Abstract
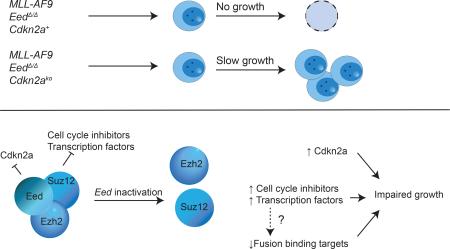
Polycomb Repressive Complex 2 (PRC2) is a chromatin regulator with central roles in development and cancer. The canonical function of PRC2 is the tri-methylation of histone 3 on lysine residue 27. This epigenetic modification is associated with gene silencing. Both tumor suppressor and oncogenic functions have been reported for PRC2, depending on cellular context. In leukemia mediated by the leukemogenic fusion MLL-AF9, complete ablation of canonical PRC2 function by genetic inactivation of the core component Embryonic Ectoderm Development (Eed) or by combined pharmacological inhibition of the PRC2 methyltransferases EZH2 and EZH1 has a strong antileukemic effect, and this effect has been linked to derepression of the PRC2 target locus Cdkn2a. We asked whether inactivation of Cdkn2a is sufficient to restore leukemic activity of Eed-inactivated MLL-AF9 leukemia cells, using combined genetic inactivation of Cdkn2a and Eed. We found that Cdkn2a inactivation partially rescues in vitro and in vivo growth of Eed-inactivated MLL-AF9 cells. However, the growth of Eed-null Cdkn2a-null MLL-AF9 cells in the absence of Cdkn2a remained severely compromised in vitro and in vivo, compared to Eed-floxed Cdkn2a-null counterparts. RNAseq analysis revealed that several genes previously implicated in inefficient growth of MLL-AF9 transformed cells, including Gata2, Egr1 and Cdkn2b were derepressed as a consequence of Eed-inactivation. Furthermore, we found that direct binding targets of MLL-fusion proteins are negatively enriched in Eed-inactivated Cdkn2a-null MLL-AF9-transformed cells. Our data show that interference with PRC2 function affects MLL-AF9 mediated leukemogenesis by both Cdkn2a-dependent and Cdkn2a-independent mechanisms.
Keywords: Stem Cells, Leukemia, Epigenetics, Polycomb Repressive Complex, MLL
Graphical Abstract
Introduction
Hyperactivity and impairment of the Polycomb Repressive Complex 2 (PRC2) have been described in patients with hematological malignancies, but the underlying mechanisms are incompletely understood. In mice, genetic inactivation of Ezh2 leads to T-ALL [1] and MDS/MPN [2], which is in keeping with PRC2 alterations in human T-lineage ALL [3, 4] and in myeloid neoplasms [5-7]. Inactivation of the PRC2 methyltransferase Ezh2 is partially compensated by the less well characterized methyltransferase Ezh1, whereas inactivation of Eed leads to complete loss of the canonical PRC2 function, i.e. the di- and tri-methylation of Lysine 27 on Histone 3 [8]. In human MLL-rearranged leukemia, to our knowledge PRC2 mutations have not been described. In genetic mouse models of MLL-rearranged leukemia, inactivation of Ezh2 impedes leukemia growth [9, 10] and knockout [9] or knockdown [11] of Eed leads to severe compromise of leukemic growth. Pharmacological interference with PRC2 function via stapled peptides targeting Eed [12] and via combined pharmacological inhibition of Ezh2 and Ezh1 [13] has also recently been demonstrated to interfere with growth of MLL-AF9 transformed cells. Derepression of the PRC2 silencing target locus Cdkn2a, which encodes the tumor suppressors p16ink4a and p19Arf, has been implicated as the major mechanism mediating the growth inhibitory effect of impaired PRC2 function in MLL-rearranged leukemia [9, 11, 13]. We here test the interplay between PRC2 and Cdkn2a stringently in a combined genetic model, asking if genetic inactivation of Cdkn2a is indeed able to restore leukemogenic activity in Eed-null MLL-AF9 leukemia.
Results and Discussion
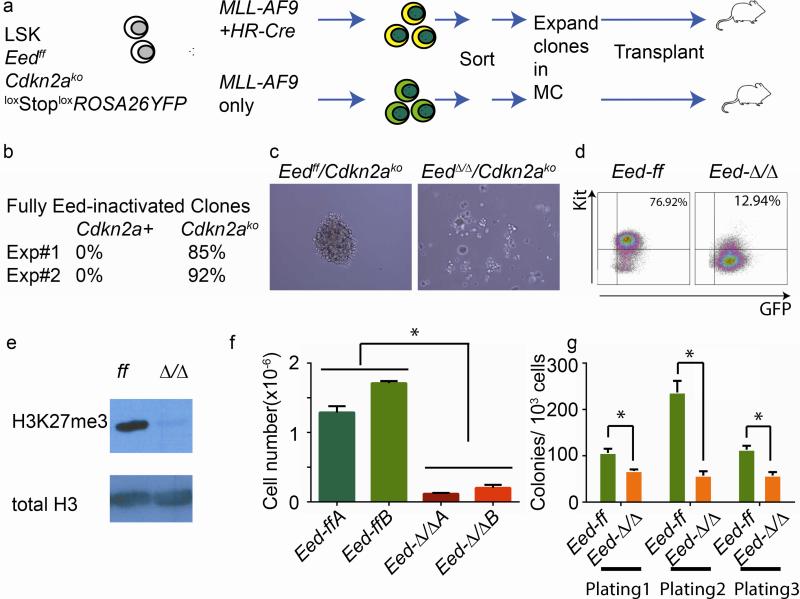
We used lineage negative, Sca-1 positive, Kit-positive (LSK-) cells harvested from mice with a homozygously floxed Eed locus crossed onto a background of wild type, or hetero- or homozygously inactivated Cdkn2a. The mice also carry a loxStoploxROSA26YFP Cre-reporter allele [14] permanently activated upon Cre-expression (Fig. 1a). We transduced LSK-cells with MSCV-MLL-AF9-ires-GFP alone or in combination with a self-excising Cre-expressing retroviral vector, thus limiting expression of the Cre-recombinase to a short time window [15]. In 2 independent experiments using 2 separate sets of donor mice (Cdkn2ako vs Cdkn2a+/+ in experiment 1 and Cdkn2ako vs Cdkn2a+/− in experiment 2) we isolated >85 % fully Eed-inactivated clones in the Cdkn2ako group (Fig. 1b). We were unable to identify any fully Eed-inactivated clones in the presence of either one or two intact Cdkn2a loci. These data confirm previous findings [9, 11, 13] that Eed-inactivated MLL-AF9 cells with an intact Cdkn2a-locus are incapable of in vitro growth and that Cdkn2a is a critically important downstream target of PRC2 in leukemia. We went on to further characterize the effects of Eed inactivation in MLL-AF9/Cdkn2ako clones. We expanded MLL-AF9/Eedff/Cdkn2ako control clones and MLL-AF9/EedΔ/Δ/Cdkn2ako clones with confirmed full excision of floxed Eed sequences (Figure 1a-b). MLL-AF9/Eedff/Cdkn2ako cells formed typical tightly packed colonies, whereas MLL-AF9/EedΔ/Δ/Cdkn2ako cells showed a more dispersed growth pattern (Fig. 1c). MLL-AF9/EedΔ/Δ/Cdkn2ako clones were also noted to have a lower proportion of Kit positive cells by flow cytometry (Fig. 1d). Western blotting confirmed global loss of H3K27me3 as a consequence of Eed inactivation (Fig. 1e).. The growth rate of MLL-AF9/EedΔ/Δ/Cdkn2ako cells in methylcellulose was reduced to approximately 10% of MLL-AF9/Eedff/Cdkn2ako control cells (Fig. 1f). While clearly compromised in their growth, MLL-AF9/EedΔ/Δ/Cdkn2ako cells were able to serially replate with a replating efficiency of approximately 50% of control (Fig. 1g and Fig. S1c) and grow at a stable rate for several weeks.
Fig. 1. Genetic inactivation of Cdkn2a partially rescues genetic inactivation of Eed in MLL-AF9 cells in vitro.
a) Schematic depiction of experimental design. Co-transduction with an MLL-AF9 encoding vector and retroviral self-excising Hit-and Run Cre vector (HR-Cre) was used to excise floxed Eed sequences. MC; methylcellulose. Cre expression leads to activation of the ROSA26-YFP reporter which was used for sorting of Eed-inactivated cells. b) Fully deleted EedΔ/Δ MLL-AF9 clones can be established in the absence, but not presence of an intact homozygous or heterozygous Cdkn2a-locus. c) MLL-AF9/Eedff/Cdkn2ako clones have the typical packed colony morphology while MLL-AF9/EedΔ/Δ/Cdkn2ako cells grow more dispersed. d) Lower proportion of Kit positive cells in MLL-AF9/EedΔ/Δ/Cdkn2ako compared to MLL-AF9/Eedff/Cdkn2ako cells. A separate pair of clones showed 89.1% Kit-positive cells for Eedff cells versus 53.6% for EedΔ/Δ cells. e) complete absence of H3K27me3 in MLL-AF9/EedΔ/Δ/Cdkn2ako cells (Western blot). f) MLL-AF9/EedΔ/Δ/Cdkn2ako cells grow more slowly in methylcellulose compared to MLL-AF9/Eedff/Cdkn2ako cells. 5000 cells were plated per well in Methylcellulose and growth factors SCF, IL3 and IL6 and total cell number was determined 4 days later. Error bars indicate standard deviation (SD). g) Approximately 50% average reduction in plating efficiency in MLL-AF9/EedΔ/Δ/Cdkn2ako clones compared to MLL-AF9/EedΔ/Δ/Cdkn2ako clones. 2 MLL-AF9/EedΔ/Δ/Cdkn2ako clones and 3 MLL-AF9/EedΔ/Δ/Cdkn2ako clones were plated in triplicate day 0, and replated in triplicate on day 5 and day 10. Error bars indicate SEM. * p<0.05 (t-test).
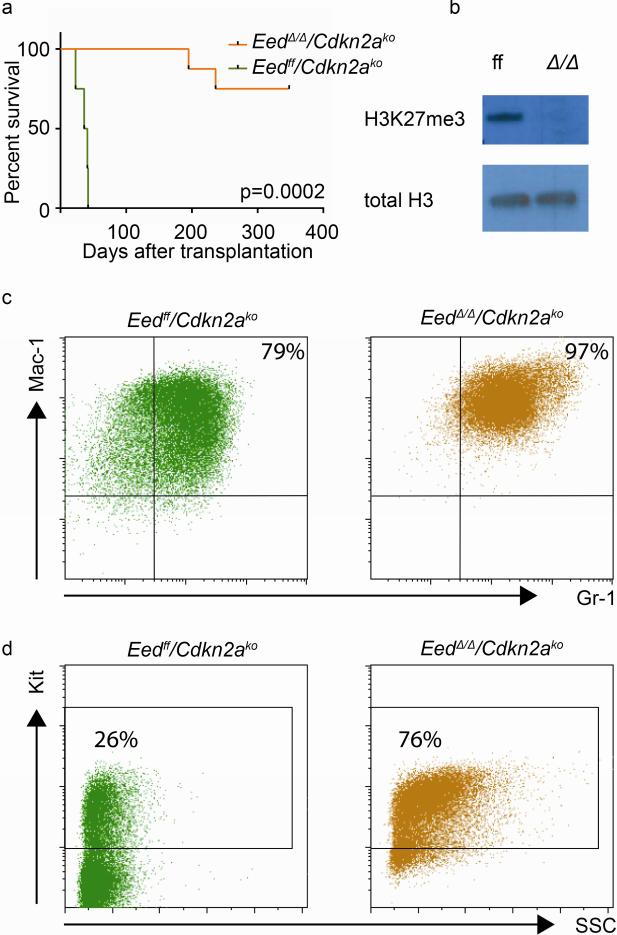
We therefore asked whether MLL-AF9/EedΔ/Δ/Cdkn2ako cells cause in vivo leukemia. In order to avoid outgrowth of non-deleted escapees, we established clonal lines, and confirmed complete loss of H3K27me3 in clones prior to injection. Complete excision of Eed sequences was also evident in the RNAseq gene expression data (Fig. S1a), and was confirmed by RT-qPCR (Fig. S1b). Cells from two MLL-AF9/Eedff/Cdkn2ako control clones and four confirmed MLL-AF9/EedΔ/Δ/Cdkn2ako clones where transplanted into syngeneic sublethally irradiated recipients (two recipients per clone). While all the recipients of control cells developed leukemia within an expected time frame, only 2/8 recipients of MLL-AF9/EedΔ/Δ/Cdkn2ako cells (derived from two separate clones) developed leukemia (one additional mouse died from unrelated reasons with low (<1%) but detectable engraftment of YFP positive cells in spleen and bone marrow). The onset of leukemia in the MLL-AF9/EedΔ/Δ/Cdknko group was significantly delayed (Fig. 2a). We confirmed complete inactivation of Eed in cells recovered from moribund animals by Western blot for H3K27me3 (Fig. 2b). Immunophenotypic analysis of both Eedff/Cdknko and EedΔ/Δ/Cdknko leukemias was consistent with AML (Fig. 2c and d), without evidence of lineage infidelity with respect to lymphoid surface markers (Fig S2a and b). Interestingly, MLL-AF9/Cdkn2ako/EedΔ/Δ leukemias showed an increased proportion of Gr-1 and Kit positive cells in the transition from pre-leukemic cell to in vivo leukemia (Fig. 2c-d and Fig. S2c-f). These data demonstrate that in the context of Cdkn2a inactivation, Eed is not strictly required for MLL-AF9 leukemia in vitro and in vivo. However, growth of MLL-AF9/Cdkn2ako/EedΔ/Δ cells remains severely compromised, suggesting that Eed-inactivation results in functionally important gene expression changes other than Cdkn2a derepression that adversely affect MLL-AF9 leukemia growth.
Fig. 2. Genetic inactivation of Cdkn2a partially rescues genetic inactivation of Eed in MLL-AF9 cells in vivo.
a) MLL-AF9/EedΔ/Δ leukemia can be established on a Cdkn2ako genetic background in vivo with incomplete penetrance and significantly prolonged latency compared to MLL-AF9/Eedff/Cdkn2ako controls. b) complete absence of H3K27me3 in MLL-AF9/EedΔ/Δ/Cdkn2ako cells in vivo (Western blot). c) Gr-1 and Mac-1 expression in vivo in MLL-AF9/Eedff/Cdkn2ako (green, gated on live GFP+ cells) and MLL-AF9/EedΔ/Δ/Cdkn2ako (yellow, gated on live GFP+YFP+ cells) blasts. d) Kit expression in vivo in MLL-AF9/Eedff/Cdkn2ako blasts (green, gated on live GFP+ cells) and MLL-AF9/EedΔ/Δ/Cdkn2ako blasts (yellow, gated on live GFP+YFP+ cells).
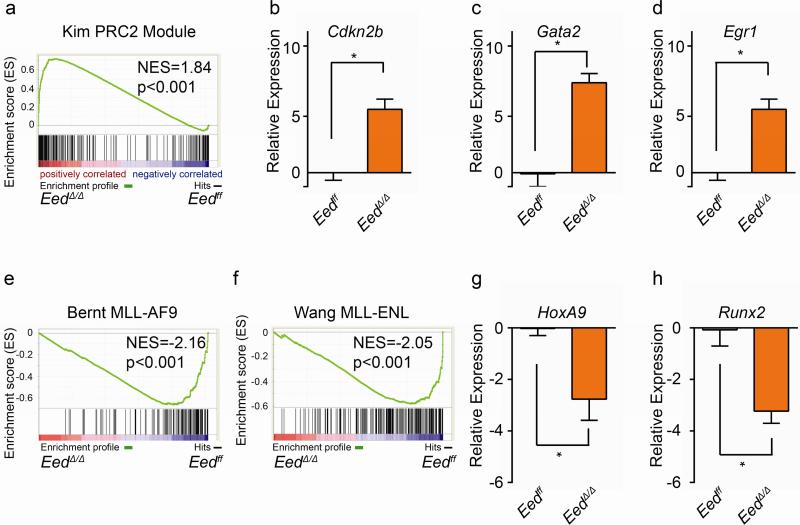
We therefore assessed additional transcriptional consequences of Eed-inactivation in MLL-AF9/EedΔ/Δ/Cdkn2ako cells. RNA-seq was performed on confirmed fully excised EedΔ/Δ/Cdkn2ako and control Eedff/Cdkn2ako cells prior to injection. Gene set enrichment analysis [16] (GSEA) revealed enrichment of canonical PRC2 targets [17] (Fig. 3a). The cell cycle inhibitor Cdkn2b, a PRC2 target (Fig. S3a) was found to be upregulated in EedΔ/Δ/Cdkn2ako cells (Fig. 3b). The stem cell associated genes Gata2 [18] (Fig. 3c) and Egr1 (Fig 3d) which are PRC2 targets in murine MLL-AF9 L-GMP and murine bulk MLL-AF9 blasts (Fig. S3a), were also upregulated. We also observed negative enrichment of primary MLL-AF9 binding targets as previously defined by Bernt et al [19] (Fig. 3e) and MLL-ENL binding targets as described by Wang et al [20] (Fig. 3f). Downregulation of HoxA9 (Fig. 3g) as a key downstream target of MLL-fusions, as well as the MLL-AF9 -fusion target Runx2 (Fig. 3h) were also confirmed by RT-qPCR.
Fig. 3. Transcriptional changes in response to Eed-inactivation.
a) Polycomb Repressive Complex 2 (PRC2) binding targets are enriched in MLL-AF9/EedΔ/Δ/Cdkn2ako cells compared to MLL-AF9/Eedff/Cdkn2ako cells. b)-d) Increased expression as measured by RT-qPCR of PRC2 binding targets Cdkn2b (b), Gata2 (c) and Egr1 (d) in MLL-AF9/EedΔ/Δ/Cdkn2ako clones. (n=2, Eedff and n=4 for Eedko). e) Negative enrichment (i.e. decreased expression) of MLL-AF9 fusion binding targets in MLL-AF9/EedΔ/Δ/Cdkn2ako cells. d) Negative enrichment (i.e. decreased expression) of MLL-ENL fusion binding targets in MLL-AF9/EedΔ/Δ/Cdkn2ako cells. g) Decreased expression of HoxA9 in MLL-AF9/EedΔ/Δ/Cdkn2ako clones (Eedff, n=2, and EedΔ/Δ n=4). h) Decreased expression of MLL-AF9 binding target Runx2 in MLL-AF9/EedΔ/Δ/Cdkn2ako clones (Eedff, n=2, and Eedko n=3). Relative expression values are expressed as log2 of MLL-AF9/EedΔ/Δ/Cdkn2ako compared to MLL-AF9/Eedff/Cdkn2ako. *p<0.05.
Recent data have suggested combined inhibition of EZH2 and EZH1 as a therapeutic strategy for MLL-leukemia [13]. We compared the gene expression changes observed in our MLL-AF9/EedΔ/Δ/Cdkn2ako model to the data described by Xu et al. [13]. We found that the genesets behaved analogously in cells treated with a combined EZH2/EZH1 inhibitor, UNC1999 (Fig. S3). Specifically, UNC1999-treated cells showed significant enrichment in PRC2 targets and negative enrichment for MLL-fusion binding targets. Genetic and pharmacologic impairment of PRC2 function is detrimental to MLL-rearranged leukemia cells. This effect has been ascribed to de-repression of the tumor suppressor locus Cdkn2a, a documented PRC2 target [9, 11, 13]. We here evaluated the effects of genetic PRC2 inactivation on MLL-AF9 leukemogenesis in a Cdkn2a-null background. Our data demonstrate that the growth disadvantage of EedΔ/Δ MLL-AF9 cells is not entirely dependent on Cdkn2a but is rather multifactorial. As expected, in response to genetic inactivation of the PRC2 core component Eed canonical PRC2 targets are derepressed, including the known negative regulator of cell cycle Cdkn2b encoding p15. We also found inappropriate expression of developmental transcription factors Gata2 and Egr1 which are normally highly expressed in stem-/early progenitor cells. The derepression of stem cell-associated genes may appear to be compatible with, or possibly supportive of a leukemic phenotype. However, the functional relevance of Gata2 and Egr1 has been validated by others, and forced expression of Gata2 [21] and Egr1 [10] has been linked to impaired growth of MLL-AF9 leukemia. In keeping with prior reports [21-23] we found that high-level forced expression of GATA2 alters colony formation and impedes growth of MLL-AF9 transformed Eedff/Cdkn2ako LSK-cells (Fig. S4), thus partially phenocopying inactivation of Eed. MLL-AF9 leukemic stem cells have been suggested to display transcriptional programs partially characteristic of adult hematopoietic stem-cells and partially resembling myeloid progenitors [24]. Our data along with published data suggest that inappropriately high expression of at least some stem cell related genes has adverse effects in MLL-AF9 leukemia, suggesting that MLL-AF9 leukemia requires a precisely regulated balance of stem- and progenitor associated genes. Finally, we observed significant loss of direct binding targets of the leukemogenic MLL-AF9 and MLL-ENL fusion. This decreased expression of binding targets was also observed in published data in response to UNC1999, a combined EZH1/EZH2 inhibitor [13], suggesting that cellular loss of H3K27me3 is responsible for the effect. In summary, impaired PRC2 severely impairs but does not abrogate MLL-AF9 leukemia growth. The growth inhibitory effect is not only mediated by dysregulation of Cdkn2a but also by dysregulation of other genes. Inhibition of PRC2 may be a strategy for the treatment of MLL-rearranged leukemia, especially if synergistic therapeutic interventions can be identified.
Experimental Procedures
For primer sequences, antibodies and detailed experimental procedures please refer to the supplemental materials.
Mice
Animals were maintained at the Animal Research Facility at the University of Colorado Anschutz Medical Campus. Animal experiments were approved by the Internal Animal Care and Use Committee. All mice were maintained on a fully backcrossed C57BL/6 background.
Generation of transformed murine cells and leukemia
See supplement for description of retroviral vectors. (Lin−Sca-1+Kit+ cells were prestimulated in liquid culture in the presence of murine SCF, Flt3L, IL6 and Tpo (Peprotec, Rocky Hill, NJ) and then transduced in the presence on Retronectin (Takara, Madison, WI). Cells were sorted for expression of GFP or coexpression of GFP and YFP and plated in methylcellulose in the presence of IL3, IL6 and SCF (Peprotech). Individual colonies were picked and expanded in 96-well plates. Cells expanded for 18 days and were then injected into syngeneic sublethally (600cGy) irradiated recipients (1 Million cells per recipient). Cell growth and viability were followed by serial cell counts. Antibodies used for flow cytometry and immunoblot detection and qPCR primers are detailed in supplementary methods.
Supplementary Material
We established MLL-AF9 leukemia with combined genetic inactivation of Cdkn2a and Eed
Cdkn2a inactivation partially rescues the previously reported growth inhibitory effects of Eed-inactivation in MLL-AF9 cells in vitro and in vivo
Cdkn2a-null Eed-null MLL-AF9 cells remain severely compromised in their growth compared to Cdkn2a-null Eed-ff MLL-AF9 cells
RNA-seq in Eed-null cells demonstrates derepression of Polycomb targets with documented adverse effect on growth of MLL-AF9 cells
RNA-seq demonstrate negative enrichment of primary MLL-fusion binding targets in Eed-null MLL-AF9 cells
Acknowledgements
We thank Karen Helm and the staff at the flow cytometry core for assistance with FACS and cell sorting. We thank Katrina Diener and Ted Shade at the Genomics core of the University of Colorado AMC. We thank Ken Jones at the Bioinformatics core for help with Illumina data analysis. We thank Tracy Haney and Holly Goold for animal Husbandry. We thank Patricia Ernst for critically reading the manuscript. The GATA2-cDNA was obtained from Gokhan Hotamisligil (Addgene plasmid 1287).This work was supported by start-up funds from Children's Hospital Colorado Research Institute (TN), as well as NHLBI K08HL102264 (KMB), NCI K08CA154777 (TN), NCI P30CA046934 (Shared Resource of the University of Colorado Cancer Center) and the Cancer League Colorado (TN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Simon C, Chagraoui J, Krosl J, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes & development. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muto T, Sashida G, Oshima M, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. The Journal of experimental medicine. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ntziachristos P, Tsirigos A, Van Vlierberghe P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine. 2012 doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 6.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature genetics. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 7.Guglielmelli P, Biamonte F, Score J, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118:5227–5234. doi: 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 8.Shen X, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neff T, Sinha AU, Kluk MJ, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka S, Miyagi S, Sashida G, et al. Ezh2 augments leukemogenecity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012 doi: 10.1182/blood-2011-11-394932. [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Wang E, Zuber J, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2012 doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim W, Bird GH, Neff T, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2- dependent cancer. Nature chemical biology. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu B, On DM, Ma A, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2014 doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Molecular cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddell J, Gazit R, Garrison BS, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernt KM, Zhu N, Sinha AU, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang QF, Wu G, Mi S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonadies N, Foster SD, Chan WI, et al. Genome-wide analysis of transcriptional reprogramming in mouse models of acute myeloid leukaemia. PloS one. 2011;6:e16330. doi: 10.1371/journal.pone.0016330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer cell. 2015;27:502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tipping AJ, Pina C, Castor A, et al. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 2009;113:2661–2672. doi: 10.1182/blood-2008-06-161117. [DOI] [PubMed] [Google Scholar]
- 24.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.