with consequent neuronal spine retraction and associated memory deficits (Figure 4B). Mice heterozygous for CBS are relatively resistant to the memory-impairing effects of the inflammatory cytokines due to lower levels of CBS [73]. In Down’s Syndrome, caused by the trisomy of chromosome 21, overexpression of CBS, which is present on the chromosome 21, leads to excess H2S production [74]. Interestingly, the expression levels of CBS in Down’s Syndrome is far greater than that expected from a trisomy. The dementia of Down’s syndrome may possibly relate to the CBS localized to astrocytes adjacent to the senile plaques [75].
Sulfhydration in the plant kingdom
Sulfhydration is not restricted to the animal kingdom. Plant proteins also undergo this posttranslational modification [76]. Cysteine metabolizing enzymes have been identified in plants, which catalyze the production of H2S from L-cysteine [77, 78, 79]. Similar to animals, H2S plays important roles in regulating responses of plants to oxidative stress, heavy metal exposure, salt, osmotic and water stress.
Reciprocity of sulfhydration and nitrosylation
As sulfhydration and nitrosylation occur on reactive cysteine residues, these modifications frequently involve the same residue [2,5]. Sulfhydration and nitrosylation usually exert opposite effects. While nitrosylation typically reduces the reactivity of the cysteine thiols, sulfhydration enhances their reactivity, rendering them more nucleophilic. For example, nitrosylation of GAPDH inhibits its glycolytic activity [80], whereas sulfhydration of the enzyme increases its activity seven-fold [25]. Similarly, nitrosylation of parkin decreases its activity [81], whereas sulfhydration stimulates it [69]. In several instances, sulfhydration precedes nitrosylation on the same cysteine residue. For example, the p65 subunit of NF-κB is sulfhydrated within the first two hours of TNF-α treatment and is then nitrosylated (Figure 5) [6]. In the context of PD, parkin is first sulfhydrated and then nitrosylated. Brains of PD patients display diminished sulfhydration and increased nitrosylation of parkin [69]. Thus, it appears that sequential modifications by sulfhydration and nitrosylation fine tune responses to physiological stimuli.
Figure 5. Reciprocity of sulfhydration and nitrosylation.
Sulfhydration and nitrosylation occur on reactive cysteines and as a result frequently modify the same residue. In general, sulfhydration and nitrosylation are functionally antagonistic although there are examples where both modifications elicit the same outcome. In several instances, sulfhydration precedes nitrosylation. During inflammatory conditions (left), cystathionine γ-lyase (CSE) expression is stimulated to produce hydrogen sulfide (H2S). H2S sulfhydrates the p65 subunit of the transcription factor NF-κB and promotes its association with its coactivator, the ribosomal protein S3 (rps3), to enhance expression of cytoprotective genes. If the inflammatory signals persist, the cells produce nitric oxide (NO), which nitrosylates p65 at the same residue and inhibits its DNA binding activity and cytoprotective functions.
Concluding remarks
With the discovery of endogenously synthesized H2S and its role in physiologic pathways, this ancient gaseous signaling molecule has been ascribed the status of a gasotransmitter, which may be its principal function. Unlike other neurotranmitters, such as acetylcholine, H2S--being a gas--cannot be stored in vesicles and thus exists in various bound forms such as sulfane sulfur and acid labile pools, which can be utilized when the need arises. H2S signaling via sulfhydration modulates a wide variety of cellular functions; perhaps more varied and extensive than those regulated by nitric oxide, given that sulfhydration appears substantially more prevalent than nitrosylation [25]. In several instances, reciprocity between sulfhydration and another posttranslational modification such as nitrosylation occurs, which orchestrates responses to physiological stimuli. As the field of H2S signaling expands, so does the need to develop reagents with high specificity and sensitivity to detect the presence and dynamics of this gasotransmitter in vivo. Several questions remain to be answered that would shed light on the in vivo role of the gasotransmitter (Outstanding questions) H2S is generated from cysteine, which itself is a cytoprotective molecule. Like H2S, cysteine is toxic at high doses and cytoprotective at low doses. Cysteine is the precursor of metabolites such as taurine, glutathione and coenzyme A. Thus, distinguishing effects mediated by H2S rather than cysteine and its metabolites may be challenging. Nevertheless, modulation of pathways governing H2S biosynthesis provides therapeutic opportunities. Diminished formation and/or actions of H2S in several neurodegenerative diseases imply that novel drugs that release the gas with minimal side effects may prove beneficial.
Outstanding questions.
What are the relative contributions of the H2S generating enzymes in various tissues?
What are the dynamics and kinetics of sulfhydration in response to different physiological stimuli?
What molecular mechanisms govern the switch between sulfhydration and nitrosylation? Is the same residue first sulfhydrated and then nitrosylated, or are there different pools of nitrosylated and sulfhydrated proteins?
What are the relative contributions of the microbiota vs. the host to H2S content in the gut?
Trends.
Hydrogen sulfide (H2S) is a gasotransmitter that signals via sulfhydration, a posttranslational modification.
Sulfhydration occurs on reactive cysteine residues and converts the Cys –SH group to an –SSH group.
Sulfhydration modulates diverse physiological processes ranging from regulation of blood pressure to signaling in the nervous system.
An emerging theme is the interplay of sulfhydration and nitrosylation which fine tunes signaling pathways.
A variety of detection agents for H2S and sulfhydration have been developed to study the role of this modification in physiological systems.
Acknowledgments
This work was supported by USPHS grants DA000266 and MH18501 to S.H.S.
Glossary
- Biotin switch assay
An assay that detcts modifications on cysteine residues of proteins by replacing the modification by a biotin moiety
- Endothelium-derived relaxation factor (EDRF)
Factors released from the endothelium of vasculature that mediate vasorelaxation
- Gasotransmitter
gaseous molecule that can act as a messenger in signaling pathways
- Ischemia
A condition where a a tissue does not receive sufficient blood supply and therefore oxygen leading to damage
- Sulfhydration
a post-translational modification on cysteine residues, wherein the –SH group is converted to a persulfide or –SSH group
- Nitrosylation
post-translational modification of cysteine residues, where the –SH group is converted to an –SNO group
- pKa
acid dissociation constant, a quantitative measure of the tendency of an acid to dissociate in solution
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yang G, et al. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science. 2008;322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul BD, Snyder SH. H2S signalling through protein sulfhydration and beyond. Nature reviews. Mol Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- 3.Shibuya N, et al. A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat Commun. 2013;4:1366. doi: 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
- 4.Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 5.Paul BD, Snyder SH. Modes of physiologic H2S signaling in the brain and peripheral tissues. Antiox Redox Signal. 2015;22:411–423. doi: 10.1089/ars.2014.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen N, et al. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu XY, et al. Glucocorticoids suppress cystathionine gamma-lyase expression and H2S production in lipopolysaccharide-treated macrophages. Cell Mol Life Sci. 2010;67:1119–1132. doi: 10.1007/s00018-009-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko Y, et al. Glucose-induced production of hydrogen sulfide may protect the pancreatic beta-cells from apoptotic cell death by high glucose. FEBS letters. 2009;583:377–382. doi: 10.1016/j.febslet.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 9.Hine C, et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160:132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan N, et al. H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein JD, et al. Activation of cystathionine synthase by adenosylmethionine and adenosylethionine. Biochem Biophys Res Commun. 1975;66:81–87. doi: 10.1016/s0006-291x(75)80297-x. [DOI] [PubMed] [Google Scholar]
- 12.Ereno-Orbea J, et al. Structural insight into the molecular mechanism of allosteric activation of human cystathionine beta-synthase by S-adenosylmethionine. Proc Nat Acad Sci USA. 2014;111:E3845–3852. doi: 10.1073/pnas.1414545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morikawa T, et al. Hypoxic regulation of the cerebral microcirculation is mediated by a carbon monoxide-sensitive hydrogen sulfide pathway. Proc Nat Acad Sci USA. 2012;109:1293–1298. doi: 10.1073/pnas.1119658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hishiki T, et al. Carbon monoxide: impact on remethylation/transsulfuration metabolism and its pathophysiologic implications. J Mol Med. 2012;90:245–254. doi: 10.1007/s00109-012-0875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vicente JB, et al. NO* binds human cystathionine beta-synthase quickly and tightly. J Biol Chem. 2014;289:8579–8587. doi: 10.1074/jbc.M113.507533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabil O, et al. Reversible heme-dependent regulation of human cystathionine beta-synthase by a flavoprotein oxidoreductase. Biochemistry. 2011;50:8261–8263. doi: 10.1021/bi201270q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enokido Y, et al. Cystathionine beta-synthase, a key enzyme for homocysteine metabolism, is preferentially expressed in the radial glia/astrocyte lineage of developing mouse CNS. FASEB J. 2005;19:1854–1856. doi: 10.1096/fj.05-3724fje. [DOI] [PubMed] [Google Scholar]
- 18.Shibuya N, et al. 3-Mercaptopyruvate sulfurtransferase produces hydrogen sulfide and bound sulfane sulfur in the brain. Antioxid Redox Signal. 2009;11:703–714. doi: 10.1089/ars.2008.2253. [DOI] [PubMed] [Google Scholar]
- 19.Kimura H. Hydrogen sulfide: its production, release and functions. Amino acids. 2011;41:113–121. doi: 10.1007/s00726-010-0510-x. [DOI] [PubMed] [Google Scholar]
- 20.Ishigami M, et al. A source of hydrogen sulfide and a mechanism of its release in the brain. Antioxid Redox Signal. 2009;11:205–214. doi: 10.1089/ars.2008.2132. [DOI] [PubMed] [Google Scholar]
- 21.Mikami Y, et al. Thioredoxin and dihydrolipoic acid are required for 3-mercaptopyruvate sulfurtransferase to produce hydrogen sulfide. Biochem J. 2011;439:479–485. doi: 10.1042/BJ20110841. [DOI] [PubMed] [Google Scholar]
- 22.Carbonero F, et al. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfennig N, Widdel F. The bacteria of the sulphur cycle. Philos Trans R Soc Lond B Biol Sci. 1982;298:433–441. doi: 10.1098/rstb.1982.0090. [DOI] [PubMed] [Google Scholar]
- 24.Linden DR. Hydrogen sulfide signaling in the gastrointestinal tract. Antioxid Redox Signal. 2014;20:818–830. doi: 10.1089/ars.2013.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mustafa AK, et al. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamler JS, et al. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Nat Acad Sci USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marino SM, Gladyshev VN. Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid Redox Signal. 2011;15:135–146. doi: 10.1089/ars.2010.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greiner R, et al. Polysulfides link H2S to protein thiol oxidation. Antioxid Redox Signal. 2013;19:1749–1765. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klomsiri C, et al. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkel T. From sulfenylation to sulfhydration: what a thiolate needs to tolerate. Sci Signal. 2012;5:pe10. doi: 10.1126/scisignal.2002943. [DOI] [PubMed] [Google Scholar]
- 32.Ohno K, et al. Endogenous S-sulfhydration of PTEN helps protect against modification by nitric oxide. Biochem Biophys Res Commun. 2015;456:245–249. doi: 10.1016/j.bbrc.2014.11.066. [DOI] [PubMed] [Google Scholar]
- 33.Francoleon NE, Carrington SJ, Fukuto JM. The reaction of H(2)S with oxidized thiols: generation of persulfides and implications to H(2)S biology. Arch Biochem Biophys. 2011;516:146–153. doi: 10.1016/j.abb.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Zhang D, et al. Detection of protein S-sulfhydration by a tag-switch technique. Angew Chem. 2014;53:575–581. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy P, Winterbourn CC. Rapid reaction of hydrogen sulfide with the neutrophil oxidant hypochlorous acid to generate polysulfides. Chem Res Toxicol. 2010;23:1541–1543. doi: 10.1021/tx100266a. [DOI] [PubMed] [Google Scholar]
- 36.Kimura Y, et al. Polysulfides are possible H2S-derived signaling molecules in rat brain. FASEB J. 2013;27(6):2451–2457. doi: 10.1096/fj.12-226415. [DOI] [PubMed] [Google Scholar]
- 37.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 38.Jaffrey SR, et al. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 39.Paul BD, Snyder SH. Protein sulfhydration. Methods Enzymol. 2015;555:79–90. doi: 10.1016/bs.mie.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 40.Pan J, Carroll KS. Persulfide reactivity in the detection of protein s-sulfhydration. ACS Chem Biol. 2013;8:1110–1116. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisz JA, et al. Thiol-blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J. 2013;280:6150–6161. doi: 10.1111/febs.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park CM, et al. Use of the “tag-switch” method for the detection of protein s-sulfhydration. Meth Enzymol. 2015;555:39–56. doi: 10.1016/bs.mie.2014.11.033. [DOI] [PubMed] [Google Scholar]
- 43.Pietri R, et al. Hydrogen sulfide and hemeproteins: knowledge and mysteries. Antioxid Redox Signal. 2011;15:393–404. doi: 10.1089/ars.2010.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishii I, et al. Cystathionine gamma-Lyase-deficient mice require dietary cysteine to protect against acute lethal myopathy and oxidative injury. J Biol Chem. 2010;285:26358–26368. doi: 10.1074/jbc.M110.147439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mustafa AK, et al. Hydrogen sulfide as endothelium-derived hyperpolarizing factor sulfhydrates potassium channels. Circ Res. 2011;109:1259–1268. doi: 10.1161/CIRCRESAHA.111.240242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes MN, et al. Making and working with hydrogen sulfide: The chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: a review. Free Radic Biol Med. 2009;47:1346–1353. doi: 10.1016/j.freeradbiomed.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Nishida M, et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat Chem Biol. 2012;8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ida T, et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Nat Acad Sci USA. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blackstone E, et al. H2S induces a suspended animation-like state in mice. Science. 2005;308:518. doi: 10.1126/science.1108581. [DOI] [PubMed] [Google Scholar]
- 50.Modis K, et al. Intramitochondrial hydrogen sulfide production by 3-mercaptopyruvate sulfurtransferase maintains mitochondrial electron flow and supports cellular bioenergetics. FASEB J. 2013;27:601–611. doi: 10.1096/fj.12-216507. [DOI] [PubMed] [Google Scholar]
- 51.Szabo C, et al. Tumor-derived hydrogen sulfide, produced by cystathionine-beta-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Nat Acad Sci USA. 2013;110:12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modis K, et al. Hydrogen sulfide-mediated stimulation of mitochondrial electron transport involves inhibition of the mitochondrial phosphodiesterase 2A, elevation of cAMP and activation of protein kinase A. Biochem Pharmacol. 2013;86:1311–1319. doi: 10.1016/j.bcp.2013.08.064. [DOI] [PubMed] [Google Scholar]
- 53.Fu M, et al. Hydrogen sulfide (H2S) metabolism in mitochondria and its regulatory role in energy production. Proc Nat Acad Sci USA. 2012;109:2943–2948. doi: 10.1073/pnas.1115634109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teng H, et al. Oxygen-sensitive mitochondrial accumulation of cystathionine beta-synthase mediated by Lon protease. Proc Nat Acad Sci USA. 2013;110:12679–12684. doi: 10.1073/pnas.1308487110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Yang G. Hydrogen sulfide maintains mitochondrial DNA replication via demethylation of TFAM. Antioxid Redox Signal. 2015 Mar 10; doi: 10.1089/ars.2014.6186. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008;275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 57.Bartholomew TC, Powell GM, Dodgson KS, Curtis CG. Oxidation of sodium sulphide by rat liver, lungs and kidney. Biochem Pharmacol. 1980;29:2431–2437. doi: 10.1016/0006-2952(80)90346-9. [DOI] [PubMed] [Google Scholar]
- 58.Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharmacol. 2001;62:255–259. doi: 10.1016/s0006-2952(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 59.Curtis CG, Bartholomew TC, Rose FA, Dodgson KS. Detoxication of sodium 35 S-sulphide in the rat. Biochem Pharmacol. 1972;21:2313–2321. doi: 10.1016/0006-2952(72)90382-6. [DOI] [PubMed] [Google Scholar]
- 60.Libiad M, Yadav PK, Vitvitsky V, Martinov M, Banerjee R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J Biol Chem. 2014;289:30901–30910. doi: 10.1074/jbc.M114.602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dickhout JG, et al. Integrated stress response modulates cellular redox state via induction of cystathionine gamma-lyase: cross-talk between integrated stress response and thiol metabolism. J Biol Chem. 2012;287:7603–7614. doi: 10.1074/jbc.M111.304576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calvert JW, et al. Hydrogen sulfide mediates cardioprotection through Nrf2 signaling. Circ Res. 2009;105:365–374. doi: 10.1161/CIRCRESAHA.109.199919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 64.Yang G, et al. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid Redox Signal. 2013;18:1906–1919. doi: 10.1089/ars.2012.4645. [DOI] [PubMed] [Google Scholar]
- 65.Kabil H, et al. Increased transsulfuration mediates longevity and dietary restriction in Drosophila. Proc Nat Acad Sci USA. 2011;108:16831–16836. doi: 10.1073/pnas.1102008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abe K, Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xuan A, et al. Hydrogen sulfide attenuates spatial memory impairment and hippocampal neuroinflammation in beta-amyloid rat model of Alzheimer’s disease. J Neuroinflamm. 2012;9:202. doi: 10.1186/1742-2094-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura Y, Kimura H. Hydrogen sulfide protects neurons from oxidative stress. FASEB J. 2004;18:1165–1167. doi: 10.1096/fj.04-1815fje. [DOI] [PubMed] [Google Scholar]
- 69.Vandiver MS, et al. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paul BD, et al. Cystathionine gamma-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature. 2014;509:96–100. doi: 10.1038/nature13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paul BD, Snyder SH. Neurodegeneration in Huntington’s disease involves loss of cystathionine gamma-lyase. Cell cycle. 2014;13:2491–2493. doi: 10.4161/15384101.2014.950538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishii I, et al. Murine cystathionine gamma-lyase: complete cDNA and genomic sequences, promoter activity, tissue distribution and developmental expression. Biochem J. 2004;381:113–123. doi: 10.1042/BJ20040243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mir S, et al. Cytokine-induced GAPDH sulfhydration affects PSD95 degradation and memory. Mol Cell. 2014;56:786–795. doi: 10.1016/j.molcel.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 74.Kamoun P, Belardinelli MC, Chabli A, Lallouchi K, Chadefaux-Vekemans B. Endogenous hydrogen sulfide overproduction in Down syndrome. Am J Med Genet A. 2003;116A:310–311. doi: 10.1002/ajmg.a.10847. [DOI] [PubMed] [Google Scholar]
- 75.Ichinohe A, Kanaumi T, Takashima S, Enokido Y, Nagai Y, Kimura H. Cystathionine beta-synthase is enriched in the brains of Down’s patients. Biochem Biophys Res Commun. 2005;338:1547–1550. doi: 10.1016/j.bbrc.2005.10.118. [DOI] [PubMed] [Google Scholar]
- 76.Aroca A, et al. S-sulfhydration: a new post-translational modification in plant systems. Plant Physiol. 2015 Mar 25; epub ahead of print. [Google Scholar]
- 77.Alvarez C, et al. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010;152:656–669. doi: 10.1104/pp.109.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calderwood A, Kopriva S. Hydrogen sulfide in plants: from dissipation of excess sulfur to signaling molecule. Nitric oxide. 2014;41:72–78. doi: 10.1016/j.niox.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 79.Hancock JT, Whiteman M. Hydrogen sulfide and cell signaling: team player or referee? Plant Physiol. 2014;78:37–42. doi: 10.1016/j.plaphy.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 80.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 81.Chung KK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 82.Zhao K, et al. S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair. EMBO Rep. 2014;15:792–800. doi: 10.1002/embr.201338213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Beus MD, et al. Modification of cysteine 111 in Cu/Zn superoxide dismutase results in altered spectroscopic and biophysical properties. Protein Sci. 2004;13:1347–1355. doi: 10.1110/ps.03576904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie ZZ, et al. Sulfhydration of p66Shc at cysteine59 mediates the antioxidant effect of hydrogen sulfide. Antioxid Redox Signal. 2014;21:2531–2542. doi: 10.1089/ars.2013.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao K, et al. Hydrogen sulfide represses androgen receptor transactivation by targeting at the second zinc finger module. J Biol Chem. 2014;289:20824–20835. doi: 10.1074/jbc.M114.559518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y, et al. Hydrogen sulfide maintains mesenchymal stem cell function and bone homeostasis via regulation of Ca(2+) channel sulfhydration. Cell Stem Cell. 2014;15:66–78. doi: 10.1016/j.stem.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]