Abstract
The detection of point mutations is required in the diagnosis of many human diseases. The conformal specificity of DNA ligases was elegantly used to distinguish single-nucleotide mismatches. However, to detect point mutations in RNA retroviruses, conventional ligase-mediated approaches require the reverse transcription of viral genomes before separate ligation and amplification steps. We developed one-step ligation on RNA amplification (LRA) for the direct detection of RNA point mutations. The process combines the ligase-mediated joining of two oligonucleotides and subsequent hot start amplification into a single-tube reaction. We report that modifications to the structure of the oligonucleotide ligation probes improve the rate of ligation and the specificity of mutation detection on RNA. We applied LRA to the detection of a common, clinically relevant HIV-1 reverse transcriptase drug-resistant point mutation, K103N, and compared it with allele-specific PCR and pyrosequencing. LRA achieved a limit of specific quantitation of 1:100 (1%), and a limit of specific detection for mutant K103N RNA transcripts among excess wild-type strands of 1:10,000 (0.01%). LRA also exhibited good detection threshold of 5 × 102 copies/μL K103N RNA transcripts. LRA is a novel point mutation detection method, with potential utilization in HIV drug resistance detection and early diagnostics of genetic disorders associated with other infectious diseases and cancer.
Rapid and sensitive diagnosis of infectious diseases and detection of drug resistance that causes point mutations can transform global health.1, 2, 3, 4, 5, 6 Specifically, personalized medicine such as the detection of point mutations in HIV-1 genes that are associated with drug resistance, has become essential in prescribing the optimal antiretroviral regimens in resource-rich settings.7, 8, 9 Consequently, many methods have been developed over the years for the detection of HIV single-point mutations.10, 11 The availability of drug-resistance testing for individual patient care in global, resource-limited settings, where HIV predominates and multiple subtypes circulate, would be invaluable.12
In their pioneering work, Landegren et al13 found that two oligonucleotides, annealed immediately adjacent to each other on a complementary target DNA molecule, can be joined covalently by the action of a DNA ligase and can distinguish single-nucleotide substitutions, provided that the nucleotides at the junction are correctly base-paired. Various ligation-mediated methods were consequently developed to detect point mutations in infectious and noninfectious circumstances, including Oligonucleotide Ligation Assay14, 15 and Ligation Amplification assay.16, 17 Other quantitative PCR-based detection techniques, such as allele-specific PCR (ASPCR)18 and variations of multiplexing PCR assays,19 depend on the retarded activity of DNA polymerase to extend on templates when there is a primer mismatch at the 3′ end. However, all ligation-mediated methods and ASPCR are DNA based, requiring initial reverse transcription of RNA to cDNA. The low-intrinsic fidelity of reverse transcription enzymes increases the likelihood of mis-incorporation of nucleotides, as occurs with the commercially available reverse transcriptase derived from Moloney Murine Leukemia Virus, which has an error rate that ranges from 4.0 × 10−6 to 0.7 × 10−4 per incorporated nucleotide for different mispairs.20 Therefore, the fidelity of the reverse transcription step limits the required specificity (ie, the ability to distinguish a target mutation from wild types, as determined by the rate of reaction on the mutant sequences compared with that on the wild types) of point mutation assays performed on synthesized cDNA.18 Ligase-mediated detection was previously applied to miRNA21, 22 in a two-step process initiated by a ligation reaction and followed by a cyclic amplification reaction. Although the use of ligase chain reaction in those previous studies enabled an exponential amplification detection scheme,23 the two-buffer process increases the complexity and overall assay time.
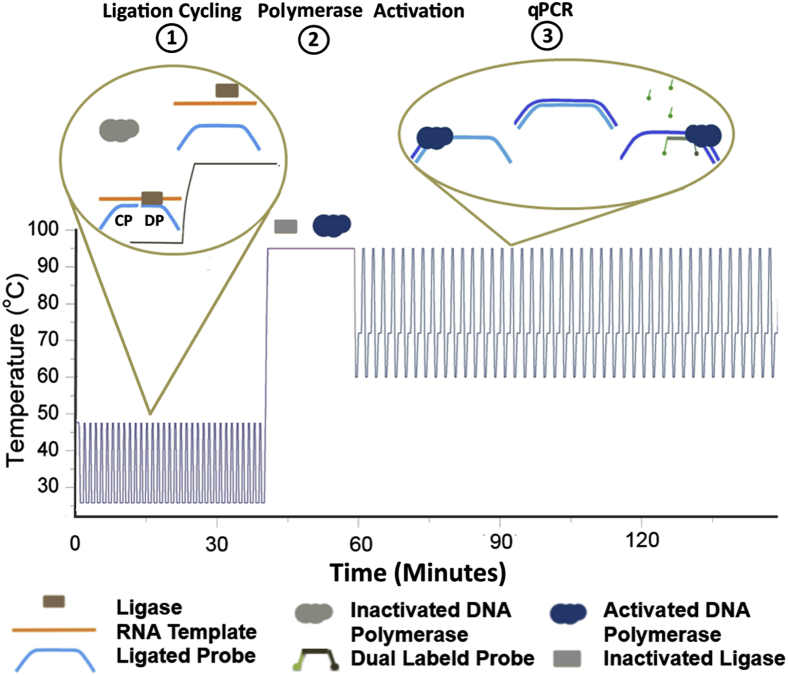
We developed LRA, a one-step, single-buffer method for the detection of point mutations from RNA templates. LRA contains three stages: ligation, polymerase activation, and quantitative PCR (Figure 1). This single-tube assay is achieved by combining a ligase, hot start DNA polymerase, and oligonucleotide probes in an optimized buffer. In the first ligation stage, a common probe (CP) fully complementary to the RNA target and a detector probe (DP) with the 3′ end nucleotide complementary only to the variant to be detected are hybridized adjacently to the RNA template. If both CP and DP are fully complementary to the RNA, they will be joined by the T4 DNA ligase. The rate of ligation will be retarded if the DP has a single-nucleotide mismatch on the 3′ end with the target RNA. The DNA polymerase remains in this stage in its chemically inactivated state, which prevents it from extending the forward primer along the CP and cleaving the RNA template during ligation cycling. In the second polymerase activation stage, the T4 DNA ligase becomes heat inactivated, and DNA polymerase extension and endonuclease activities are restored. In the third quantitative PCR stage, the ligated probes are amplified with a sequence-specific dual-labeled probe for detection.
Figure 1.
A schematic overview of the three stages of ligation on RNA amplification. The temperature cycling during the ligation stage 1 allows for a linear increase of ligated products and off sets the lower hybridization rate of low copies of mutant in high wild-type concentration mixtures. The ligase is heat inactivated, whereas the polymerase becomes activated during stage 2. The polymerase initiates quantitative PCR amplification of the ligated products during stage 3.
We report here that the ligation probe length affects the ligation specificity and its efficiency on RNA and investigate the effects of probe chemical structure modification on LRA specificity and reaction rate. We applied this new method to the detection of K103N (lysine to asparagine change at the 103rd position of the HIV-1 reverse transcriptase), a common mutation that confers high-level resistance to first-generation nonnucleoside reverse transcriptase inhibitors8 and found that it outperforms ASPCR and pyrosequencing in K103N detection specificity.
Materials and Methods
Design of Ligation Probes, PCR Primers, and Dual-Labeled PCR Probes
All custom-designed probes and primers were purchased from Integrated DNA Technologies (Coralville, IA) and Eurofins Operon (Huntsville, AL). PAGE purification was performed on the oligonucleotides when possible, otherwise high-performance liquid chromatography purification was used. The sequences of the probes and primers used are summarized in Table 1.
Table 1.
List of Sequences of Oligonucleotide Probes Used for LRA, Pyrosequencing, and ASPCR Reactions
| Name | Sequence |
|---|---|
| K103 wild type | 5′-ACAUCCUGCAGGGUUAAAACAGAAAAAAUCAGUAACAGUACUGGAUGU-3′ |
| K103N mutant | 5′-ACAUCCUGCAGGGUUAAAACAGAACAAAUCAGUAACAGUACUGGAUGU-3′ |
| K103N DP | 5′-CACGAGTAGACACCTGCCATCCAAGCACAAGCTCTCCTGCTGAGTTACTGATC(fluoro-U)G-3′ |
| CP | 5′-/5phos/TTCTGTTTTAACCAATCGTCTTCTAGCACCACTGC/6Spacer/-3′ |
| LRA reverse primer | 5′-CACGAGTAGACACCTGCCA-3′ |
| LRA forward primer | 5′-GCAGTGGTGCTAGAAGACGA-3′ |
| Dual-labeled probe | 5′-CCAAGCACAAGCTCTCCTGCTG-3′ |
| Pyrosequencing primer | 5′-CCATCACCTACATCC-3′ |
| Pyrosequencing amplicon | 5′-TCCTGCAGGGTTAAAACAGAAAAAATCAGTAACAGTACTGGATGTAGGTGATGGAAGTACGG-3′ |
| Pyrosequencing PCR forward primer | 5′-/biotin/TCCTGCAGGGTTAAAACAGAA-3′ |
| Pyrosequencing PCR reverse primer | 5′-CCGTACTTCCATCACCTACATC-3′ |
| ASPCR forward primer | 5′-CCTACATCCAGTACTGTTACTGA-3′ |
| ASPCR reverse primer | 5′-CCTGCAGGGTTAAAACAGAAC-3′ |
ASPCR, allele-specific PCR; CP, common probe; DP, detector probe; LRA, ligation on RNA amplification.
Pre-Adenylation of the CP
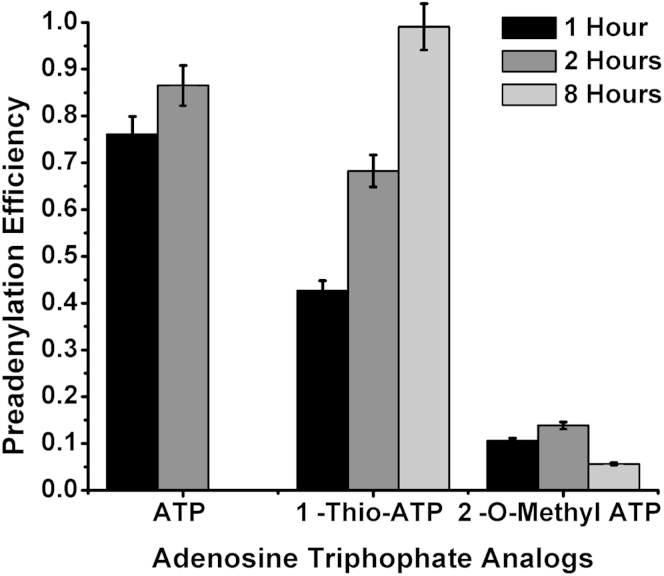
The common ligation probe was chemically phosphorylated by Integrated DNA Technologies at the 5′ end and has a 3′ six-carbon glycol spacer to prevent circularization. For ligation reactions performed with pre-adenylated probes, the 5′ phosphorylated ends of the probes were enzymatically adenylated with a 5′ DNA Adenylation Kit from New England Biolabs (Ipswich, MA). Two ATP analogs, 1-Thio-ATP and 2-O-Methyl-ATP, purchased from Trilink Biotechnologies (San Diego, CA), were also used with the pre-adenylation kit. The reaction was performed according to the manufacturer's guidelines with the use of equimolar concentrations of Mth RNA Ligase and probe for 1, 2, or 8 hours at 65°C. After 10 minutes of heat inactivation at 85°C, the pre-adenylated probes were purified with a QIAquick Nucleotide Removal Kit from Qiagen (Germantown, MD) to remove all salts, enzymes, and residual ATP from the reaction. The concentration of the eluted probes was measured with Nanodrop ND-1000 Spectrophotometer from Thermo Scientific (Waltham, MA). The adenylation efficiency (Supplemental Figure S1) was determined with Bioanalyzer Gel Electrophoresis (Agilent Technologies, Santa Clara, CA).
LRA Reactions
T4 DNA ligase was purchased from New England Biolabs. AmpliTaq Gold 360 DNA Polymerase was purchased from Life Technologies (Grand Island, NY), and dNTPs were purchased from TriLink Biotechnologies (San Diego, CA). EvaGreen was purchased from Biotium (Hayward, CA). Reactions were performed on PikoReal quantitative PCR cycler from Thermo Scientific (Waltham, MA) for 10-μL volume and a custom droplet-based real-time PCR platform for 5-μL reactions.24 Reactions were performed in buffer with pH of 8.3, containing 20 mmol/L Tris-HCl, 50 mmol/L KCl, 10 mmol/L dithiothreitol, and 2.5 mmol/L MgCl2 (Supplemental Figure S2). CP (0.05 pmol) and DP (0.03 pmol) were used with 1 unit of polymerase and 5 units of ligase. Five units of SUPERase•In RNase Inhibitor from Life Technologies were used in each reaction. In reactions that contained ATP, it was present at 20 μmol/L. The normalized specificity is determined by the ratio of specific reaction (mutant DP on mutant K103N RNA) to unspecific reaction (mutant DP on wild-type RNA) for each set of modifications normalized to the no modification control. LRA reactions were performed with 10 rounds of ligation at 25°C for 120 seconds and 43°C for 5 seconds, then 10 minutes of heat activation at 95°C, and followed by 50 cycles of PCR at 95°C for 5 seconds, 63°C for 5 seconds, and 72°C for 10 seconds. LRA reactions were run in triplicates. For reaction determining the effects of modifications, 109 copies of both wild-type and mutant RNA were used. For reaction determining the effects of probe length and mismatch at the n and n–1 sites, 108 copies of both wild-type and mutant RNA were used. For reactions determining the specificity of mutant in an excess of wild types, 109 copies of RNA were used. For reactions determining the detection threshold, 1 μL of each different concentration of mutant RNA was used. The relative specificity and efficiency were calculated from quantitative PCR results by running different sets of conditions for both mutant and wild-type RNA against a control. Sample calculations are shown in Supplemental Tables S1–S3.
Pyrosequencing
Pyrosequencing primers were designed in house (Table 1). Samples were prepared in house according to the manufacturer's guidelines. Sequencing runs were performed on a PyroMark Q24 machine from Qiagen at the Protein and Nucleic Acids facility, Stanford University.
ASPCR
Reactions were performed in 50-μL volumes with the use of SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit from Invitrogen (Grand Island, NY). ASPCR primers are included in Table 1. Reactions were performed on PikoReal quantitative PCR cycler from Thermo Scientific. ASPCRs were performed with a reverse transcription step of 3 minutes at 50°C, then a heat activation step of 95°C for 5 minutes, followed by 60 cycles of 95°C for 15 seconds and 60°C for 30 seconds.
Preparation of RNA Transcripts
The K103N mutation was introduced into the polymerase gene of the HIV-1 genome with the use of site-directed mutagenesis (Agilent Technologies, Palo Alto, CA). Wild-type (K103, aaa codon) and mutant (K103N, aac codon) fragments were in vitro transcribed into RNA with the use of T7 RNA Polymerase (Promega, Madison, WI) and DNase treated per the manufacturer's instructions. RNA fragments were purified with RNeasy MinElute Cleanup Kit from Qiagen, and concentration was determined with the Qubit RNA Assay Kit from Life Technologies.
Results
Optimization of LRA Buffer Conditions
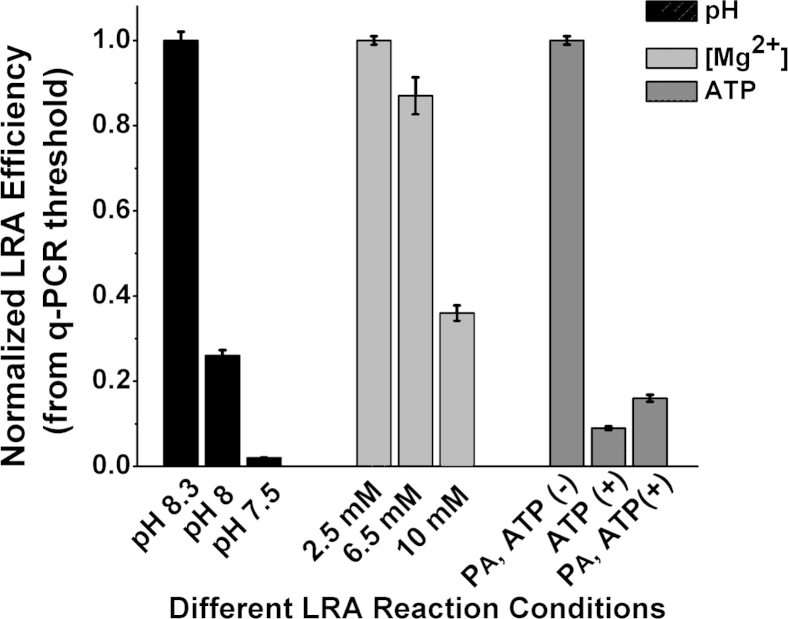
To investigate the effects of ATP concentration on the rate of ligation, we performed enzymatic pre-adenylation of the CP. During pre-adenylation, an AMP group is transferred onto the 5′ phosphate of the CP before the LRA reaction. Supplemental Figure S1 shows the efficiency of enzymatic pre-adenylation of CPs with the use of unmodified ATP and two ATP analogs. After 2 hours of pre-adenylation with unmodified ATP, 87% of CPs is charged with an AMP group. After 8 hours of incubation with 1-Thio-ATP, 99% of CPs is charged with AMP. The percentage of CPs charged with AMP group remains <15% after 8 hours of incubation with 2-O-Methyl-ATP, indicating low pre-adenylation efficiency. When pre-adenylated CPs are used, no ATP is required for the ligation step of the assay. Supplemental Figure S2 shows that pre-adenylation increased LRA efficiency more than six times compared with when uncharged CPs were used. When ATP is present and pre-adenylated CPs are used, the rate of the reactions slows down by almost 10-fold compared with reactions without ATP (Supplemental Figure S2).
Ligation reactions that used T4 DNA ligase require a buffer with pH of 7.5, whereas PCR is usually performed in a buffer with pH ≥8. To determine the optimum pH for one-step LRA, different buffers with pH 7.5, 8, and 8.3 were tested. The most efficient reaction, as determined by the normalized threshold value from the quantitative PCR amplification, resulted from using a buffer with pH 8.3 (Supplemental Figure S2). The efficiency increased 50-fold as the pH increased from 7.5 to 8.3, implying that the ligase is more tolerant of pH changes than the polymerase.
Although ligation reactions have an optimal magnesium concentration of 10 mmol/L, PCR requires a much lower concentration of Mg2+. LRA reactions performed with 2.5 mmol/L Mg2+ resulted in the most efficient reaction (Supplemental Figure S2).
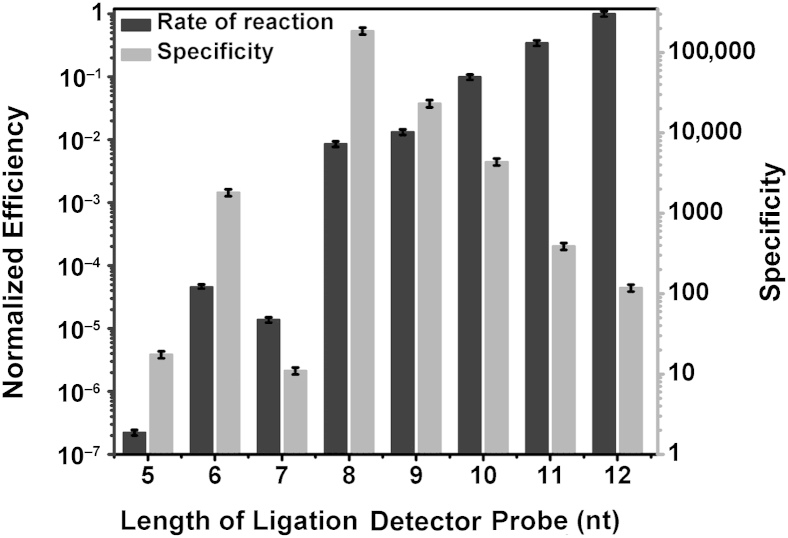
Effect of Ligation Probe Length on RNA Ligation Efficiency and Specificity
To understand the effect of the length of ligation probes on LRA efficiency and specificity, we performed experiments with the use of DP with different hybridization region length while keeping that of the CP constant at 12 nucleotides (nt) (Figure 2). The specificity of LRA is measured by how much faster its rate is on the target mutant templates compared with the nontarget wild-type templates. The efficiency is determined by the rate of reaction of different lengths of DP on mutant templates normalized to the optimal (fastest) rate. Both the specificity and the efficiency are obtained with the relative threshold values from the quantitative PCR amplification (1/2ΔCt). As the length of DP increases, the normalized rate of the LRA reaction increases, and the specificity of LRA decreases. The efficiency of ligation on RNA approaches its maximum as the length of DP is increased to 12 nt, suggesting that a minimum probe length of 12 nt is required for efficient ligation on RNA molecules (Figure 2). At that DP length, the reaction on mutant templates is 117-fold more efficient than on wild-type templates.
Figure 2.
The effects of length of ligation probes on the rate and the specificity of ligation on RNA. The right y axis indicates the specificity expressed as rate of reaction on mutant templates divided by the rate of reaction on wild-type templates. The left y axis indicates the efficiency expressed as rate of reaction with the use of different detector probe lengths divided by the optimum rate. Data are expressed as means ± SD. nt, nucleotide.
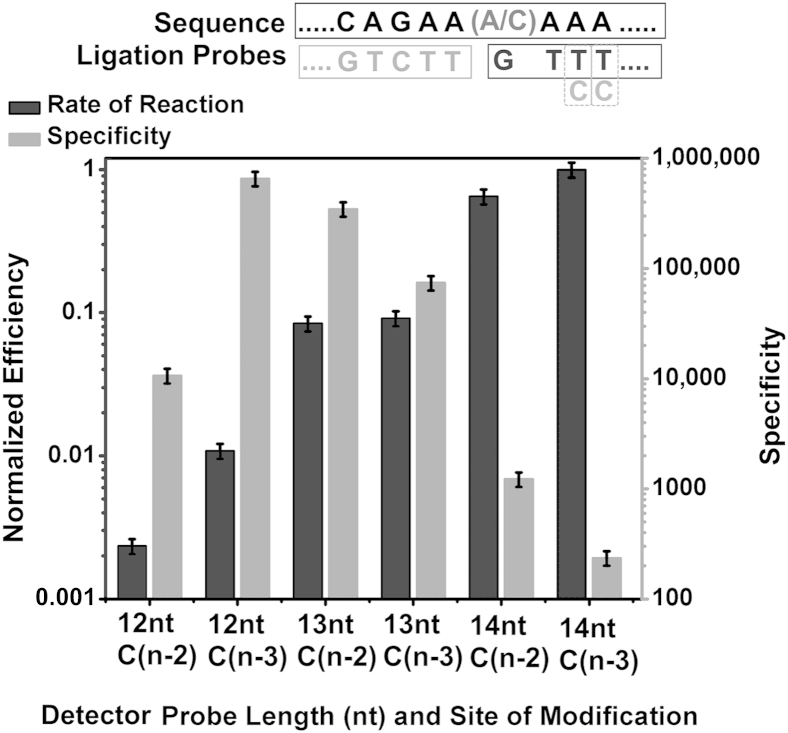
Effect of a Single Mismatch on the DP on RNA Ligation Efficiency and Specificity
When a mismatch is introduced at the n–2 or n–3 positions on the DP, the specificity decreases and the efficiency increases as the probe length is increased, and the required probe length for maximum ligation efficiency increases to 14 nt (Figure 3). Within the same DP length, a mismatch at the n–3 position has less of an effect on reducing the ligation efficiency than a mismatch at the n–2 position.
Figure 3.
The effect of a single DP nucleotide mismatch on RNA ligation efficiency and specificity. The right y axis indicates the specificity expressed as rate of reaction on mutant templates divided by the rate of reaction on wild-type templates. The left y axis indicates the efficiency expressed as rate of reaction using different DP lengths divided by the optimum rate. Shown above the graph are the target sequence (top) with wild-type/mutant nucleic acids; the CP sequence (bottom left); and the DP sequence (bottom right) with mismatches (Cs instead of Ts) at positions 2 and 3. Data are expressed as means ± SD. CP, common probe; DP, detector probe; nt, nucleotide.
Chemical Modifications to Ligation Probes
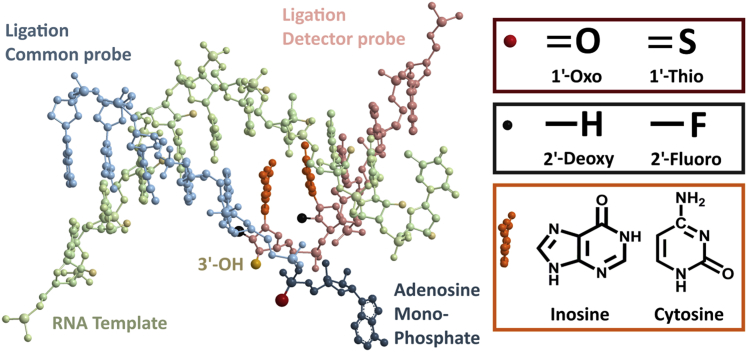
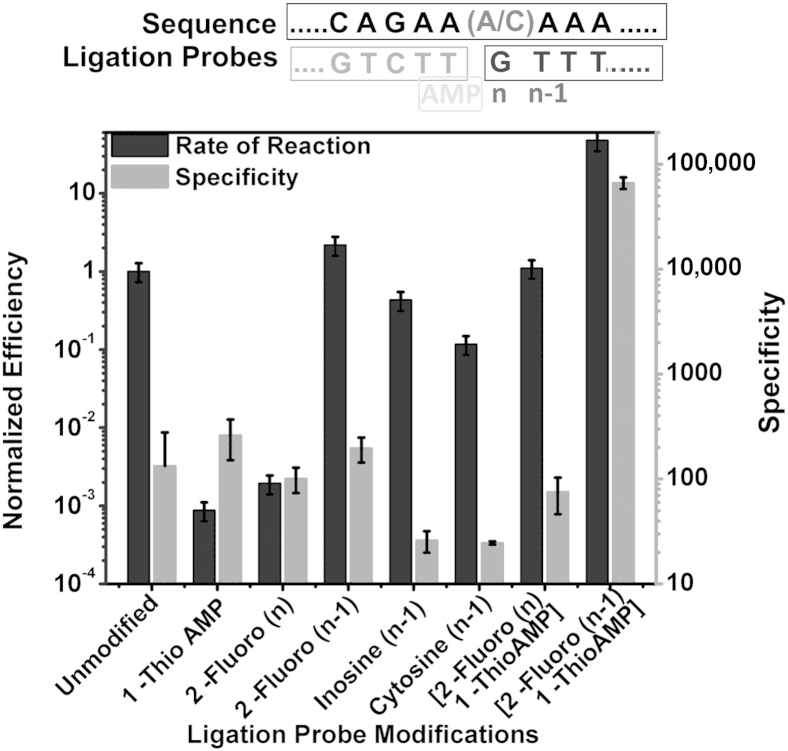
To improve LRA performance we evaluated the effects of chemical modifications to the structure of the ligation probes on LRA specificity and efficiency (rate of reaction). We performed LRA with the use of 12-nucleotide CPs with modified AMP groups and 12-nucleotide DPs with modified nitrogenous base and deoxyribose sugar groups (Figure 4). Nitrogenous base modifications tested include inosine and cytosine. Deoxyribose sugar modifications include the substitution of the 2′-OH group for a fluoride ion on various nucleotides near the 3′ end of the DP.
Figure 4.
Chemical modifications used on the CP and the DP. Adenosine monophosphate (navy) modifications used include 1-Thio-AMP (the red denotes the sulfur on the phosphate group). Deoxyribose sugar modifications used include 2-Fluoro analogs (black). Nitrogenous base modifications used include inosine and cytosine (orange). RNA template is in lime, CP is in cyan, and DP is in pink. The 3′-OH group on DP is depicted in yellow. CP, common probe; DP, detector probe.
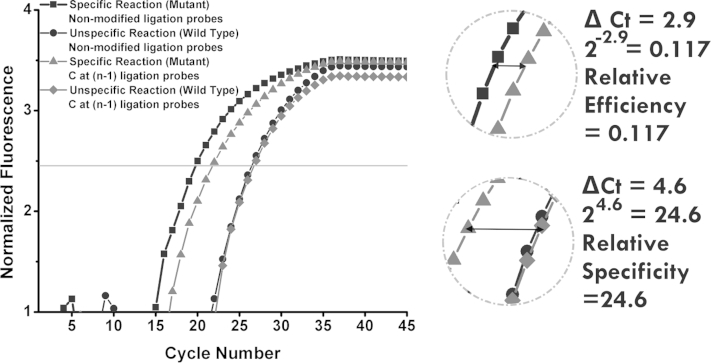
As shown in Supplemental Figure S3, when a cytosine modification is introduced to the structure of the DP at the n–1 nucleotide, the difference in Ct values between LRA reactions on mutant and wild type was 4.6 cycles. This shows that, when this cytosine modification is used, the mutant reaction is 24.6 (24.6) times more efficient than the wild-type reaction. This is the relative specificity for the substitution of cytosine at the n–1 position on the DP. For mutant template reactions, the difference in Ct of when nonmodified probes are used and that of when the cytosine-modified probes are used, is 2.9 cycles. The relative efficiency of using this modification is 0.117 (2−2.9).
When the three categories of modifications were tested individually, only reactions that used 1-Thio-AMP CP and (n–1) 2-Fluoro DP found an increase in specificity (Figure 5). Although 1-Thio-AMP CP found an improvement in specificity, the slowdown in reaction rate needed to be compensated by a longer reaction time. A combination of the 1-Thio-AMP CP with nitrogenous base and deoxyribose modifications were also evaluated. The combination of 1-Thio-AMP CP and (n–1) 2-Fluoro DP found the highest (190-fold) specificity increase compared with a control of no modifications. In combination, they increased the rate of specific ligation and ligation efficiency.
Figure 5.
The effects of chemical modifications of CP and DP on the specificity of LRA. The right y axis indicates the specificity expressed as rate of the reaction on mutant templates divided by the rate of reaction on wild-type templates. The left y axis indicates the efficiency expressed as rate of the reaction using different lengths of the DP divided by the optimum rate. For each modification, the rates of specific and unspecific reactions are determined by their difference in the Cts with that of no modification control. Shown above the graph are the target sequence (top) with wild-type/mutant nucleic acids being differentiated by the A/C nucleotide; the CP sequence (bottom left) with AMP modification (yellow); and the DP sequence (blue) with modifications (bottom right) with various modifications. Data are expressed as means ± SD. CP, common probe; DP, detector probe; LRA, ligation on RNA amplification.
Application of LRA to K103N Detection
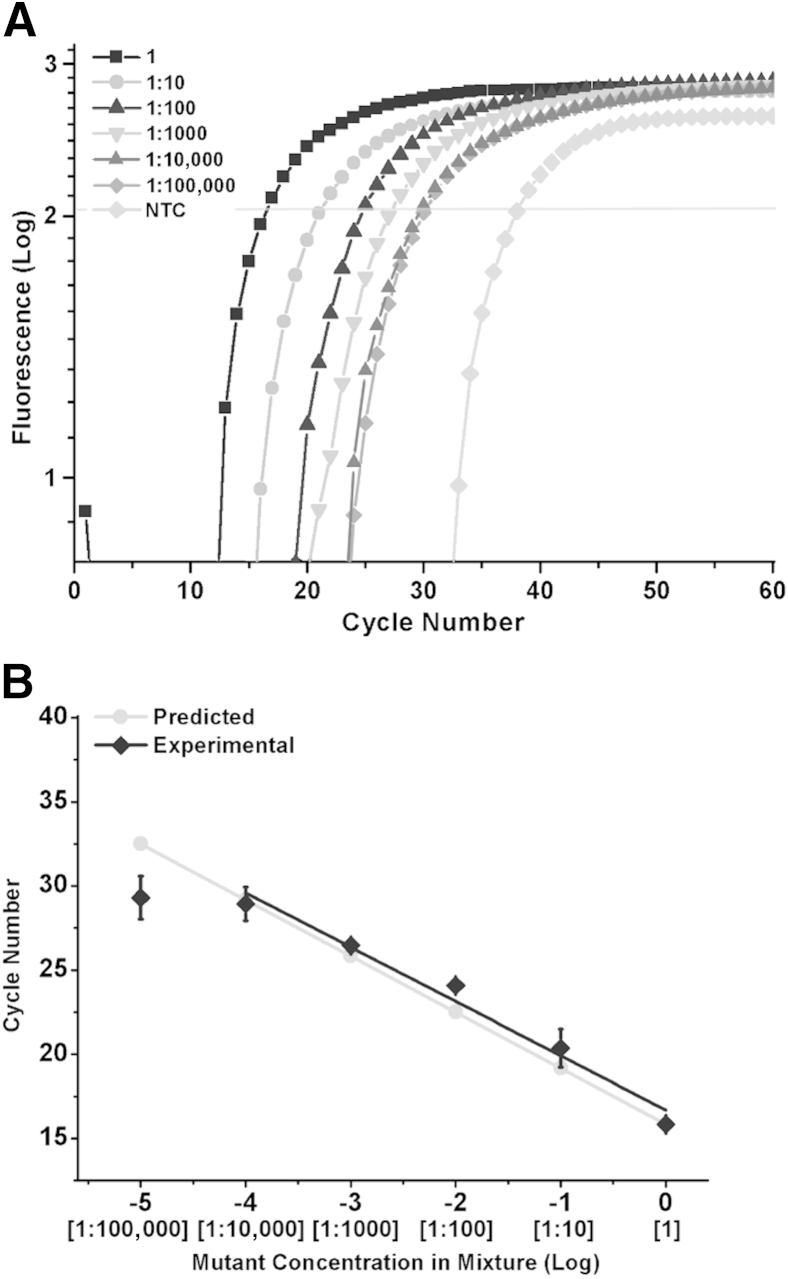
In vitro-transcribed K103N (aac codon) mutant RNA transcripts were serially diluted into the wild-type K103 (aaa codon) RNA transcripts. Reactions were performed with the optimum combination of ligation probes, 12 nt CP pre-adenylated with 1-Thio-AMP and 12 nt DP with a 2-Fluoro modification on the n–1 nucleotide and a cytosine substitution on the n–2 nucleotide. Figure 6A shows LRA signal as a function of cycle number, with a threshold line for calculations of Ct. The Ct values are plotted against the concentration of mutant in wild type. Ideally, a separation of 3.32 cycles should be observed between 10-fold dilutions of the target template, assuming perfect efficiency. The separation between the dilutions reduces from the canonical 3.32 as the concentration of mutant RNA decreases (Figure 6, A and B). This indicates the reactions become less specific toward lower concentrations of mutant RNA. However, we identified the 1:10,000 ratio of mutant to wild-type RNA as our limit of (specific) detection (LoSD). Here, we define LoSD as the lowest concentration in which we are able to identify and distinguish between two different dilutions. We defined limit of specific quantitation (LoSQ) as the lowest concentration in which we are able to identify and distinguish between two different dilutions that have an accurate separation of 3.32 Cts between them. We achieved the LoSQ of 1:100 (Figure 6, A and B).
Figure 6.
Detection of K103N-containing RNA transcripts in wild-type mixtures by ligation on RNA amplification. A: Quantitative PCR amplification curves of the RNA mixtures. B: Ratios of mutant to wild-type RNA mixtures plotted against their cycle number. The slope of the plot is −3.2. Error bars represent 1 SD. NTC, no template control.
Formation of DP Dimer through PCR Amplification
When no-template control (NTC) reactions were performed with no ligase and no CP, a false-positive PCR signal was observed (not shown). Two potential mechanisms for this signal were considered: undesired annealing and extension during the quantitative PCR amplification stage (Supplemental Figure S4, A–D) and the formation of DP dimers during amplification (Supplemental Figure S4E). Because the extension on RNA sequences by thermostable DNA polymerases (Taq) is intrinsically limited, preventing the formation of the RNA reverse complement, it is unlikely that the false-positive signal arose from the first proposed mechanism. During the heat activation step between the ligation and the amplification stages, the RNA is susceptible to degradation. This further limits the possibility of the formation of unspecific product because of the first mechanism. In the second mechanism considered, the formed DP dimers are amplifiable by the reverse primers and also contain whole or parts of the dual-labeled probe-binding sequence. However, because of the effectively halved primer concentration, the amount of PCR-amplified DP dimer should be less than one-half of that of a positive reaction. Because the 3′ end of the DPs depends on the template sequence, the degree of DP dimer formation depends on the particular point mutation and the sequence of target genome. The presence of DP dimers will affect the sensitivity of LRA. However, as with any dimer formation, through adjustments to the DP concentration and reaction conditions during the amplification stage, such as annealing temperature and time, the formation of DP dimers can be minimized.
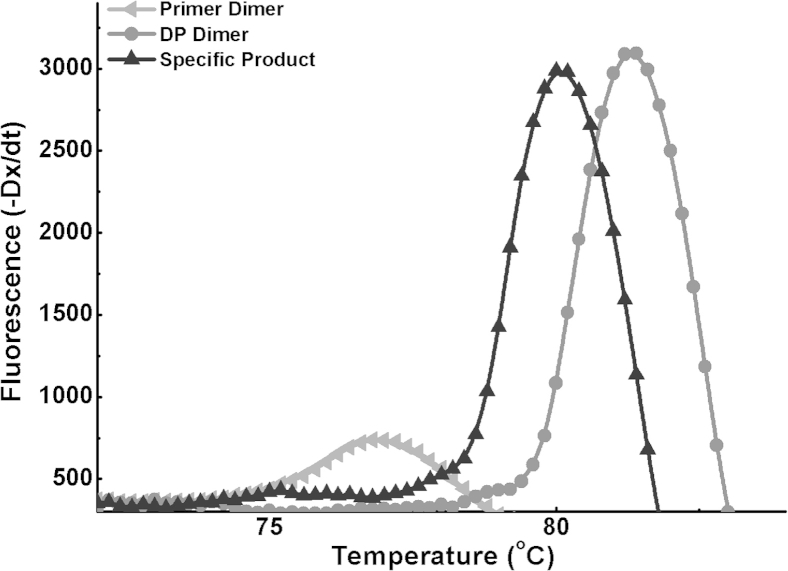
Melting curve analysis was also performed on NTC and positive control reactions with 108 copies/μL mutant K103N RNA. EvaGreen intercalating dye was used instead of dual-labeled probes in these reactions. For the NTC reaction, the melting curve shows a peak that is 1.3°C higher than the specific product peak (Supplemental Figure S5). Because the DP is longer than the CP, the produced DP dimer could be longer than the specific ligated product. Therefore, a higher melting point is observed with the NTC reaction. In addition to DP dimers, primer dimers were also observed when low concentrations of mutant RNA samples were used.
Comparison of LRA Specificity with ASPCR and Pyrosequencing
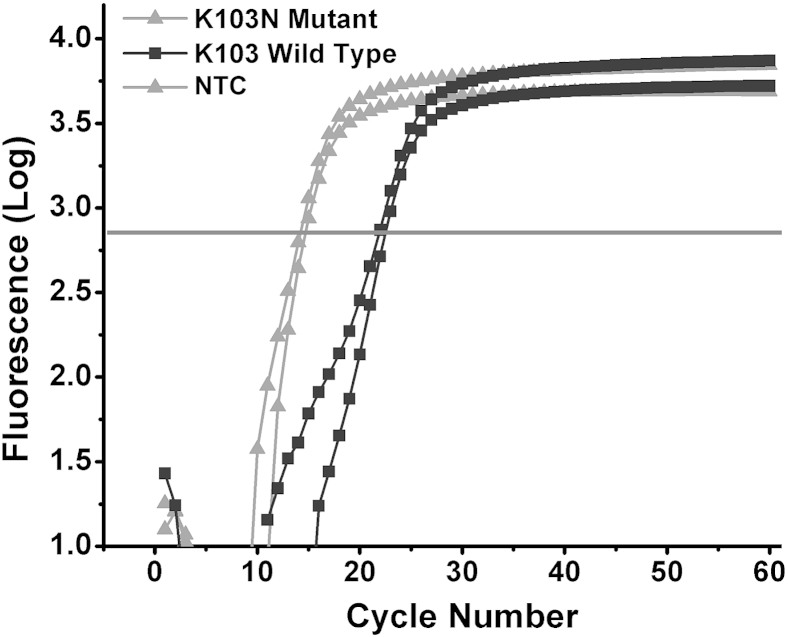
To evaluate the performance of LRA, we compared it with two currently used assays for the detection of HIV drug resistance mutations. First, we performed one-step reverse transcription ASPCR on K103N mutant and K103 wild-type in vitro-transcribed RNA fragments with the use of the same concentrations as for LRA reactions. We found the rate of reaction on mutant RNA to be 208 times faster than the rate on wild-type RNA, as determined by their relative Ct values (Figure 7).
Figure 7.
Quantitative PCR amplification curves of the detection of K103N mutant and wild-type RNA with the use of reverse transcription ASPCR. The rate of ASPCR on mutant RNA is 208 times faster than that on wild-type RNA (as determined by the difference in threshold values during quantitative PCR, 2−ΔCt). ASPCR, allele-specific PCR; NTC, no template control.
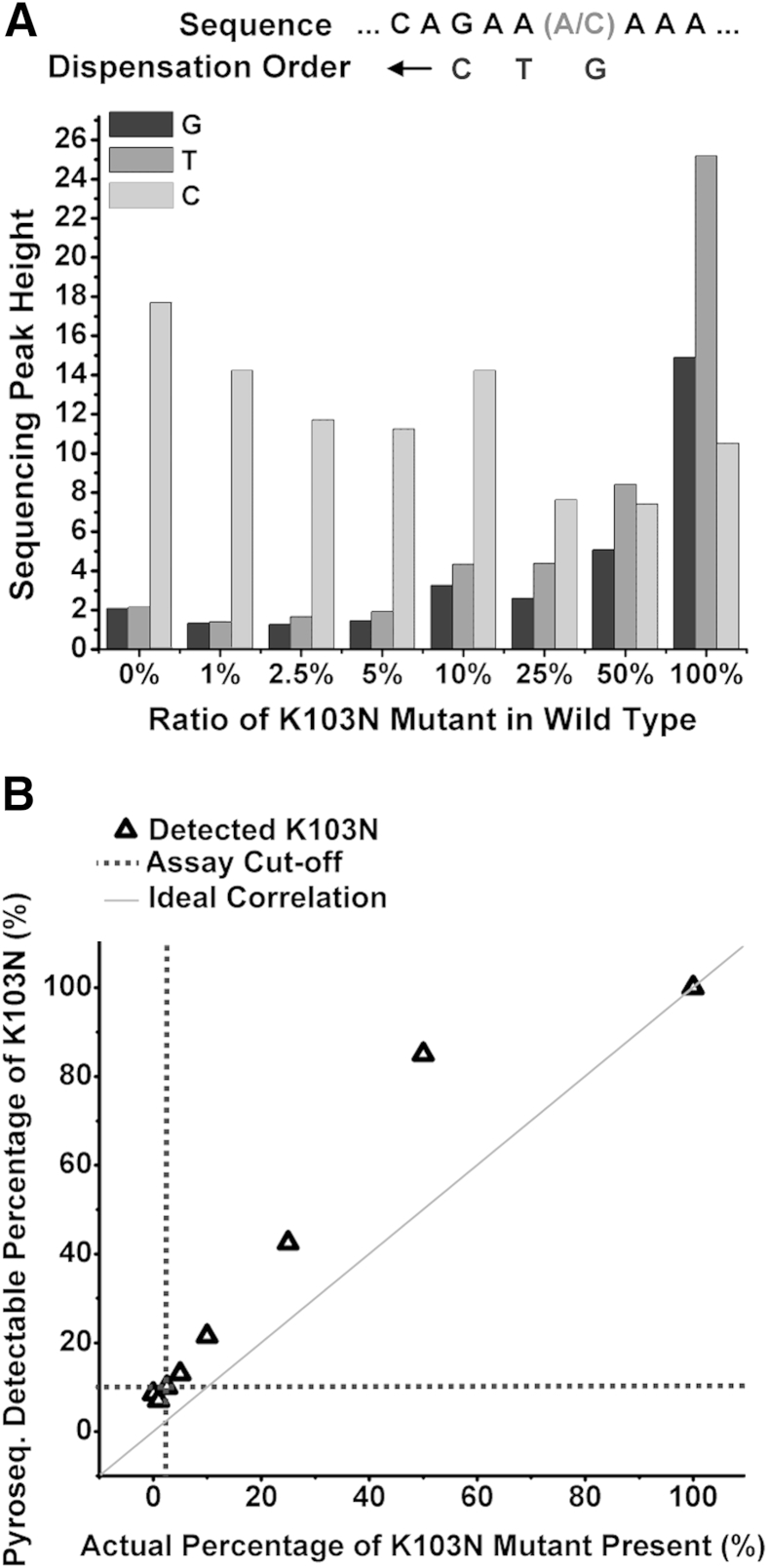
We subsequently performed pyrosequencing on different concentrations of mutant K103N (aac) DNA spiked into wild-type K103 (aaa). Figure 8A shows the sequencing peak reads of the three dispensed nucleotides next to the target mutation site. Because the mutant template has a cytosine at the mutation site, the signal strength corresponding to the dispensed guanosine nucleotide will determine the relative concentrations of the mutant. As the concentration of mutant K103N in the sample mixture decreases, the sequencing peak height corresponding to the dispensed guanosine nucleotide decreases. Figure 8B compares percentages of K103N mutant present, as detected by pyrosequencing, with the actual percentages of mutant templates spiked into wild-type templates. Because the ratio for the 0% mutant K103N sample is lower than the ratio for the 1% mutant sample, we infer that pyrosequencing could not resolve between 0% mutant concentration and 1% mutant concentration. The extent at which pyrosequencing was able to detect the presence of a target point mutation in an excess of wild type (and therefore its specificity) is determined to be 2.5% (1:40 ratio of mutant to wild-type template).
Figure 8.
Detection of concentration of K103N templates diluted into K103 wild-type templates by pyrosequencing. A: Pyrosequencing reads of K103N mutant in wild-type templates. B: Percentage of K103N detected by pyrosequencing compared with the actual percentage of K103N mutant present.
Sensitivity of LRA
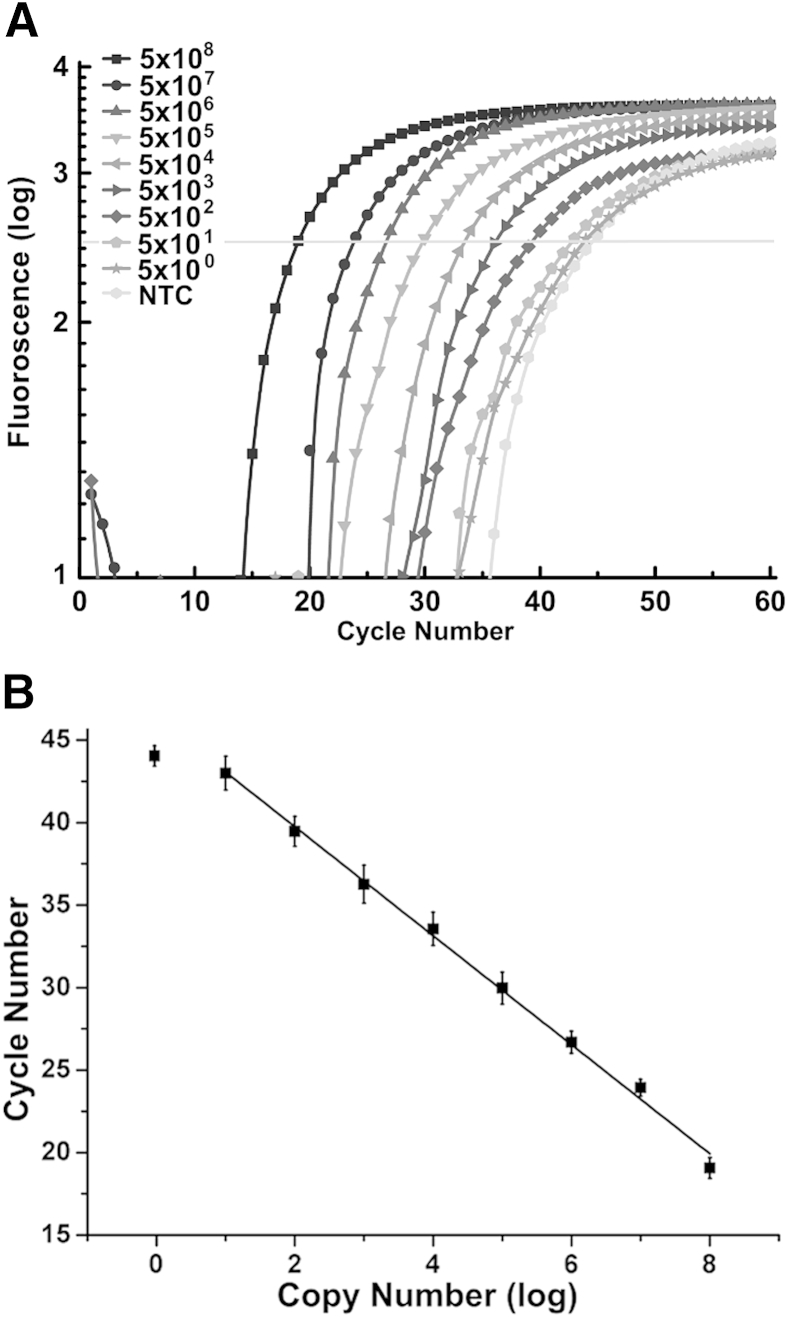
To determine assay sensitivity, defined as the minimal concentration in which mutations can be detected, mutant DPs and CPs were used, and 5 × 102 copies/μL mutant RNA were detected (Figure 9). Sequence-specific dual-labeled probes were used for the quantitative PCR amplification stage of LRA.
Figure 9.
Detection of HIV-1 K103N RNA transcripts with the use of ligation on RNA amplification. The sensitivity of the assay is evaluated by quantitative PCR amplification. A: Amplification curves of different concentrations of the K103N-containing RNA and NTC. The samples were prepared by 10-fold serial dilutions into nuclease-free water, and water is used in the NTC. The limit of detection is determined to be 5 × 102 copies/μL. B: The concentrations of K103N mutant RNA, in log (copy number), plotted against their cycle number. The slope is −3.2. Error bars represent 1 SD. NTC, no template control.
Discussion
The work presented here proposes and demonstrates a novel method for the rapid and specific detection of point mutations directly on RNA. LRA uses the heat activation properties of hot start thermostable polymerases to cleanly separate a RNA-templated ligation process and a subsequent quantitative PCR reaction. Much akin to the now staple one-step reverse transcription-PCR reaction, LRA combines its constituent stages in a simple one-step and single-buffer system. During the low-temperature ligation phase, the ligase is the only active enzyme, whereas the DNA polymerase remains inactivated. After 10 minutes of incubation at 95°C, the ligase enzyme is inactivated, the DNA polymerase becomes activated, and the amplification phase of the assay begins. This single-buffer, two-phase reaction system is only possible because the ligation oligonucleotide probes do not interfere with the primers for amplification. Because only one reaction is active at a time, maximum sensitivity for each phase of LRA can be achieved. Although there are advantages to incorporating a cyclic exponential amplification method such as ligase chain reaction to the mutation detection assays, it comes at the cost of relying on a two-buffer system, which increases the complexity and time required for the assays. Because T4 DNA ligase is capable of blunt-end ligation, if the probes required to initiate ligase chain reaction are present at the same time as the RNA ligation probes, a false-positive result will occur even when no target RNA template is present. This renders ligase chain reaction unsuitable as a detection method in a single-buffer reaction.
Ligases are sequence-independent enzymes that derivate their specificity from the structural complementarities of double-stranded duplexes. There is some degree of nonspecificity in ligation reactions, which leads to ligation typically occurring on a small number of wild-type RNA. As the ratio of mutant RNA in this mixture decreases, the overall effects of the wild-type RNA template nonspecific reaction will become more prominent. This effect is observed in our data. In point-of-care diagnostics, LoSD is often a sufficient measurement of specificity. We have shown that LRA can be used to detect K103N mutant RNA in an excess of wild-type RNA with a LoSQ of up to 1:100 (1%) and a LoSD of up to 1:10,000 (0.01%). Compared with ASPCR and pyrosequencing, which resulted in LoSDs of 1:208 (0.5%) and 1:40 (2.5%), respectively, LRA exhibited good specificity to distinguish single-nucleotide mismatches on synthetic HIV-1 RNA that contained the K103N mutation.
We found that increasing the length of the ligation probes has opposite effects on the efficiency and specificity of the ligation on RNA. As DP length is increased, the specificity of the reaction decreases, whereas the rate of the reaction increases. The minimum length of the DP required for an efficient ligation rate is 12 nt. This could be an intrinsic property of the T4 ligase on RNA. As previously shown,17 a mismatch on the DP at the n–2 position from the 3′ end of the probe increases the ligation specificity on DNA. We also found that introducing a mismatch on the n–2 or n–3 positions on the DP increased the specificity of the ligation reaction at the cost of reduced ligation efficiency on RNA. The reduced efficiency can be compensated by increasing the DP length. The rate of the reaction on target mutant RNA is 117 times faster than the rate on wild-type RNA when both unmodified DP and CP (at 12 nt) are used. This corresponds to the limit that the T4 DNA ligase is able to natively distinguish between single-nucleotide mismatches. As the DP length is decreased, the specificity increases because the rate of ligation on mismatched RNA templates decreases at a faster rate than the rate of ligation on the target RNA template. Previously,25 the use of [γ-thio]-triphosphate as a substrate for T4 DNA ligase in DNA array profiling found a delayed rate of ligation reactions. We show that CPs with pre-adenylated 1-Thio-AMP groups improve the specificity of ligation reactions on RNA when mismatches are present. By incorporating a number of chemical modifications to the design of the oligonucleotide probes, we found that the combination of 1-Thio-AMP CP and (n–1) 2-Fluoro DP produced the highest increase in reaction specificity compared with LRA performed with nonmodified ligation probes.
In all NTC reactions a late-positive signal was detected. In traditional DNA-based ligation assays the dimerization of the complement of the CP and the extended DP could produce an amplifiable product during the quantitative PCR cycling phase (Supplemental Figures S4 and S5). For ligation on RNA, this method of forming a NTC signal is not possible. However, we determined that the DP could by itself dimerize and become amplified. Because of the design of ligation-mediated molecular point mutation assays (in which the sequences to be amplified are always present even if they are not ligated together) and the nature of quantitative PCR amplification, random dimerization and amplification are difficult to avoid. In most cases, however, this does not affect the interpretation of the results because of the lateness of the signal.
The RNA transcripts used in this study are shorter than the full-length HIV-1 viral genome; therefore, they have limited secondary structures. The presence of additional secondary structures on full-length HIV-1 RNA could pose problems to ligation probe hybridization. Although the RNA transcripts used in this study are heated to 70°C for 2 minutes to open up any secondary structures, further optimization on clinical samples from HIV-infected patients will be required. Recombinant RNase inhibitors were included in all reaction mixtures to limit RNA degradation which could decrease the detection threshold and specificity of LRA.
Although we have successfully explored the feasibility of the LRA concept, it still needs further optimization in cases when accurate determination of concentration of the target is required, rather than just being able to detect the presence of targets at a certain concentration. The LoSD and LoSQ observed in our study need further evaluation for real-patient samples.
Conclusions
In summary, this work represents proof of principle of a novel assay for direct ligation and amplification on RNA, which may have a revolutionary impact in diagnosing point mutations. Particularly, the potential of LRA to quickly and efficiently detect the presence of minority variant mutations make it highly appropriate for inclusion in a point-of-care device for HIV drug resistance detection in resource-limited settings. Apart from HIV, it can have other relevant applications such as in other infectious diseases and cancer diagnostics.4
Acknowledgments
R.K. and A.T. conceived and oversaw the project; L.Z. designed and performed experiments, analyzed the data, and wrote the manuscript; J.W. provided technical advice and edited the manuscript; M.C. generated the RNA transcripts and provided technical advice; M.C. and R.K. edited the manuscript and provided the RNA transcripts; S.A. participated in technical discussions; A.T. edited the manuscript and provided technical advice.
Footnotes
Supported in part by a developmental grant from the Lifespan/Tufts/Brown Center for AIDS Research (R.K., A.T.), NIH National Institute of Allergy and Infectious Diseases grant P30AI042853, and a Seed Award for Translational Research from Brown University (R.K., A.T.).
Disclosures: None declared.
R.K. and A.T. contributed equally to this work as senior authors.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2015.07.001.
Supplemental Data
Supplemental Figure S1.
The pre-adenylation efficiency of the common probe with the use of ATP and ATP analogs. The pre-adenylation efficiency was evaluated over a period of 1, 2, and 8 hours. Data are expressed as means ± SD.
Supplemental Figure S2.
Effects of reaction conditions on rates of the LRA reaction. Rates (y axis) are shown by the relative Ct values normalized to the lowest Ct within each set of conditions (1/2ΔCt). The sets of conditions evaluated are shown on the x axis and include the pH, Mg2+, and the presence of ATP with and without PA. Data are expressed as means ± SD. LRA, ligation on RNA amplification; PA, pre-adenylation.
Supplemental Figure S3.
Derivation of relative efficiency and specificity of using cytosine-modified ligation probe for ligation on RNA amplification. The relative efficiency and specificity are calculated from Ct values obtained from quantitative PCR reactions.
Supplemental Figure S4.
Illustration of potential undesired production of ligated products for traditional ligation amplification on DNA. A: Besides the desired reaction, side reactions can produce the ligated product. B: The FP can be extended along the CP to produce a complimentary copy of the CP. C: In the event, the CP and the DP are not enzymatically ligated because of a single-nucleotide mismatch on the 3′ end of the DP, extension of the DP can still occur on the DNA target sequence. D: The DP′ and the FP′ on hybridization can dimerize and produce the ligated product. E: The formation of DP dimers. CP, common probe; DP, detector probe; DP′, extended detector probe; FP, forward primer; FP′, extended forward primer.
Supplemental Figure S5.
Melting curve analysis of one-step ligation on RNA amplification with the sequence-specific probe replaced with EvaGreen intercalating dye. The graph shows the formation of primer dimers (lower peak) in a nonoptimized NTC reaction, the formation of the specific amplicon with 107 copies of mutant RNA (middle peak), and the formation of DP dimer (higher peak) in a NTC reaction with no CP. Both the specific product and the DP dimer will be detectable when sequence-specific probes are used. CP, common probe; DP, detector probe; NTC, no-template control.
References
- 1.Hiatt J.B., Pritchard C.C., Salipante S.J., O'Roak B.J., Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milbury C.A., Li J., Makrigiorgos G.M. PCR-based methods for the enrichment of minority alleles and mutations. Clin Chem. 2009;55:632–640. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewandowska M.A., Jozwicki W., Jochymski C., Kowalewski J. Application of PCR methods to evaluate EGFR, KRAS and BRAF mutations in a small number of tumor cells in cytological material from lung cancer patients. Oncol Rep. 2013;30:1045–1052. doi: 10.3892/or.2013.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lun F.M., Tsui N.B., Chan K.C., Leung T.Y., Lau T.K., Charoenkwan P., Chow K.C., Lo W.Y., Wanapirak C., Sanguansermsri T., Cantor C.R., Chiu R.W., Lo Y.M. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:19920–19925. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jani I.V., Peter T.F. How point-of-care testing could drive innovation in global health. N Engl J Med. 2013;368:2319–2324. doi: 10.1056/NEJMsb1214197. [DOI] [PubMed] [Google Scholar]
- 7.Thompson M.A., Aberg J.A., Hoy J.F., Telenti A., Benson C., Cahn P., Eron J.J., Gunthard H.F., Hammer S.M., Reiss P., Richman D.D., Rizzardini G., Thomas D.L., Jacobsen D.M., Volberding P.A. Antiretroviral treatment of adult HIV infection 2012 recommendations of the International Antiviral Society-USA Panel. JAMA. 2012;308:387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 8.Wensing A.M., Calvez V., Gunthard H.F., Johnson V.A., Paredes R., Pillay D., Shafer R.W., Richman D.D. 2014 Update of the drug resistance mutations in HIV-1. Top Antivir Med. 2014;22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 9.Department of Health and Human Services Panel on Antiretroviral Guidelines for Adults and Adolescents – A Working Group of the Office of AIDS Research Advisory Council (OARAC): Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents [Internet]. 2015. Available at http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf
- 10.Wittek M., Sturmer M., Doerr H.W., Berger A. Molecular assays for monitoring HIV infection and antiretroviral therapy. Expert Rev Mol Diagn. 2007;7:237–246. doi: 10.1586/14737159.7.3.237. [DOI] [PubMed] [Google Scholar]
- 11.Gianella S., Richman D.D. Minority variants of drug-resistant HIV. J Infect Dis. 2010;202:657–666. doi: 10.1086/655397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kantor R., Katzenstein D.A., Efron B., Carvalho A.P., Wynhoven B., Cane P., Clarke J., Sirivichayakul S., Soares M.A., Snoeck J., Pillay C., Rudich H., Rodrigues R., Holguin A., Ariyoshi K., Bouzas M.B., Cahn P., Sugiura W., Soriano V., Brigido L.F., Grossman Z., Morris L., Vandamme A.M., Tanuri A., Phanuphak P., Weber J.N., Pillay D., Harrigan P.R., Camacho R., Schapiro J.M., Shafer R.W. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landegren U., Kaiser R., Sanders J., Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 14.Beck I.A., Mahalanabis M., Pepper G., Wright A., Hamilton S., Langston E., Frenkel L.M. Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J Clin Microbiol. 2002;40:1413–1419. doi: 10.1128/JCM.40.4.1413-1419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner T.A., Kress C.M., Beck I., Techapornroong M., Wittayapraparat P., Tansuphasawasdikul S., Jourdain G., Ngo-Giang-Huong N., Lallemant M., Frenkel L.M. Detection of HIV-1 drug resistance in women following administration of a single dose of nevirapine: comparison of plasma RNA to cellular DNA by consensus sequencing and by oligonucleotide ligation assay. J Clin Microbiol. 2010;48:1555–1561. doi: 10.1128/JCM.02062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poeta M.L., Manola J., Goldenberg D., Forastiere A., Califano J.A., Ridge J.A., Goodwin J., Kenady D., Saunders J., Westra W., Sidransky D., Koch W.M. The Ligamp TP53 assay for detection of minimal residual disease in head and neck squamous cell carcinoma surgical margins. Clin Cancer Res. 2009;15:7658–7665. doi: 10.1158/1078-0432.CCR-09-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi C.J., Eshleman S.H., Jones D., Fukushima N., Hua L., Parker A.R., Yeo C.J., Hruban R.H., Goggins M.G., Eshleman J.R. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 18.Loubser S., Balfe P., Sherman G., Hammer S., Kuhn L., Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006;20:995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson J.A., Li J.F., Wei X.R., Lipscomb J., Bennett D., Brant A., Cong M.E., Spira T., Shafer R.W., Heneine W. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS One. 2007;2:e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varela-Echavarria A., Garvey N., Preston B.D., Dougherty J.P. Comparison of Moloney murine leukemia virus mutation rate with the fidelity of its reverse transcriptase in vitro. J Biol Chem. 1992;267:24681–24688. [PubMed] [Google Scholar]
- 21.Nilsson M., Antson D.O., Barbany G., Landegren U. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 2001;29:578–581. doi: 10.1093/nar/29.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J., Li Z., Liu C., Cheng Y. Simple and sensitive detection of microRNAs with ligase chain reaction. Chem Commun (Camb) 2010;46:2432–2434. doi: 10.1039/b923521c. [DOI] [PubMed] [Google Scholar]
- 23.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci U S A. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angione S.L., Chauhan A., Tripathi A. Real-time droplet DNA amplification with a new tablet platform. Anal Chem. 2012;84:2654–2661. doi: 10.1021/ac202532a. [DOI] [PubMed] [Google Scholar]
- 25.Kim J., Mrksich M. Profiling the selectivity of DNA ligases in an array format with mass spectrometry. Nucleic Acids Res. 2010;38:e2. doi: 10.1093/nar/gkp827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.