Abstract
Fertilization, the union of male and female gametes to create offspring, is an intricate biological process dependent upon several biochemical and physiological events. Our understanding of the functions of protein constituents of the outer acrosomal membrane-associated matrix complex (OMC) is limited. A highly purified OMC fraction isolated from bovine cauda sperm heads is comprised of 54, 50, 45, and 38–19kDa polypeptides. The objective of this study is to identify and to characterize the 45kDa (OMC45) polypeptide and to define its role in binding acrosomal hydrolases and to examine the fate of OMC45 polypeptide during the acrosome reaction. We isolated OMC45 polypeptide from the high-pH insoluble fraction of OMC. Proteomic analysis of OMC45 by MALDI–TOF–TOF yielded 8 peptides that matched the NCBI database sequence of Tektin 3 (TEKT3). Triton X–100–permeabilized cauda sperm exhibited intense staining of the acrosomal segment with anti–OMC45 and anti–TEKT3. The OMC45 polypeptide was solubilized by RIPA (radio-immunoprecipitation assay) buffer extraction. The solubilized fraction was subjected to immunoprecipitation analysis. The OMC45 polypeptide was recovered in the anti–OMC45 immunoprecipitation pellet. An identical blot stained with anti–TEKT3 exhibited the presence of TEKT3 polypeptide in the anti–OMC45 pellet. Our immunofluorescence and biochemical studies confirm the proteomics identification of OMC45 polypeptide; that it exhibits a sequence similarity to TEKT3. OMC45 glycoprotein possesses both N–linked and O–linked oligosaccharides. Deglycosylated OMC45 revealed a significant reduction in both acrosin and N–acetylglucosaminidase (NAGA) binding in comparison with acrosin and NAGA binding to a native OMC45 polypeptide, demonstrating the important role of oligosaccharides in hydrolase binding. OMC45 polypeptide is not released during the acrosome reaction but remains in the particulate cell subfraction, associated with the hybrid membrane complex.
Keywords: Bovine sperm, Acrosome, Glycoproteins, Tektin 3, Acrosomal Hydrolases
Introduction
Mammalian fertilization leads to the formation of a new organism when the exchange of the nuclear components of opposite gametes occurs [1–3]. The formation of a fertilization competent sperm cell is a complex, multifaceted process initiated in the testis (spermatogenesis) and completed in the female reproductive tract. Testicular spermatozoa must undergo epididymal maturation, capacitation, and the acrosome reaction to be able to fertilize the oocyte [1]. Both male and female gametes are equipped with specialized cell-type specific proteins needed for the completion of fertilization. The interaction of sperm proteins and glycoproteins of the egg’s zona pellucida activates signal transduction pathways and calcium uptake, leading to the acrosome reaction. The acrosome reaction is an extensive fusion between the outer acrosomal membrane and plasma membrane located at the anterior of a sperm head permitting the release of acrosomal hydrolases. A well-organized secretory event from sperm (i.e., the acrosome reaction) is requisite for fertilization [4].
The mammalian sperm acrosome is a membrane-bounded, exocytotic vesicle located over the anterior portion of the nucleus and contains hydrolytic enzymes which are required for fertilization. Some hydrolases are compartmentalized within morphologically distinct domains of the acrosomal matrix [5–11]. The insoluble matrix assemblies define acrosomal shape, but they also bind specific hydrolases [5, 8, 10, 12–14]. The association of acrosomal enzymes with domain-specific matrix proteins has been suggested to function in enzyme stabilization prior to the acrosome reaction and to regulate sequential enzyme release during the acrosome reaction [7, 9, 11, 13, 15–17]. Thus, several studies suggest that mammalian fertilization is indeed a complicated process in which multiple hydrolases are involved.
We have previously isolated and characterized a stable acrosomal matrix assembly from the bovine acrosome termed the outer acrosomal membrane-associated matrix complex (OMC) [13, 18]. Our understanding of the functions of protein constituents of the outer acrosomal membrane–associated matrix complex (OMC) is limited. This stable matrix assembly exhibits specific binding activity for acrosin [13] and N-acetylglucosaminidase (NAGA) [19]. A highly purified OMC fraction is comprised of three major (54, 50, and 45kDa) and several minor (38-19kDa) polypeptides. The set of 38-19kDa polypeptides termed “rpf” (related polypeptide family) is selectively solubilized by extraction of OMC in 0.1M CAPS buffer (3-[cyclohexylamino]-1-propane sulfonic acid), pH 10.5, while the remaining three major polypeptides (55, 50 and 45kDa) remain associated with the sedimentable “stripped” OMC [20]. We published the purification of a 45kDa polypeptide (OMC45) from the high–pH insoluble fraction by continuous-elution SDS-PAGE. Anti-OMC45 polyclonal antibody reacts strongly on immunoblots with the OMC45 band. Using immunofluorescence, anti-OMC45 localizes specifically to the acrosomal cap [19]. Acrosin and NAGA bound the OMC32 polypeptide (one of the major polypeptides in rpf) in a concentration-dependent fashion. In contrast, OMC45 polypeptide exhibited stronger affinity to acrosin than NAGA [19]. The binding specificity of acrosomal matrix proteins to hydrolases strongly suggests that the matrix polypeptides play an important role in the regulation of hydrolases released during the acrosome reaction. However, the molecular characterization of OMC45 polypeptide is not known. The objective of this study is to identify and to characterize the 45kDa (OMC45) polypeptide and to define its role in binding acrosomal hydrolases and to investigate the fate of OMC45 polypeptide during the acrosome reaction. Our immunofluorescence and biochemical studies strongly confirm the proteomic identification of OMC45 polypeptide; that it exhibits a sequence similarity to Tektin 3 (TEKT3). Our results suggest that oligosaccharide moieties of OMC45 polypeptide play an important role in hydrolase binding.
Materials and Methods
Isolation of Outer Acrosomal Membrane-Associated Matrix Complex (OMC)
Bovine epididymides were obtained from Randloph Packing Co., Inc., Asheboro, NC. Heads from bovine cauda epididymal spermatozoa were isolated following our published procedure [13]. The OMC fraction was isolated by centrifugation on a Percoll gradient [13]. Isolated OMC fractions were fixed with 4% glutaraldehyde in 0.1M sodium cacodylate buffer, pH 7.4. The samples were then postfixed with OsO4, dehydrated, and embedded in epon [13]. Thin sections were stained with uranyl acetate and lead citrate to examine the purity of OMC by electron microscopic analysis. The purification of the 45kDa polypeptide (OMC45) from the high-pH insoluble fraction was performed by continuous-elution SDS-PAGE on 7.5% acrylamide gels using a Model 491 Prep Cell (Bio-Rad Laboratories, Hercules, CA) following the method of Nagdas et al. [19].
Proteomic Analysis
Proteomic identification of OMC45 polypeptide was performed at the Mass Spectrometry Facility of UNC School of Medicine Proteomic Center, Chapel Hill, NC. The 45kDa band was subjected to MALDI-TOF-TOF analysis to obtain internal amino acid sequences of several tryptic peptides. Derived peptide sequences was analyzed in the National Center for Biotechnology Information (NCBI) database to determine if a full length sequence has been reported and to identify potential functional motifs such as a transmembrane hydrophobic domain, an extracellular domain with consensus glycosylation sites, and to define potential phosphorylation sites as well as protein interaction domains on its cytoplasmic segment.
Gel Electrophoresis and Western Blotting
SDS-PAGE was performed on 12% or 7.5% continuous or 7.5% to 15% gradient polyacrylamide gels [21]. Polypeptides were then electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes [22] or stained with Coomassie blue [23] or silver [24]. Western blots were stained either with affinity-purified anti-OMC45 IgG fraction, anti-TEKT3, anti-phosphotyrosine antibodies (BD Transduction Laboratories, Lexington, KY), or preimmune serum. We received an aliquot of TEKT3 antibody from the laboratory of Prof. Hiroshi Iida, Laboratory of Zoology, Kyushu University, Fukuoka, Japan. Lectin blot analysis was used to identify glycosylated polypeptides following the method of Nagdas et al. [25] using biotinylated lectins (Vector Laboratories, Burlingame, CA). The specificity of lectin sugar interactions was examined with incubations containing a proper saccharide inhibitor in lectin-containing buffer, including 0.2M N-acetylglucosamine for Wheat Germ Agglutinin (WGA) and 0.2M D-galactose for Ricinus Communis Agglutinin (RCA). Lectin binding polypeptides were visualized by color development using H2O2 and diaminobenzidine.
Immunofluorescence Microscopy
Spermatozoa were fixed in 4% formalin in PBS (0.1M sodium phosphate buffer, pH 7.4) at 4°C for 30 min and plated on poly-L-lysine-coated coverslips. Cells were then washed in PBS and permeabilized in PBS containing 0.1% Triton X-100 for 30 min at 4°C. After three rinses in PBS, samples were blocked with 1% normal goat serum in PBS (PBS-NGS). Coverslips were then incubated with equal dilutions of immune or preimmune serum in PBS-NGS for 1 hr and washed three times with PBS. Cells were then incubated with Cy3-conjugated goat anti-rabbit IgG (KPL) in PBS-NGS for 1 hr. Following incubation, cells were then washed three times with PBS to remove unbound antibody molecules. Coverslips were examined by phase contrast and epifluorescence microscopy.
Immunoprecipitation
For immunoprecipitation experiments, the particulate fraction of OMC obtained after high-pH extraction was extracted in RIPA buffer composed of 1% NP-40, 1% Na-deoxycholate, 0.1% SDS, 0.15M NaCl, 10mM sodium phosphate, pH 7.2, at 4°C for 1 hr and then centrifuged at 12,000 ×g for 15 min. The supernatant fraction (lysate) was then precleared of endogenous IgG by incubation with Protein A Sepharose beads for 15 min at 4°C and centrifugation at 12,000 ×g for 1–2 min. Immunoprecipitation was performed using Pierce Immunoprecipitation Kit following the manufacturer’s protocol. The supernatant obtained after the removal of endogenous IgG was immunoprecipitated with affinity-purified anti-OMC45 IgG fraction covalently conjugated to Protein A-Sepharose. Control immunoprecipitations utilized preimmune IgG conjugated to Protein A-Sepharose. The beads containing bound proteins were washed two or three times by centrifugation in RIPA buffer. Immunoprecipitates were fractionated by SDS-PAGE, transferred to PVDF membranes, and stained with anti-OMC45 or anti-TEKT3 antibodies.
Deglycosylation of OMC45 Polypeptide and Hydrolases Binding to Deglycosylated OMC45
OMC45 polypeptide was treated with N-glycanase and O-glycanase separately following the manufacturer’s instructions. Both N-glycanase and O-glycanase were purchased from New England Biolabs., MA. After enzymatic digestion, deglycosylated OMC45 polypeptide was analzed by Western blot stained with anti-OMC45.
For the hydrolase binding experiments, the homogenous fraction of OMC45 polypeptide was first treated with N-glycanase (to remove all N-linked oligosaccharide moieties) and the resulting OMC45 polypeptide was treated with O-glycanase (an endoenzyme known to cleave O-linked OS chains). Both incubations were done overnight at 37°C according to the New England Biolabs’ instructions. After enzymatic digestion, deglycosylated OMC45 polypeptide and native OMC45 polypeptide were conjugated to AminoLink Plus coupling gel (Pierce Chemical Co.) separately at pH 10.0 according to the manufacturer’s instructions. As a control, same units of N-glycanase and O-glycanase were conjugated to AminoLink Plus coupling gel. A centrifugation assay was performed to determine the binding efficacy of hydrolases to the deglysosylated OMC45 polypeptide following the method of Nagdas et al. [13].
Sperm Capacitation and Acrosome Reaction
Sperm (4–6 × 107/ml) were capacitated in the presence of heparin (10 mg/ml) in a modified Tyrode’s medium (pH 7.4) for 4 hours at 39°C with a 95% air: 5% CO2 atmosphere [26]. To initiate the acrosome reaction, sperm were incubated with 100 µg/mL lysophosphatidyl choline (LPC) for 15 min following the end of the 4 hr incubation. Samples were either fixed for immunofluorescence staining or centrifuged at 10,000 ×g for 10 min at 4°C. The pellets and supernatants were adjusted to equal volumes and used for SDS-PAGE and acrosin determination.
Hydrolase Assays
Acrosin activity was measured spectrophotometrically at 410nm in 0.05M Tris-HCl buffer (pH 8.0), 0.1% Triton X-100, 0.05M CaCl2, and 0.1mM N-p-Tosyl-Gly-Pro-Arg-p-niroanilide (Sigma Chemical Co., St. Louis, MO) (27). β-N-acetylglucosaminidase (NAGA) activity was also measured spectrophotometrically at 410nm using 2mM p-nitrophenyl N-acetyl-β-D-glucosaminide as a substrate [28]. Protein was estimated by the procedure of Bradford [29].
Results
Purification and Proteomic Identification of OMC45 Polypeptide
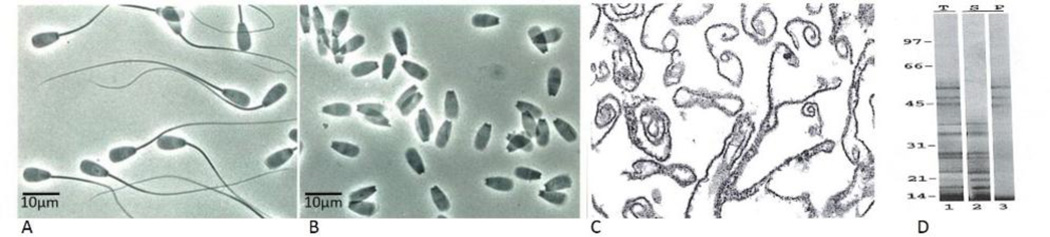
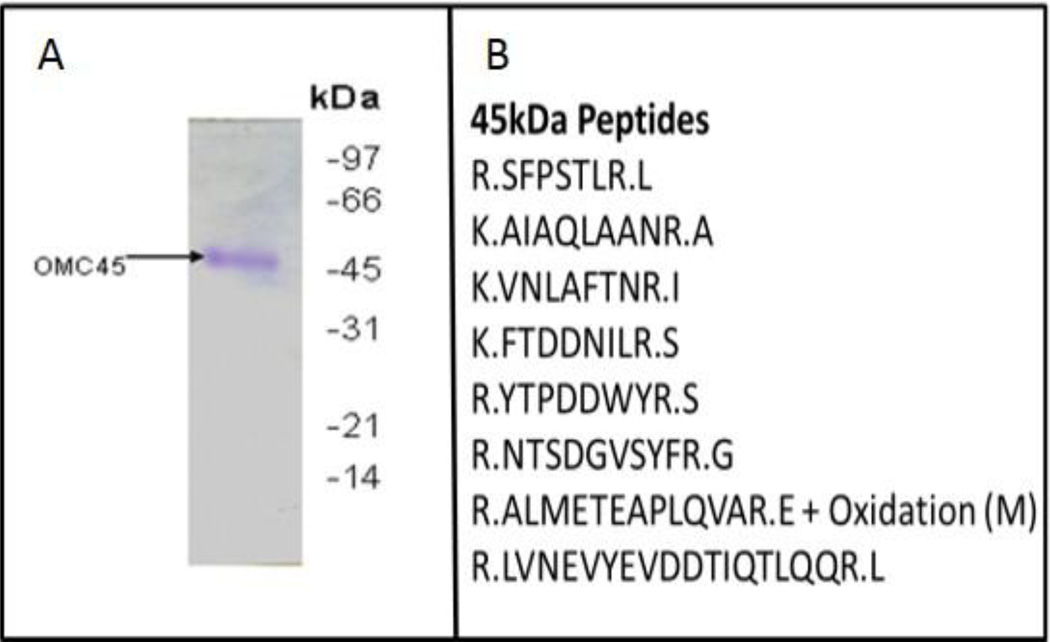
We have previously identified a set of acrosomal components from bovine cauda epididymal spermatozoa (Fig. 1A) that resist disruption by sonication and detergent treatment [18, 30]. Sperm heads (Fig. 1B) were purified from suspensions of sonicated cauda spermatozoa following the method of Nagdas et al. [13]. Then, the OMC (Fig. 1C) was purified from Triton X-100 treated heads by centrifugation on Percoll gradients following our published procedure [13]. SDS-PAGE of the OMC fraction reveals a distinct set of major polypeptides (Fig. 1D, lane 1) that includes protein bands of 54, 50, and 45 kDa and a family of polypeptides between 38 and 19 kDa, termed as “rpf”. High-pH extraction of OMC solubilized rpf polypeptides (Fig. 1D, lane 2), whereas the 54, 50, and 45 kDa polypeptides remained associated with the high-pH insoluble particulate fraction (Fig. 1D, lane 3). The homogeneous fraction of OMC45 polypeptide (Fig. 2A) was isolated from the high-pH insoluble fraction by continuous-elution SDS-PAGE following the method of Nagdas et al. [19] for the proteomic identification. Proteomic identification of the OMC45 polypeptide by MALDI-TOF-TOF analysis yielded 8 peptides (Fig. 2B) that matched the NCBI database sequence of Tektin 3 protein (Bos Taurus).
Figure 1.
Phase contrast photomicrograph showing intact ejaculated bovine sperm (A) and enriched sperm head fraction isolated from sonicated spermatozoa (B), employed in OMC isolation. Electron micrograph demonstrating purity of isolated OMC fraction (C) X26000. Silver-stained SDS-polyacrylamide gel (D). Lane 1 shows the pattern of polypeptides in the total OMC fraction (T). Lane 2 shows that the 38-19kDa family of polypeptides is released to the supernatant fraction (S) following high-pH extraction. Lane 3 shows that the high molecular weight polypeptides of the OMC complex remain in the pellet (P) fraction after high-pH extraction.
Figure 2.
OMC45 polypeptide was identified by Coomassie blue staining (A) and was subjected to MALDI-TOF-TOF analysis at the UNC School of Medicine Proteomic Center, Chapel Hill, NC. Proteomic analysis of OMC45 by MALDI-TOF-TOF yielded 8 peptides (B) that matched the NCBI database sequences of Tektin 3.
Immunofluorescence Localization of OMC45 Polypeptide
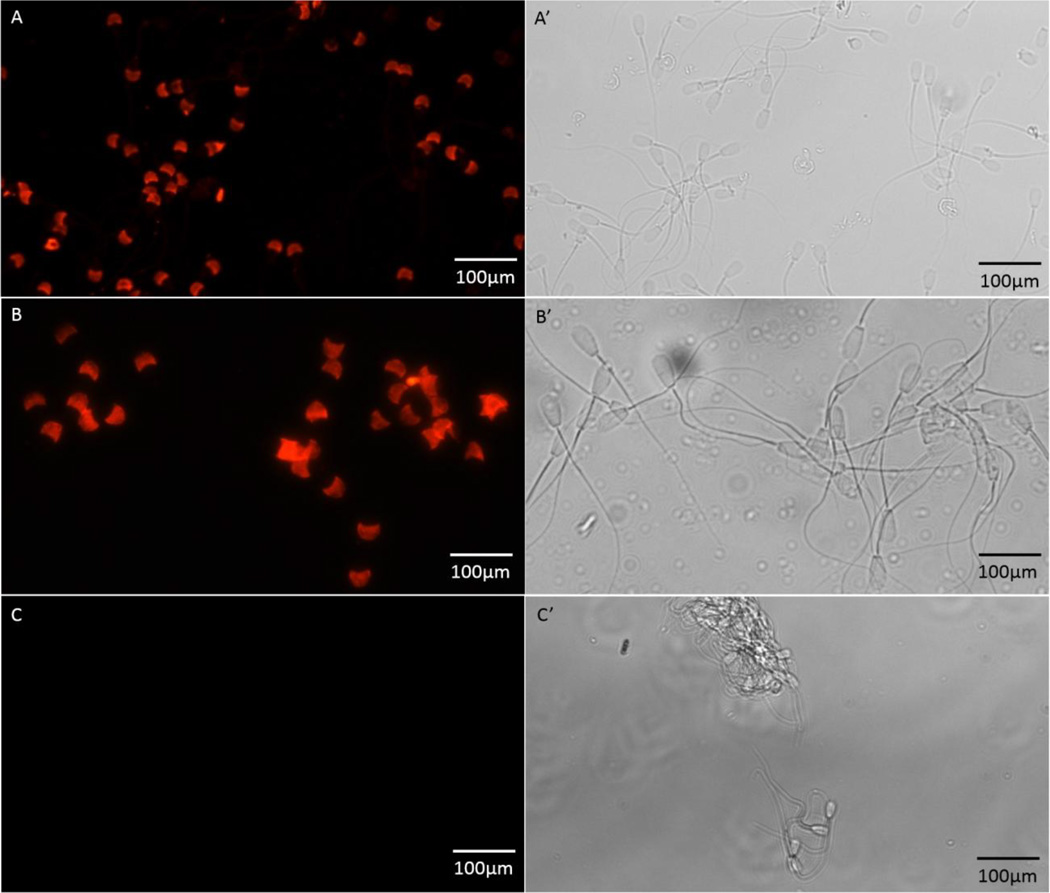
Takiguchi et al. [31] showed that TEKT3 is a constituent of the rat sperm midpiece as well as the acrosomal membrane and proposed that TEKT3 may be involved in acrosome-related events. Using both TEKT3 and OMC45 antibodies, we examined the immunological localization of OMC45 polypeptide. Triton X-100-permeabilized cauda sperm exhibited intense staining of the acrosomal segment both with anti-OMC45 (Fig. 3 A and A’) and anti-TEKT3 (Fig. 3 B and B’). No staining of the equatorial segment or postacrosomal segment of the head was observed. No stain was evident with preimmune serum (Fig. 3 C and C’). This study demonstrates that both anti-TEKT3 and anti-OMC45 antibodies localize OMC45 polypeptide and TEKT3 polypeptide to the anterior segment of the acrosome.
Figure 3.
Paired phase-contrast (A’, B’, and C’) and fluorescence images (A, B, and C) of bovine cauda sperm immunostained with anti-OMC45 (A and A’), anti-TEKT3 (B and B’) antibodies and preimmune serum (C and C’). Cauda spermatozoa were permeabilized with Triton X-100 and then immunostained with antibodies. They show intense fluorescence of the acrosomal segment. Both the equatorial segment and the postacrosomal segment are negative.
Solubilization of OMC45 Polypeptide
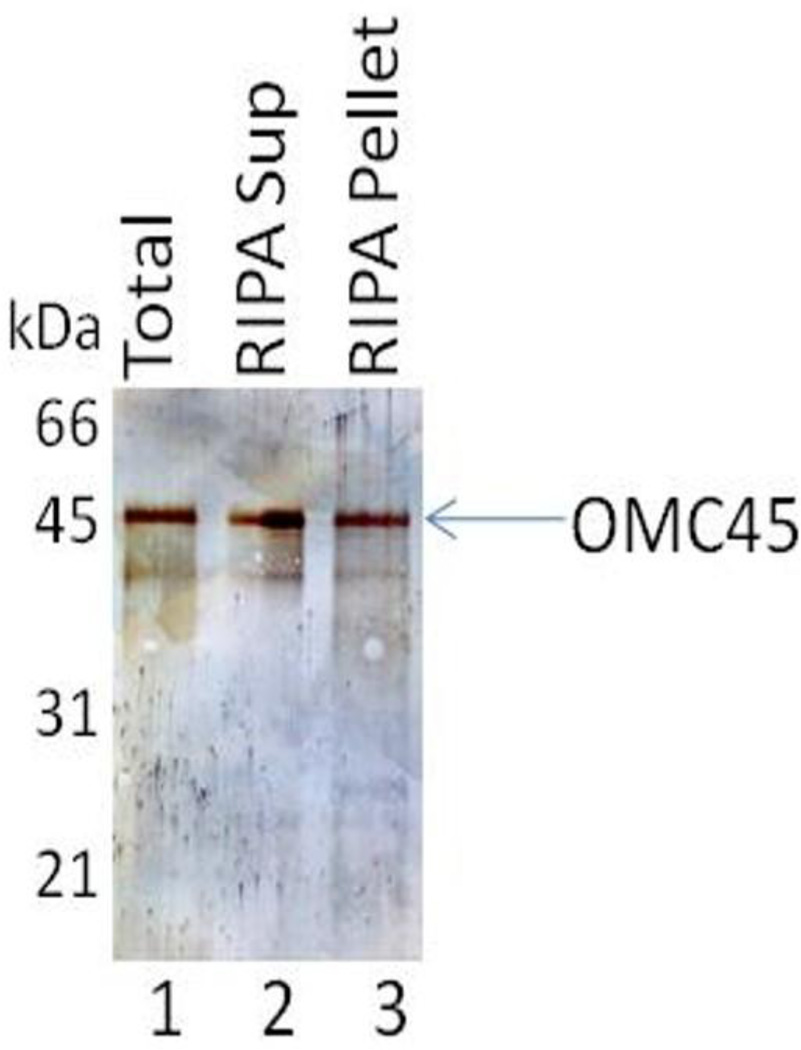
Since OMC45 polypeptide remains insoluble in the particulate fraction after high-pH extraction (20), we tested the solubility properties of OMC45 polypeptide in the high-pH pellet fraction (Fig. 4, lane 1) in RIPA buffer. Our data revealed that a portion of the OMC45 polypeptide was solubilized by RIPA buffer (Fig. 4, lane 2) and the remaining OMC45 polypeptide was present in the particulate fraction (Fig. 4, lane 3).
Figure 4.
Immunoblot showing the solubility properties of OMC45 polypeptide (lane 1) in RIPA buffer. A portion of the OMC45 polypeptide was solubilized by RIPA buffer (lane 2) and the remaining OMC45 polypeptide was present in the particulate fraction (lane 3).
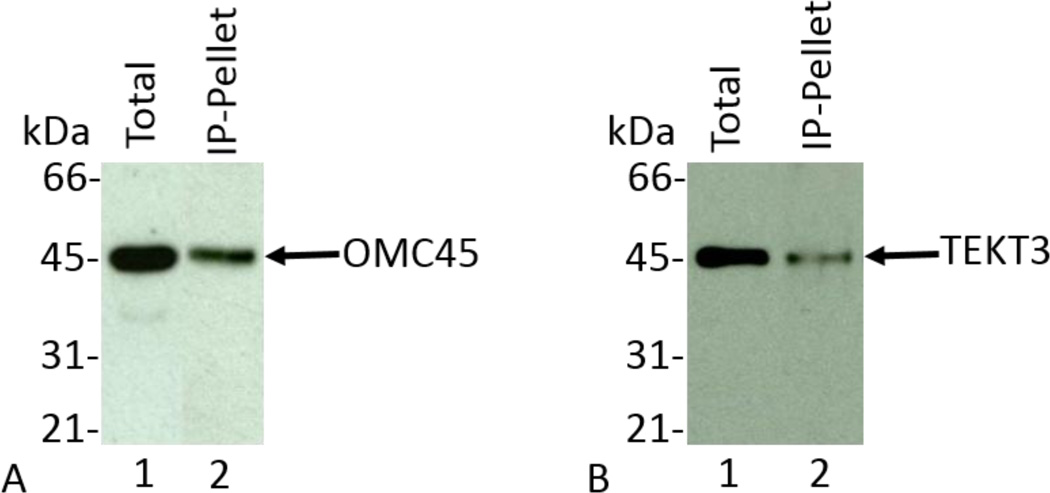
Immunoprecipitation Analysis of OMC45 Polypeptide
Immunoprecipitation analyses were performed to examine the biochemical co-localization of TEKT3 utilizing anti-OMC45 antibody. The supernatant obtained after RIPA buffer extraction (lane 1 of Figs. 5A and 5B) was subjected to immunoprecipitation analysis. As shown in figure 5A, lane 2, the recovery of OMC45 polypeptide was achieved in the anti-OMC45 immunoprecipitation pellet. When an identical blot was stained with anti-TEKT3 antibody, OMC45 polypeptide was immunologically detected in the anti-OMC45 immunoprecipitation pellet (Fig. 5B, lane 2). Our immunofluorescence and biochemical studies strongly confirm the proteomics identification of OMC45 polypeptide, which exhibits the sequence similarity to TEKT3 protein.
Figure 5.
The supernatant obtained after RIPA buffer extraction was subjected to immunoprecipitation analysis. Total (Fig. 5A, lane 1), and IP- Pellet (Fig. 5A, lane 2) fractions were analyzed by Western blot analyses and immunostained with anti-OMC45 antibody. As shown in figure 5A lane 2, OMC45 polypeptide was recovered in the anti-OMC45 immunoprecipitation pellet. Another immunoblot having the same total (Fig. 5B, lane 1), and IP-Pellet (Fig. 5B, lane 2) fractions were stained with anti-TEKT3 antibody. The presence of a TEKT3 band was exhibited in the anti-OMC45 IP-Pellet (Fig. 5B, lane 2).
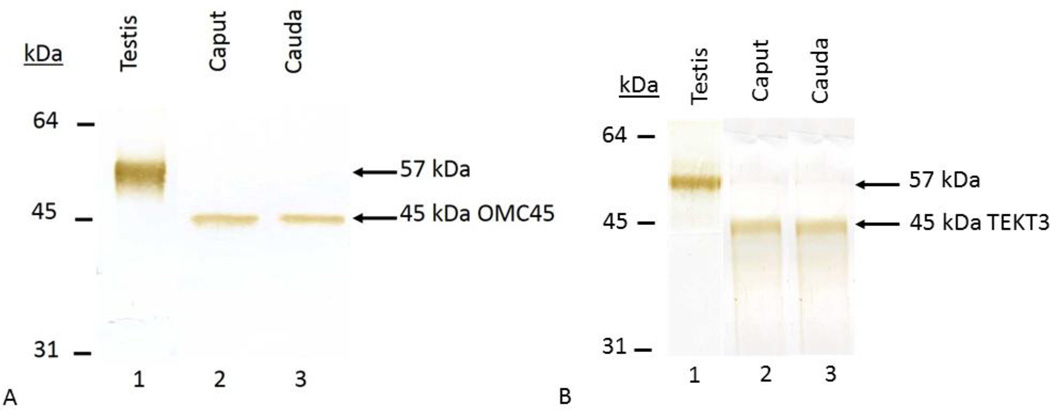
Biochemical Localization of OMC45 Polypeptide in Testis, Caput and Cauda Epididymal Spermatozoa
Bovine testes, caput and cauda spermatozoa were homogenized in a Tris-saline-protease inhibitor solution (TNI), composed of 150mM NaCl, 25mM Tris-HCl (pH 7.5), 2mM benzamidine, 1µg/ml leupeptin, 1µg/ml pepstatin, 1mM NaF, 1mM sodium orthovanadate and 0.05% sodium azide. An equal amount of protein of each homogenized fraction was examined by Western blot analysis stained with anti-OMC45 and anti-TEKT3 antibodies. The testicular fraction possessed a 57kDa (apparent molecular weight) polypeptide (Fig. 6A, lane 1) that reacted with anti-OMC45 polyclonal antibody. Total lysates of caput (Fig. 6A, lane 2) and cauda (Fig. 6A, lane 3) spermatozoa exhibited the presence of a 45kDa immunoreactive polypeptide. When an identical blot was stained with anti-TEKT3 antibody, a 57kDa band was observed in the testicular fraction (Fig. 6B, lane 1) and a 45kDa TEKT3 immunorecative band was present both in caput (Fig. 6B, lane 2) and cauda (Fig. 6B, lane 3) sperm lysates. No band was seen when an identical blot was stained with preimmune serum (data not shown). These data suggest the 57kDa polypeptide of the testis represent a precursor polypeptide that is processed during spermatogenesis into a mature OMC45/TEKT3 polypeptide. It is also noted that no change in molecular form of OMC45 polypeptide was observed during epididymal transit.
Figure 6.
Immunoblot showing total testis lysate (lane 1), total caput (lane 2) and cauda (lane 3) sperm lysates stained with anti-OMC45 (Fig. 6A) and anti-TEKT3 (Fig. 6B) antibodies. Each lane was loaded with 2.5µg of protein.
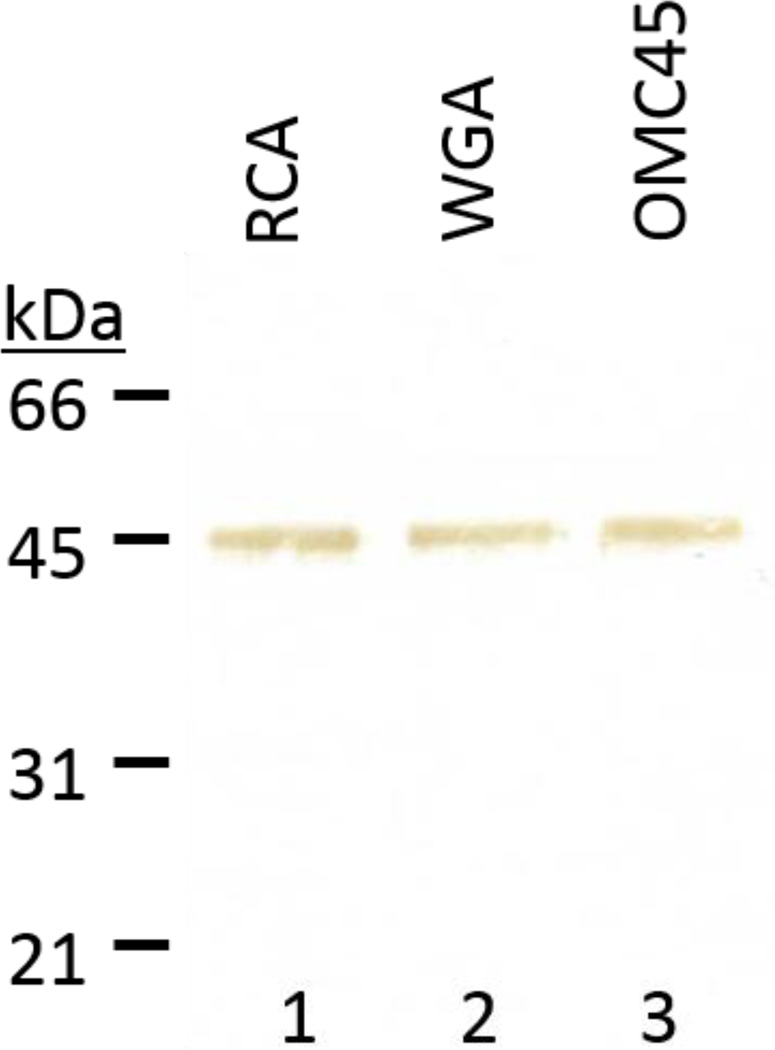
OMC45 Polypeptide is a Glycoprotein
Western blot of purified OMC45 polypeptide was stained with biotinylated RCA and WGA to examine whether OMC45 polypeptide is a glycoconjugated protein. OMC45 polypeptide stained intensely with both RCA (Fig. 7, lane 1) and WGA (Fig. 7, lane 2). No staining was observed when biotinylated RCA and WGA were pre-incubated with D-galactose and N-acetylglucosamine, respectively (data not shown) demonstrating the specificity of the RCA and WGA glycoproteins staining.
Figure 7.
The OMC45 polypeptide was analyzed by Western blot analyses and stained with RCA and WGA biotinylated lectins and with anti-OMC45 antibody. OMC45 polypeptide binds both RCA (lane 1) and WGA (lane 2). Lane 3 immunostained with anti-OMC45 antibody showed the presence of a 45kDa band. Lectin blot analyses demonstrate that OMC45 is a glycoprotein. Each lane was loaded with 3µg of protein.
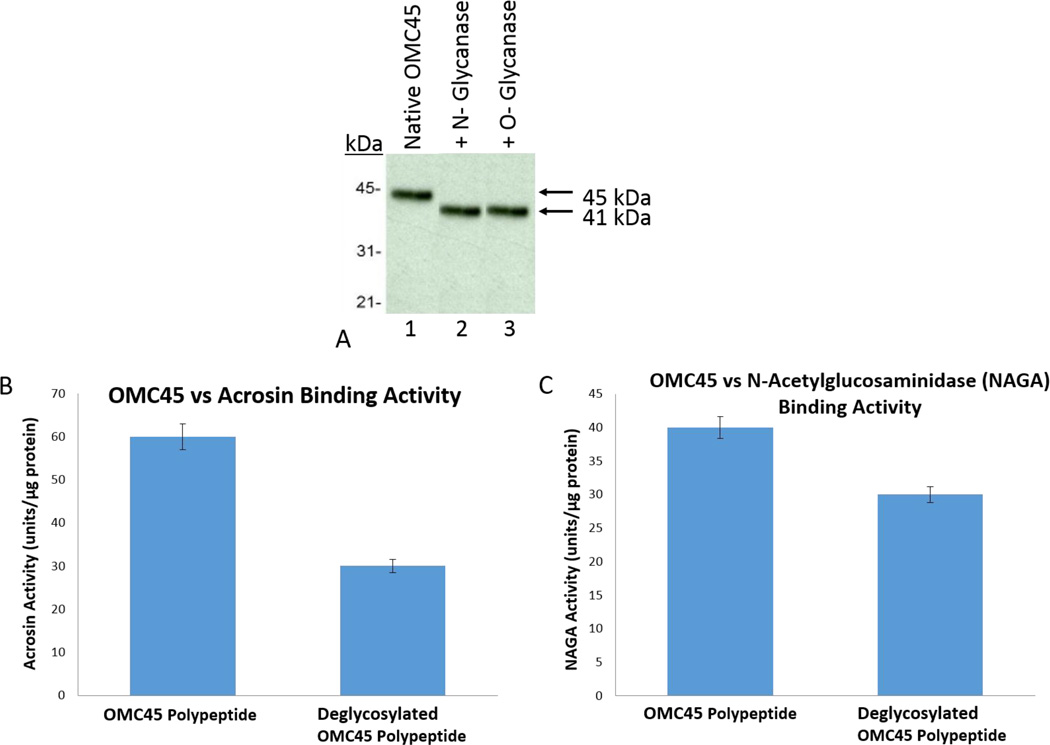
Evidence for the Presence of N-linked and O-linked Sugars on OMC45 Polypeptide
To identify N-linked and O-linked oligosaccharides, OMC45 polypeptide was treated with N-Glycanase and O-Glycanase separately, followed by SDS-PAGE and immunoblot analysis. Compared to native OMC45 polypeptide (Fig. 8A, lane 1), both N-Glycanase (Fig. 8A, lane 2) and O-Glycanase (Fig. 8, lane 3) treated deglycosylated OMC45 polypeptide migrated with an apparent molecular weight of 41kDa. Our results reveal that OMC45 possesses approximately 4kDa of N-linked and approximately 4kDa of O-linked oligosaccharides.
Figure 8.
A: Solubilized OMC45 polypeptide (lane 1) was treated with N-glycanase (lane 2) and O-glycanase (lane 3) separately following the manufacturer’s instructions. After enzymatic digestion, deglycosylated OMC45 polypeptide was analzed by Western blot stained with anti-OMC45. Our results reveal that OMC45 possesses ~4kDa N-linked and ~4kDa O-linked oligosaccharides.
B and C: After enzymatic digestion, deglycosylated OMC45 polypeptide and native OMC 45 polypeptide were conjugated to AminoLink Plus coupling gel (Pierce Chemical Co.) separately at pH 10.0, according to the manufacturer’s instructions. As a control, same units of N-glycanase and O-glycanase were conjugated to AminoLink Plus coupling gel. Bovine epididymal cauda spermatozoa were extracted in high-salt Triton X-100 solution. The supernatant obtained after centrifugation, that contained hydrolase activities, was dialyzed against 25mM Tris-HCl buffer, pH 7.5, incubated with deglycosylated and native OMC45 polypeptide, and N-Glycanase/O-Glycanase conjugated beads separately overnight at 4°C. After centrifugation at 1000 ×g for 5 min at 4°C, all three pellet fractions were assayed for acrosin (Fig. 8B) and NAGA (Fig. 8C). Values represent mean ± SD of three experiments. One unit of acrosin is the quantity of enzyme required to hydrolyze one µM of substrate per min at 25°C. One unit of NAGA is the quantity of enzyme required to release one mM of p-nitrophenol per hr at 25°C.
Demonstration of Acrosin and NAGA Binding to Deglycosylated and Native OMC45 Polypeptide
A sedimentation assay was employed to determine the binding efficiency of deglycosylated OMC45 to acrosin (Fig. 8B) and NAGA (Fig. 8C). Deglycosylated OMC45 revealed a noticeable reduction in both acrosin (~50% reduction) and NAGA (~25% reduction) binding in comparison to native OMC45 polypeptide binding to acrosin and NAGA. In contrast, N-Glycanase/O-Glycanase conjugated beads, used as control for specificity, did not bind acrosin and NAGA (data not shown), demonstrating the specificity of binding of hydrolases to native and deglycosylated OMC45 polypeptide. Our results suggest that carbohydrate moieties play an important role in hydrolase binding.
Biochemical and Immunocytochemical Localization of OMC45 Polypeptide during Capacitation and the Acrosome Reaction
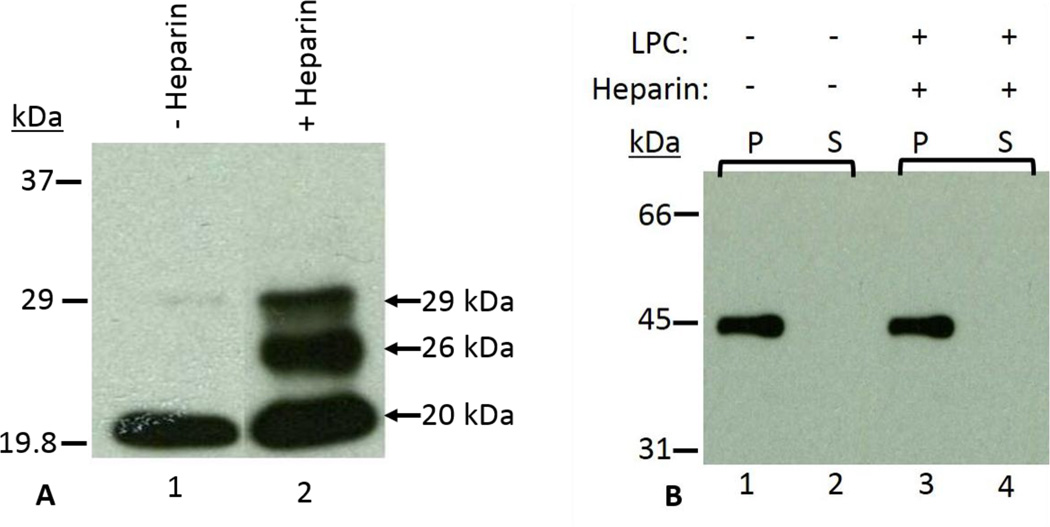
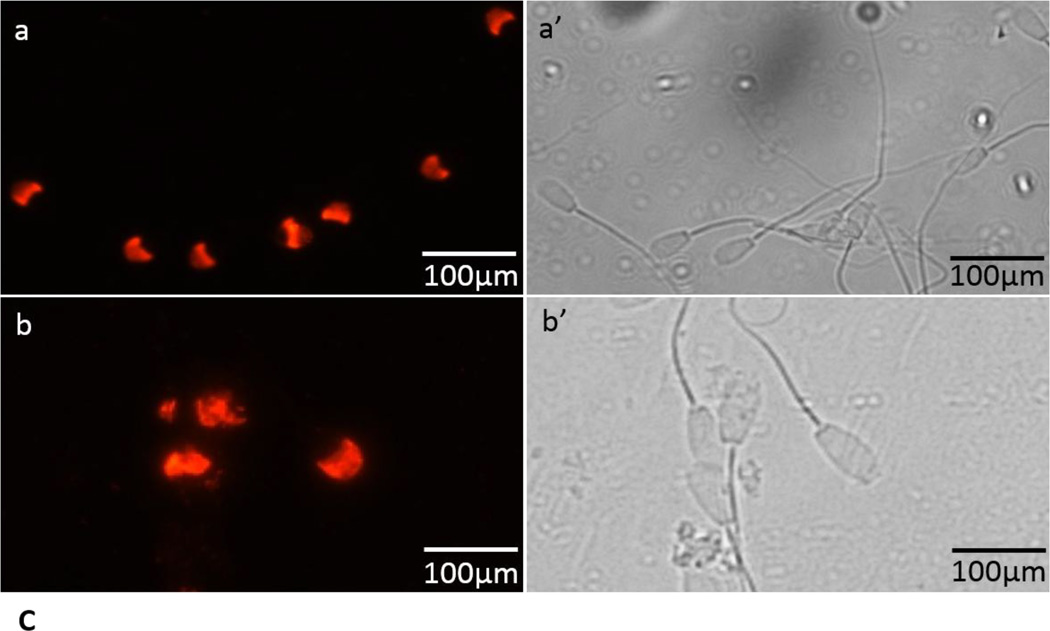
Protein tyrosine phosphorylation is a key biochemical event accompanying sperm capacitation [32–34]. To examine whether capacitation successfully occurred under our current conditions, we analyzed our capacitated sperm fraction with immunoblots stained with anti-phosphotyrosine antibody (Fig. 9A). We observed that the 29 and 26kDa polypeptides were phosphorylated during capacitation (Fig. 9A, lane 2). Since the 20kDa phosphorylated polypeptide was present in both the control (Fig. 9A, lane 1) and capacitated (Fig. 9A, lane 2) sperm fractions, it suggests that the 20kDa is a non-specific band. Phosphorylation of the bovine cauda sperm polypeptide during capacitation confirms the efficacy of our capacitation protocol. Acrosin levels were analyzed in the pellet and supernatant fractions of non-capacitated and acrosome reacted spermatozoa to confirm the occurrence of an LPC-induced acrosome reaction. As shown in Table-1, almost 90% acrosin activity remained in the pellet fraction of non-capacitated spermatozoa whereas the addition of LPC to heparin-capacitated sperm exhibited the release of ~81% of total acrosin into the supernatant fraction. All results demonstrate the competency of our acrosome reaction model. Previously, comparable results were also reported in bovine ejaculated and cauda epididymal spermatozoa (20, 35). Immunoblot analysis was employed to examine whether OMC45 polypeptide is retained or released following capacitation and the acrosome reaction. As demonstrated in figure 9B, the OMC45 polypeptide was retained in the sperm pellets (Fig. 9B, lanes 1 and 3) after LPC-induced acrosome reaction; no immunoreactive polypeptides were detected in the supernatant fractions (Fig. 9B, lanes 2 and 4). LPC induced acrosome reacted sperm were permeabilized with Triton X-100 and immunostained with anti-OMC45 antibody to delineate the fate of OMC45 polypeptide by immunofluorescence localization. All positive staining remained associated with the hybrid membrane complex over the apical and principal segments of the acrosome (Fig. 9C, panel b). It is also noted that non-capacitated spermatozoa exhibited intense staining of the acrosomal segment (Fig. 9C, panel a). These studies suggest that the OMC45 polypeptide remains associated to the particulate fraction even after the release of acrosomal contents (acrosomal exocytosis).
Figure 9.
A: Demonstates the detection of phosphorylation in capacitated sperm fractions. X-ray film of capacitated sperm fractions incubated in the absence and presence of heparin. Sperm fractions incubated in the presence of heparin (lane 2) underwent phosphorylation, as opposed to those incubated in the absence of heparin (lane 1). These results provide biochemical evidence that the capacitation reaction was properly executed.
B: Immunoblot analysis of the pattern of OMC45 polypeptide in the particulate (P) and supernatant (S) fractions of non-capacitated (lanes1 and 2) and LPC-induced capacitated (lanes 3 and 4) spermatozoa. The immunoblot was stained with anti-OMC45 antibody. Note that under each treatment condition the OMC45 polypeptide remains in the pellet fraction (lanes 1 and 3), no stained band was observed in the supernatant (lanes 2 and 4).
C: Matched phase contrast (panels a’ and b’) and fluorescence (panels a and b) photomicrographs of non-capacitated sperm (a and a’) and sperm capacitated 4 hrs in the presence of heparin and exposed to LPC for 15 min (b and b’). Note that acrosome-reacted sperm show specific staining of the acrosomal segment.
Table-1.
Distribution of Total Acrosin of Non-Capacitated and Acrosome-Reacted Spermatozoa
| Non-Capacitated Sperm | Acrosome-Reacted Sperm | |
|---|---|---|
| Pellet | 90 ± 8 | 19 ± 10 |
| Supernatant | 10 ± 6 | 81 ± 7 |
The distribution of acrosin between pellet and supernatant fractions is shown in Table-1. Only acrosome-reacted sperm exhibit substantial acrosin release to the supernatant fraction. Values represent mean ± SD of three experiments.
Discussion
The acrosomal segment of spermatozoon plays key roles in the recognition, binding, and penetration of the egg investments [1]. The outer acrosomal membrane possesses the restricted domain that fuses with the overlying plasma membrane during the acrosome reaction. It has been reported that neither the outer acrosomal membrane of the equatorial segment nor the inner acrosomal membrane participate in the membrane fusion event. Our previously published studies suggested that the OMC provides the maintenance of acrosome assembly, the segregation of hydrolases within the acrosome, and the regulation of acrosomal hydrolases during the acrosome reaction [13,19, 20]. First, we isolated and characterized a localized, stable acrosomal matrix assembly from the bovine acrosome termed the outer acrosomal membrane-associated matrix complex (OMC) [13, 18] and this stable matrix is restricted to the apical and principal segments of the acrosome where it is associated with the luminal surface of the outer acrosomal membrane [13, 18]. Second, we demonstrated that OMC exhibits specific binding activity for acrosin [13] and NAGA [19]. Third, a highly purified OMC fraction is comprised of three major (54, 50, and 45kDa) and several minor (38-19kDa) polypeptides. The set of polypeptides (38-19kDa) termed “rpf” is selectively solubilized by extraction of OMC at pH 10.5, while the remaining three major polypeptides (55, 50 and 45kDa) remain associated with the sedimentable “stripped” OMC [20]. Fourth, we purified a 45kDa polypeptide from the high-pH insoluble fraction by continuous-elution SDS-PAGE. Anti-OMC45 polyclonal antibody reacts strongly on immunoblots with the OMC45 band. Using immunofluorescence, anti-OMC45 localizes specifically to the acrosomal cap [19]. Acrosin and NAGA bound OMC32 polypeptide in a concentration-dependent fashion. In contrast, OMC45 polypeptide exhibited stronger affinity to acrosin than NAGA [19]. Using biochemical, immunological, and proteomic analyses we demonstrated the molecular characterization of OMC45 polypeptide. Proteomic analysis of OMC45 polypeptide by MALDI-TOF-TOF yielded 8 peptides that matched the NCBI database sequence of Tektin3 (TEKT3).
Tektins are intermediate filament (IF) proteins [36–38] and nuclear lamins [39–41] localized in cilia and flagella. Five Tektin proteins (TEKT1-5) have been identified in mammals [42–49]. In situ hybridization analyses of mice testes revealed the expression of TEKT3 mRNA particularly in late pachytene and early round spermatids [46]. It has been observed that TEKT3-null mice generate sperm with reduced motility and forward progression as well as increased flagellar structural bending defects. They proposed that sperm motility is regulated by TEKT3 via a distinct mechanism [50]. Takiguchi et al. [31] investigated the sub-cellular localization of TEKT3 in rat sperm at the ultra-structure level using a polyclonal antibody that is raised against an unique hydrophobic amino acid sequence of rat TEKT3 (RKTQADSTQN), and this specific amino acid sequence is not present in other Tektin family proteins. All ultra-structural and biochemical results demonstrate the association of TEKT3 with the surface of mitochondria and outer dense fibers in the middle piece, and not directly associated with the flagellar axoneme. In addition, they found that TEKT3 was present at the equatorial region of the acrosomal membrane in rat sperm heads. They also proposed that TEKT3 may be involved in acrosome-related events. The presence and the functions of other Tektin family member proteins in sperm have also been illustrated. In bull and mouse sperm, TEKT1 was localized in the tail as well as the apical region of the acrosomal cap. In mice, loss of acrosome-associated TEKT1 was observed after the in vitro acrosome reaction [51]. TEKT2 is present at the surface of outer dense fibers (ODFs) and may be involved in flagellum stability and sperm motility [52]. TEKT2-null sperm exhibited flagella bending and reduced motility []). Recently, Yamaguchi et al. [54] identified a 36kDa TEKT2-binding protein1 associated with mitochondria of rat sperm flagella. It has been reported that Tektin 2 of hamster sperm becomes tyrosine phosphorylated during in vitro capacitation [55]. TEKT4 is associated with outer dense fibers, not with axonemal tubulins of rat and mouse spermatozoa [48]. Roy et al. [50] found significant reduction of forward progressive velocity in TEKT4-null mouse sperm. Murayama et al. [49] reported the localization of TEKT5 at the surface of mitochondrial sheaths in rat sperm flagella. The high expression of TEKT5 mRNA was observed during late stages of spermiogenesis and TEKT5 protein was localized throughout the sperm tail [56]. It has been suggested that TEKT5 plays an important role in flagella formation during spermiogenesis and involved in sperm motility. Thus, several studies have proposed the localization and putative role of each Tektin protein in mammalian spermatozoa. However, the precise function of each Tektin protein is not clearly elucidated. In the current study, using the rat TEKT3 polyclonal antibody, we observed the localization of anti-TEKT3 specifically to the acrosomal cap of bovine cauda epididymal sperm using immunofluorescence microscopy. No staining of the tail was observed. Thus, our biochemical and immunofluorescence studies strongly confirm the proteomic identification of OMC45 polypeptide; that it exhibits a sequence similarity to TEKT3. Only two TEKT family members (TEKT1 and TEKT3) are reported in mammalian sperm acrosome [31]. We are the first to report the presence of Tektin 3 in detergent-resistant acrosomal matrix in mammalian spermatozoa. We propose potential roles of TEKT3 as being a detergent (Triton X-100) resistant acrosomal matrix structural element in the bovine sperm acrosome. Our data also suggest the participation of TEKT3 in the regulation of hydrolases released during the acrosome reaction. In addition, it may be possible that the acrosomal matrix TEKT3 may be involved in the segregation of acrosomal hydrolases and other matrix polypeptides within the acrosome interior.
We now demonstrate the presence of a precursor form (~57kDa) of the OMC45 in the total testicular lysate. This result allows us to suggest that the OMC45 polypeptide is synthesized in the testis in a high molecular weight precursor form which undergoes processing, presumably in the testicular germ cells. Previously, it has been shown that two major acrosomal matrix proteins (29kDa and 22kDa) in hamster caput and cauda epidiydmal spermatozoa are structurally related and appear to arise from a common 40kDa precursor protein in round spermatids [57]. In other species, including the baboon, human, and mouse, the testicular forms of SP10 or SP10-related polypeptides are high-molecular weight precursors of the mature polypeptides found in epididymal spermatozoa [58–61]. In bovine sperm, Olson et al. [20] demonstrated that a 32kDa acrosomal protein, one of the members of “rpf”, exhibits sequence homology to SP-10 proteins and the Western blot analysis of total testicular lysate stained with anti-OMC32 antibody showed two (50.5 and 48kDa) high molecular weight precursor forms (unpublished data). Although several high molecular weight precursor forms of mSP10 of mouse testis [58] and “rpf” family of bovine testis have been identified, Olson et al. [57] found only a single immunologically reactive band of 40KDa in hamster testis. In the present study, a 57kDa (apparent molecular weight) precursor form of OMC45/TEKT3 polypeptide was observed in bovine testis. This may reflect species specific differences of polypeptides. Other intermediate forms of OMC45/TEKT3 polypeptide in the bovine testis could be identified as more mature spermatids or testicular spermatozoa are examined. Additional studies with the isolated germ cells will be needed to determine the stage-specific synthesis and processing of the acrosomal matrix polypetide. Moreover, it is possible that size processing of the precursor polypeptide is required for the assembly of the insoluble matrix elements and for the binding of the acrosomal matrix elements to the outer acrosomal membrane. Future studies will address these issues. The deduced molecular weight of Bos Taurus Tektin 3 having 490 amino acids is 56607, in close agreement to the noted apparent molecular weight (57,000) of the testicular precursor form of OMC45 polypeptide.
Our lectin blot and glycohydrolases-treated results revealed that OMC45 is a glycoprotein. Previously, we have shown by two-dimensional PAGE that OMC45 polypeptide possesses a charge-variant pattern with 5–6 isoforms which may represent glycosylation variants [19]. Thus, our current observation that OMC45 polypeptide contains ~4kDa of each N-linked and O-linked oligosaccharides strongly supports that the presence of charge-variants of OMC45 polypeptide are due to the post-translational glycosylation of apoprotein. Whether other post-translational modifications such as sulfation, nitration, or phosphorylation also contribute to charge heterogenetity of OMC45 remains to be investigated. Previously, we have demonstrated that the OMC45 polypeptide exhibited stronger binding affinity to acrosin than NAGA [19]. In the current study, we show that the oligosaccharide moieties of OMC45 polypeptide play important roles in acrosomal hydrolase binding. A significant reduction of acrosin binding (~50% reduction) to deglycosylated OMC45 polypeptide was observed in comparison to native OMC45 polypeptide. NAGA’s lower binding affinity to native OMC45 polypeptide [19] could be the plausible explanation for less binding reduction of NAGA (~25% reduction) in comparison to acrosin with carbohydrate-stripped OMC45 polypeptide. Although we have shown that the carbohydrate moieties of OMC45 polypeptide are involved in acrosin/NAGA binding, the current study will not rule out whether N-linked or O-linked oligosaccharides alone are involved in hydrolase binding. Even if both oligosaccharide moieties of OMC45 polypeptide are involved in binding, would there be any differential binding affinity of N-linked and O-linked oligosaccharides to acrosin/NAGA? Future studies will better define the role of both N-linked and O-linked oligosaccharides of OMC45 polypeptide in hydrolase binding.
Following capacitation and the acrosome reaction, some matrix polypeptides are proteolytically processed and released along with the acrosomal contents [62–64], whereas the retention of other acrosomal matrix proteins with the hybrid membrane complex was previously noted [65, 66]. Human SP-10 polypeptide fraction remains associated with various acrosomal structures following the ionophore-induced acrosome reaction [67]. Previously, we have shown that bovine OMCrpf polypeptides [20] and PRDX5 polypeptide [35] remained associated with the hybrid membrane complex. Our current data clearly reveal that the bovine acrosomal matrix OMC45 polypeptide is exclusively localized to the hybrid membrane complex and remains in the particulate following the acrosome reaction. The association of OMC45 polypeptide to a stable acrosomal matrix assembly suggests that this polypeptide plays a significant role in maintaining the structural integrity of the hybrid membrane complex generated by the membrane fusion process of the acrosome reaction. We propose that OMC45 polypeptide having sequence similarity to TEKT3 polypeptide is one of the constituents of stable intra-acrosomal framework of the bovine sperm acrosome and it plays an important role in the regulation of hydrolases released during the acrosome reaction. It may have a potential role in the segregation of acrosomal hydrolases and other matrix polypeptides.
Acknowledgements
Supported by NIH/NIGMS/ 1SC3GM096875-04, NSF HBCU-UP #1036257, and FSU RISE Grant
References
- 1.Yanagimachi R. Mammalian Fertilization. In: Knobil E, Neill JD, editors. The physiology of reproduction. New York: Raven Press; 1993. pp. 189–317. [Google Scholar]
- 2.Wassarman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat. Cell Biol. 2001;3:E59–E64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- 3.Primakoff P, Myles DG. Penetration, adhesion, and fusion in mammalian sperm-egg interaction. Science. 2002;296:2183–2185. doi: 10.1126/science.1072029. [DOI] [PubMed] [Google Scholar]
- 4.Kopf GS, Gerton GL. The mammalian sperm acrosome and the acrosome reaction. In: Wassarman PM, editor. Elements of Mammalian Fertilization. Boca Raton, Ann Arbor, Boston: CRC Press; 1991. pp. 153–203. [Google Scholar]
- 5.Huang TTF, Hardy DM, Yanagimachi H, Teuscher C, Tung K, Wild G, Yanagimachi R. pH and protease control of acrosomal stasis and release during the guinea pig sperm acrosome reaction. Biol. Reprod. 1985;32:451–462. doi: 10.1095/biolreprod32.2.451. [DOI] [PubMed] [Google Scholar]
- 6.Talbot P, DiCarlantonio G. Cytochemical localization of dipeptidyl peptidase II (DPP-II) in mature guinea pig sperm. J. Histochem. Cytochem. 1985;33:1169–1172. doi: 10.1177/33.11.4056380. [DOI] [PubMed] [Google Scholar]
- 7.DiCarlantonio G, Talbot P. Evidence for sequential deployment of secretory enzymes during the normal acrosome reaction of guinea pig sperm in vitro. Gamete Res. 1988;21:425–438. doi: 10.1002/mrd.1120210410. [DOI] [PubMed] [Google Scholar]
- 8.Hyatt H, Gwatkin RBL. Characterization of isolated acrosomal matrices from hamster spermatozoa. J. Reprod. Fert. 1988;83:419–429. doi: 10.1530/jrf.0.0830419. [DOI] [PubMed] [Google Scholar]
- 9.Olson GE, Winfrey VP, Davenport GR. Characterization of matrix domains of the hamster acrosome. Biol. Reprod. 1988;39:1145–1158. doi: 10.1095/biolreprod39.5.1145. [DOI] [PubMed] [Google Scholar]
- 10.Noland TD, Davis LS, Olson GE. Regulation of proacrosin conversion in isolated guinea pig sperm acrosomal apical segments. J. Biol. Chem. 1989;264:13586–13590. [PubMed] [Google Scholar]
- 11.Hardy DM, Oda MN, Friend DS, Huang TTF. A mechanism for differential release of acrosomal enzymes during the acrosome reaction. Biochem. J. 1991;275:759–766. doi: 10.1042/bj2750759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NagDas SK, Winfrey VP, Olson GE. Identification of hydrolase binding activities of the acrosomal matrix of hamster spermatozoa. Biol. Reprod. 1996;55:1405–1414. doi: 10.1095/biolreprod55.6.1405. [DOI] [PubMed] [Google Scholar]
- 13.NagDas SK, Winfrey VP, Olson GE. Proacrosin-acrosomal matrix binding interactions in ejaculated bovine spermatozoa. Biol. Reprod. 1996;54:111–121. doi: 10.1095/biolreprod54.1.111. [DOI] [PubMed] [Google Scholar]
- 14.Baba T, Niida Y, Michikawa Y, Kasiwabara S, Kodaira K, Takenaka M, Kohno N, Gerton GL, Arai Y. An acrosomal protein, sp32, in mammalian sperm is a binding protein specific for two proacrosins and an acrosin intermediate. J. Biol. Chem. 1994;269:10133–10140. [PubMed] [Google Scholar]
- 15.Holt WV. Development and maturation of the mammalian acrosome. A cytochemical study using phosphotungstic acid staining. J. Ultrastruct. Res. 1979;68:58–71. doi: 10.1016/s0022-5320(79)90142-4. [DOI] [PubMed] [Google Scholar]
- 16.Green DPL. The activation of proteolysis in the acrosome reaction of guinea pig sperm. J. Cell Sci. 1978;32:153–164. doi: 10.1242/jcs.32.1.153. [DOI] [PubMed] [Google Scholar]
- 17.Nuzzo NA, Anderson RA, Zaneveld LJD. Proacrosin activation and acrosin release during the guinea pig acrosome reaction. Mol. Reprod. Dev. 1990;25:52–60. doi: 10.1002/mrd.1080250110. [DOI] [PubMed] [Google Scholar]
- 18.Olson GE, Winfrey VP, Garbers DL, Noland TD. Isolation and characterization of a macromolecular complex associated with the outer acrosomal membrane of bovine spermatozoa. Biol. Reprod. 1985;33:761–779. doi: 10.1095/biolreprod33.3.761. [DOI] [PubMed] [Google Scholar]
- 19.Nagdas SK, Hamilton SL, Raychoudhury S. Identification of acrosomal matrix-specific hydrolases binding proteins of bovine cauda epididymal spermatozoa. J Androl. 2010;31:177–187. doi: 10.2164/jandrol.108.007146. [DOI] [PubMed] [Google Scholar]
- 20.Olson GE, Winfrey VP, Neff JC, Lukas TJ, NagDas SK. An antigenically related polypeptide family is a major structural constituent of a stable acrosomal matrix assembly in bovine spermatozoa. Biol. Reprod. 1997;57:325–334. doi: 10.1095/biolreprod57.2.325. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairbanks G, Steck TL, Wallach DFH. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 24.Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- 25.NagDas SK, Winfrey VP, Olson GE. Identification of a hamster epididymal region-specific secretory glycoprotein that binds nonviable spermatozoa. Biol. Reprod. 2000;63:1428–1436. doi: 10.1095/biolreprod63.5.1428. [DOI] [PubMed] [Google Scholar]
- 26.Parrish JJ, Susko-Parrish J, Winer MA, First NL. Capacitation of bovine sperm by heparin. Biol. Reprod. 1988;38:1171–1180. doi: 10.1095/biolreprod38.5.1171. [DOI] [PubMed] [Google Scholar]
- 27.Lottenberg R, Christensen U, Jackson CM, Coleman PL. Assay of coagulation proteases using peptide chromogenic and fluorogenic substrates. Meth. Enzymol. 1981;80:341–361. doi: 10.1016/s0076-6879(81)80030-4. [DOI] [PubMed] [Google Scholar]
- 28.Miller DJ, Gong X, Shur BD. Sperm require B-N-acetylglucosaminidase to penetrate through the egg zona pellucida. Development. 1993;118:1279–1289. doi: 10.1242/dev.118.4.1279. [DOI] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Olson GE, Winfrey VP. Structure of membrane domains and matrix components of the bovine acrosome. J. Ultrastruct. Res. 1985;90:9–25. doi: 10.1016/0889-1605(85)90113-2. [DOI] [PubMed] [Google Scholar]
- 31.Takiguchi H, Murayama E, Kaneko T, Kurio H, Toshimori K, Iida H. Characterization and subcellular localization of Tektin 3 in rat spermatozoa. Mol. Reprod. Dev. 2011;78:611–620. doi: 10.1002/mrd.21352. [DOI] [PubMed] [Google Scholar]
- 32.Visconti PE, Bailey JL, Moore GD, Pan D, Olds-Clarke P, Kopf GS. Capacitation of mouse spermatozoa. I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development. 1995;121:1129–1137. doi: 10.1242/dev.121.4.1129. [DOI] [PubMed] [Google Scholar]
- 33.Porambo JR, Salicioni AM, Visconti PE, Platt MD. Sperm phosphoproteomics: historical perspectives and current methodologies. Expert Rev Proteomics. 2012;9:533–548. doi: 10.1586/epr.12.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saccary L, She YM, Oko R, Kan FW. Hamster oviductin regulates tyrosine phosphorylation of sperm proteins during in vitro capacitation. Biol Reprod. 2013;89:1–11. doi: 10.1095/biolreprod.113.109314. [DOI] [PubMed] [Google Scholar]
- 35.Nagdas SK, Buchanan T, Raychoudhury S. Identification of peroxiredoxin-5 in bovine cauda epididymal sperm. Mol. Cell. Biochem. 2014;387:113–121. doi: 10.1007/s11010-013-1876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang XJ, Piperno G. Cross-reactivity of antibodies specific for flagellar tektin and intermediate filament subunits. J Cell Biol. 1987;104:1563–1568. doi: 10.1083/jcb.104.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steffen W, Linck RW. Relationship between tektins and intermediate filament proteins: an immunological study. Cell Motil. Cytoskeleton. 1989;14:359–371. doi: 10.1002/cm.970140306. [DOI] [PubMed] [Google Scholar]
- 39.McLachlan AD, Stewart M. Periodic charge distribution in the intermediate filament proteins desmin and vimentin. J Mol. Biol. 1982;162:693–698. doi: 10.1016/0022-2836(82)90396-5. [DOI] [PubMed] [Google Scholar]
- 40.Parry DA, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp. Cell Res. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Goldie KN, Wedig T, Mitra AK, Aebi U, Herrmann H, Hoenger A. Dissecting the 3-D structure of vimentin intermediate filaments by cryo-electron tomography. J Struct. Biol. 2007;158:378–385. doi: 10.1016/j.jsb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Iguchi N, Tanaka H, Fujii T, Tamura K, Kaneko Y, Nojima H, Nishimune Y. Molecular cloning of haploid germ cell-specific tektin cDNA and analysis of the protein in mouse testis. FEBS Lett. 1999;456:315–321. doi: 10.1016/s0014-5793(99)00967-9. [DOI] [PubMed] [Google Scholar]
- 43.Larsson M, Norrander J, Gräslund S, Brundell E, Linck R, Ståhl S, Höög C. The spatial and temporal expression of Tekt1, a mouse tektin C homologue, during spermatogenesis suggest that it is involved in the development of the sperm tail basal body and axoneme. Eur. J Cell Biol. 2000;79:718–725. doi: 10.1078/0171-9335-00097. [DOI] [PubMed] [Google Scholar]
- 44.Xu M, Zhou Z, Cheng C, Zhao W, Tang R, Huang Y, Wang W, Xu J, Zeng L, Xie Y, Mao Y. Cloning and characterization of a novel human TEKTIN1 gene. Int. J Biochem. Cell Biol. 2001;33:1172–1182. doi: 10.1016/s1357-2725(01)00089-9. [DOI] [PubMed] [Google Scholar]
- 45.Wolkowicz MJ, Naaby-Hansen S, Gamble AR, Reddi PP, Flickinger CJ, Herr JC. Tektin B1 demonstrates flagellar localization in human sperm. Biol. Reprod. 2002;66:241–250. doi: 10.1095/biolreprod66.1.241. [DOI] [PubMed] [Google Scholar]
- 46.Roy A, Yan W, Burns KH, Matzuk MM. Tektin3 encodes an evolutionarily conserved putative testicular microtubules-related protein expressed preferentially in male germ cells. Mol. Reprod. Dev. 2004;67:295–302. doi: 10.1002/mrd.20025. [DOI] [PubMed] [Google Scholar]
- 47.Matsuyama T, Honda Y, Doiguchi M, Iida H. Molecular cloning of a new member of TEKTIN family, Tektin4, located to the flagella of rat spermatozoa. Mol. Reprod. Dev. 2005;72:120–128. doi: 10.1002/mrd.20331. [DOI] [PubMed] [Google Scholar]
- 48.Iida H, Honda Y, Matsuyama T, Shibata Y, Inai T. Tektin 4 is located on outer dense fibers, not associated with axonemal tubulins of flagella in rodent spermatozoa. Mol. Reprod. Dev. 2006;73:929–936. doi: 10.1002/mrd.20486. [DOI] [PubMed] [Google Scholar]
- 49.Murayama E, Yamamoto E, Kaneko T, Shibata Y, Inai T, Iida H. Tektin5, a new Tektin family member, is a component of the middle piece of flagella in rat spermatozoa. Mol. Reprod. Dev. 2008;75:650–658. doi: 10.1002/mrd.20804. [DOI] [PubMed] [Google Scholar]
- 50.Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Tektin 3 is required for progressive sperm motility in mice. Mol. Reprod. Dev. 2009;76:453–459. doi: 10.1002/mrd.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oiki S, Hiyama E, Gotoh T, Iida H. Localization of Tektin 1 at both acrosome and flagella of mouse and bull spermatozoa. Zoolog. Sci. 2014;31:101–107. doi: 10.2108/zsj.31.101. [DOI] [PubMed] [Google Scholar]
- 52.Shimasaki S, Yamamoto E, Murayama E, Kurio H, Kaneko T, Shibata Y, Inai T, Iida H. Subcellular localization of Tektin2 in rat sperm flagellum. Zoolog. Sci. 2010;27:755–761. doi: 10.2108/zsj.27.755. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol. Cell. Biol. 2004;24:7958–7964. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi A, Kaneko T, Inai T, Iida H. Molecular cloning and subcellular localization of Tektin2-binding protein 1 (Ccdc 172) in rat spermatozoa. J. Histochem. Cytochem. 2014;62:286–297. doi: 10.1369/0022155413520607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mariappa D, Aladakatti RH, Dasari SK, Sreekumar A, Wolkowicz M, Van Der Hoorn F, Seshagiri PB. Inhibition of tyrosine phosphorylation of sperm flagellar proteins, outer dense fiber protein-2 and tektin-2, is associated with impaired motility during capacitation of hamster spermatozoa. Mol. Reprod. Dev. 2010;77:182–193. doi: 10.1002/mrd.21131. [DOI] [PubMed] [Google Scholar]
- 56.Cao W, Ijiri TW, Huang AP, Gerton GL. Characterization of a novel tektin member, TEKT5, in mouse sperm. J. Androl. 2011;32:55–69. doi: 10.2164/jandrol.109.009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olson GE, Winfrey VP, NagDas SK. Acrosome biogenesis in the hamster: Ultrastructurally distinct matrix regions are assembled from a common precursor polypeptide. Biol. Reprod. 1998;58:361–370. doi: 10.1095/biolreprod58.2.361. [DOI] [PubMed] [Google Scholar]
- 58.Reddi PP, Naaby-Hansen S, Aguolnik I, Tsai J-Y, Silver LM, Flickinger CJ, Herr JC. Complementary deoxyribonucleic acid cloning and characterization of mSP-10: The mouse homologue of human acrosomal protein SP-10. Biol. Reprod. 1995;53:873–881. doi: 10.1095/biolreprod53.4.873. [DOI] [PubMed] [Google Scholar]
- 59.Liu MS, Aebersold R, Fann CH, Lee CG. Molecular and developmental studies of a sperm acrosome antigen recognized by HS-63 monoclonal antibody. Biol. Reprod. 1992;46:937–948. doi: 10.1095/biolreprod46.5.937. [DOI] [PubMed] [Google Scholar]
- 60.Freemerman AJ, Wright RM, Flickinger CJ, Herr JC. Cloning and sequencing of baboon and cynomologus monkey intraacrosomal protein SP-10: Homology with human SP-10 and a mouse sperm antigen (MSA-63) Mol. Reprod. Dev. 1993;34:140–148. doi: 10.1002/mrd.1080340205. [DOI] [PubMed] [Google Scholar]
- 61.Herr JC, Klotz K, Shannon J, Wright RM, Flickinger CJ. Purification and microsequencing of the intra-acrosomal protein SP-10. Evidence that SP-10 heterogeneity results from endoproteolytic processes. Biol. Reprod. 1992;47:11–20. doi: 10.1095/biolreprod47.1.11. [DOI] [PubMed] [Google Scholar]
- 62.Westbrook-Case VA, Winfrey VP, Olson GE. A domain-specific 50-kilodalton structural protein of the acrosomal matrix is processed and released during the acrosome reaction in guinea pig. Biol. Reprod. 1994;51:1–13. doi: 10.1095/biolreprod51.1.1. [DOI] [PubMed] [Google Scholar]
- 63.Noland TD, Friday BB, Maulit MT, Gerton GL. The sperm acrosomal matrix contains a novel member of the pentaxin family of calcium-dependent binding proteins. J. Biol. Chem. 1994;269:32607–32614. [PubMed] [Google Scholar]
- 64.Reid MS, Blobel CP. Apexin, an acrosomal pentaxin. J. Biol. Chem. 1994;269:32615–32620. [PubMed] [Google Scholar]
- 65.Barros C, Bedford JM, Franklin LE, Austin CR. Membrane vesiculation as a feature of the mammalian acrosome reaction. J. Cell Biol. 1967;34:C1–C5. doi: 10.1083/jcb.34.3.c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yanagimachi R, Phillips DM. The status of acrosomal caps immediately before fertilization in vivo. Gamete Res. 1984;9:1–19. [Google Scholar]
- 67.Foster JA, Klotz KL, Flickinger CJ, Thomas TS, Wright RM, Castillo JR, Herr JC. Human SP-10: Acrosomal distribution, processing, and fate after the acrosome reaction. Biol. Reprod. 1994;51:1222–1231. doi: 10.1095/biolreprod51.6.1222. [DOI] [PubMed] [Google Scholar]