Abstract
Studies in myeloid neoplasms have described recurrent IDH1 and IDH2 mutations as primarily mutually exclusive. Over a 6-month period of clinical testing with a targeted next-generation sequencing assay, we evaluated 92 patients with acute myeloid leukemia, myelodysplastic syndrome, and chronic myelomonocytic leukemia and identified a subset of 21 patients (23%) who harbored mutations in either IDH1 or IDH2. Of the 21 patients with IDH mutations, 4 (19%) were found to have single nucleotide variants in both IDH1 and IDH2. An additional patient included in the study was found to have two different IDH2 mutations. The mutations were typically present at different variant allelic frequencies, with one predominating over the other, consistent with the presence of multiple subclones in a single patient. In one case, the variant allelic frequencies in both IDH1 and IDH2 were equally low in the setting of a high percentage of blasts, suggesting that the IDH mutations were unlikely to be present in the founding clone. Given these data, we conclude that dual IDH1/2 mutations likely were previously underestimated, a finding that may carry important treatment implications.
In recent years, whole-genome sequencing of acute myeloid leukemias (AMLs) led to the identification of frequent heterozygous mutations in isocitrate dehydrogenase 1 gene (IDH1).1, 2 Subsequent exome sequencing and targeted re-sequencing studies in AML also identified frequent IDH2 mutations.3, 4 Overall, approximately 15% to 30% of AML have mutations in IDH1 or IDH2, almost exclusively at codon Arg132 for IDH1 and at codons Arg140 and Arg172 for IDH2.1, 3, 4, 5, 6 Patients with cytogenetically normal AML are enriched for these mutations. The mutations confer neomorphic enzyme activity through the NADPH-dependent reduction of the normal end-product α-ketoglutarate to the putative oncometabolite 2-hydroxyglutarate. The accumulation of high levels of 2-hydroxyglutarate in the IDH1/2-mutant tumor provides an important mechanism of cellular transformation through the targeting of epigenetic regulators.7
IDH mutations were also identified in preleukemic clonal malignancies, including myelodysplastic syndromes (MDSs) and myeloproliferative neoplasms (approximately 5% of MDS/myeloproliferative neoplasm and approximately 20% of AML arising from MDS or myeloproliferative neoplasm).1, 4, 8 They occur early in disease pathogenesis in the founding clone9 and act as major initiating mutations.10 Mutations in the two IDH genes are likely mutually exclusive,11 with only rare reports of concurrent IDH1 and IDH2 mutations12 at unknown allelic frequency (AFs).
Testing for these mutations is becoming increasingly important to patient care, given that first-generation IDH inhibitors were shown to suppress the growth of 2-hydroxyglutarate-producing IDH-mutant tumor cells both in vitro and in vivo.13, 14, 15 Clinical trials with IDH1 and IDH2 inhibitors for patients with IDH1/2 mutations are ongoing.16, 17, 18, 19, 20
Because our laboratory launched a next-generation sequencing (NGS)-based tumor genotyping assay in early 2014, we found that IDH1 and IDH2 mutations co-occur in the same tumors more frequently than was reported. This study is a detailed description of five cases of dual mutations we have thus far identified and of the potential implications of these results.
Materials and Methods
Identification of Cases
The Partners HealthCare Institutional Review Board granted approval for this study before its initiation. The electronic files of the Massachusetts General Hospital Pathology Department were queried for all cases run with the use of the NGS assay (SNAPSHOT NGS) since its launch in April 2014 until October 3, 2014, with a primary diagnosis of AML, MDS, or chronic myelomonocytic leukemia. A subsequent query was run to identify the subset of cases with either IDH1 or IDH2 single nucleotide variants (SNVs). Unique patients were enumerated such that multiple specimens sent on a single patient were not individually counted.
Targeted DNA-Seq Using Anchored Multiplex PCR
The SNAPSHOT NGS assay uses a multiplex PCR technology called Anchored Multiplex PCR for SNVs and insertion/deletion detection in genomic DNA with the use of NGS.21 Briefly, genomic DNA was isolated from blood or bone marrow aspirates (QIAcube; Qiagen, Valencia, CA). The genomic DNA was sheared with the Covaris (Woburn, MA) M220 instrument followed by end-repair, adenylation, and ligation with an adapter. A sequencing library that targeted hotspots and exons in 39 genes (Supplemental Table S1) was generated with two hemi-nested PCR reactions with the use of one primer specific to a sequence in the gene of interest and one specific to a universal sequence in the adapter, for each PCR reaction.21 Illumina (San Diego, CA) MiSeq 2 × 151 bp paired-end sequencing results were aligned to the hg19 human genome reference with the use of BWA-MEM.22 PCR/optical duplicates were removed on the basis of unique start sites of sequenced molecules. MuTect23 was used for SNV detection and Oncotator was used for mutation annotation (http://www.broadinstitute.org/oncotator, last accessed May 6, 2015). A minimum threshold of five unique reads that supported the canonical mutation and visualization in JBrowse24 were required to make a variant call.
Focused Ultra-Deep Amplicon Sequencing Using the Ion Torrent PGM Platform
Barcoded and tagged unidirectional PCR primers targeting codons 132 of IDH1 and 140 of IDH2 were designed to generate amplicons approximately 100 bp in length (Table 1). Genomic DNA samples were quantitated with the Qubit high-sensitivity DNA assay kit (Thermo Fisher Scientific, Waltham, MA), individually amplified with Platinum Taq HiFi, purified with Ampure XL beads (Agencourt, Brea, CA), and quantitated with High-Sensitivity DNA chip on the Agilent Technologies BioAnalyzer (Santa Clara, CA). The amplicons were normalized and pooled to generate emulsion PCR libraries on the Ion Torrent OneTouch2 platform with the use of the Ion PGM Template OT2 400 kit (Thermo Fisher Scientific). The libraries were enriched with the Ion Torrent ES platform, and all sequencing was performed with the Ion PGM Hi-Q Sequencing kit on the Ion Torrent PGM platform, analyzed, and visualized with the IGV version 2.3. The average base pair coverage of the amplicon was approximately 100,000 times. Relative frequencies of the mutant alleles were derived from dividing the number of mutant calls by total calls of the relevant position (Supplemental Table S2).
Table 1.
PCR Primers for Focused Ultra-Deep Amplicon Sequencing Using the Ion Torrent PGM Platform
| Primers | Sequence |
|---|---|
| IDH1 primers | |
| IDH1_R132_1F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGGCATGTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_2F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGCGATCTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_3F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGAGCTATCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_4F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGACACATCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_5F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCTCTTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_6F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCAGTTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_7F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTATATTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_8F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGAGTCATCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_9F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTAGCTTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_10F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTGTGTTCACATTATTGCCAACATGACT-3′ |
| IDH1_R132_R | 5′-CCTCTCTATGGGCAGTCGGTGATGCATGCGGTCTTCAGAGAAGCCATT-3′ |
| IDH2 primers | |
| IDH2_R140_1F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGGCATGCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_2F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGCGATCCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_3F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGAGCTACTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_4F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGACACACTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_5F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCTCTCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_6F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCAGTCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_7F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTATATCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_8F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGAGTCACTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_9F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTAGCTCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_10F | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTGTGTCTAGGCGTGGGATGTTTTTG-3′ |
| IDH2_R140_R | 5′-CCTCTCTATGGGCAGTCGGTGATGCATGTCTGTCCTCACAGAGTTCAAGC-3′ |
Bold text indicates sequence of the adapter and barcode.
Droplet Digital PCR for IDH1 c.395G>A, p.Arg132His
Genomic DNA was used as input for digital PCR at 10 ng per reaction. Primers spanned an intron/exon junction and were as follows: IDH1 forward primer, 5′-CTGCAAAAATATCCCCCGGC-3′; IDH1 reverse primer, 5′-CAAGTTGGAAATTTCTGGGCCAT-3′; mutant probe, 5′-ATCATAGGTCGTCATGCTTAT-3′; and wild-type probe, 5′-ATCATAGGTCATCATGCTTAT-3′. Reactions were performed in a 20-μL reaction with the use of the droplet digital PCR Supermix for Probes (no dUTP; Bio-Rad, Hercules, CA). Final concentration of primers and probes were 900 nmol/L and 250 nmol/L, respectively. The relation between the presence or absence of mutant or wild-type molecules in a droplet is defined by the Poisson distribution and allows robust, digital quantification of the two molecules relative to one another (Supplemental Table S3). Droplet generation was performed according to the manufacturer's instructions. Cycling conditions were 95°C for 10 minutes, followed by 40 cycles of 94°C for 30 seconds and 64°C for 2 minutes, and then a final 10-minute incubation at 98°C. The temperature ramp rate was 2.5°C/second. Droplet reading was performed on a QX200 droplet digital PCR droplet reader (Bio-Rad), and the analysis was performed with manual mode on a QuantaSoft Analysis software version 1.4 (Bio-Rad) by adjusting the gates to the no template control samples.
FLT3, NPM1, and CEBPA Mutation Analysis
Specimens were sent to LabCorp (Research Triangle Park, NC) for FLT3, NPM1, and CEBPA mutation analysis. Briefly, PCR amplification for the detection of FLT3 internal tandem duplication mutations (FLT3-ITDs) was performed, and the products were run on the ABI 3500xl genetic analyzer (Thermo Fisher Scientific) for size determination. The ITD wild-type produces a fragment that is approximately 327 bp, whereas the presence of an insertion produces a fragment that is ≥330 bp. PCR and size determination was also performed for NPM1 analysis to detect a 4-bp insertion at nucleotide position 959 (exon 12). For CEBPA analysis, the CEBPA coding region was PCR-amplified and was Sanger sequenced to identify sequence variations.
Immunohistochemistry
Two micron-thick formalin-fixed, paraffin-embedded bone marrow (BM) core biopsy sections were stained with hematoxylin and eosin. Immunohistochemistry for IDH1 was performed on 5-μm–thick formalin-fixed, paraffin-embedded BM biopsy sections with an antibody specific for the mutant IDH1 p.Arg132His protein (dilution 1:150; H09; Dianova, Hamburg, Germany). A labeled streptavidin biotin kit was used as a detection system (Leica Biosystem, Nussloch, Germany). Combined cytoplasmic and nuclear staining was interpreted as immunopositive. Currently, antibodies are only available against IDH1 p.Arg132His mutant protein; no antibodies specific for any of the IDH2 mutated proteins are available for use in immunohistochemistry.
Results
Clinical and Cytogenetic Characteristics
Clinical, cytogenetic, and molecular characteristics of the patients with dual mutations in IDH1 and IDH2 are listed in Table 2. Three of the five patients, all men, exhibited diagnoses of AML. Two additional patients (one female and one male) were diagnosed with MDS (both refractory anemia with excess blasts-2; RAEB-2). The age at diagnosis ranged from 61 to 73 years. The AML patients had BM aspirate or (when not available) peripheral blood blast counts that ranged from 22% to 33% at diagnosis. Blast counts from the RAEB-2 patients were not available for the sequenced specimens. All five patients had normal karyotypes.
Table 2.
Patient Characteristics and IDH1 and IDH2 Single Nucleotide Variants
| Case No. | Dx | Age, years/sex | Speci-men type | Blast, % | IDH1 mutation (allelic frequency, %) | No. of Reads (ALT/REF) | IDH2 mutation (allelic frequency, %) | No. of Reads (ALT/REF) | Other (allelic frequency, %) | Karyo-type | FLT3-ITD, NPM1, CEBPA status |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AML | 63/M | BM | 22 | p.Arg132His (c.395G>A) (12) | 588/4228 | p.Arg140Gln (c.419G>A) (2) | 232/12310 |
SMAD4 p.Ala458Val (c.1373C>T) (45), CDH1 p.Ala592Thr (c.1774G>A) (46) |
46,XY |
FLT3 wild-type NPM1 wild-type CEBPA wild-type |
| 2 | AML | 73/M | BM | 30∗ (PB) |
p.Arg132His (c.395G>A) (44) | 191/256 | p.Arg140Trp (c.418C>T) (5) | 50/1044 | 46,XY |
FLT3 wild-type NPM1 mutated (4 bp insertion at c.959) CEBPA wild-type |
|
| 3 | AML | 61/M | BM | 33 | p.Arg132Cys (c.394C>T) (3) | 7/237 | p.Arg140Gln (c.419G>A) (3) | 19/589 |
NRAS p.Gly12Asp (c.35G>A) (34), NRAS p.Gly12Ser (c.34G>A) (2) |
46,XY |
FLT3-ITD mutated [PCR product of 429 bp (wild-type ∼327 bp)] NPM1 wild-type CEBPA double mutant [p.Asn74X (c.219insT), p.Ala295Thr (c.883G>A)] |
| 4 | RAEB-2 | 68/F | PB | 13† | p.Arg132His (c.395G>A) (4) | 8/199 | p.Arg140Gln (c.419G>A) (12) | 60/482 | 46,XX | NA | |
| 5 | RAEB-2 | 64/M | BM | 13† | p.Arg140Gln (c.419G>A) (27); p.Arg140Trp (c.418C>T) (13) |
337/943, 169/1138 |
46,XY |
FLT3 wild-type NPM1 wild-type CEBPA wild-type |
F, female; M, male; ALT, alternative allele; AML, acute myeloid leukemia; BM, bone marrow; Dx, diagnosis; ITD, internal duplication mutation; NA, not applicable; PB, peripheral blood; RAEB-2, refractory anemia with excess blasts-2; REF, reference allele.
BM was unavailable for use to determine blast percentage by structural characteristics because of extensive necrosis. Instead, the blast percentage from PB is shown.
The blast count was unavailable for the specimen submitted for sequencing. The blast percentage shown is from a different specimen on a different day (from PB 16 days before the sequencing sample for patient 4, and from BM 13 days before the sequencing sample for patient 5.).
Of note, patient 2 had a history of bladder cancer, treated with surgery and Bacille Calmette-Guérin. None of the patients had any reported environmental exposures or a family history of blood disorders or hematologic neoplasms.
Frequency of IDH1/2 Mutations
This study included all AML (n = 53), MDS (n = 34), and chronic myelomonocytic leukemia (n = 5) patients with specimens submitted for the SNAPSHOT NGS assay during the 6-month period from the assay's launch in April 2014 through October 2014 (total n = 92). Of these 92 patients, 21 (23%) harbored IDH mutations (14 AML, 6 MDS, 1 chronic myelomonocytic leukemia). Of these 21 patients, 4 (19%) had coexisting IDH1 and IDH2 mutations. A fifth patient (with MDS) who was included in our study was previously identified as harboring two different IDH2 mutations on a previously used hotspot profiling platform, which were confirmed by the current SNAPSHOT NGS assay. The dual IDH1 and IDH2 variants identified were missense mutations at the canonically mutated codons, Arg132 and Arg140, respectively (Table 2).
Although several IDH mutations were observed at low AF, our confidence in these calls was based on a high depth of coverage (average target region coverage for each of the five samples, 3453; range, 690 to 8909), with a high number of unique molecules supporting the calls (well above background), the mutations' location at canonical sites, and the identification of the mutations on multiple samples from the same patient on repeat testing in some instances (data not shown). In addition, the mutations were verified by an orthogonal sequencing method (see Focused Ultra-Deep Amplicon Sequencing Using the Ion Torrent PGM Platform) (Supplemental Table S2). The IDH1/2 mutational burden for each case (as represented by the variant AF) is described in the sections below along with non-IDH mutations.
Patient 1
Patient 1 had AML with normal karyotype and without FLT3, NPM1, or CEBPA mutations and showed an IDH1 p.Arg132His (c.395G>A) mutation at 12% AF and an IDH2 p.Arg140Gln (c.419G>A) mutation at 2%, in the setting of a marrow blast percentage of 22% (Table 2 and Figure 1). Although the blast count does not necessarily reflect the size of the clonal population, if we assumed a heterozygous IDH1 mutation were present in every cell of the blast population, the marrow involvement by 22% blasts would predict a variant AF of approximately 11%. The calculated AF of 12% by SNAPSHOT NGS, combined with the observation of mutant IDH protein by immunohistochemistry in nearly all of the blasts, is consistent with this assumption (Supplemental Figure S1). SNVs in SMAD family member 4 and CDH1 genes were also detected by the SNAPSHOT NGS assay. The variant AFs of the SNVs in SMAD family member 4 and cadherin 1, type 1 genes, however, were close to 50% (45% and 46%, respectively), suggesting these were not somatic tumor mutations.
Figure 1.
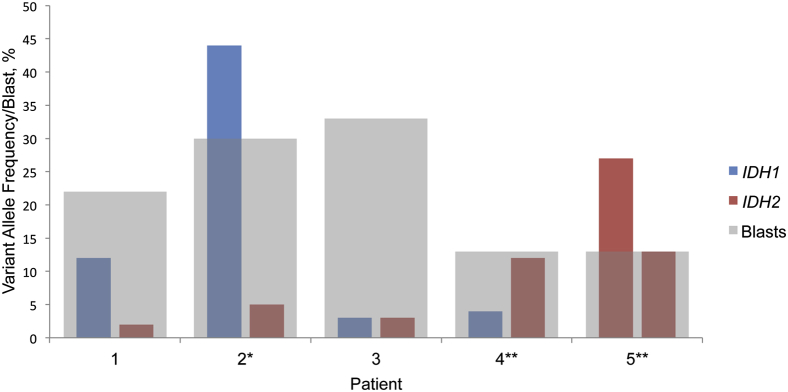
Relation between variant allelic frequency and blast percentage. The histogram displays the variant allelic frequency in the context of the blast count for each of the five patients. The asterisk affixed to Patient 2 indicates blast count was obtained from PB. The double asterisk affixed to Patients 4 and 5 indicate blast count was obtained from a different specimen or date (PB 16 days and BM 13 days before the sequencing sample, respectively). BM, bone marrow; IDH, isocitrate dehydrogenase; PB, peripheral blood.
Patient 2
Patient 2 had AML with normal karyotype and mutated NPM1, but wild-type FLT3 and CEBPA, and showed a predominant IDH1 p.Arg132His (c.395G>A) mutation on the BM aspirate with a 44% AF, whereas an IDH2 p.Arg140Trp (c.418C>T) mutation was present at 5% AF. The marrow blast percentage could not be determined because of extensive necrosis, but a concurrent peripheral blood sample showed a blast percentage of 30% (Table 2 and Figure 1). No other SNVs were detected by SNAPSHOT NGS.
Patient 3
Patient 3 had AML with normal karyotype, a FLT3 ITD mutation, two CEBPA mutations, and no NPM1 mutation. Both an IDH1 p.Arg132Cys (c.394C>T) and an IDH2 p.Arg140Gln (c.419G>A) mutation were present, each at an equally low AF of 3%, in the setting of 33% blasts in the marrow (Table 2 and Figure 1). SNAPSHOT NGS also detected two neuroblastoma RAS viral (v-ras) oncogene homolog (NRAS) gene mutations that were present in trans, both at the hotspot codon 12 [p.Gly12Asp (c.35G>A) and p.Gly12Ser (c.34G>A)] at AFs of 34% and 2%, respectively (Supplemental Figure S2).
Patient 4
Patient 4, diagnosed with RAEB-2 with normal karyotype, harbored a predominant IDH2 p.Arg140Gln (c.419G>A) mutation at 12% AF and an IDH1 p.Arg132His (c.395G>A) mutation at 4% AF (Table 2 and Figure 1). The peripheral blood for this patient had 13% blasts 16 days before the NGS testing. No other SNVs were detected by SNAPSHOT NGS. Mutational analyses for FLT3, NPM1, and CEBPA mutations were not performed.
Patient 5
Patient 5, diagnosed with RAEB-2 with normal karyotype, had two separate IDH2 mutations with AFs of 27% and 13%. The BM aspirate for this patient had 13% blasts 13 days before the NGS testing. The two mutations represented different SNVs that affected the same hot spot codon 140 [p.Arg140Gln (c.419G>A) and p.Arg140Trp (c.418C>T), respectively] and were shown to be in trans, confirmed by visualization on independent sequencing reads (Figure 2). No other SNVs were detected by SNAPSHOT NGS. Mutational analyses by PCR for FLT3, NPM1, and CEBPA did not identify mutations in these genes.
Figure 2.
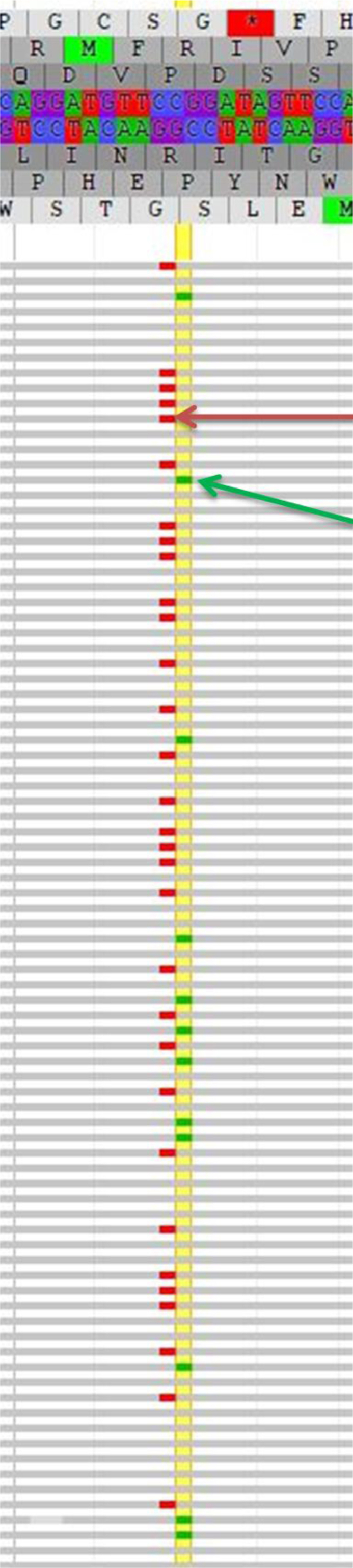
Screenshot of the dual mutations visualized in JBrowse. In the upper portion of the figure is the hg19 reference genome sequence (and corresponding amino acid translation) for codons 137 to 143. The horizontal gray rows indicate individual reads (ie, individual molecules) sequenced in the forward direction. The small red bars indicate a nucleotide change (SNV) from the reference nucleotide G to A (c.419G>A) [the red arrow points to example IDH2 p.Arg140Trp (c.418C>T)]; the small green bars indicate a nucleotide change (SNV) from the reference nucleotide C to T (c.418C>T) [the green arrow points to example IDH2 p.Arg140Gln (c.419G>A)]. The yellow vertical highlight line marks the nucleotides at position 419 in the coding sequence where an SNV was detected. IDH, isocitrate dehydrogenase; SNV, single nucleotide variant.
Validation of Low Allelic Frequency Mutations
The four patient samples harboring mutations at ≤5% AF were resequenced with a second NGS assay that uses a different platform, the Ion Torrent PGM platform. Ultra-deep sequencing (average base coverage, approximately 100,000 times) verified the presence of the mutations at similar AF to the original method (Supplemental Table S2). For patients with the IDH1 Arg132His mutation (patients 1, 2, and 4), droplet digital PCR, an additional orthogonal method, was used to confirm the mutations, one of which was ≤5% AF (Supplemental Table S3).
Discussion
Recurrent IDH mutations were described in a variety of malignancies, including gliomas,25, 26 chondrosarcomas,27, 28, 29 cholangiocarcinomas,30 and breast carcinomas,31 and also in a number of myeloid neoplasms such as AML, MDS, and myeloproliferative neoplasm.1, 32 Both IDH1 and IDH2 encode IDH enzymes that normally function to convert isocitrate to α-ketoglutarate. Mutant enzymes, however, reduce α-ketoglutarate to the oncometabolite 2-hydroxyglutarate, which in turn goes on to inhibit α-ketoglutarate–dependent enzymes, such as TET2, resulting in aberrant epigenetic modifications.11 Mutant IDH1 and IDH2 enzymes thus serve as promising therapeutic targets, and specific inhibitors are currently in various stages of development.13, 14, 15, 16, 17, 18, 19, 20, 33, 34
With the use of a sensitive NGS mutation panel, we identified dual mutations in IDH1 and IDH2 that occur at greater frequency than previously described. Among the IDH-mutated myeloid neoplasms (n = 21), nearly one-quarter were found to have dual IDH mutations (5 of 21 patients, including one patient who harbored two IDH2 mutations.) The previous under-recognition of dual mutations is likely because of the lack of highly sensitive sequencing assays with the ability to routinely detect variants that are only present at an AF of 1% to 5% of reads. Although in some cases the detected IDH1/2 mutations were present at an AF as high as 44%, we were also able to identify low-level mutations at 2% to 3% AF that would have otherwise been missed by less-sensitive assays. Importantly, we verified the mutations with ultra-deep sequencing with the use of an alternative NGS platform, an orthogonal method, to help rule out the possibility that the low-level mutations were a result of sequencing error.35 Furthermore, dual IDH mutations were not detected in 1390 other tumor samples that concurrently underwent SNAPSHOT NGS sequencing between April and October 2014 (data not shown).
In addition to identifying low-level mutations, we also found that multiple IDH1/2 mutations may be present at substantially different AFs. In fact, in four of the five patients, one IDH1/2 mutation predominated compared with the other, with a twofold to ninefold difference between the mutations' AFs (patient 1, IDH1 12% versus IDH2 2%; patient 2, 44% versus 5%; patient 4, 4% versus 12%; patient 5, IDH2 p.Arg140Gln 27% versus IDH2 p.Arg140Trp 13%) (Table 2). Recognition of the high frequency of concomitant mutations in IDH1 and IDH2 underscores the utility of panel testing and of sufficient read coverage for accurate and comprehensive molecular diagnosis. It may also affect decisions about conventional or targeted therapy.
The identification of mutations at low AFs, often in the setting of relatively extensive myeloblast marrow infiltration, may suggest that IDH1/2 mutations do not always occur in the founding neoplastic clone. This may have important implications about the role of mutant IDH1 and IDH2 in the clonal evolution of myeloid neoplasms and their targeted inhibition. Of note, in our study, two of the five patients had co-occurring non-IDH mutations. Patient 2 harbored an NPM1 mutation (AF unknown), and patient 3 had two canonical mutations in neuroblastoma RAS viral (v-ras) oncogene homolog gene (p.Gly12Asp, p.Gly12Ser), of which one was at much higher AF (34%) than the concurrent IDH mutations (3% each), as well as FLT3-ITD and dual CEBPA mutations (AF unknown) (Table 2). Although the remaining three patients did not harbor other somatic mutations among the 39 genes tested in our NGS panel, the panel did not assess for a number of genes relevant to myeloid neoplasms such as DNMT3A, TET2, or ASXL1 (Supplemental Table S1). Overall, our findings suggest that mutations in IDH1/2 may not always be present in the founding clone and rather may represent simultaneous or sequential events throughout clonal evolution.
In summary, we report that concomitant mutations in IDH1 and IDH2 are not infrequent events in IDH-mutated myeloid neoplasms. The use of panel testing that has sufficient depth of coverage, along with the reporting of allele frequencies of identified mutations, may have considerable impact on both the initial diagnostics and subsequent clinical decision making about therapy. Finally, although it remains to be definitively demonstrated whether IDH1/2 mutations co-occur in the same tumor cells or in discrete tumor cell subsets, our data support the co-existence of multiple tumor subclones within individual patients. This finding may have practical consequences; although the initial clinical data with the use of IDH-specific inhibitors may be encouraging, other strategies such as simultaneous inhibition of both IDH1 and IDH2 activity, in addition to other antileukemic therapies, may be necessary for patients with both IDH1- and IDH2-mutation–positive subclones.
Footnotes
Supported by T32 training/NIH grant T32 CA 71345-18 (A.M.B.) and Vancouver Coastal Health Research Institute Mentored Clinician Scientist Award, BrainCare BC (S.Y.).
Disclosures: A.T.F. has participated in advisory boards for Agios Pharmaceuticals; Z.Z., L.P.L., and A.J.I. hold patents for the Anchored Multiplex PCR technology, licensed to ArcherDx; Z.Z., L.P.L., and A.J.I. hold stocks in ArcherDx; and L.P.L. and A.J.I. were consultants for Enzymatics, which had acquired ArcherDx.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2015.06.004.
Supplemental Data
Representative image of blasts from a high-grade myelodysplastic syndrome case stained with the mutant IDH1 antibody. A: Hematoxylin and eosin staining. B: IDH1 p.Arg132His immunohistochemical staining. Nearly all of the blasts stain positive for the mutant IDH protein. IDH, isocitrate dehydrogenase.
Screenshot of the dual NRAS mutations visualized in JBrowse. In the upper portion of the figure is the hg19 reference genome sequence (and corresponding amino acid translation) for NRAS codons 9 to 14. The horizontal gray rows indicate individual reads (ie, individual molecules) sequenced in the forward direction; the horizontal light blue rows indicate individual reads sequenced in the reverse direction. The small red bars indicate a nucleotide change (SNV) from the reference nucleotide G to A. The dark blue ticks indicate soft clips. The yellow vertical line marks the NRAS nucleotides at position 34 in the coding sequence where an SNV was detected. Labeled arrows are pointing to examples of each of the two SNVs.
References
- 1.Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward P.S., Patel J., Wise D.R., Abdel-Wahab O., Bennett B.D., Coller H.A., Cross J.R., Fantin V.R., Hedvat C.V., Perl A.E., Rabinowitz J.D., Carroll M., Su S.M., Sharp K.A., Levine R.L., Thompson C.B. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcucci G., Maharry K., Wu Y.Z., Radmacher M.D., Mrozek K., Margeson D., Holland K.B., Whitman S.P., Becker H., Schwind S., Metzeler K.H., Powell B.L., Carter T.H., Kolitz J.E., Wetzler M., Carroll A.J., Baer M.R., Caligiuri M.A., Larson R.A., Bloomfield C.D. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green C.L., Evans C.M., Zhao L., Hills R.K., Burnett A.K., Linch D.C., Gale R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118:409–412. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- 6.Patel J.P., Gonen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., Huberman K., Cheng J., Viale A., Socci N.D., Heguy A., Cherry A., Vance G., Higgins R.R., Ketterling R.P., Gallagher R.E., Litzow M., van den Brink M.R., Lazarus H.M., Rowe J.M., Luger S., Ferrando A., Paietta E., Tallman M.S., Melnick A., Abdel-Wahab O., Levine R.L. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losman J.A., Looper R.E., Koivunen P., Lee S., Schneider R.K., McMahon C., Cowley G.S., Root D.E., Ebert B.L., Kaelin W.G., Jr. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chotirat S., Thongnoppakhun W., Wanachiwanawin W., Auewarakul C.U. Acquired somatic mutations of isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) in preleukemic disorders. Blood Cells Mol Dis. 2015;54:286–291. doi: 10.1016/j.bcmd.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Lin T.L., Nagata Y., Kao H.W., Sanada M., Okuno Y., Huang C.F., Liang D.C., Kuo M.C., Lai C.L., Lee E.H., Shih Y.S., Tanaka H., Shiraishi Y., Chiba K., Lin T.H., Wu J.H., Miyano S., Ogawa S., Shih L.Y. Clonal leukemic evolution in myelodysplastic syndromes with TET2 and IDH1/2 mutations. Haematologica. 2014;99:28–36. doi: 10.3324/haematol.2013.091249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., Tallman M.S., Sun Z., Wolniak K., Peeters J.K., Liu W., Choe S.E., Fantin V.R., Paietta E., Lowenberg B., Licht J.D., Godley L.A., Delwel R., Valk P.J., Thompson C.B., Levine R.L., Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paschka P., Schlenk R.F., Gaidzik V.I., Habdank M., Kronke J., Bullinger L., Spath D., Kayser S., Zucknick M., Gotze K., Horst H.A., Germing U., Dohner H., Dohner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 13.Popovici-Muller J., Saunders J.O., Salituro F.G., Travins J.M., Yan S., Zhao F., Gross S., Dang L., Yen K.E., Yang H., Straley K.S., Jin S., Kunii K., Fantin V.R., Zhang S., Pan Q., Shi D., Biller S.A., Su S.M. Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett. 2012;3:850–855. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohle D., Popovici-Muller J., Palaskas N., Turcan S., Grommes C., Campos C., Tsoi J., Clark O., Oldrini B., Komisopoulou E., Kunii K., Pedraza A., Schalm S., Silverman L., Miller A., Wang F., Yang H., Chen Y., Kernytsky A., Rosenblum M.K., Liu W., Biller S.A., Su S.M., Brennan C.W., Chan T.A., Graeber T.G., Yen K.E., Mellinghoff I.K. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., Travins J., DeLaBarre B., Penard-Lacronique V., Schalm S., Hansen E., Straley K., Kernytsky A., Liu W., Gliser C., Yang H., Gross S., Artin E., Saada V., Mylonas E., Quivoron C., Popovici-Muller J., Saunders J.O., Salituro F.G., Yan S., Murray S., Wei W., Gao Y., Dang L., Dorsch M., Agresta S., Schenkein D.P., Biller S.A., Su S.M., de Botton S., Yen K.E. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 16.Hansen E., Quivoron C., Straley K., Lemieux R.M., Popovici-Muller J., Sadrzadeh H., Fathi A.T., Gliser C., David M., Saada V., Micol J.-B., Bernard O., Dorsch M., Yang H., Su M., Agresta S., de Botton S., Penard-Lacronique V., Yen K. AG-120, an oral, selective, first-in-class, potent inhibitor of mutant IDH1, reduces intracellular 2HG and induces cellular differentiation in TF-1 R132H cells and primary human IDH1 mutant AML patient samples treated ex vivo. Blood. 2014;124:3734. [Google Scholar]
- 17.Shih A.H., Shank K.R., Meydan C., Intlekofer A.M., Ward P., Thompson C.B., Melnick A.M., Travins J., Straley K., Gliser C., Yen K., Levine R.L. AG-221, a small molecule mutant IDH2 inhibitor, remodels the epigenetic state of IDH2-mutant cells and induces alterations in self-renewal/differentiation in IDH2-mutant AML model in vivo. Blood. 2014;124:437. [Google Scholar]
- 18.Quivoron C., David M., Straley K., Travins J., Kim H., Chen Y., Zhu D., Saada V., Bawa O., Opolon P., Polrot M., Micol J.-B., Willekens C., Bernard O., Yang H., Agresta S., de Botton S., Yen K., Penard-Lacronique V. AG-221, an oral, selective, first-in-class, potent IDH2-R140Q mutant inhibitor, induces differentiation in a xenotransplant model. Blood. 2014;124:3735. [Google Scholar]
- 19.Stein E.M., Altman J.K., Collins R., DeAngelo D.J., Fathi A.T., Flinn I., Frankel A., Levine R.L., Medeiros B.C., Patel M., Pollyea D.A., Roboz G.J., Stone R.M., Swords R.T., Tallman M.S., Agresta S., Fan B., Yang H., Yen K., de Botton S. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant metabolic enzyme, induces durable remissions in a phase I study in patients with IDH2 mutation positive advanced hematologic malignancies. Blood. 2014;124:115. [Google Scholar]
- 20.Fan B., Chen Y., Wang F., Yen K., Utley L., Almon C., Biller S., Agresta S., Yang H. Evaluation of pharmacokinetic-pharmacodynamic (PKPD) relationship of an oral, selective, first-in-class, potent IDH2 inhibitor, AG-221, from a phase 1 trial in patients with advanced IDH2 mutant positive hematologic malignancies. Blood. 2014;124:3737. [Google Scholar]
- 21.Zheng Z., Liebers M., Zhelyazkova B., Cao Y., Panditi D., Lynch K.D., Chen J., Robinson H.E., Shim H.S., Chmielecki J., Pao W., Engelman J.A., Iafrate A.J., Le L.P. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westesson O., Skinner M., Holmes I. Visualizing next-generation sequencing data with JBrowse. Brief Bioinform. 2013;14:172–177. doi: 10.1093/bib/bbr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K.W., Velculescu V.E., Vogelstein B., Bigner D.D. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., Olivi A., McLendon R., Rasheed B.A., Keir S., Nikolskaya T., Nikolsky Y., Busam D.A., Tekleab H., Diaz L.A., Jr., Hartigan J., Smith D.R., Strausberg R.L., Marie S.K., Shinjo S.M., Yan H., Riggins G.J., Bigner D.D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr D.A., Lopez H.U., Deshpande V., Hornicek F.J., Duan Z., Zhang Y., Rosenberg A.E., Borger D.R., Nielsen G.P. Molecular distinction of chondrosarcoma from chondroblastic osteosarcoma through IDH1/2 mutations. Am J Surg Pathol. 2013;37:787–795. doi: 10.1097/PAS.0b013e31827ab703. [DOI] [PubMed] [Google Scholar]
- 28.Arai M., Nobusawa S., Ikota H., Takemura S., Nakazato Y. Frequent IDH1/2 mutations in intracranial chondrosarcoma: a possible diagnostic clue for its differentiation from chordoma. Brain Tumor Pathol. 2012;29:201–206. doi: 10.1007/s10014-012-0085-1. [DOI] [PubMed] [Google Scholar]
- 29.Amary M.F., Bacsi K., Maggiani F., Damato S., Halai D., Berisha F., Pollock R., O'Donnell P., Grigoriadis A., Diss T., Eskandarpour M., Presneau N., Hogendoorn P.C., Futreal A., Tirabosco R., Flanagan A.M. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 30.Borger D.R., Tanabe K.K., Fan K.C., Lopez H.U., Fantin V.R., Straley K.S., Schenkein D.P., Hezel A.F., Ancukiewicz M., Liebman H.M., Kwak E.L., Clark J.W., Ryan D.P., Deshpande V., Dias-Santagata D., Ellisen L.W., Zhu A.X., Iafrate A.J. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fathi A.T., Sadrzadeh H., Comander A.H., Higgins M.J., Bardia A., Perry A., Burke M., Silver R., Matulis C.R., Straley K.S., Yen K.E., Agresta S., Kim H., Schenkein D.P., Borger D.R. Isocitrate dehydrogenase 1 (IDH1) mutation in breast adenocarcinoma is associated with elevated levels of serum and urine 2-hydroxyglutarate. Oncologist. 2014;19:602–607. doi: 10.1634/theoncologist.2013-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosmider O., Gelsi-Boyer V., Slama L., Dreyfus F., Beyne-Rauzy O., Quesnel B., Hunault-Berger M., Slama B., Vey N., Lacombe C., Solary E., Birnbaum D., Bernard O.A., Fontenay M. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–1096. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- 33.Chaturvedi A., Araujo Cruz M.M., Jyotsana N., Sharma A., Yun H., Gorlich K., Wichmann M., Schwarzer A., Preller M., Thol F., Meyer J., Haemmerle R., Struys E.A., Jansen E.E., Modlich U., Li Z., Sly L.M., Geffers R., Lindner R., Manstein D.J., Lehmann U., Krauter J., Ganser A., Heuser M. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122:2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 34.Pirozzi C.J., Reitman Z.J., Yan H. Releasing the block: setting differentiation free with mutant IDH inhibitors. Cancer Cell. 2013;23:570–572. doi: 10.1016/j.ccr.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R., Bertoni A., Swerdlow H.P., Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative image of blasts from a high-grade myelodysplastic syndrome case stained with the mutant IDH1 antibody. A: Hematoxylin and eosin staining. B: IDH1 p.Arg132His immunohistochemical staining. Nearly all of the blasts stain positive for the mutant IDH protein. IDH, isocitrate dehydrogenase.
Screenshot of the dual NRAS mutations visualized in JBrowse. In the upper portion of the figure is the hg19 reference genome sequence (and corresponding amino acid translation) for NRAS codons 9 to 14. The horizontal gray rows indicate individual reads (ie, individual molecules) sequenced in the forward direction; the horizontal light blue rows indicate individual reads sequenced in the reverse direction. The small red bars indicate a nucleotide change (SNV) from the reference nucleotide G to A. The dark blue ticks indicate soft clips. The yellow vertical line marks the NRAS nucleotides at position 34 in the coding sequence where an SNV was detected. Labeled arrows are pointing to examples of each of the two SNVs.


