Abstract
Human experts can annotate peaks in MALDI-TOF profiles of detached N-glycans with some degree of accuracy. Even though MALDI-TOF profiles give only intact masses without any fragmentation information, expert knowledge of the most common glycans and biosynthetic pathways in the biological system can point to a small set of most likely glycan structures at the “cartoon” level of detail. Cartoonist is a recently developed, fully automatic annotation tool for MALDI-TOF glycan profiles. Here we benchmark Cartoonist’s automatic annotations against human expert annotations on human and mouse N-glycan data from the Consortium for Functional Glycomics. We find that Cartoonist and expert annotations largely agree, but the expert tends to annotate more specifically, meaning fewer suggested structures per peak, and Cartoonist more comprehensively, meaning more annotated peaks. On peaks for which both Cartoonist and the expert give unique cartoons, the two cartoons agree in over 90% of all cases.
Keywords: Mass spectrometry, glycosylation, glycomics, bioinformatics
Graphical Abstract
1. Introduction
Glycosylation is one of the most common and important post-translational modifications of proteins, involved in crucial biological processes such as sperm-egg binding, immune system recognition and evasion, virus cell entry and exit, and a variety of other diseases including cancer. Glycan modifications are much more complex than low-mass protein modifications such as methylation, acetylation, and phosphorylation, because glycans are built enzymatically and have various oligosaccharide compositions and numerous isomers for most compositions. Glycosidic and peptide bonds also exhibit different fragmentation properties in mass spectrometry, so that it is difficult to simultaneously fragment both the glycan and peptide of a glycopeptide. Hence for ease of data acquisition and data analysis, glycans are most often studied after release from their protein carriers. MALDI-TOF mass spectrometry has proven to be one of the most successful methods [1, 2] to study detached N- and O-linked glycans, because of its efficient ionization of permethylated glycans, extensive m/z range, and large dynamic range in measured intensity. A MALDI-TOF profile of detached N-glycans from a single tissue sample often contains 50 or more easily discernible peak series, each representing a distinct glycan mass and its natural isotopic variants. Assigning these peaks to likely glycan structures is an important data analysis step, essential for forming hypotheses or inferences about biosynthetic pathways, enzyme activity, cell signaling, or biomarkers.
Glycan structures can be specified at various levels of detail. Oligosaccharide composition, meaning the number of pentose units, hexose units, N-acetyl hexosamines, deoxyhexoses, etc., is relatively easy to obtain; an accurate glycan mass alone is often sufficient to determine the composition, or to determine it except for the ambiguity left by the fact that the mass of deoxyhexose and N-acetyl neuraminic acid (NeuAc) equals the mass of hexose and N-glycolyl neuraminic acid (NeuGc). The cartoon (or topology) level of detail specifies the tree of connections among the monosaccharides, but not the “linkage information”, that is, the exact types of glycosidic bonds between linked monosaccharides. This level cannot be obtained from glycan mass alone, but expert knowledge can often supply one or at most a few most likely cartoons for low-mass glycans, say those below 4000 Da. High-mass glycans may have 10s or even 100s of plausible cartoons. The complete structure specifies the topology and also the “linkage information”, for example, “beta 1,4”, specifying the positions of the carbons and the stereochemistry of the glycosidic bond between each pair of linked monosaccharides. Topology can be obtained with minor ambiguities from MS2 (tandem mass spectrometry), but for linkage information extensive MSn [3] and/or exoglycosidase digestions [4] may be necessary to obtain structures that are well supported by observations.
Glycan structure assignment, however, need not be complete nor error-free to be useful. For example, elevated levels of high-mannose N-glycans have been linked to breast cancer progression [5, 6], and increased levels of HexNAc(4)Hex(3)Fuc(1) have been linked to ovarian cancer [7, 8], and gross observations such as these can be derived from approximate glycan annotations that leave many details unspecified.
In this paper, we describe software for annotating MALDI-TOF glycan profiles to the level of cartoons. This software was initially developed by Dr. David Goldberg in collaboration with Core C of the Consortium for Functional Glycomics under the direction of Prof. Anne Dell, and has recently undergone a series of improvements by the present group of authors. We benchmarked the improved software by comparing its annotations to manual annotations produced by Core C’s experts.
2. Materials and Methods
Cartoonist is a fully automatic annotation tool for MALDI glycan profiles [9]. In brief, Cartoonist detects significant peaks in a MALDI mass spectrum, identifies their likely oligosaccharide compositions, and finds a set of cartoons to match the compositions. Since its initial development [9] and applications [10, 11], the tool, implemented as platform-independent Java software, has gone through a series of improvements in its spectrum viewer (front end) and core detection, identification, and annotation algorithms (back end), including improvements in speed and accuracy as well as added functionality. Cartoonist now supports both sodiated and protonated MALDI systems, both native (aldose or ketose) and alditol reducing ends, and both native and permethylated glycans.
As described previously [9], the bulk of the code in Cartoonist is devoted to peak detection. Cartoonist rates each series of peaks in the spectrum for signal-to-noise ratio, closeness of the match to the expected isotope ratios for an average glycan elemental composition, and mass error after recalibration based on an initial set of identifications. Peak series are then matched to possible oligosaccharide compositions using an absolute mass tolerance (1.5 Da); a relative mass tolerance computed from matched peaks (for example, 50 ppm) is used in viewing annotated spectra. The user can scale the viewing tolerance on the Options Advanced tab. Peak series that cannot be matched to oligosaccharide compositions do not go on to annotation.
As shown in Figure 1, Cartoonist now has two annotation modes: automatic glycan generation and database search. Automatic is the tool’s default mode. This mode generates N-glycan cartoons according to an expert system. As described previously [9], Cartoonist uses a graph grammar to generate a large library of plausible cartoons (only topology without linkage information). The graph grammar, which encodes a model of common biosynthetic pathways, starts from the trimannosyl core with four antenna “slots”. Each antenna is extended with zero or more lactosamine units (GlcNAc-Gal going up the antenna), each of which may carry a fucose on GlcNAc. Each antenna is then capped with one of ten capping groups: GlcNAc, GlcNAc-GalNAc, GlcNAc-(Fuc)-GalNAc, GlcNAc-(Fuc)-Gal-(Fuc), GlcNAc-(Fuc)-Gal-NeuAc, GlcNAc-Gal-NeuAc, GlcNAc-Gal-Gal, GlcNAc-Gal-(GalNAc)-NeuAc, GlcNAc-GalNAc-NeuAc, GlcNAc-Gal-NeuAc-NeuAc. Finally the grammar allows the optional addition of bisecting GlcNAc and/or core fucose, and the substitution of one or more NeuGc’s for NeuAc’s.
Figure 1.
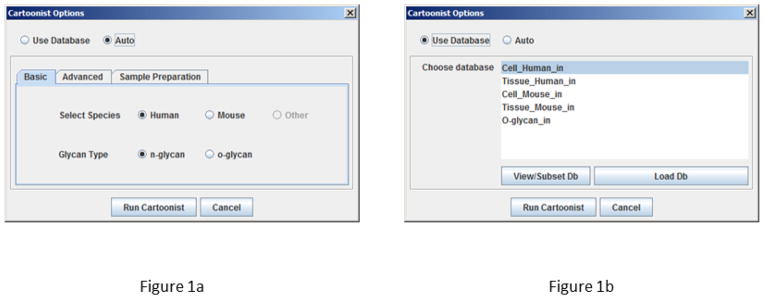
Cartoonist inputs for (a) automatic glycan generation and (b) glycan database search. Automatic mode generates glycan cartoons according to a model of biosynthetic pathways, giving in effect a database with tens of thousands of possibilities; glycan databases are generally much smaller and can be customized for each project.
Cartoonist’s automatic mode then applies score “demerits” to the generated cartoons in order to penalize unlikely glycan substructures. There are mild demerits (−1 score point) for bisecting GlcNAc, more than one fucose on an antenna, or uncapped terminal GlcNAc; these demerits apply no matter which organism (Mouse, Human, or Other) the user chooses. For human samples, there are medium demerits (−3 score points) for one-antenna glycans or presence of LacdiNAc (GalNAc-GlcNAc) and large demerits (−5 score points) for the presence of NeuGc or Gal-alpha-Gal. Cartoonist reports all topologies that tie for the top (least negative) score. As shown in Supplementary Table S1, about 20 different substructures are included in the tables of demerits. Cartoonist is designed for extensibility; the eventual goal is to have tables of demerits for a large number of organisms and tissues.
Cartoonist’s database-search mode matches a predetermined list of cartoons, which may be N- or O-linked glycans, to MS peaks with oligosaccharide compositions. In this mode, Cartoonist rates MS peaks but does not score cartoons, and it outputs all cartoons with mass matching within 1.5 Da. This functionality was used to digitize the manually annotated MALDI-TOF profiles on the CFG website. Using a text editor, we made a small custom database containing the manual annotations for each spectrum, and then let Cartoonist find the matching peaks. For this application, we programmed Cartoonist to print a warning message if there was no peak to match a database cartoon. Such a warning message is not desired for most database-search applications, but in this case Cartoonist’s warnings were useful for checking the manual annotations. The manual annotations were made with PowerPoint, and initially included fairly frequent copy-and-paste errors that were found and corrected by Cartoonist. Right-clicking on a cartoon in a MALDI-TOF profile on the CFG website takes the user to a page giving more complete information, including possible structures with linkage information.
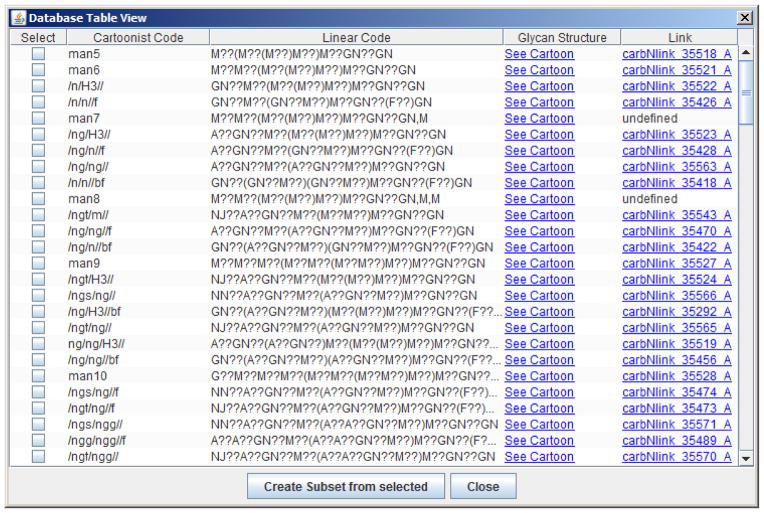
Cartoonist offers five preset databases, one O-glycan database and four N-glycan databases, corresponding to the top-level classification on the CFG web site: mouse cells, mouse tissue, human cells, and human tissue. As shown in Figure 2, Cartoonist uses two different encodings of cartoons: its own compact encoding that specifies cartoon level only or linear codes developed by GlycoMinds (Maccabim, Israel). Cartoonist’s compact codes use one-letter abbreviations for monosaccharides (n = GlcNAc, o = GalNAc, g = Gal, l = Glu, s = NeuAc, t = NeuGc, f = Fuc, x = Xyl) and specifies only the antennae, separated by slashes, for N-glycans containing the trimannosyl core. A string of letters indicates a linear chain of monosaccharides, for example, ngngs means GlcNAc-Gal-GlcNAc-Gal-NeuAc going “up” the antenna. An f after a monosaccharide indicates a fucose side branch. For example, nfgs indicates GlcNAc-Gal-NeuAc with Fuc branching off the GlcNAc, which could be either sialyl Lewis X or sialyl Lewis A. An f after all four slashes ending antennae indicates core fucose. For example, /ng/ng//f in Figure 2 specifies an empty leftmost antenna, GlcNAc-Gal on each of the two middle antenna, an empty rightmost antenna, and core fucosylation, that is, the bi-antennary N-glycan usually called G2F in the context of monoclonal antibodies. A b after all the antennae specifies bisecting GlcNAc, for example, /ng/ng//bf specifies /ng/ng//f with the addition of bisecting GlcNAc. H3 specifies a hybrid structure with three mannose residues on one “arm” of the trimannosyl core; for example, ng/ng/H3// specifies GlcNAc-Gal antennae on the two leftmost positions, and mannoses on the two rightmost positions. (Perhaps a more logical code for ng/ng/H3// would be ng/ng/m/m/ with m meaning mannose.) Cartoonist’s compact codes are not fully general—they cannot currently encode glycans with 5 antennae—but they cover the vast majority of cases and provide an especially quick and convenient way to build databases.
Figure 2.
View of a glycan database inside Cartoonist. Glycans are given by compact Cartoonist codes, linear codes, links to images, and links to pages on the Consortium for Functional Glycomics web site. The user can create a customized glycan database by selecting a subset of glycans from a larger database.
Linear codes (see http://web.mit.edu/glycomics/cbp/linearcode.html) are fully general and expressive; they use upper-case abbreviations for monosaccharides (GN = GlcNAc, AN = GalNAc, A = Gal, G = Glu, M = Man, NN = NeuAc, NJ= NeuGc, F = Fuc), parentheses for branching, and lower-case letters and numbers for linkages, such as a3 for alpha 1,3. Cartoonist, however, does not display linkage information, so the linear codes in Cartoonist glycan databases generally have ?? (unknown anomericity and position) for each linkage as seen in Figure 2.
In order to evaluate Cartoonist’s performance, we compared Cartoonist’s automatic annotations with manual expert annotations of human and mouse N-glycan MALDI-TOF profiles from the Consortium for Functional Glycomics. The set of 87 MALDI-TOF spectra, containing a total of 3627 manually annotated peaks, used in this benchmark is given in Supplemental Information Table 1 and includes a wide variety of cell and tissue types. Manual annotations are not ground truth, but they are generally fairly accurate, because they are informed by knowledge of biosynthetic pathways, and for many of the samples, by MS2 data and even glycosidase analysis.
Cartoonist automatic annotations were obtained with the following parameter settings: sodiation and not protonation, permethylation, maximum number of annotations per MS1 scan set to 200, peak smoothing filter width set to 0.25 Thomsons, and organism set to Mouse for mouse samples and Human for human samples. Quality and intensity thresholds and mass tolerance scaling were set to their default values.
3. Results and Discussion
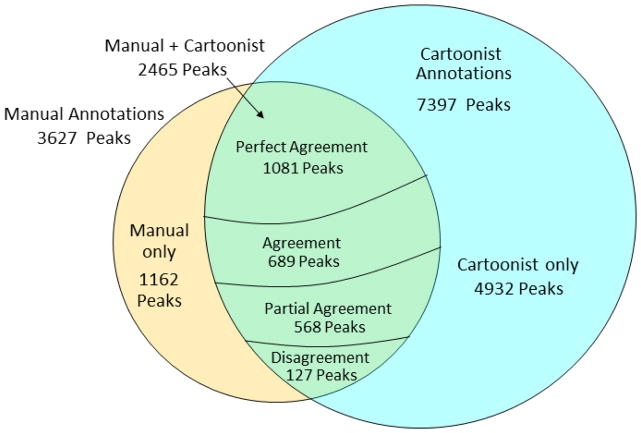
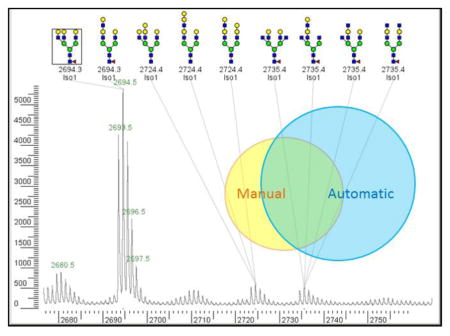
Figure 3 gives a summary of the agreement between the manual and automatic annotations. It is useful to distinguish between “peak agreement”, meaning which peaks to annotate, and “cartoon agreement”, the similarity between the manual and automatic cartoons.
Figure 3.
Venn diagram showing agreement of manual and automatic peak annotations over 87 MALDI-TOF profiles from the Consortium for Functional Glycomics web site (http://www.functionalglycomics.org/). Perfect Agreement means that manual and automatic annotations give the same unique peptide; Agreement means that the manual annotations are a subset of the automatic annotations; Partial Agreement means that there is nonempty intersection of the manual and automatic annotations; and Disagreement means empty intersection of two nonempty sets.
Peak Agreement
Cartoonist annotated 7397 peaks in the 87 MALDI-TOF spectra, including 2465 (~68%) of the 3627 manually annotated peaks. For many peaks, mass implies oligosaccharide composition, for example, a peak at 1579.8 Da is almost surely man5, but for many others, the oligosaccharide composition may be ambiguous, because GalNAc and GlcNAc have exactly the same mass, one Fuc plus one NeuGc have exactly the same mass as one Gal plus one NeuAc, and two fucoses have mass only 1 Da larger than one NeuAc. Most of the 4932 peaks annotated by Cartoonist but not by the human expert are low-intensity peaks in the 3000 – 6000 Da mass range, but there are also a large number of lower mass peaks as clear as the peak at 2327.2 Da in Figure 4. There are various reasons why an expert may skip a clear peak: low intensity, cartoon uncertainty, or even crowding in the PowerPoint slide. Almost all of the 1162 manually annotated peaks annotated by the human expert but not by Cartoonist were skipped by Cartoonist due to deviations from the expected m/z or the expected isotope envelopes. An example with unexpected isotope envelope is the peak series starting at 1620.9 Da in Figure 5. This peak series overlaps with lower mass peak series, most likely one starting at 1617.9 and another at 1615.8. Cartoonist includes sliders allowing the user to adjust peak intensity and isotope ratio (“quality”) thresholds. Cartoonist adaptively sets its mass measurement model (how error scales with measured mass) and mass matching threshold based on initial peak identifications, but the user can adjust the tolerance on the Options Advanced tab. We used the default settings for all MS profiles in our computational experiments rather than adjust settings for each spectrum.
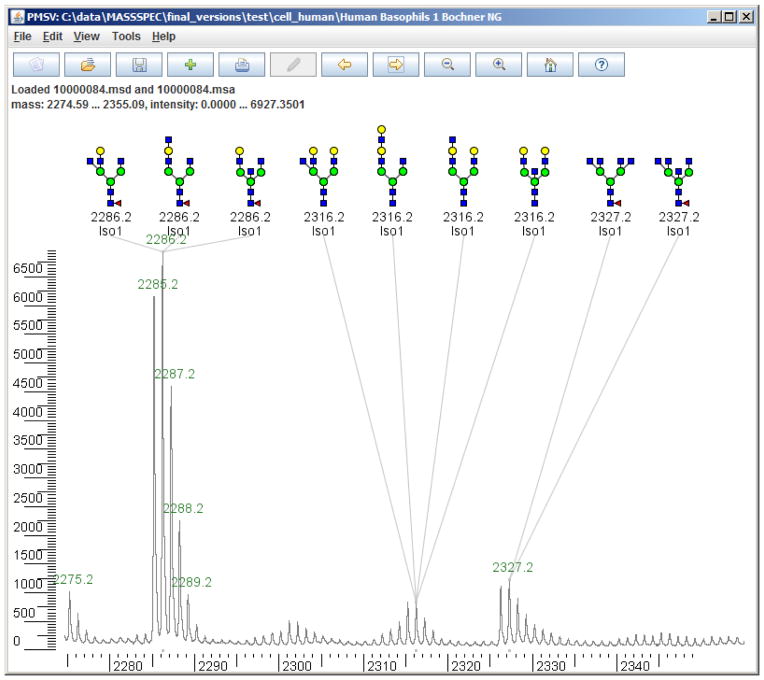
Figure 4.
In this example, Cartoonist annotates the peak at 2327.2 Da, which is not included in the manual annotations. Cartoonist offers several alternatives for 2286.2 and 2316.2 Da, which are annotated with single cartoons in the manual annotations. Cartoonist annotates the isotope peak that it expects to be most intense; here “Iso1” means the first isotope peak, that is, one Dalton heavier than the monoisotopic form.
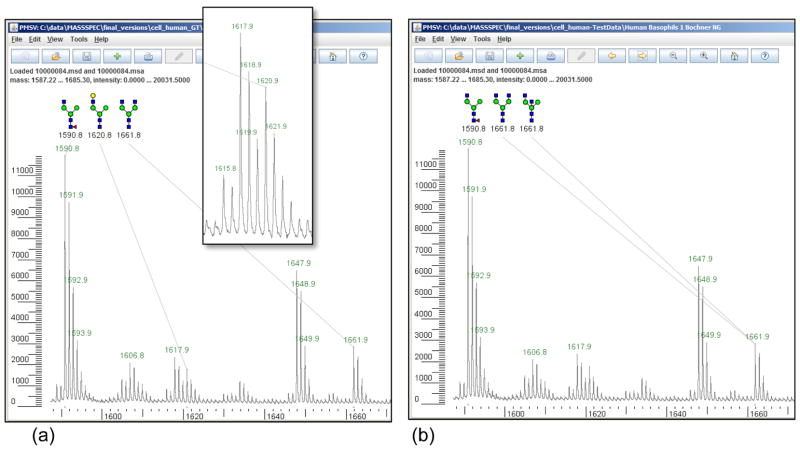
Figure 5.
(a) Manual annotations on m/z 1580 – 1670 from human basophil cells. (b) Cartoonist’s automatic annotations on the same data. The annotations for the peak at 1590.8 are in “perfect agreement”, because the human expert and Cartoonist gave the same unambiguous annotation; the annotations for the peak at 1661.8 are in “agreement” because Cartoonist’s annotations include all the expert annotations. Cartoonist did not annotate the small peak series (inset) starting at 1620.8, which matches sodiated permethylated HexNAc(3)Hex(4) with calculated monoisotopic mass 1620.8097, because of a poor match to the expected isotope distribution due to interfering peaks. Neither the expert nor Cartoonist annotated the peaks at 1606.8 or 1647.9, which are incompletely permethylated forms of the glycans at 1620.8 and 1661.9.
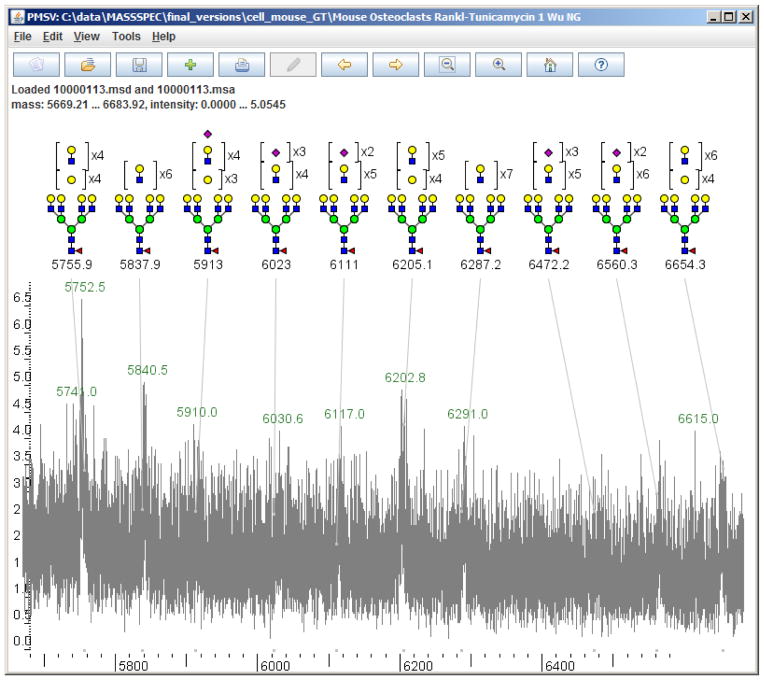
There are also 95 manually annotated peaks that were skipped by Cartoonist, because their masses were too large or their oligosaccharide compositions exceeded limits assumed in Cartoonist’s automatic mode. Cartoonist currently allows at most 12 HexNAc residues, at most 12 Hex residues, at most 6 Fuc, at most 4 NeuAc, and at most 4 NeuGc. Figure 6 shows a manual annotation with a peak at 6654.3 Da with a composition of HexNAc=12, Hexose=17, and Fucose=1. When we relaxed the limits on the numbers of monosaccharides, Cartoonist still missed most of the peaks in Figure 6, because Cartoonist is designed to pick peaks that stand out from the background noise. We have devised more sensitive peak-picking algorithms [12] that can find N-glycan peaks up to 12,000 Da in spectra acquired on the latest MALDI-TOF instruments, but have not yet incorporated them into Cartoonist.
Figure 6.
Manual annotations from the CFG web site of peaks with high mass and low signal-to-noise ratios. Cartoonist missed these peaks for two reasons: it has limits on the sizes of the glycans, and it does not annotate peaks with signal-to-noise ratio below about four.
Cartoon Agreement
When N-glycans are shown with linkage information, the left-to-right order of the antennae has a conventional meaning, for example, the alpha-3-linked mannose in the trimannosyl core is shown on the left and the alpha-6-linked mannose on the right. When glycans are shown without linkage information as in Cartoonist and the CFG web site, however, the left-to-right order of the antennae is not generally meaningful. Thus we define two cartoons to be the identical if their unordered set of antennae agree, along with the presence/absence of bisecting GlcNAc and presence/absence of core fucose. Since both manual and automatic annotations can offer multiple cartoons for a given MS peak, we define several types of cartoon agreement:
Perfect agreement: Manual and automatic annotations each give a single cartoon and the two cartoons are identical.
Agreement: Manual annotation gives a single cartoon that is identical to one of the more than one cartoons given by automatic annotation.
Partial agreement: Manual annotation gives more than one cartoon (usually due to unattached substructures) and at least one of the manual cartoons is identical to one of the cartoons given by automatic annotation.
Disagreement: None of the manual cartoons is identical to a cartoon given by automatic annotation.
Figure 5 shows examples of agreement and perfect agreement. We did not need to consider “reverse agreement” in which automatic annotation gives a single cartoon that is one of the multiple manual cartoons, because automatic annotation always gave multiple cartoons when the human expert gave multiple cartoons.
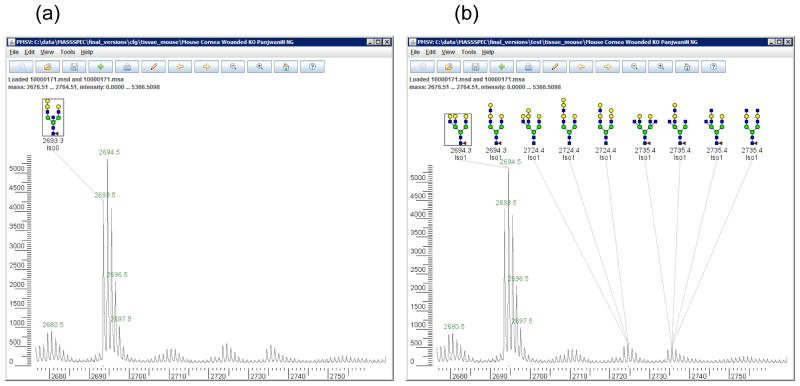
Figure 3 gives the breakdown of the 2465 peaks with both manual and automatic annotations: 1081 perfect agreements, 689 agreements, 568 partial agreements, and 127 disagreements. On peaks for which both Cartoonist and the expert give unique cartoons, the two cartoons agree on 1081 and disagree on 102 peak assignments, for a success rate of over 90%. Figure 7 shows a case of disagreement in which Cartoonist’s demerit system preferred a structure without bisecting GlcNAc.
Figure 7.
(a) Manual annotations on m/z 2680 – 2760 from wounded mouse cornea. (b) Cartoonist’s automatic annotations on the same data. The annotations for the peak at 2694 Da are in “disagreement”, because the human expert and Cartoonist gave disjoint sets of glycans. Neither the expert nor the computer program annotated the peaks at 2680, which most likely represent incompletely permethylated forms of the glycan at 2694.
4. Conclusion
We have described software improvements and an objective evaluation for Cartoonist, a fully automatic annotation tool for MALDI-TOF glycan profiles. We benchmarked Cartoonist’s automatic annotations against human expert annotations on human and mouse N-glycan data from the Consortium for Functional Glycomics using glycan oligosaccharide composition and topology. We found that Cartoonist and expert annotations largely agree, but the expert tends to annotate more specifically, meaning fewer suggested structures per peak, and Cartoonist more comprehensively, meaning more annotated peaks.
There are of course numerous avenues for future improvements: integration with MS2 annotation tools, improved support for modified (phosphorylated, acetylated, and sulfated) glycans, generalization to electrospray ionization methods, support for glycolipids, expert-system knowledge for non-mammalian glycosylation, differential quantification, and so forth. Glycomics is clearly underserved relative to proteomics, because proteomics tools that can handle both MS1 and MS2, both MALDI and electrospray, and so forth, have existed for years. We believe, however, that as high-throughput research efforts incorporate ever higher levels of integration and complexity, there will be greater demand for automation in the analysis of glycosylation.
Cartoonist’s database-search mode annotates and displays the interactive glycan profiles on the Consortium for Functional Glycomics Web site http://www.functionalglycomics.org/. Cartoonist is available by request from the corresponding author; it will be posted for public download when ready.
Supplementary Material
Biological Significance.
N-linked glycosylation is a nearly ubiquitous modification on secreted and membrane proteins, and protein / glycan binding plays numerous biological roles, including endogenous cell signaling and exogenous pathogen recognition. Alterations in glycosylation are a hallmark of many diseases including cancer and auto-immune diseases, and many of these alterations, for example, increases or decreases in the rate of fucosylation or sialylation, are observable from MALDI-TOF profiles. The software described here will enable researchers, even those without specialized training in glycomics, to discover and evaluate potential glycan biomarkers.
Highlights.
Cartoon-level annotation of MALDI-TOF mass spectra of detached glycans
Benchmark comparison of automatic and manual annotations on N-linked glycans
Automatic annotations are more comprehensive but less specific than manual
Cartoonist software provides fast objective analysis of glycan population
Acknowledgments
This work was supported by NIH grant R01GM085718.
Footnotes
- We declare no conflict of interest. Cartoonist is freeware, available upon request.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wada Y, Dell A, Haslam SM, Tissot B, Canis K, Azadi P, et al. Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1. Molecular & cellular proteomics: MCP. 2010;9:719–27. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, et al. Comparison of the methods for profiling glycoprotein glycans--HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17:411–22. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 3.Reinhold V, Zhang H, Hanneman A, Ashline D. Toward a platform for comprehensive glycan sequencing. Molecular & cellular proteomics: MCP. 2013;12:866–73. doi: 10.1074/mcp.R112.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royle L, Radcliffe CM, Dwek RA, Rudd PM. Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. Methods in molecular biology. 2006;347:125–43. doi: 10.1385/1-59745-167-3:125. [DOI] [PubMed] [Google Scholar]
- 5.de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, et al. High-mannose glycans are elevated during breast cancer progression. Molecular & cellular proteomics: MCP. 2011;10:M110 002717. doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruhaak LR, Miyamoto S, Lebrilla CB. Developments in the identification of glycan biomarkers for the detection of cancer. Molecular & cellular proteomics: MCP. 2013;12:846–55. doi: 10.1074/mcp.R112.026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kronewitter SR, De Leoz ML, Strum JS, An HJ, Dimapasoc LM, Guerrero A, et al. The glycolyzer: automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12:2523–38. doi: 10.1002/pmic.201100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–56. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg D, Sutton-Smith M, Paulson J, Dell A. Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics. 2005;5:865–75. doi: 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- 10.Comelli EM, Sutton-Smith M, Yan Q, Amado M, Panico M, Gilmartin T, et al. Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. Journal of immunology. 2006;177:2431–40. doi: 10.4049/jimmunol.177.4.2431. [DOI] [PubMed] [Google Scholar]
- 11.Burlak C, Bern M, Brito AE, Isailovic D, Wang ZY, Estrada JL, et al. N-linked glycan profiling of GGTA1/CMAH knockout pigs identifies new potential carbohydrate xenoantigens. Xenotransplantation. 2013;20:277–91. doi: 10.1111/xen.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bern M, Brito AE, Pang PC, Rekhi A, Dell A, Haslam SM. Polylactosaminoglycan glycomics: enhancing the detection of high-molecular-weight N-glycans in matrix-assisted laser desorption ionization time-of-flight profiles by matched filtering. Molecular & cellular proteomics: MCP. 2013;12:996–1004. doi: 10.1074/mcp.O112.026377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.