Abstract
Experimental tools to model cell-tissue interactions will likely lead the way to new ways to both understand and treat cancer. While the mechanical properties and regulation of invasion have been recently studied for tumor cells, these have received less attention in the context of tumor vascular dynamics. In this article, we have investigated the interaction between the surfaces of structures encountered by endothelial cells invading their surrounding extracellular matrix (ECM) during angiogenesis. For this purpose, we have fabricated round and sharp geometries with various curvature and sharpness indices in collagen hydrogel at a wide range of stiffness to mimic different microenvironments varying from normal to tumor tissues. We have then cultured endothelial cells on these structures to investigate the bi-directional interaction between the cells and ECM. We have observed that cell invasion frequency is higher from the structures with the highest sharpness and curvature index, while interestingly the dependence of invasion on local micro-geometry is strongest for the highest density matrices. Notably, structures with the highest invasion length are linked with higher deformation of side structures, which may be related to traction force-activated signaling suggesting further investigation. We have noted that round structures are more favorable for cell adhesion and in some cases round structures drive cell invasion faster than sharp ones. These results highlight the ability of endothelial cells to sense small variations in ECM geometry, and to respond with a balance of matrix invasion as well as deformation, with potential implications for feedback mechanisms that may enhance vascular abnormality in response to tumor-induced ECM alterations.
Introduction
In tissue engineering, collagen hydrogels are used extensively as scaffolds to model the extracellular matrix (ECM) due to the abundance of collagen as the most prevalent ECM protein in the human body. Moreover, it is convenient to adjust the chemical and mechanical characteristics of collagen hydrogel 1, 2 and to investigate cell invasion in physiologically relevant 3D microenvironments 3, 4. Many studies have investigated the role of molecular signaling driven by soluble proteins such as vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), and platelet-derived growth factor (PDGF), which have underpinned the standard paradigm of tumor angiogenesis and the angiogenic switch 5–8. However, recent evidence suggests that mechanical cues likely play an equally important role in several aspects of tumor progression. Mechanical cues such as matrix density, pore size, and fiber thickness regulate many cell behaviors. A dense matrix of fibrous collagen indicates a high risk for breast cancer metastasis 9, 10. Small pores permit cell attachment and permit faster but limited cell motility whereas large pores are better suited for long-range cell motility 11, 12. Furthermore, the role of matrix stiffness and pore size have previously been studied independently. It has been shown that decreasing the confinement size by decreasing the channel width for a given ECM stiffness enhances the cell migration while increasing the ECM stiffness increases the cell migration through the same confinement effect 13. However the independent effects are a challenge to decouple, and the exact role of more complex aspects of the ECM structural microenvironment remain unclear.
In terms of substrate topography, studies have shown that cell migration speed and direction depend on the curvature of the surface 14. Geometrical cues play a role in determining the position of new branching sites from pre-existing mammary epithelial cells 15 and tumor cell invasion has been investigated from different positions within mammary ducts 4; however, the relevance to tumor vascular networks, which are characterized by complex branching structures, has not been demonstrated clearly. Moreover, blood vessel diameters in tumor environments are uneven and vary in size abruptly and abnormally compared to those in healthy blood vessels 16. In our previous work, we have observed that local ECM cues in certain contexts may override more traditional chemical drivers of endothelial cell (EC) invasion, however, analyzing the mechanisms has remained a challenge due to limited ability to control the ECM topography presented to cells 17 Additionally, the mechanism by which the curvature index, which represents the topography of the basement membrane of blood vessels, impacts EC invasion has rarely been studied.
The traditional methods to study EC-tissue interactions have involved seeding cells within or on top of scaffolds made from natural ECM hydrogels (e.g. matrigel, collagen, fibronectin), or synthetic polymeric scaffolds (e.g. poly(lactic-co-glycolic acid (PLGA) or poly(ethylene glycol) (PEG)) 18, 19. These platforms typically do not contain complex gradients or heterogeneity that is essential to regulate relevant cellular phenotypes such as migration or invasion. While in vivo studies provide a higher degree of physiological relevance, dynamic experiments are challenging. Also, generally in vivo models provide correlations rather than causal information regarding the role of the microenvironment in regulating invasion dynamics, due to an inability to define or tune key microenvironment parameters in living systems. Microfluidic blood vessel models can provide a high degree of physiological relevance as well as the potential for dynamic, causal studies. These have been developed using either lithographic top-down or sacrificial methods to define template channels coated with ECs as biomimetic vessels; however, these studies have been mainly focused on simple structures including rectangular geometries and straight vessels 20–23.
In this paper, we have investigated the effect of different microscale topographies formed in collagen hydrogel on EC migration and capillary formation. We have fabricated a set of sharp and round structures that mimic the geometries in blood vessels such as the tube and branching sites over a wide variety of stiffness to model tissues with different mechanical properties. Our approach takes full advantage of the ability to control, with high precision, the topological microenvironment of tissues. We have furthermore characterized the cell invasion process by measuring the average sprout length and invasion frequency. Finally, we have investigated the cell-ECM interaction of ECs with sharp and round structures revealing new bi-directional regulation between the ECM microstructure and EC invasion. Such technology will enable in vitro definition, at an unprecedented spatial resolution, of 3D tissue geometries that mimic the complex structures found in vivo. These platforms will enable detailed studies of the interaction between mechanical and chemical cues in tumor processes such as angiogenesis and metastasis, as well as the effect of cancer drugs in targeting these processes in a physiologically relevant context.
Materials and methods
Collagen hydrogel
Collagen extraction
Collagen type I was obtained from rat tail tendon under sterile conditions as described previously 24. In brief, rat tails were dissected and the tendons were extracted and processed in 0.1% acetic acid at 4°C for 48 hours. The collagen solution was purified by high-speed centrifugation (Avanti J-25, Beckman Coulter, CA, USA) at ×15,000g for 90 minutes. The solution was then placed at −20°C overnight and subsequently lyophilized for 48 hours until complete dryness and kept at −20°C until reconstitution for 3D cell culture experiments.
Collagen hydrogel preparation
Collagen hydrogel was made at a final concentration of 5, 7.5, and 10mg/ml from the stock solution of 7.5, 11.25, and 15mg/ml, respectively. This range of collagen hydrogel concentration spans a mechanical stiffness including both normal and tumor tissues according to number of publications 25–27. Collagen solution and the diluent media consisting of Dulbecco’s Modified Eagle’s medium (DMEM) 10× (0.1 × final volume neutralized collagen), NaOH (volume of collagen × 0.02), and DMEM 1× (as much as necessary to adjust the final desired collagen mass fraction) were separately prepared on ice. The diluent media was pipetted in the collagen solution and gently mixed with a small, sterilized spatula to avoid the formation of bubbles. Once the collagen solution was neutralized, as determined roughly by the color of the phenol red dye in the media, and confirmed with pH paper, it was dispensed on the cover glass with the polydimethylsiloxane (PDMS) supports on the side, and cast against a PDMS stamp which imprinted the microstructures into the hydrogel. First, the neutralized collagen solutions were pre-incubated at 4°C to facilitate the assembly of fiber molecules and then incubated at 37°C to cross-link the fibers. All the hardware and accessories including the PDMS stamp were kept on ice prior to pre-incubation to prevent immature collagen cross-linking. As the density of collagen fibers increased, the pre-incubation time was also increased to allow enough time for the molecular assembly of fibers.28, 29 We pre-incubated 5mg/ml collagen solutions for 1 hour and both 7.5mg/ml and 10mg/ml collagen solutions for 3 hours.
Cell culture
Human umbilical vein endothelial cells (HUVECs) (Lonza, MD, USA) were cultured in EGM2 basal media supplemented with a growth factor bullet kit (Lonza). HUVECs were seeded at 200cells/mm2 on the collagen hydrogel devices between passages 3–4 to yield a rapid monolayer 25. After formation of a confluent monolayer on the hydrogel surface (~48 hours), hVEGF165 (10ng/ml) (Cell Signaling Technology, MA, USA) and 12-O-Tetradecanoylphorbol-13-Acetate (TPA) (40ng/ml) (Cell Signaling Technology) were added as previously reported 17. Cell media and growth factors were changed every 48 hours during the experiment. 5mg/ml hydrogel devices were fixed 48 hours after the addition of the growth factors (VEGF and TPA). This time for 7.5mg/ml and 10mg/ml hydrogel devices was increased to 72 hours.
Immunofluorescence staining
Cells and collagen fibers were fixed at room temperature for 20 minutes with 10% buffered formalin (Fisher Scientific). Between each step, the samples were washed 3 times with PBS for 5 minutes each rinse. They were permeabilized by submerging in Superblock blocking buffer in phosphate buffered saline (PBS) (Thermo Scientific) at room temperature for 2 hours while gently shaking on an orbital shaker. In the first stage, they were incubated with primary mouse monoclonal anti-collagen I (Abcam, MA, USA) (1:1000) and rabbit monoclonal VE-Cadherin antibody (Cell Signaling Technology) (1:250) at 4°C overnight. In the second stage, they were incubated with anti-mouse Alexa Flour 546 (Life Technologies) (1:250) to stain the fibers, DAPI (Life Technologies) (1:5000) to stain the nuclei, and anti-rabbit Alexa Flour 488 (Life Technologies) (1:250) to stain cell-cell junctions at room temperature for 1 hour. To stain the actin filaments, in the second stage, cells were alternately incubated with Phalloidin Alexa Flour 488 (Life Technologies) (1:250) excluding the anti VE-Cadherin stain.
Hydrogel structure fabrication
The structures were first fabricated in silicon utilizing deep reactive ion etching (DRIE) (AMS 100, France) as we have previously published 30. Utilizing soft lithography, the PDMS stamps with the negative replica of structures were fabricated. The stamps were treated with oxygen plasma (Harrick-Plasma, NY, USA) and coated with 1% bovine serum albumin (BSA) for 30 minutes at room temperature and kept on ice under sterile conditions in a biosafety cabinet. This makes the PDMS surface non-adhesive to collagen hydrogel. Neutralized collagen solutions with a final concentration of 5mg/ml, 7.5mg/ml, and 10mg/ml were made from concentrated collagen stock solutions and kept in separate microtubes on ice. The BSA from the PDMS stamps was aspirated and replaced with a ~50μl drop of neutralized collagen. As 7.5mg/ml and 10mg/ml collagen solutions are highly viscous, when the collagen drops are poured on the PDMS structures, bubbles may form between the PDMS structures and the collagen hydrogel. To remove these potential bubbles from the structures and hydrogel drops, 7.5mg/ml and 10mg/ml hydrogel devices were placed on a beaker filled with ice. The beakers were sealed with perforated parafilm and placed under house vacuum for about 2 minutes. Then, the neutralized collagen solution is pipetted on to a coverslip with two strips of PDMS support on both sides to hold the collagen solution, and the PDMS stamps with structures were cast against the collagen solution.
Cell invasion characterization and analysis
The cell invasion progress was monitored and captured by live imaging utilizing an inverted optical microscope (Axio Observer.Z1, Carl-Zeiss) equipped with an incubation chamber with a 10× objective lens. Each hydrogel device was imaged right before cell seeding and then every 24 hours until fixation. In order to characterize the cell invasion progress from the structures of interest, we measured cell invasion frequency and cell invasion length utilizing Zen Lite 2012 software (Carl-Zeiss). The number of sprouts was counted from each structure (from the tip to the edge of structures). Structures with no sprouts (zero count) were also included in the measurement. Cell invasion length was measured by the ruler feature of the Zen Lite software. Because the cells are lying on the sidewall of the microchannel, the aforementioned parameters could be simply measured as they were imaged from the top utilizing the inverted optical microscope.
Statistical analysis
Cell invasion frequency and average length were expressed as mean with the standard error of the mean (SEM). One-way ANOVA with Tukey’s honest significant difference (JMP Software) was performed to find significant difference between the cell invasion length results. Chi-square test with Poisson distribution (JMP Software) was performed to find significant differences between the cell invasion frequency results. Differences were considered statistically significant for (*** p < 0.001, ** p<0.01, and * p<0.05) with level of significance α=0.05.
Results
Collagen hydrogel structure fabrication
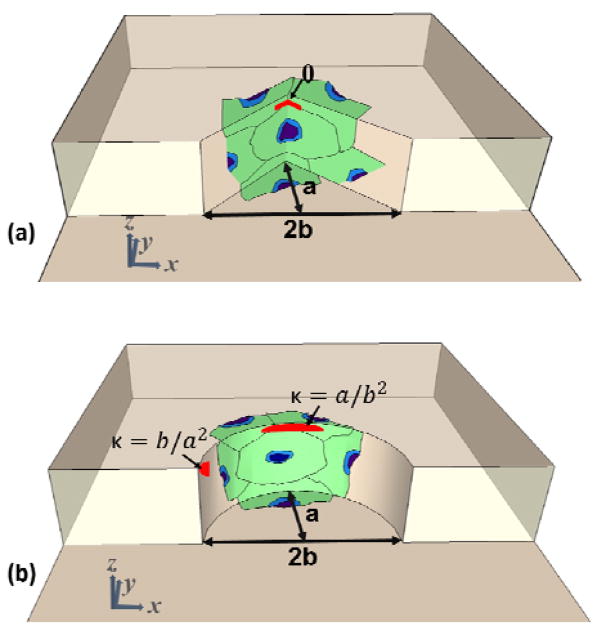
Three sets of round and sharp structures with different curvature and sharpness indices were designed and fabricated in collagen hydrogel. The curved structures are comprised of semi-elliptic structures with minor radius (b) and major radius (a), with corresponding curvature index of b/a2 and a/b2 on the tip and side of the structures, as shown in Figure 1.a. On the other hand, the sharp structures are comprised of flat surfaces on the sides while intersecting each other with angle (θ) on the tip as shown in Figure 1.b. As shown in Figure 1.a–b, cells were seeded on the structures, along with the top and bottom of the channels. Cells on the curved structures sense the curvature index between b/a2 and a/b2, and on the sharp structures sense the flat surfaces with angle (θ) on the tip. Table 1 provides the dimension, curvature index, and angle of the fabricated structures. Topography a contains round (Ra) and sharp (Sa) structures with x=100μm and y=50μm, Topography b contains round (Rb) and sharp (Sb) structures with x= 100μm and y=125μm, and Topography c contains round (Rc) and sharp (Sc) structures with x= 100μm and y=250μm. Spacing between adjacent structures in all topographies is 150μm. Each device contains five topographies from each set of round and sharp structures docked along a 500μm-wide and 95μm-deep microchannel (Figure 2.a–c).
Figure 1.
Orientation of seeded cells with respect to and dimension of fabricated structures on (a) sharp and (b) curved structures.
Table 1.
Dimension of fabricated structures and the curvature and sharpness index of round and sharp structures.
| a | 2b | axial ratio (a/b) | angle (θ)/curvature index (C.I.) | ||
|---|---|---|---|---|---|
| Topography a | Sharp a (Sa) | 100μm | 50μm | 4 | θ = 28° |
| Round a (Ra) | C.I. min= 0.0025, max=0.16 | ||||
| Topography b | Sharp b (Sb) | 125μm | 1.6 | θ = 65° | |
| Round b (Rb) | C.I. min= 0.0062, max=0.025 | ||||
| Topography c | Sharp c (Sc) | 250μm | 0.8 | θ = 100° | |
| Round c (Rc) | C.I. min= 0.0064, max=0.0125 | ||||
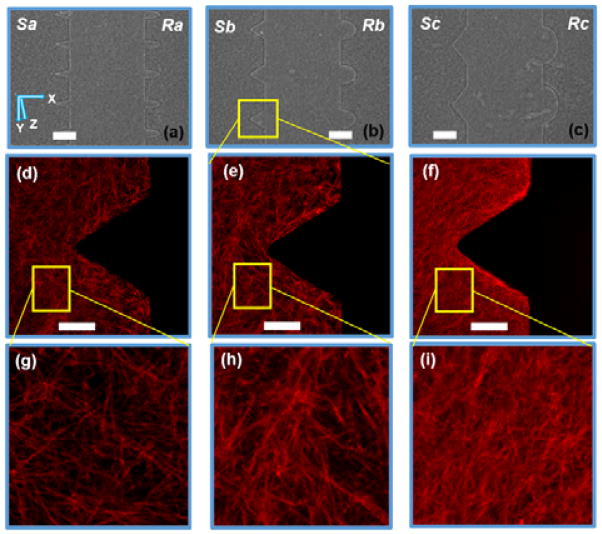
Figure 2.
Micrographs of fabricated structures in 7.5mg/ml collagen hydrogel for structures (a) Sa and Ra, (b) Sb and Rb, and (c) Sc and Rc, and the confocal images of collagen fiber orientation around Sb for (d) 5mg/ml, (e) 7.5mg/ml, and (f) 10mg/ml hydrogel devices utilizing an 25× objective lens, and zoomed-in view of collagen fibers of (g) 5mg/ml, (h) 7.5mg/ml, and (i) 10mg/ml hydrogel devices. scale bar: 150μm for (ac) and 50μm for (d–f). Frame size approx. 70μm×70μm for (g–i).
Furthermore, Table 2 shows the polymerization conditions of hydrogel devices with different concentrations, as well as the depth of the hydrogel microchannel, and fabrication yield. The depth of the hydrogel channel was calculated by seeding fluorescent micro-beads on the hydrogel devices, and measuring the difference between the point of focus of microbeads on the top and the bottom of microchannels by using an inverted microscope (Axio Observer.Z1, Carl-Zeiss, Germany). For cell seeding experiments, we used collagen hydrogel topographies whose curvature and sharpness indices were within 10% of those of the silicon template. As shown in Table 1, the yield for 5mg/ml concentration was 70% while yields for 7.5mg/ml and 10mg/ml were more than 90%.
Table 2.
Summary of fabrication parameters and conditions of hydrogel structures for different stiffness. Yield was determined as the percentage of hydrogel devices fabricated within 90% pattern transfer accuracy from the original silicon structures.
| 5mg/ml | 7.5mg/ml | 10mg/ml | |
|---|---|---|---|
|
| |||
| Mold treatment | No treatment | Under vacuum | |
|
| |||
| Polymerization | |||
| Pre-incubation | 4°C for 1 hour | 4°C for 3 hours | |
| Incubation | 37°C for 1 hour | 37°C for 1 hour | |
|
| |||
| z (depth) = (measured D, SD) | 67.7μm, 6.8 μm | 71.7μm, 3.9 μm | 68.8μm, 3.1 μm |
|
| |||
| Yield (within 10% limit) | ~70% | ~90% | ~90% |
Cell sensing and remodeling of collagen hydrogel
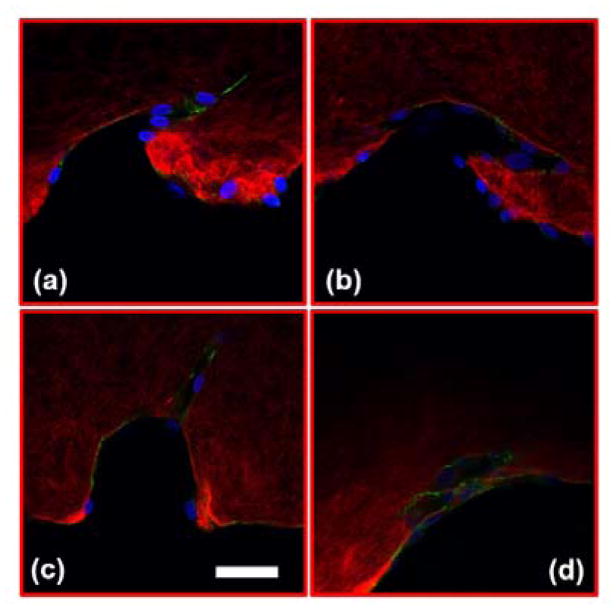
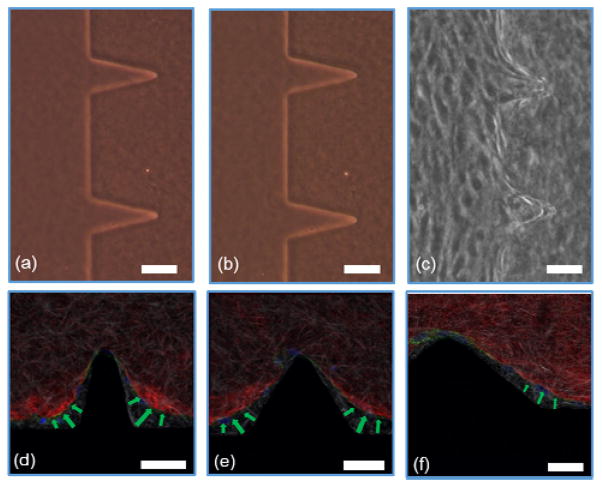
Figure 3 shows the confocal images of structures Sa, Sc, Ra, and Rc with developed neo-vessels from the tip of the structures. We note that in all four images, the neo-vessels were formed from the regions having higher curvature or sharpness indices. These parameters were chosen so that cells sense structures with a wide range of sharpness (from flat surfaces to sharp angels) and curvature (semi- ellipse structures with various curvature indices). Figure 4.a–b shows the micrograph of structure Sa right after fabrication and 3 days after fabrication without the presence of cells. As evident, the structures have not shown any sign of degradation. Figure 4.c shows the micrograph of the same structures with cell seeding after 3 days (1 day after the addition of growth factor). As evident, the cell-cell attachment has started to deform the junctions and side of the structures. In more details, Figure 4.d–f shows this phenoneman with the help of the confocal microscopy. These images show the structures Sa, Sb, and Sc (collagen fibers shown in red) 1 day after the addition of the growth factors to the cell media with overlay images of the hydrogel structures before cell culturing (collagen fibers shown in white). As the images suggest, the ECM has been deformed from the end sides which has resulted in an increase in the collagen fiber density at those locations (saturated red color). The structures with the highest shaprness and curvature index have suffered larger alteration of collagen micropatterned structures (shown with green arrows).
Figure 3.
Confocal images of cell invasion through collagen hydrogel structures for (a) Sa, (b) Sc, (c) Ra, and (d) Rc. [red: collagen fibers, blue: nuclei, and green: VE-Cadherin]. scale bar: 50μm.
Figure 4.
Micrograph of 7.5mg/ml collagen hydrogel structures (a) after fabrication, (b) 3 days after fabrication, (c) 3 days after cell seeding (1 day after the addition of growth factors). The confocal images of hydrogel structure deformation by cell-cell traction forces on the edge of structures for (d) Sa, (e) Sb, and (f) Sc. [red: collagen fibers after cell seeding, white: collagen fibers before cell seeding, blue: nuclei, and green: Actin]. scale bar: 50μm.
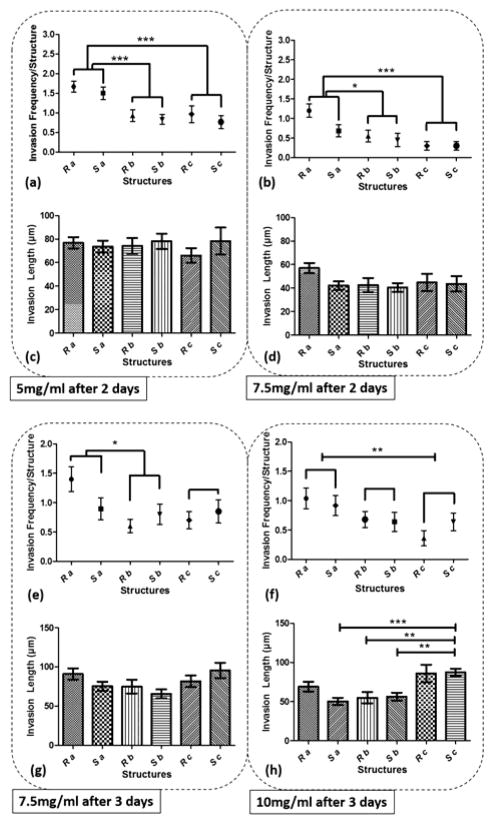
Collagen hydrogel structures mediate cell invasion
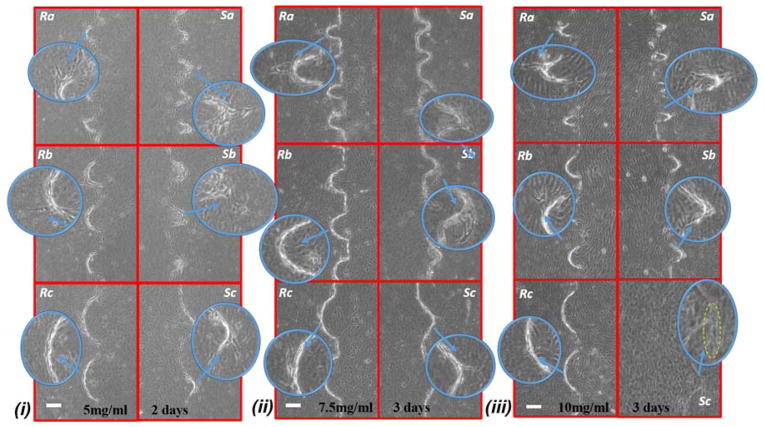
Figure 5 shows the graph of endothelial cell invasion frequency and length for the hydrogel devices. Invasion length and frequency were measured after 2 days for 5mg/ml and 7.5mg/ml devices, and 3 days for 7.5mg/ml and 10mg/ml devices. For all cases, we did not find any significant difference between the invasion frequency within the same sharpness index category (e.g, within Topography a, b or c). However, the more common trend (which was not statistically significant) was towards a higher invasion frequency for the round as opposed to the sharp structures. To facilitate the comparison between index groups, the results from the same index category group were merged and compared with other groups (e.g., Topography a vs Topography b). For instance, in 5mg/ml hydrogel devices, we did not find any significant difference between Sa and Ra, but the invasion frequency of (Sa and Ra) was significantly higher compared to (Sb and Rb). For 5mg/ml devices (6 devices; 30 samples for each structure), we observed that cell invasion frequency was significantly higher for Topography a compared to Topography b and c (Fig 5.a). For 7.5mg/ml devices (4 devices; 20 structures from each), after 2 days, cell invasion frequency was significantly higher for Topography a compared to Topography b and Topography c (Fig 5.b), and after 3 days, cell invasion frequency remained significantly higher for Topography a compared to Topography b but not Topography c (Fig 5.e). For 10mg/ml devices (5 devices; 25 structures from each), cell invasion frequency was significantly higher for Topography a compared to Topography c (Fig 5.f) but not Topography b. The invasion length for 5mg/ml and 7.5mg/ml devices did not result in any significant difference between various structures (Fig 5.c–d,g), however, these differences were significant at 10 mg/ml, with the wider structures driving longer invasions (Fig 5.h). Furthermore, it can be observed that the cell invasion length for the corresponding structures is higher from the softer hydrogel devices while they were measured at the same time point (e.g, 5mg/ml and 7.5mg/ml devices after 2 days, and 7.5mg/ml and 10mg/ml devices after 3 days) (Fig 5.c–d, g–h). As it has been shown already that softer hydrogel substrates lead to increased EC invasion 25, 31, 32, we will focus the following result and analysis sections on the EC invasion from 5mg/ml devices fixed after 2 days, and 7.5mg/ml and 10mg/ml devices fixed after 3 days. Where we have cross-compared the invasion frequency or length, we have done so using the same time-points. Figure 6 shows the micrograph of collagen hydrogel structures with HUVECs at the final concentration of 5mg/ml, 7.5mg/ml, and 10mg/ml. Each image includes a zoom-in inset that shows the structure of interest along with new sprouts.
Figure 5.
The graph of cell invasion frequency measured after 2 days for structures Sa, Sb, Sc, Ra, Rb, and Rc for (a) 5mg/ml, (b) 7.5mg/ml, and cell invasion length for the same time period and structures for (c) 5mg/ml, (d) 7.5mg/ml, and cell invasion frequency measured after 3 days for (e) 7.5mg/ml (f) 10mg/ml hydrogel devices, and cell invasion length for the same time period for (g) 7.5mg/ml (h) 10mg/ml hydrogel devices. (*** p < 0.001, ** p<0.01, and * p<0.05)
Figure 6.
Micrograph images of cell invasion from structures Sa, Sb, Sc, Ra, Rb, and Rc. for (i) 5mg/ml hydrogel devices, fixed 48 hours (ii) 7.5mg/ml hydrogel devices, fixed 72 hours, and (iii) 10mg/ml hydrogel devices fixed 72 hours after addition of the growth factor. In most cases cell invasion is developed from the sharp or curved tip of the structures. scale bar: 100μm.
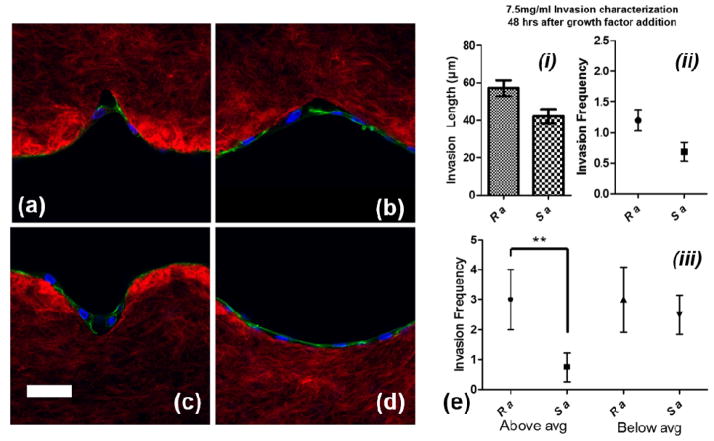
Structure sensing versus cell invasion
As the confocal images show in Figure 7.a–d, the round surfaces (Ra and Rc) are more favorable than sharp structures (Sa and Sc) for cell attachment. Cells on the wall of sharp structures form an arc to bypass the sharp tip whereas the cells fully sense the round structure during the initial phase of growth and proliferation. We compared the cell invasion length and invasion frequency from Sa and Ra for 7.5mg/ml devices, 2 days after the addition of growth factors, before the cell sprouts become so long that there would be no significant difference between them for comparison (because of saturation of average length) (Fig 7.e (i) & (ii)). While there was no significant difference for invasion length or frequency between the two structures, the trend was towards Ra maintaining a higher average invasion length and frequency. In order to evaluate which structures promote cell invasion faster in terms of producing longer neo-vessels, we measured the number of sprouts which were above and below average (51.8μm) for both groups (each 20 structures) since smaller neo-vessels are continuously being produced after the addition of growth factors. As shown in the graph in Figure 7.e (iii), when analyzed in this way the round structures (Ra) produced longer neo-vessels than the sharp structures (Sa), with a statistically significant difference (p<0.01).
Figure 7.
Confocal images of cell attachment on round and sharp structures on 7.5mg/ml hydrogel devices for structures (a) Sa, (b) Sc, (c) Ra, and (d) Rc 24 hours after cell culturing. scale bar: 50μm, (e) invasion frequency and length for Sa and Ra 48 hours after addition of growth factors, and (f) invasion frequency for Sa and Ra for below and above average cell invasion length. [red: collagen fibers, blue: nuclei, and green: actin filaments].
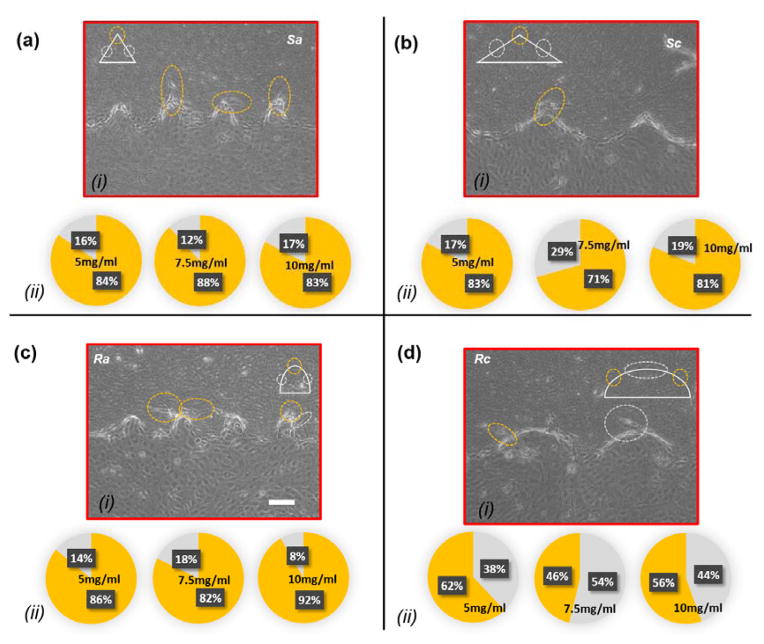
Cells sensing local curvature and sharpness indices
Each structure within a hydrogel device consists of multiple sub-regions with different local curvature or sharpness indices. Curved structures are in fact a semi-elliptical structure with variable curvature indices along their perimeter. In Topography a, the sub-region with the highest curvature region is located on the tip whereas in structures c, the subregions with the highest curvature are located on the sides. On the other hand, the sharp structures consist of side flat subregions and a sharp turn on the tip. Figure 8.a–d (i) shows the sites of low- and high-index curved and sharp regions for Topography a and Topography c, along with the neo-vessels, which are marked with white and orange dash circles, respectively. For structures Sa, Ra, Sc, and Rc, we assumed the radius of the yellow circles centered on the tip of the structures, which represent the sub-regions of high curvature/sharpness indices to be 40μm, 40μm, 50μm, and 70μm. Outside these sub-regions, we considered that the invasion was initiated from the sub-regions with lower curvature/sharpness index. Figure 8.a–d (ii) shows pie charts of neo-vessel distribution across different sub-regions of curved and sharp structures for 5mg/ml, 7.5mg/ml, and 10mg/ml hydrogel devices for Sa, Ra, Sc, and Rc, respectively. In Figure 5.a–b and 5.e–f, it was shown that Topography a (Sa and Ra), which includes the structures with the highest curvature/sharpness index, produces more neo-vessels compared to the other topographies. Here, within the same structure group, it is shown that cell invasion is observed more from the sharper sub-regions or sub-regions with the higher curvature index. As evident in Figure 8.a (ii) and Figure 8.b (ii), on the structures Sa and Sc, the majority of neo-vessels were formed from the sharp tip of the structures. Likewise, as shown in Figure 8.c (ii), on the structures Ra, the majority of neo-vessels were formed from the tip of the structures where the curvature index is the highest. However, in structures Rc the cell invasion was distributed equally across the perimeter as shown in Figure 8.d (ii).
Figure 8.
Cell invasion micrographs and confocal images with the distribution pie chart of cell invasion from different sites within each structure for (a) Sa, (b) Sc, (c) Ra, and (d) Rc. The pie chart indicates the percentile of cell invasion from high-index curved/sharp sub-regions (yellow) and low-index curved/sharp sub-regions (gray). scale bar: 100μm.
Discussion
In this study, we have explored the interaction of ECs with sharp and curved geometries in terms of ECM sensing, remodeling and invasion. We observed the highest invasion frequency for the narrower ECM structures over all of our experiments, while this frequency was reduced at higher collagen concentrations. Both of these observations are as expected based on our prior observations of enhanced EC invasion in the sharp corners of microchannels 17, as well as observations by ourselves and others on the inhibition of EC invasion in denser hydrogel 25, 31, 32. However, we observed a trend towards higher invasion rates for the round versus sharp structures of the same width, although this was not statistically significant. We believe this may be related to our observation that during the initial phase of cell adhesion, round structures were more favorable for cell attachment than sharp structures. We furthermore, interestingly, observed the strongest dependence of invasion length on micro-geometry for the highest density collagen. Additionally, cell-cell attachment force on the edges of the collagen hydrogel structures deformed those structures locally. All of these latter observations suggest a balance between collagen restructuring versus invasion resulting in a complex dependence of invasion length on ECM density and structure.
Previous studies have shown that regions of tumor cell invasion were correlated with high mechanical stress from the tumor sites 4. Traction forces at the invasion site have also been demonstrated to be associated with local proteolytic activity, facilitating the cancer cell invasion via matrix degradation 33, 34. In our current work, this cell-cell and cell-ECM attachment might induce such traction forces that enhance the cell invasion process in nearby structures where higher local cell density mediated by sharpness and curvature index results in an ECM that is more amenable to invasion. As noted previously, we observed the higher traction force adjacent to regions where we observed the higher cell invasion. In future work, the impact of such forces can be investigated by varying the geometry on the edges of the structures while maintaining the curvature and sharpness indices constant where cells have previously been observed to initiate invasion.
One factor that might affect the cell invasion length among different structures is that the growth factors might not be equally distributed if the cell invasion density is not the same across all structures. For instance, it was observed that there are more cell sprouts from Topography a than Topography c, while Topography c maintain even larger surface area. This means that in Topography c, a smaller number of cell sprouts have more room to grow while there will be less competition for growth factors among the sprouts. Furthermore, the progress of cell invasion and growth of the sprouts is a continuous phenomenon that starts right after the addition of the growth factors until the cells are fixed. Therefore, the average invasion length is biased downwards by the newly formed vessels. However, the structures that promote cell invasion should contain longer neo-vessels. At early time-points, this difference might be very small, yet as time passes and the number of neo-vessels increases, there might not be any noticeable difference between the length of neo-vessels. Our preliminary attempts to remove this bias by binning of above versus below average length sprouts in one particular case (7.5mg/ml hydrogel devices fixed 48 hours after addition of growth factors) suggested that within one curvature and sharpness group, round structures promote cell invasion earlier in the process than the sharp structures, resulting in longer than average invasion lengths persisting at later times. The slower invasion process from this sharp structure might be attributed to the additional time and energy cells spend on remodeling and attaching to the sharp structures, and also may help explain the observed trend of higher invasion frequency for round versus sharp structures.
We have demonstrated experimentally that cell invasion frequency is dependent on geometrical cues, and geometries with the highest curvature/sharpness index produced more neo-vessels. For 7.5mg/ml devices, cell invasion frequency was significantly higher from Topography a (narrowest) than Topography b (intermediate width) and Topography c (widest) (Fig 5.b) after 2 days, and from Topography a (narrowest) than just Topography b (intermediate width) after 3 days (Fig 5.e). For 10mg/ml devices, cell invasion frequency was significantly higher for Topography a compared to Topography c (widest) after 3 days (Fig 5.f). These results suggest that with increasing time, the cell invasion dependency on curvature/sharpness index decreases (Fig 5.b, 5.e), therefore these differences may be most relevant to early stages of sprouting and neovessel formation. Furthermore, we can argue that the curvature/sharpness index is playing a larger role than surface area in driving cell invasion from the stiffer substrate (Fig 5.e, 5.f) since Topography c has higher surface area but lower surface curvature than Topography b. Furthermore, as noted, we observe the largest variation in invasion length with geometry for the stiffest ECM, e.g. for 10mg/ml devices the average invasion length from Topography c was higher compared to other structures (Fig 5.h). These observations indicate that by increasing the mechanical stiffness of the ECM, the cell invasion dependency on curvature and sharpness configuration increases. This might be partially attributed to the low number of cell sprouts competing for cell media and growth factors within Topography c., however additional mechanisms related to effects such as traction forces and matrix restructuring should be examined in future work, while microfluidic channel feeding may be used to remove compounding variations in nutrition levels.
Overall our present work provides evidence that ECs balance mechanical re-organization of ECM structure with cellular invasion, depending on curvature index and matrix compliance. Such dynamic regulation of ECM by vascular structures has not been demonstrated previously, and suggests that there may be complex feedback loops between ECM structure and geometry and vascular dynamics that could provide therapeutic targets, or else lead to uncontrolled tumor growth if left unchecked. Our observation of enhanced dependence of sprout length on local ECM micro-geometry at higher versus lower tissue density may also have important implications for our understanding of the leaky, tortuous, branching, and generally highly disordered vasculature observed in dense tumor tissue. Our platforms will enable a unique exploration of the transition in EC dynamics from 2D to 3D geometries in terms of alteration in cell morphology and focal adhesion dynamics as well as studies of the molecular signaling regulating the observed cell dynamics
Conclusions
In this work, we fabricated lithographically defined complex surfaces in collagen hydrogel to mimic the microstructures that vascular networks may encounter in natural ECM. The mechanical properties of the hydrogel structures were tuned to mimic a wide range of tissues from normal to tumor. We studied the interaction of cells with such microenvironments, which greatly impact the processes of cell proliferation, migration and development of capillary networks, all of which are significant factors in tumor neo-vascularization and growth. We found that ECs are able to fully interact with round microstructures at earlier time-points as compared with sharp ones. Furthermore, cells remodeled the microstructures with the higher sharpness/curvature index to a greater degree, and cell invasion frequency was the highest in most cases from the microstructures with the highest sharpness/curvature index. Finally we found that dependence of EC invasion distance on local ECM micro-geometry was the strongest at higher matrix density. In summary, as stiffness increases from a softer to stiffer substrate, cell invasion frequency and length decrease within the same structures. While, keeping the stiffness fixed, as the sharpness and curvature index increase, these parameters increase. Furthermore, we showed that curved structures enhance cell invasion at early time-points. Such exploration of cell-ECM interactions, and elucidation of the basic mechanisms may provide novel designs for vascular-targeting cancer treatments, and potentially improve the performance of existing anti-vascular drugs which have been a major research and investment focus.
Supplementary Material
Acknowledgments
We would like to thank the Institute for Critical Technology and Applied Science (ICTAS), the National Science Foundation (CBET-1403304 to M. Agah), and the National Institutes of Health (NIBIB-R21EB019123 to S. Verbridge) for financially supporting this project. Furthermore, we would like to thank Mr. Donald Leber at the Micron Cleanroom Facility in Virginia Tech and Dr. Bob Geil at the Institute for Advanced Materials, NanoScience and Technology in the University of North Carolina at Chapel Hill for their assistance with fabrication process, and Dr. Kristi DeCourcy at the Fralin Biotechnology Center in Virginia Tech for her assistance with the confocal imaging, respectively. We also would like to thank Dr. Elizabeth Antoine, Dr. Celeste Nelson, and Mr. Aboozar Monavarfeshani for providing helpful discussions as well.
Notes and references
- 1.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Advanced Materials. 2009;21:3307–3329. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, Camci-Unal G, Dokmeci MR, Peppas NA, Khademhosseini A. Adv Mater. 2014;26:85–123. doi: 10.1002/adma.201303233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD. Proc Natl Acad Sci U S A. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boghaert E, Gleghorn JP, Lee K, Gjorevski N, Radisky DC, Nelson CM. Proc Natl Acad Sci U S A. 2012;109:19632–19637. doi: 10.1073/pnas.1118872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergers G, Benjamin LE. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P, Jain RK. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang SY, Bu X, Zhao H, Yu JT, Wang YM, Li D, Zhu CC, Zhu T, Ren TT, Liu XP, Yao LB, Su J. J Pathol. 2014;232:436–448. doi: 10.1002/path.4311. [DOI] [PubMed] [Google Scholar]
- 8.Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA. Pharmacol Rev. 2004;56:549–580. doi: 10.1124/pr.56.4.3. [DOI] [PubMed] [Google Scholar]
- 9.Kakkad SM, Solaiyappan M, Argani P, Sukumar S, Jacobs LK, Leibfritz D, Bhujwalla ZM, Glunde K. Journal of biomedical optics. 2012;17:116017. doi: 10.1117/1.JBO.17.11.116017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. The Journal of biological chemistry. 2013;288:12722–12732. doi: 10.1074/jbc.M112.447631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy CM, O’Brien FJ. Cell adhesion & migration. 2010;4:377–381. doi: 10.4161/cam.4.3.11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harley BA, Kim HD, Zaman MH, Yannas IV, Lauffenburger DA, Gibson LJ. Biophysical journal. 2008;95:4013–4024. doi: 10.1529/biophysj.107.122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak A, Kumar S. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MC, Kim C, Wood L, Neal D, Kamm RD, Asada HH. Integrative Biology. 2012;4:1386–1397. doi: 10.1039/c2ib20159c. [DOI] [PubMed] [Google Scholar]
- 15.Nelson CM, VanDuijn MM, Inman JL, Fletcher DA, Bissell MJ. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukumura D, Duda DG, Munn LL, Jain RK. Microcirculation. 2010;17:206–225. doi: 10.1111/j.1549-8719.2010.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verbridge SS, Chakrabarti A, DelNero P, Kwee B, Varner JD, Stroock AD, Fischbach C. Journal of Biomedical Materials Research Part A. 2013 doi: 10.1002/jbm.a.34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Rodrigues J, Tomas H. Chemical Society reviews. 2012;41:2193–2221. doi: 10.1039/c1cs15203c. [DOI] [PubMed] [Google Scholar]
- 20.Bian W, Liau B, Badie N, Bursac N. Nature protocols. 2009;4:1522–1534. doi: 10.1038/nprot.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden A, Tien J. Lab on a Chip. 2007;7:720–725. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- 22.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Integrative biology : quantitative biosciences from nano to macro. 2013;5:606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghavan S, Nelson CM, Baranski JD, Lim E, Chen CS. Tissue Engineering; Parts A. 2010;16:2255–2263. doi: 10.1089/ten.tea.2009.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajan N, Habermehl J, Cote MF, Doillon CJ, Mantovani D. Nature protocols. 2006;1:2753–2758. doi: 10.1038/nprot.2006.430. [DOI] [PubMed] [Google Scholar]
- 25.Cross VL, Zheng Y, Won Choi N, Verbridge SS, Sutermaster BA, Bonassar LJ, Fischbach C, Stroock AD. Biomaterials. 2010;31:8596–8607. doi: 10.1016/j.biomaterials.2010.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Ramanujan S, Pluen A, McKee TD, Brown EB, Boucher Y, Jain RK. Biophys J. 2002;83:1650–1660. doi: 10.1016/S0006-3495(02)73933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang YL, Motte S, Kaufman LJ. Biomaterials. 2010;31:5678–5688. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Wood GC. The Biochemical journal. 1960;75:598–605. doi: 10.1042/bj0750598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosseini Y, Verbridge SS, Agah M. Journal of biomedical materials research. Part A. 2014 doi: 10.1002/jbm.a.35352. [DOI] [PubMed] [Google Scholar]
- 31.Ghajar CM, Chen X, Harris JW, Suresh V, Hughes CC, Jeon NL, Putnam AJ, George SC. Biophys J. 2008;94:1930–1941. doi: 10.1529/biophysj.107.120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis-Sedlak ME, Moya ML, Huang JJ, Lucas SA, Chandrasekharan N, Larson JC, Cheng MH, Brey EM. Microvascular research. 2010;80:3–9. doi: 10.1016/j.mvr.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Aung A, Seo YN, Lu S, Wang Y, Jamora C, del Alamo JC, Varghese S. Biophys J. 2014;107:2528–2537. doi: 10.1016/j.bpj.2014.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jerrell RJ, Parekh A. Acta biomaterialia. 2014;10:1886–1896. doi: 10.1016/j.actbio.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.