Abstract
Celiac disease (CD) is an immune-mediated enteropathy triggered by gluten in genetically susceptible individuals. The recent increase in CD incidence suggests that additional environmental factors, such as intestinal microbiota alterations, are involved in its pathogenesis. However, there is no direct evidence of modulation of gluten-induced immunopathology by the microbiota. We investigated whether specific microbiota compositions influence immune responses to gluten in mice expressing the human DQ8 gene, which confers moderate CD genetic susceptibility. Germ-free mice, clean specific-pathogen-free (SPF) mice colonized with a microbiota devoid of opportunistic pathogens and Proteobacteria, and conventional SPF mice that harbor a complex microbiota that includes opportunistic pathogens were used. Clean SPF mice had attenuated responses to gluten compared to germ-free and conventional SPF mice. Germ-free mice developed increased intraepithelial lymphocytes, markers of intraepithelial lymphocyte cytotoxicity, gliadin-specific antibodies, and a proinflammatory gliadin-specific T-cell response. Antibiotic treatment, leading to Proteobacteria expansion, further enhanced gluten-induced immunopathology in conventional SPF mice. Protection against gluten-induced immunopathology in clean SPF mice was reversed after supplementation with a member of the Proteobacteria phylum, an enteroadherent Escherichia coli isolated from a CD patient. The intestinal microbiota can both positively and negatively modulate gluten-induced immunopathology in mice. In subjects with moderate genetic susceptibility, intestinal microbiota changes may be a factor that increases CD risk.
Celiac disease (CD) is an immune-mediated enteropathy triggered by gluten in individuals with genetic risk. Proteolytic-resistant gluten peptides are deamidated by transglutaminase 2 (TG2) in the small-intestinal lamina propria, increasing their binding affinity to the CD-associated HLA-DQ2 or DQ8 molecules, leading to T-cell activation.1, 2 CD also requires an innate immune response, characterized by up-regulation of stress markers on epithelial cells as well as up-regulation and activation of intraepithelial lymphocytes (IELs).3, 4 There has been a rapid rise in CD prevalence over the past 50 years.5 This, in conjunction with the fact that only 2% to 5% of genetically susceptible individuals will develop CD, argues for environmental modulators of CD expression.6
The intestinal microbiota plays an important role in mucosal immune maturation and homeostasis as evidenced from seminal studies using germ-free and gnotobiotic mice.7, 8 Clinical and animal studies also suggest that altered colonization early in life increases susceptibility to chronic inflammatory diseases and food sensitivities.9, 10, 11 Indeed, alterations in intestinal microbial composition have been described in CD patients, some of which normalize after treatment with a gluten-free diet.12 Clinical studies have also proposed a link between antibiotic use and elective caesarean section and CD development.13, 14, 15 However, recent studies in families with high genetic risk for CD (positive family history or homozygous for HLA-DQ2.5) have not been able to identify an environmental determinant, including the timing and dose of gluten introduction to an infant's diet.16, 17 Microbial factors were not directly investigated, and results may not apply to the general population or individuals with moderate genetic risk for CD (HLA-DQ2 heterozygous or HLA-DQ8).
Whether altered colonization instigates CD in an individual at moderate risk of developing CD remains unclear.18 Therefore, we investigated whether microbial colonization modulates host responses to gluten using transgenic HLA-DQ8 mice on the nonobese diabetic background (NOD/DQ8).19 We used complementary strategies to investigate host responses to gluten under different microbial conditions. Clean specific-pathogen-free (SPF) mice, strictly monitored for the absence of a variety of pathobionts and Proteobacteria, were protected from gluten-induced immunopathology when compared to germ-free and conventional SPF mice. Perinatal disruption of the microbiota leading to Proteobacteria expansion in conventional SPF mice further enhanced severity of responses to gluten; whereas specific pathobiont supplementation to clean SPF mice reversed the protective effect of the benign microbiota.
Materials and Methods
Mice and Colonization Procedures
Female and male germ-free, clean SPF and conventional SPF NOD AB° DQ8 (NOD/DQ8) transgenic mice maintained on a gluten-free diet were used for experiments.20 Germ-free mice were generated by two-stage embryo transfer, as previously described,21 and bred and maintained in flexible film isolators in McMaster's Axenic Gnotobiotic Unit. Clean SPF mice originated from germ-free mice that were naturally colonized by co-housing with female mouse colonizers harboring altered Schaedler flora and bred for three generations in individually ventilated cage racks. Pathogen contamination and microbiota diversification in mouse cecum contents of clean SPF mice was evaluated every 2 weeks in cage sentinels and at the end of the study in the experimental mice by PCR for Helicobacter bilis, H. ganmani, H. hepaticus, H. mastomyrinus, H. rodentium, Helicobacter spp., H. typhlonius, and Pneumocystis murina. Mouse serum was also tested for murine viral pathogens by multiplexed fluorometric immunoassay/enzyme-linked immunosorbent assay (ELISA)/indirect fluorescent antibody tests. Germ-free status was monitored in sentinels and, at the end of the study in the experimental mice, by immunofluorescence (SYTOX Green; Invitrogen, Burlington, ON, Canada), anaerobic and aerobic culture, as well as PCR technique.
Additional experiments were performed in germ-free and clean SPF C57BL/6 mice. For pathobiont supplementation experiments, 8- to 12-week-old clean SPF NOD/DQ8 mice were orally fed with 108 colony-forming units of Escherichia coli ENT CAI:5, isolated from fecal microbiota of a CD patient,22 three times a week, 1 week before the start of sensitization and once a week during the sensitization and challenge period. Conventional SPF mice were bred and maintained in a conventional SPF facility at McMaster University. All experiments were conducted with approval from the McMaster University Animal Care Committee.
Gluten Sensitization and Challenge
NOD/DQ8 mice were sensitized with 500 μg of sterilized pepsin-trypsin digest of gliadin (PT-gliadin) and 25 μg of cholera toxin (Sigma-Aldrich, St. Louis, MO) by oral gavage once a week for 3 weeks, as previously described.19 PT-gliadin was prepared as previously described.19 In antibiotic experiments, mice were sensitized at 3 weeks of age, following weaning. For all other experiments, 8- to 12-week-old mice were used for sensitizations. Following PT-gliadin sensitization, gluten-treated mice were challenged by oral gavage with 2 mg of sterile gluten (Sigma-Aldrich) dissolved in acetic acid three times a week for 2 weeks. Nonsensitized control mice received cholera toxin alone during the sensitization phase and acetic acid alone during the challenge phase. NOD/DQ8 mice were weaned and maintained on a gluten-free diet. In additional experiments, C57BL/6 mice were sensitized with PT-zein and cholera toxin, once a week for 3 weeks, and challenged with zein dissolved in acetic acid three times a week for 2 weeks. All preparations were tested for lipopolysaccharide contamination using the E-Toxate kit (Sigma-Aldrich). Mice were sacrificed 18 to 24 hours following the final gluten or zein challenge.
Microbial Analysis
Fecal and cecal samples were collected and flash frozen on dry ice. DNA was extracted from samples as previously described.23 Extracted DNA underwent amplification for the hypervariable 16S rRNA gene v3 region and sequenced on the Illumina MiSeq platform (Illumina, San Diego, CA). Generated data were analyzed as described previously.23 Briefly, sequences were trimmed using Cutadapt software version 1.2.1,24 aligned using PANDAseq software version 2.8,25 operational taxonomic units selected via AbundantOTU,26 and taxonomy assigned against the Greengenes reference database.27 α-Diversity was calculated using Quantitative Insights Into Microbial Ecology (QIIME),28 and heat maps were generated using R (R Foundation for Statistical Computing, Vienna, Austria), clustered based on Bray-Curtis dissimilarity.29
Histology and Immunohistochemistry
Cross sections of the jejunum were fixed in 10% formalin, embedded in paraffin, and stained with hematoxylin and eosin for histological evaluation by light microscopy (Olympus, Richmond Hill, ON, Canada) using Image-Pro Plus software version 6.3 (Media Cybernetics, Rockville, MD). Enteropathy was determined by measuring villus-to-crypt (V/C) ratios in a blinded fashion, as previously described.19 Intraepithelial lymphocytosis was determined by counting CD3+ IELs per 20 enterocytes in five randomly chosen villus tips, as previously described, and expressed as IELs/100 enterocytes. CD3+ immunostaining was performed on paraffin-embedded sections of the jejunum as previously described.19
Enterocyte cell death was determined by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining using the ApopTag Peroxidase in Situ Apoptosis Detection Kit (Millipore, Billerica, MA) according to the manufacturer's instructions. Slides were viewed by light microscopy (Olympus). The percentage of TUNEL-positive enterocytes in 20 villi was determined for each mouse.
Small-Intestinal Lamina Propria and IEL Preparation
Small intestines were removed from mice, and IELs and lamina propria lymphocytes isolated by established protocols. Briefly, small intestines from mice were flushed to remove intestinal contents, Peyer's patches and mesentery were removed, intestines opened longitudinally, and cut into 3- to 5-mm pieces. Intestinal pieces were incubated five to six times in EDTA/HEPES/Dulbecco's phosphate buffered saline for 15 minutes in a 37°C shaker. After each 15-minute incubation, intestines were vigorously vortexed and the IELs were collected by passing the supernatants through a 40-μm cell strainer. Intestinal pieces were further digested with DNase I (Roche, Mississuaga, ON, Canada) and Collagenase Type VIII (Sigma-Aldrich) to collect lamina propria lymphocytes. IELs and lamina propria lymphocytes were enriched on a Percoll gradient and resuspended in fluorescence-activated cell sorting buffer for cell staining.
Single-cell suspensions of lamina propria preparations were stained with fluorochrome-labeled cell-surface antibodies including CD4-APC (RM4-5), CD8a-PerCP (53-6.7), and CD25-PE (7D4) purchased from BD Biosciences (San Jose, CA). IEL cell suspensions were stained with fluorochrome-labeled cell-surface antibodies including CD3-Alexa Fluor-700 (ebio500AZ; eBioscience, San Diego, CA), CD3-Pacific Blue (RM4-5; BD Biosciences), CD8-PerCP (53-6.7; BD Biosciences), β T-cell receptor (βTCR) (H57-597; eBioscience), TCRγδ-APC (eBioGL3; eBioscience), NKG2D-PE (CX5; eBioscience), and CD69-PE-CF594 (H1.2F3; BD Biosciences). For intracellular staining, cells were permeabilized using the Foxp3 staining buffer set (eBioscience). Lamina propria cells were incubated with fluorescein isothiocyanate–conjugated antibodies to Foxp3 (FJK-16s; eBioscience), and IELs were incubated with PE-Cy7–conjugated Granzyme-B (NGZB; eBioscience) for 90 minutes at 4°C. Stained cells were acquired using the LSR II (BD Biosciences) and analyzed with FlowJo software version 7.2.4 (TreeStar, Ashland, OR).
T-Cell Proliferation and Cytokine Analysis
Single-cell suspensions of mesenteric lymph nodes were prepared in RPMI 1640 (1% penicillin/streptomycin, 10% fetal calf serum, 2 mmol/L l-glutamine). CD4+ T cells were isolated from mesenteric lymph nodes using the EasySep Mouse CD4+ T cell Enrichment Kit (StemCell, Vancouver, BC, Canada), and labeled with carboxyfluorescein succinimidyl ester (CFSE; Life Technologies, Grand Island, NY). CD11c+ cells were isolated from spleens using the Easysep Mouse CD11c Selection Kit (StemCell). A total of 5 × 104 CD11c+ cells were co-cultured with 2 × 105 CD4+ T cells in the presence of 500 μg/mL PT-gliadin, 500 μg/mL PT-zein, or medium alone in a round-bottom 96-well plate for 3 days at 37°C, 5% CO2. Cells were resuspended in fluorescence-activated cell sorting buffer and stained with fluorochrome-conjugated antibodies to CD3 and CD4 and a viability stain. CFSE-labeled cells were acquired using the LSR II (BD Biosciences). Viable cells were gated on CD4+ T cells. CFSE intensity for this population was determined using FlowJo software (TreeStar) and the percentage of divided cells determined for each condition (PT-gliadin, PT-zein, medium). Proliferation of cells in response to PT-gliadin or PT-zein stimulation were normalized to the proliferation of medium alone and expressed as a proliferation index.
Anti-Gliadin ELISA
Serum IgA and IgG antibodies to gliadin were measured by ELISA as previously described,30, 31 with minor modifications. In addition, intestinal wash IgA antibodies to gliadin were measured similarly. Intestinal wash IgG antibody reactivity was too low to be detected reliably and was not measured. One hundred mg of the US hard red spring wheat Triticum aestivum cv Butte 86 variety flour was suspended in 1 mL of phosphate-buffered saline and mixed for 1 hour at 4°C. The suspension was centrifuged at 10,000 × g for 20 minutes. The supernatant containing mostly non-gluten proteins, was removed. The pellet was washed with phosphate-buffered saline, resuspended in 50% isopropanol, and mixed for 1 hour at room temperature. The suspension was centrifuged at 10,000 × g for 20 minutes, and the supernatant, containing gliadin and glutenin proteins, was stored at −20°C. The 96-well Maxisorp round-bottom polystyrene plates (Nunc, Roskilde, Denmark) were coated with 50 μL/well of a 0.01 mg/mL solution of the gliadin gluten extract in 0.1 mol/L carbonate buffer (pH 9.6) or were left uncoated to serve as control wells. Wells were blocked by incubation with 1% bovine serum albumin. Serum samples were diluted at 1:100, whereas intestinal wash samples were diluted at 1:10. The samples were added at 50 μL per well in duplicates and incubated for 1 hour. Each plate contained a positive control sample. After washing the wells, they were incubated with a 1:2000 dilution of either horseradish peroxidase–conjugated anti-mouse IgG (GE Healthcare, Piscataway, NJ) or IgA (Abcam, Cambridge, MA) secondary antibodies. The plates were washed, and 50 μL of developing solution was added to each well; absorbance was measured at 450 nm after 20 minutes. Absorbance values were corrected for nonspecific binding by subtraction of the mean absorbance of the associated bovine serum albumin–coated control wells. The corrected values were first normalized according to the mean value of the positive control duplicate on each plate. The mean antibody level for the clean SPF control group was then set as 1.0 arbitrary units, and all other results were normalized accordingly.
Anti-Gliadin Western Blots
Antibody reactivity to gluten in sera was confirmed by Western blot. The Butte 86 gluten extract was dissolved in sample buffer, heated for 10 minutes at 75°C, and separated by SDS-PAGE (0.66 μg of protein per lane) using NuPAGE 4% to 20% bis-tris gels (Life Technologies). Protein transfer onto nitrocellulose membranes was performed with the iBlot Dry Blotting System (Life Technologies). The membrane was incubated for 2 hours in blocking solution (5% milk + 0.5% bovine serum albumin) in tris-buffered saline containing 0.05% Tween-20 (TBS-T). Serum specimens (1:500) were incubated in dilution buffer (10% blocking solution + 10% fetal bovine serum in TBS-T) for 1 hour. The secondary antibody used was horseradish peroxidase–conjugated anti-mouse IgG or IgA. Bound antibodies were detected by the ECL system (Millipore) and autoradiography film (Fuji, Valhalla, NY).
Cytokine Measurement
Sections of the jejunum were collected 18 to 24 hours following the final gluten challenge, homogenized, and tissue supernatants collected. Supernatants were also collected from T-cell proliferation assays after 3 days of stimulation. Cytokines were measured in tissue supernatants and cell culture supernatants using the Mouse Inflammatory CBA kit (BD Biosciences), and then analyzed using FACSarray Bioanalyzer System (BD Biosciences).
Antibiotic Treatment
Pregnant conventional SPF NOD/DQ8 mice were placed on 200 mg/L vancomycin (Sigma-Aldrich) in sterile drinking water and continued after birth until pups were weaned at 3 weeks of age. Vancomycin-containing water was replaced every 3 days. Fecal pellets were collected at 3 weeks of age for microbial analysis by 16S rRNA gene sequencing. Additional mice originating from non–antibiotic-treated NOD/DQ8 mice served as controls.
Statistics
Data were evaluated by analysis of variance with the Bonferroni post-hoc test for multiple comparisons when comparing more than two groups. Unpaired t-test was used to compare two groups. For microbial analysis, the U-test was used. P < 0.05 was considered statistically significant. All statistical analysis were performed in GraphPad Prism software version 6 (GraphPad Software, San Diego CA).
Results
Colonization with a Clean Microbiota Attenuates Gluten-Induced Markers of IEL Cytotoxicity and Enterocyte Cell Death
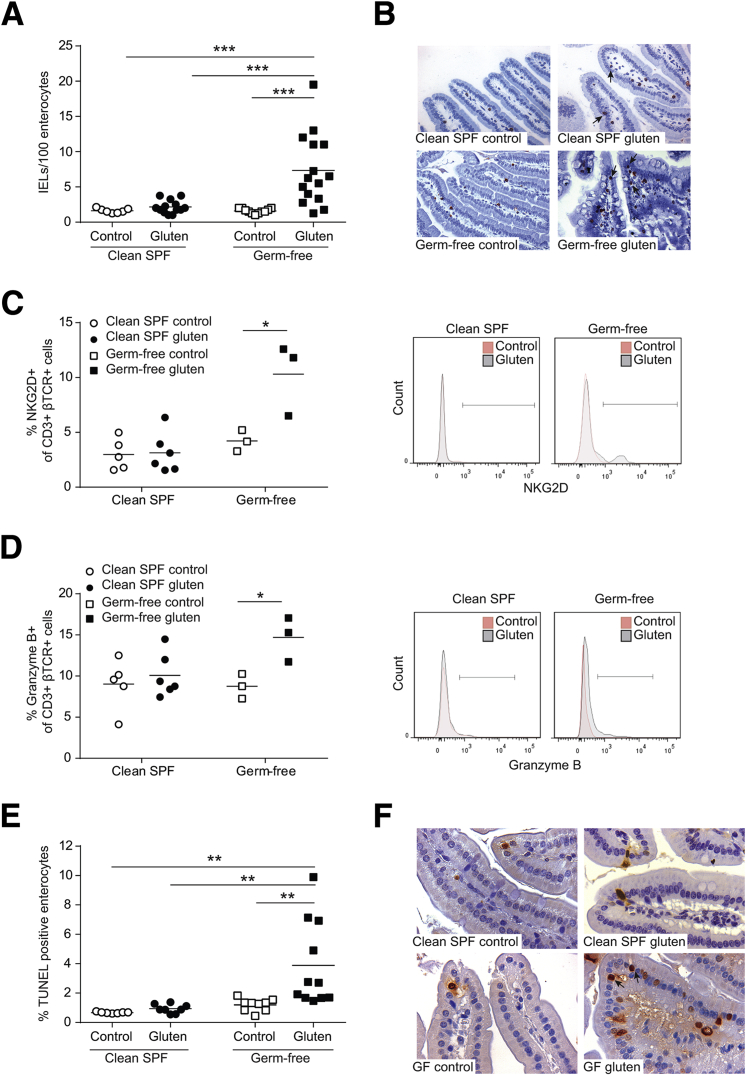
Proliferation and activation of IELs is a hallmark of CD.32 To test the hypothesis that the background microbiota modulates host responses to gluten, we first compared IEL numbers and phenotype in germ-free and clean SPF NOD/DQ8 mice following gluten sensitization and challenge (gluten treatment). The microbiota of clean SPF mice was dominated by members of the Bacteroidetes and Firmicutes phyla, and was free from any pathogens, opportunistic bacteria, or bacteria from the Proteobacteria phylum (Table 1). IEL counts increased following gluten treatment in germ-free, but not in clean SPF mice, and were greater in gluten-treated germ-free versus clean SPF mice (Figure 1, A and B). To test whether the germ-free condition enhances mucosal sensitivity to cholera toxin or to other non-gluten antigens such as zein, we determined IEL counts in germ-free and clean SPF C57BL/6 mice following sensitization and challenge with zein. No alterations in IEL counts were observed in germ-free or clean SPF C57BL/6 mice sensitized to zein (Supplemental Figure S1A).
Table 1.
Microbial Composition of Clean SPF and Conventional SPF NOD/DQ8 Mice
| Phylum | Class | Order | Family | Genus | Clean SPF, % | Conv SPF, % |
|---|---|---|---|---|---|---|
| Bacteroidetes | Bacteroidia | Bacteroidales | Unclassified | Unclassified | 0 | 3.7 |
| Bacteroidaceae | Bacteroides∗ | 0 | 62.2 | |||
| Porphyromonadaceae | Unclassified | 0 | 0.5 | |||
| Parabacteroides | 73.9 | 14.3 | ||||
| Rikenellaceae | Unclassified | 0 | 0.3 | |||
| Alistipes | 0 | 0.1 | ||||
| Deferribacteres | Deferribacteres | Deferribacterales | Deferribacteraceae | Mucispirillum | 1.9 | 0 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus∗ | 0 | 0.1 |
| Gemellales | Gemellaceae | Gemella | 0 | 0.2 | ||
| Lactobacillales | Aerococcaceae | Aerococcus | 0 | 0.1 | ||
| Lactobacillaceae | Lactobacillus | 0.1 | 1.7 | |||
| Streptococcaceae | Streptococcus∗ | 0 | 0.1 | |||
| Turicibacterales | Turicibacteraceae | Unclassified | 2.4 | 0.1 | ||
| Clostridia | Clostridiales | Unclassified | Unclassified | 0.2 | 0 | |
| Clostridiales Family XIII. Incertae Sedis | Eubacterium | 0 | 0.1 | |||
| Lachnospiraceae | Unclassified | 21 | 13.1 | |||
| Blautia | 0.2 | 0.2 | ||||
| Clostridium | 0 | 0.1 | ||||
| Ruminococcaceae | Unclassified | 0.2 | 0.2 | |||
| Anaerotruncus | 0 | 0.1 | ||||
| Proteobacteria | Betaproteobacteria | Burkholderiales | Unclassified | Unclassified∗ | 0 | 0.1 |
| Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | Helicobacter∗ | 0 | 0.1 | |
| Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Escherichia∗ | 0 | 2.0 | |
| Tenericutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Allobaculum | 0 | 0.4 |
| Coprobacillus | 0 | 0.1 |
Numbers indicate relative proportions at the genus level. Cutoff: 0.1% abundance.
Conv, conventional; SPF, specific pathogen free.
Contains pathogenic or opportunistic bacteria associated with inflammation.
Figure 1.
Colonization with a clean specific pathogen free (SPF) microbiota attenuates gluten-induced intraepithelial lymphocytosis, markers of intraepithelial lymphocyte (IEL) cytotoxicity, and enterocyte cell death in germ-free NOD/DQ8 mice. A: Quantification of CD3+ cells in villi tips in sections of jejunum, expressed as IELs per 100 enterocytes. B: Representative CD3+ stained sections of jejunum. Arrows indicate examples of IELs. C and D: Quantification and corresponding histograms of NKG2D+ (C) and granzyme B+ (D) cells gated on CD3+ β T-cell receptor (TCR)+ small-intestinal IELs. Open circles represent clean SPF controls, closed circles represent clean SPF gluten-treated mice, open squares represent germ-free controls, and closed squares represent germ-free gluten treated mice. E: Quantification of the percentage of terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL)-positive enterocytes in the villi of jejunum sections. F: Representative TUNEL-stained sections of the jejunum. Arrows indicate examples of positive cells. Each dot represents an individual mouse. ∗P < 0.05 (C and D), ∗∗P < 0.01 (E), and ∗∗∗P < 0.001 (A). Original magnification: ×20 (B); ×40 (F).
Consistent with the literature,33 naive germ-free mice had a higher proportion of γδTCR+ IELs compared to clean SPF mice (Supplemental Figure S2A). The proportions of γδTCR+ IELs or βTCR+ IELs did not change following gluten treatment in germ-free or clean SPF conditions (Supplemental Figure S2, A–C). However, gluten-treatment in germ-free mice, but not clean SPF mice, led to an increase in NKG2D and granzyme B expression in CD3+βTCR+ IELs (Figure 1, C and D). No changes in NKG2D or granzyme B were detected on γδTCR+ IELs in germ-free or clean SPF mice following gluten treatment (data not shown). The increase in IEL activation markers was accompanied by an increase in the percentage of TUNEL-positive enterocytes in gluten-treated germ-free mice, but not in gluten-treated clean SPF mice (Figure 1, E and F). IL-15 mRNA was expressed at low levels in small-intestinal tissues in all groups, and did not change following gluten treatment (Supplemental Figure S2D). IL-21 mRNA levels were undetectable in all groups (data not shown). The data suggest that in the absence of commensal colonization, gluten induces a cytotoxic IEL response associated with increased enterocyte cell death.
Colonization with a Clean Microbiota Attenuates Gluten-Induced Pathology
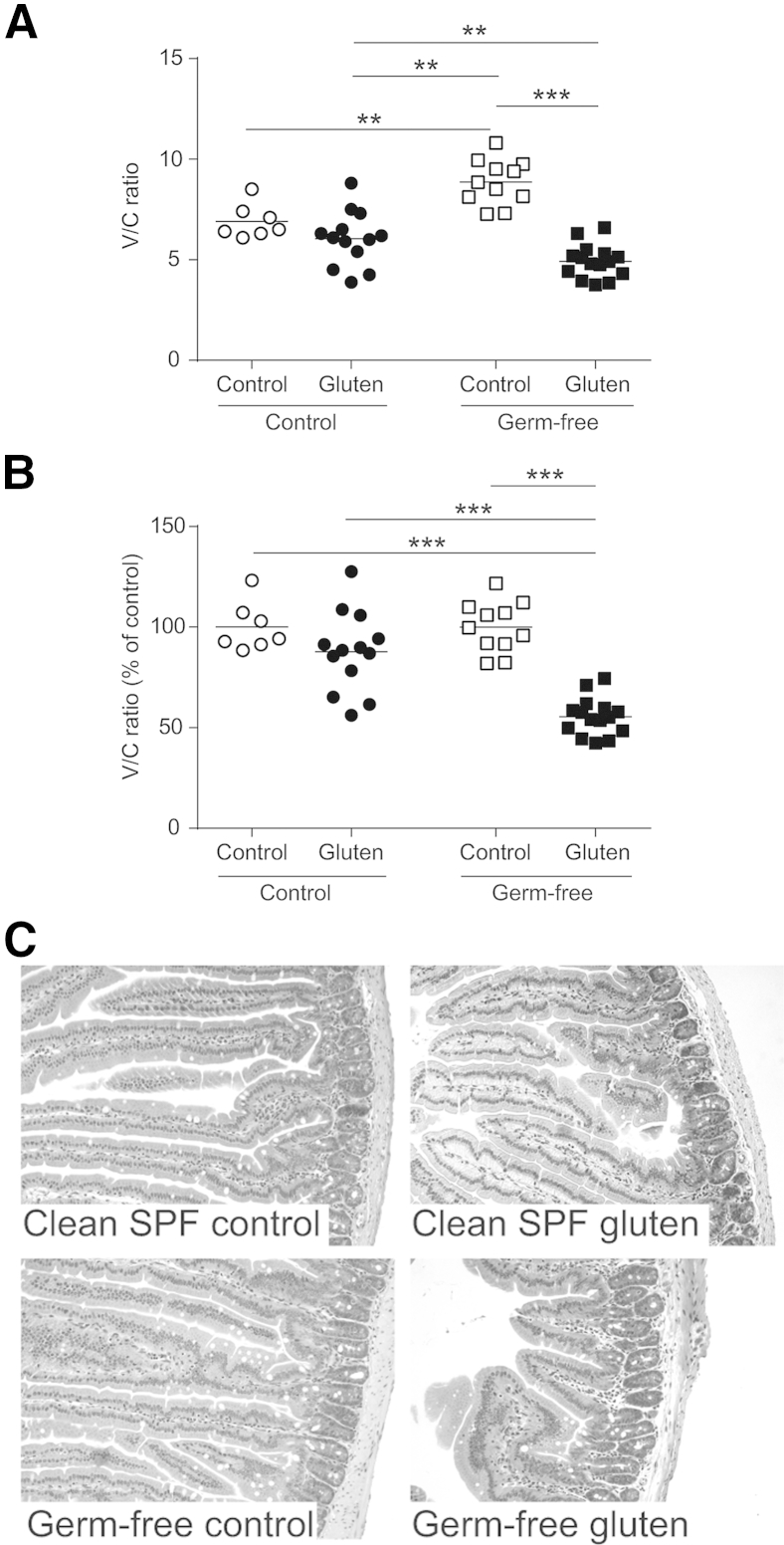
Gluten treatment in germ-free but not clean SPF NOD/DQ8 mice led to a reduction in V/C ratios (Figure 2, A and C). Under baseline conditions, germ-free mice have longer and thinner villi compared to colonized mice (Figure 2A and Supplemental Figure S3).34 Thus, to account for baseline differences under different microbial conditions, V/C ratios for gluten-treated mice were also expressed as a percentage of their respective controls for all experiments. When normalized to their controls, V/C ratios in germ-free mice were significantly lower compared to clean SPF mice following gluten treatment (Figure 2B). No alterations in V/C ratios were observed in zein-treated mice (Supplemental Figure S1, B–D). Thus, in the presence of susceptibility genes, commensal colonization with a benign microbiota attenuates gluten-induced enteropathy.
Figure 2.
Colonization with a clean specific pathogen free (SPF) microbiota attenuates gluten-induced enteropathy in germ-free NOD/DQ8 mice. A: Quantification of villus-to-crypt (V/C) ratios in jejunum sections. B: V/C ratios for clean SPF and germ-free control and gluten-treated mice, expressed as a percentage of controls. Each dot represents an individual mouse. C: Representative hematoxylin and eosin–stained sections of jejunum. ∗∗P < 0.01, ∗∗∗P < 0.001. Original magnification, ×10 (C).
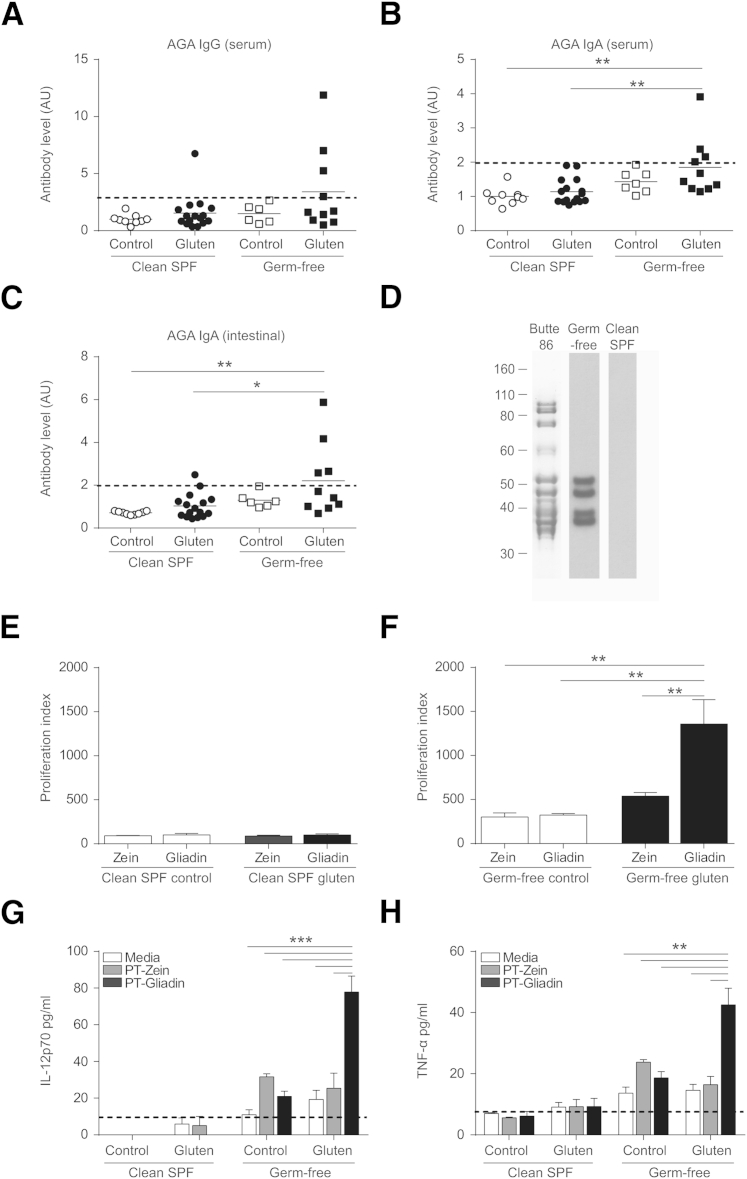
Colonization with a Clean Microbiota Attenuates Gliadin-Specific Responses
Serum and intestinal washes were tested for the presence of anti-gliadin IgA and IgG antibodies (AGA). No clean SPF or germ-free control mouse developed AGA (Figure 3, A–C). In germ-free conditions, 4 of 10 gluten-treated mice developed positive AGA in serum (IgG and IgA) and intestinal washes (IgA); by contrast, only 1 of 16 clean SPF mice developed AGA (Figure 3, A–C). Antibody reactivity to specific gluten proteins of wheat was confirmed by Western blot analysis (Figure 3D). No differences in anti-TG2 IgA antibodies were detected between groups (data not shown). To further assess the immune reactivity to gliadin, T-cell proliferation in response to PT-gliadin or PT-zein stimulation was determined using CD4+ T cells isolated from the mesenteric lymph nodes of germ-free and clean SPF control and gluten-treated mice. No response to PT-gliadin or PT-zein was observed in T cells isolated from clean SPF mice (Figure 3E). CD4+ T cells isolated from gluten-treated germ-free mice had a greater response to PT-gliadin stimulation than to PT-zein stimulation (Figure 3F). Increased reactivity to PT-gliadin in cultures from gluten-treated germ-free mice was accompanied by increased production of IL-12p70 and tumor necrosis factor-α (TNF-α) in culture supernatant (Figure 3, G and H) and mild increases in IL-6 and interferon-γ (IFN-γ) (Supplemental Figure S4, A and B). No response to gluten or zein was observed in germ-free NOD/DQ8 controls (Figure 3F) or in zein-treated mice (Supplemental Figure S1E), indicating the responses observed in germ-free mice are gluten-specific and are attenuated by colonization with clean SPF microbiota.
Figure 3.
Colonization with a clean specific pathogen free (SPF) microbiota attenuates gliadin-specific immune responses in germ-free NOD/DQ8 mice. A–C: Serum and small-intestinal washes were collected for determination of serum anti-gliadin IgG (A), serum anti-gliadin IgA (B), and intestinal anti-gliadin IgA (C). Each dot represents an individual mouse. Positive reactivity was determined by using a positive cutoff value of ≥2 standard deviations above the mean of control mice, as indicated by the dotted line. D: Confirmation of antibody reactivity to gluten proteins by Western blotting. SDS-PAGE profile of the gluten extract from the Butte 86 cultivar used for immunoblotting assays. Western blot reactivity of serum antibodies from a representative germ-free gluten-treated mouse and a representative clean SPF control mouse. Molecular weight markers on the left are in kiloDaltons. E and F: CD4+ T cells were isolated from mesenteric lymph nodes of nonsensitized control and gluten-treated clean SPF (E) and germ-free NOD/DQ8 (F) mice and stimulated with pepsin-trypsin (PT)-gliadin, PT-zein, or medium. The percentage of divided cells was determined for each stimulation condition (PT-gliadin, PT-zein), normalized to the proliferation of medium only wells, and expressed as a proliferation index. G and H: Production of IL-12p70 (G) and tumor necrosis factor (TNF)-α (H) in cell culture supernatant. Black dotted line represents the limit of detection. Data presented as means ± SEM. n = 3 (G and H, per group). ∗P < 0.05 (C), ∗∗P < 0.01 (B, C, F, and H), and ∗∗∗P < 0.001 (G). AGA, anti-gliadin antibodies.
Attenuation of Gluten-Induced Responses in Clean SPF Mice Is Independent of Increased T Regulatory Cells
Regulatory T cells (Tregs), defined as CD4+CD25+Foxp3+ T cells, were increased in the small intestine of naive germ-free NOD/DQ8 mice compared to naive clean SPF mice (Supplemental Figure S4, C and E). Furthermore, gluten treatment had no effect on Treg proportions or small-intestinal IL-10 levels (Supplemental Figure S4, D–F). In addition, no changes in TNF-α, IFN-γ, monocyte chemotactic protein 1, IL-6, or IL-12p70 cytokine levels were detected in small-intestinal tissues of clean SPF or germ-free gluten-treated mice (data not shown).
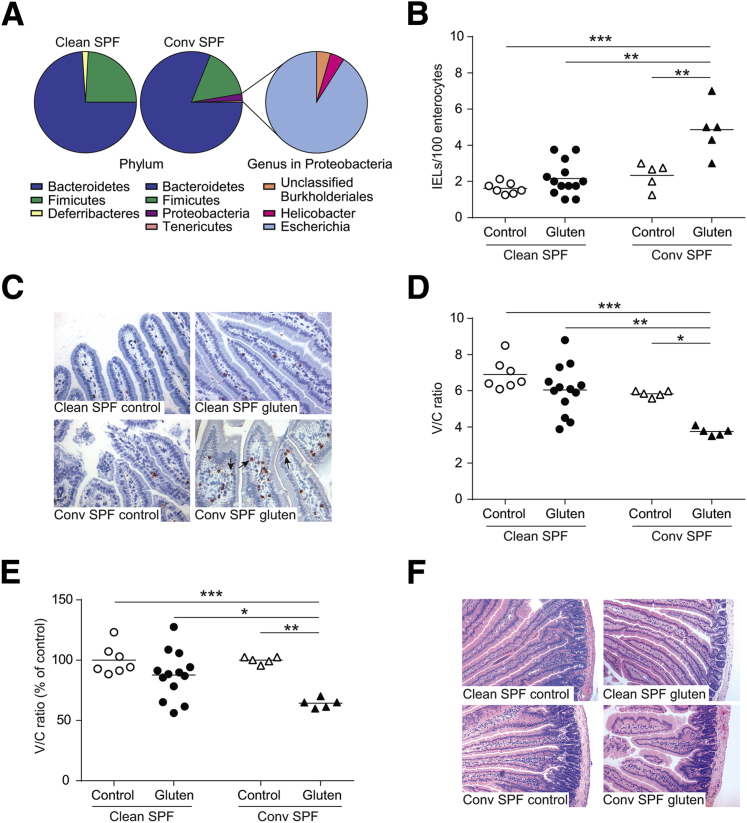
Conventional SPF Mice Develop More Severe Gluten-Induced Pathology Compared to Clean SPF NOD/DQ8 Mice
Unlike clean SPF, conventional SPF mice harbor several members from the Proteobacteria phylum such as Helicobacter and Escherichia species (Table 1 and Figure 4A). We found that gluten treatment in conventional SPF mice increased IELs in villi tips (Figure 4, B and C) and decreased V/C ratios (Figure 4, D–F) to a greater extent than in clean SPF mice.
Figure 4.
Conventional specific pathogen free (SPF) mice harbor opportunistic bacteria and develop more severe gluten-induced pathology compared to clean SPF NOD/DQ8 mice. A: Microbial composition of cecal contents from clean SPF and conventional (conv) SPF NOD/DQ8 mice by 16s rRNA sequencing at the phylum and genus level within the Proteobacteria phylum. B: Quantification of CD3+ cells in villi tips of jejunum sections, expressed as intraepithelial lymphocytes (IELs) per 100 enterocytes. C: Representative CD3+-stained sections of the jejunum. Black arrows indicate examples of IELs. D: Quantification of villus-to-crypt (V/C) ratios in jejunum sections. E: V/C ratios for clean SPF and conventional SPF control and gluten-treated mice, expressed as a percentage of controls. Each dot represents an individual mouse. F: Representative hematoxylin and eosin–stained sections of jejunum. ∗P < 0.05 (D and E), ∗∗P < 0.01 (B, D, and E), and ∗∗∗P < 0.001(B, D, and E). Original magnification: ×20 (C); ×10 (F).
Perinatal Antibiotic Treatment of Conventional SPF NOD/DQ8 Mice Worsens Gluten-Induced Pathology
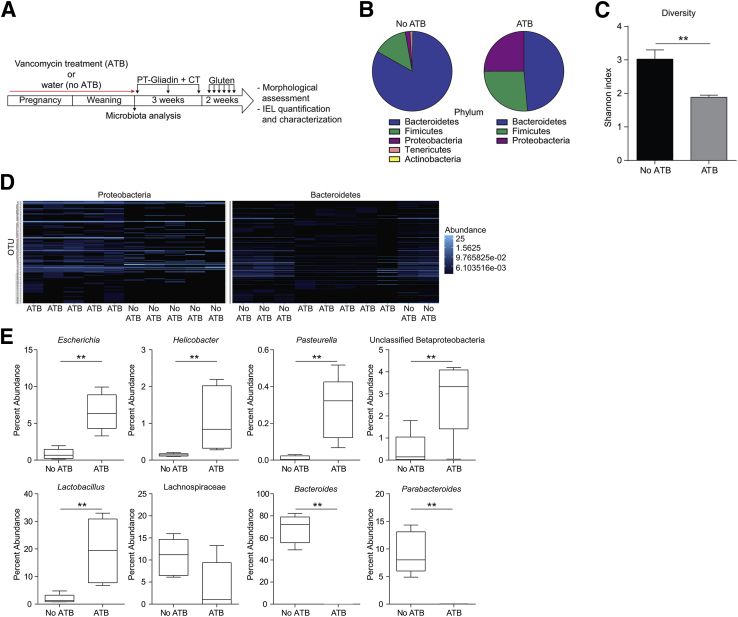
To evaluate whether expansion of the Proteobacteria phylum in conventional SPF mice further exacerbated gluten-induced pathology, we treated NOD/DQ8 mice perinatally with vancomycin (Figure 5A). Antibiotic treatment resulted in an increase in the relative abundance of Proteobacteria and Firmicutes; a decrease in the relative abundance of Actinobacteria, Bacteroidetes, and Tenericutes; and lower fecal microbiota diversity (Figure 5, B–D). At the genus level, antibiotic-treated mice had greater abundances of Escherichia, Helicobacter, Pasteurella, an unclassified Betaproteobacteria, and Lactobacillus (Figure 5E). Additionally, the family Lachnospiraceae was reduced (P = 0.056) and the genera Bacteroides and Parabacteroides were significantly reduced in antibiotic-treated mice (Figure 5E).
Figure 5.
In utero and neonatal vancomycin treatment induces changes in microbial composition in conventional specific pathogen free (SPF) NOD/DQ8 mice. A: Pregnant conventional SPF mice received antibiotics (vancomycin; ATB) in drinking water until pups were weaned at 3 weeks of age. Non–antibiotic-treated mice received water alone (no ATB). B: Fecal microbial composition by 16s rRNA sequencing, at the phylum level. C: Fecal microbial diversity, expressed via Shannon Index. D: Heat map of Proteobacteria and Bacteroidetes phyla, each band representing a unique operational taxonomic unit, generated using Bray-Curtis dissimilarity. E: Percent abundance of genera Escherichia, Helicobacter, Pasteurella, unclassified Betaproteobacteria, Bacteroides, Parabacteroidetes, Lactobacillus, and family Lachnospiraceae. Data presented as means ± SEM (C) or as medians (E), with each box extending from the 25th to 75th percentile, and whiskers extending from the minimum to maximum value. n = 5 (per group). ∗∗P < 0.01.
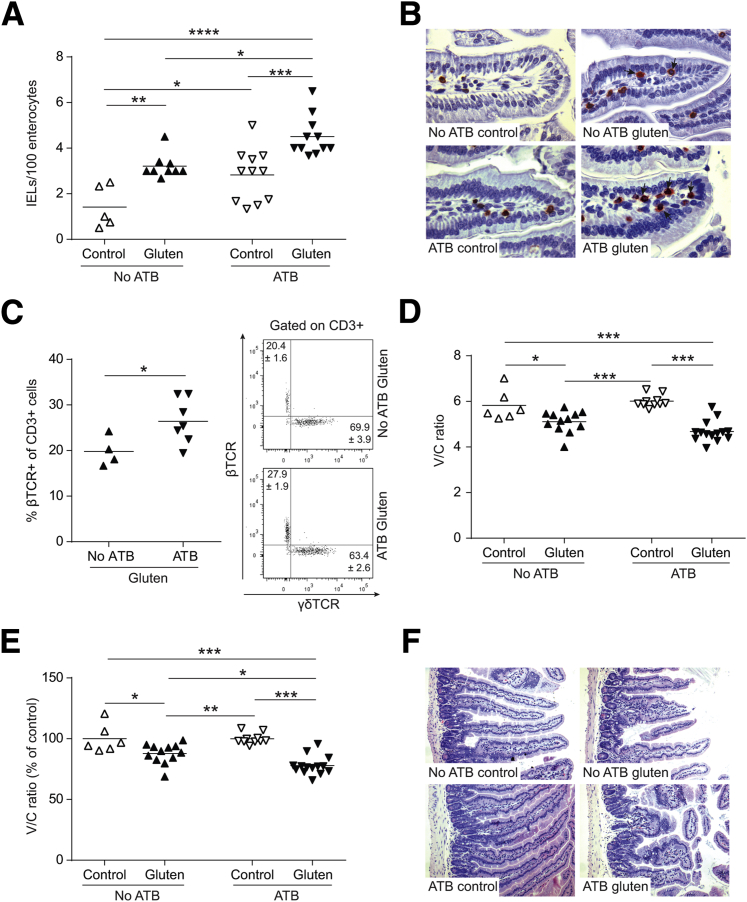
Gluten treatment increased IEL counts in mice treated with or without antibiotics. However, IEL counts were greater in gluten-treated mice that received antibiotics compared to non–antibiotic-treated mice (Figure 6, A and B). Furthermore, antibiotic treatment increased the proportion of βTCR+ IELs following gluten treatment (Figure 6C). No changes in NKG2D or granzyme B expression were detected following gluten treatment in either group (data not shown). Gluten treatment also decreased V/C ratios, with or without antibiotics (Figure 6, D–F). Together, these studies demonstrate that perturbation of early colonization and induction of dysbiosis, characterized by increased Proteobacteria, enhances the severity of gluten-induced responses in NOD/DQ8 mice.
Figure 6.
Perturbation of the colonization process in NOD/DQ8 mice increases severity of gluten-induced pathology. A: Quantification of CD3+ cells in villi tips of jejunum sections, expressed as intraepithelial lymphocytes (IELs) per 100 enterocytes. B: Representative CD3+-stained sections of jejunum. Arrows indicate examples of IELs. C: Quantification of the percentage of β T-cell receptor (TCR)+ cells gated on CD3+ small-intestinal IELs and representative flow cytometry plots for CD3+ γδTCR+ and CD3+ βTCR+ IELs are shown with the means ± SEM indicated. D: Quantification of villus-to-crypt (V/C) ratios in jejunum sections. E: V/C ratios for control and gluten-treated mice, expressed as a percentage of controls. Each dot represents an individual mouse. F: Representative hematoxylin and eosin–stained sections of the jejunum. ∗P = 0.05 (A, C, D, and E), ∗∗P < 0.01 (A and E), ∗∗∗P < 0.001 (A, D, and E), and ∗∗∗∗P < 0.0001 (A). Original magnification: ×40 (B); ×10 (F). ATB, antibiotics.
Administration of E. coli ENT CAI:5 to Clean SPF Mice Renders Mice Susceptible to Gluten-Induced Pathology
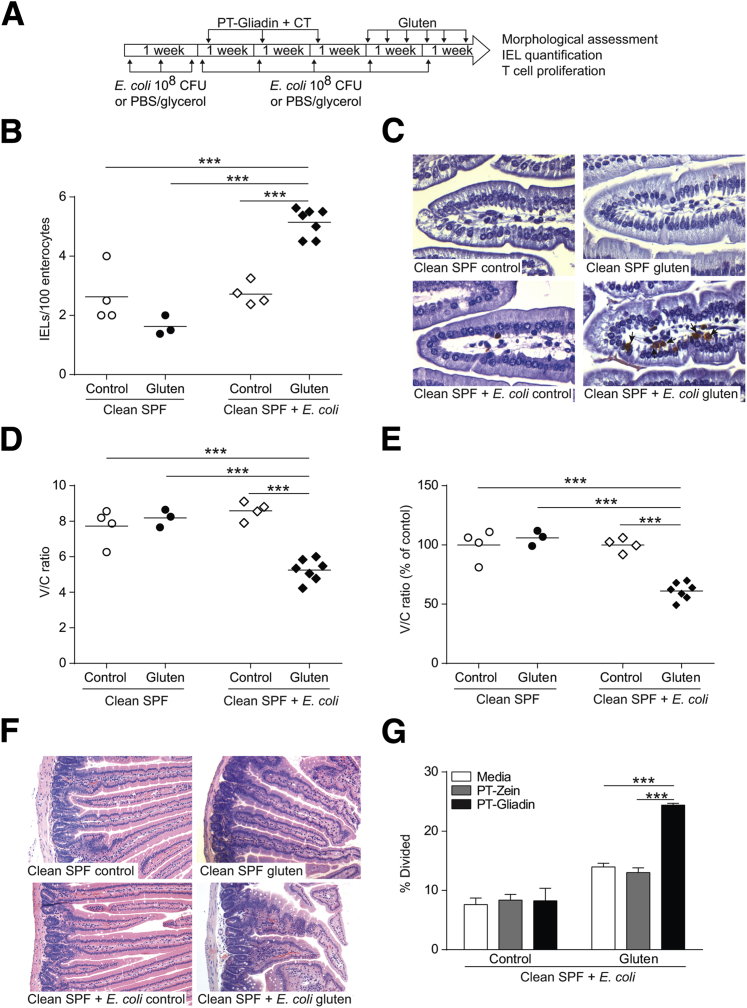
Finally, we investigated whether protection against gluten-induced immunopathology in clean SPF mice could be reversed through administration of a Proteobacteria member. Given the expansion of Escherichia associated with gluten-induced pathology in antibiotic-treated mice, and the clinical association between CD and increased abundance of Proteobacteria,35, 36, 37 we supplemented clean SPF microbiota with a noninvasive, enteroadherent E. coli ENT CAI:5 strain carrying multiple virulence genes (fimA and kfiC),22 originally isolated from a CD patient (Figure 7A). E. coli–supplemented clean SPF mice developed increased IEL counts in villi tips (Figure 7, B and C), reduced V/C ratios (Figure 7, D–F), and gliadin-specific CD4+ T-cell responses (Figure 7G) after gluten treatment. E. coli ENT CAI:5 supplementation alone had no effect (Figure 7, B–G), supporting the notion that the presence of specific groups of bacteria, combined with genetic susceptibility and gluten, influences the development of immunopathology.
Figure 7.
Supplementation of clean specific pathogen free (SPF) microbiota with Escherichia coli ENT CAI:5 increases severity of gluten-induced pathology in NOD/DQ8 mice. A:E. coli ENT CAI:5 supplementation and gluten treatment protocol. B: Quantification of CD3+ cells in villi tips of jejunum sections, expressed as intraepithelial lymphocytes (IELs) per 100 enterocytes. C: Representative CD3+-stained sections of the jejunum. Arrows indicate examples of IELs. D: Quantification of villus-to-crypt (V/C) ratios in jejunum sections. E: V/C ratios for control and gluten-treated mice, expressed as a percentage of controls. Each dot represents an individual mouse. F: Representative hematoxylin and eosin–stained sections of the jejunum. G: Mesenteric lymph node CD4+ T-cell proliferation in response to pepsin-trypsin (PT)-zein or PT-gliadin stimulation in E. coli ENT CAI:5-supplemented clean SPF mice. Data presented as means ± SEM. n = 3 per group. ∗∗∗P < 0.001. Original magnification: ×40 (C); ×10 (F). CFU, colony-forming units.
Discussion
The incidence of CD has risen dramatically over the last 5 decades, suggesting an important role for environmental factors in disease development.5, 38 Studies investigating environmental modulators of CD risk have not confirmed a protective role of feeding practices in infants at high risk.16, 17, 39 A role for the intestinal microbiota as a contributing factor to CD has been suggested in some clinical studies.13, 14 However, a modulatory role of the microbiota on gluten-induced responses has remained elusive. We used a gnotobiotic approach to test the hypothesis that the background microbiota constitutes an environmental factor that modulates host responses to gluten in NOD/DQ8 mice. Colonization with a microbiota free from any opportunistic bacteria and bred in gnotobiotic conditions (clean SPF) prevented the development of gluten-induced immunopathology compared to the germ-free status, or to conventional SPF mice that harbor a diverse microbiota containing opportunistic pathogens belonging to the Proteobacteria phylum. Perinatal antibiotic disruption of the microbiota, leading to Proteobacteria expansion, further enhanced gluten-induced immunopathology in conventional SPF mice. When clean SPF mice were supplemented with an E. coli isolated from a CD patient (E. coli ENT CAI:5), the protection conferred by the benign microbiota was suppressed.
Gluten-treated germ-free mice developed decreased V/C ratios, a cytotoxic IEL phenotype and increased enterocyte cell death compared to clean SPF mice. The finding is in agreement with an earlier study, which demonstrated that long-term gliadin feeding to germ-free wild-type rats induced moderate small-intestinal damage.40 However, interpretation of that study was limited by the lack of appropriate colonized controls. Expanding on that study, we show through several different strategies, that the composition of the microbiota can modulate gluten-induced immunopathology in the context of the HLA-DQ8 gene. Moreover, we demonstrate these effects with short-term gluten challenge. IELs from gluten-treated germ-free mice in our study had increased expression of NKG2D and granzyme B, which mediate epithelial cell death and are increased in IELs from active CD patients.4, 41 In addition to increased markers of IEL cytotoxicity, germ-free mice treated with gluten developed gliadin-specific antibodies and a proinflammatory gliadin-specific T-cell response, which were absent in clean SPF conditions. The underlying mechanisms could relate to the absence of homeostatic regulation by the commensal microbiota in germ-free mice. It is known that commensal bacterial colonization induces maturation of intestinal structure as well as immune gut function.42 The microbiota of our clean SPF mice is primarily composed of bacteria that are generally considered important for inducing maturation of the immune system7, 21 and down-regulating adverse inflammatory responses.10, 43 At the genus level, the microbiota was dominated by Parabacteroides which has been shown to protect against experimental colitis through several pathways, including induction of regulatory mechanisms.43
Tregs can be modulated by the microbiota, and they play a central role in oral tolerance.7, 44 However, the role of the microbiota in modulating oral tolerance is controversial.45, 46 Unlike what has been previously reported for colonic Tregs,7 our study suggests the proportion of small-intestinal lamina propria Tregs was higher in naive germ-free NOD/DQ8 mice compared to colonized mice, indicating that a decrease in Treg proportions does not explain the higher reactivity to gluten in germ-free mice. Although we did not detect changes in small-intestinal levels of the regulatory cytokine IL-10, the results do not rule out that differences in Treg function underlie this observation. However, studies have suggested that small-intestinal Tregs from patients with CD47 or IL-15 transgenic mice48 are functional and suppressive, and that the defective response resides in effector T cells becoming unresponsive to Tregs due to dysregulated IL-15 signaling.49 Increased expression of IL-15 has been found in a proportion of CD patients,50 and animal models have reported IL-15–mediated gluten- or ovalbumin-induced enteropathy.48, 50 There is also evidence that IL-15 can induce IEL activation.4, 51 However, we found that IL-15 mRNA expression was low, and no changes were detected between germ-free or clean SPF groups at the transcriptional level. Although methodological issues have been raised regarding IL-15 measurement,52 NOD mice have been reported to have reduced IL15 gene expression, which explains our results.53 This suggests that the increased IEL numbers and markers of cytotoxicity observed in germ-free NOD/DQ8 mice following gluten treatment is mediated through an IL-15–independent pathway.
Conventional SPF NOD/DQ8 mice have previously been shown to respond to gliadin sensitization and challenge, developing a mild decrease in V/C ratios, increased IEL counts in villi tips, increased intestinal permeability, as well as gliadin-specific antibody and T-cell responses compared to nonsensitized controls.19, 54 However, mice with distinct colonization conditions have never been compared with respect to the degree of gluten-induced immunopathology. We found more severe gluten-induced responses in conventional SPF compared to clean SPF mice. Together with the germ-free studies, the data reveal a complex modulatory role of the microbiota to gluten, which may also include exacerbation of responses due to the presence of opportunistic pathogens within the conventional SPF microbiota. To test this hypothesis, we performed experiments using perinatal vancomycin treatment of conventional SPF NOD/DQ8 mice to deliberately perturb the normal colonization process and expand the Proteobacteria phylum.55 Early-life antibiotic treatment led to significant changes in microbial profiles at 3 weeks of age with increases in Proteobacteria and Firmicutes. Interestingly, infants with a high genetic risk for CD have a higher relative abundance of Proteobacteria, including Escherichia.56 Antibiotic-treated mice had increased IELs, with or without gluten, as well as increased proportions of βTCR+ IELs in adult gluten-treated mice, an IEL subset that has been shown to be responsible for small-intestinal enteropathy associated with CD,57 whereas γδTCR IELs may play a protective role.41 Microbial signaling also modulate IEL number and phenotype,58 and thus, combinatory effects of both changes in microbial composition and presence of gluten likely explain the higher IEL numbers in antibiotic- and gluten-treated mice.
Increased abundance of Proteobacteria, including E. coli, has been reported in CD children,35, 36 and increased abundance of Proteobacteria has been associated with persistent symptoms in CD patients following a gluten-free diet.37 Furthermore, E. coli isolated from CD children have been shown to carry a higher number of virulence genes22 and induce proinflammatory cytokine production and activation markers in response to gluten stimulation in peripheral blood mononuclear cell cultures, dendritic cell cultures, and intestinal loops.59, 60, 61 To further explore the role of pathobionts in modulation of responses to gluten, we supplemented clean SPF mice with E. coli ENT CAI:5. This rendered clean SPF mice, otherwise protected, susceptible to gluten sensitization as evidenced by increased IEL counts and increased T-cell proliferation to gliadin in vitro. Although the findings highlight a potential disease-modifying role of Proteobacteria in CD, there could be additional microbial differences between clean, conventional, and antibiotic-treated mice that may contribute. Thus, additional basic and clinical studies are needed to define the exact contribution of Proteobacteria versus other microbes in the modulation of gluten-induced responses.
In summary, we show that distinct changes in microbiota structure can either ameliorate or enhance IEL and CD4+ T-cell responses to gluten in NOD/DQ8 mice. Our results support the concept that alterations in microbiota recently reported in active or symptomatic CD patients who are on a gluten-free diet could be causally related. Importantly, the data argue that the recognized increase in CD prevalence in the general population is causally driven, at least in part, by perturbations in intestinal microbial ecology. Specific microbiota-based therapies may aid in the prevention or treatment of CD in subjects with moderate genetic risk.
Acknowledgments
We thank Joe Notarangelo, Sarah Armstrong, and Sheryll Competente from McMaster's Axenic Gnotobiotic Unit for their technical support in gnotobiotic experiments and Dr. Michael G. Surette and the staff of the Farncombe Metagenomics Facility for advice on microbiota analysis.
Footnotes
See related Commentary on page 2864
Supported by Canadian Institutes of Health Research grant MOP#123282 (E.F.V.), partially by NIH grant R01 DK67189 (B.J.), The Stanley Medical Research Institute 08R-2061 (A.A. and S.H.), and MINECO grant AGL2011-25169 (Y.S.). H.J.G. and J.L.M. received a New Investigator Award from the Canadian Celiac Association, and M.M., Erwin Schrödinger Fellowship J 3418-B19 from the FWF Austrian Science Fund. E.F.V. and M.J. hold Canada Research Chairs.
H.J.G. and J.L.M. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2015.07.018.
Supplemental Data
Clean specific pathogen free (SPF) and germ-free C57BL/6 mice are protected from zein sensitization. Zein-treated clean SPF and germ-free C57BL/6 mice were orally sensitized to zein plus cholera toxin and challenged with zein three times a week for 2 weeks. Nonsensitized controls were treated with cholera toxin alone. A: Quantification of CD3+ cells in villi tips of jejunum sections, expressed as intraepithelial lymphocytes (IEL) per 100 enterocytes. B: Quantification of villus-to-crypt (V/C) ratios in jejunum sections. C: V/C ratios for clean SPF and germ-free control and zein-treated mice, expressed as a percentage of controls. Each dot represents an individual mouse. D: Representative hematoxylin and eosin–stained sections of jejunum. E: CD4+ T cells were isolated from mesenteric lymph nodes of zein-treated clean SPF and germ-free mice, labeled with carboxyfluorescein succinimidyl ester, and stimulated with pepsin-trypsin (PT)-gliadin, PT-zein, or medium. T-cell proliferation (% divided) was determined by flow cytometry. Data are presented as means ± SEM. n = 4 to 6 (per group). Original magnification, ×10 (D).
Gluten treatment does not induce changes in β T-cell receptor (TCR)+ or γδTCR+ intraepithelial lymphocyte (IEL) frequency or small-intestinal IL-15 levels in clean specific pathogen free (SPF) or germ-free NOD/DQ8 mice. A–C: IELs were isolated from the small intestine of nonsensitized controls and gluten-treated clean SPF and germ-free NOD/DQ8 mice, and the expression of βTCR and γδTCR was determined by flow cytometry. Quantification of γδTCR+ (A) and βTCR+ (B) cells gated on CD3+ lymphocytes. Each dot represents an individual mouse. Open circles represent clean SPF controls, closed circles represent clean SPF gluten-treated mice, open squares represent germ-free controls, closed squares represent germ-free gluten-treated mice. C: Representative flow cytometry plots for γδTCR+ and βTCR+ cells, gated on CD3+ IELs, are shown with the mean ± SEM indicated. D: IL-15 mRNA expression in the small intestine, normalized to GAPDH, and expressed as fold induction relative to controls. Data are presented as means ± SEM. n = 6 to 10 (per group).
Naive germ-free NOD/DQ8 mice have greater villus-to-crypt (V/C) ratios compared to naive clean specific pathogen free (SPF) NOD/DQ8 mice. A: Quantification of V/C ratios in jejunum sections from naive clean SPF and germ-free NOD/DQ8 mice. Each dot represents an individual mouse. B: Representative hematoxylin and eosin–stained jejunum sections from naive clean SPF and germ-free NOD/DQ8 mice. ∗P < 0.05. Original magnification, ×4 (B).
Gluten-treatment in germ-free NOD/DQ8 mice induces a proinflammatory gliadin-specific immune response, but is not associated with changes in small-intestinal regulator T cells (Tregs). A and B: CD4+ T cells were isolated from mesenteric lymph nodes of control and gluten-treated clean specific pathogen free (SPF) and germ-free mice and stimulated with pepsin-trypsin (PT)-gliadin, PT-zein, or medium for 3 days. Production of IL-6 (A) and IFN-γ (B) was measured in cell culture supernatant. Red line represents the limit of detection. C and D: Quantification of CD25+Foxp3+ cells gated on CD3+CD4+ lymphocytes in the small intestine of naïve mice (C) and control and gluten-treated clean SPF and germ-free mice (D). Each dot represents an individual mouse. E: Representative flow cytometry plots of small-intestinal CD25+Foxp3+ cells gated on CD3+CD4+ cells in naive nonsensitized control, and gluten-treated clean SPF and germ-free mice are shown with the mean ± SEM indicated. F: Quantification of IL-10 in small-intestinal tissue samples of nonsensitized control and gluten-treated clean SPF and germ-free mice. Data are presented as mean ± SEM. n = 3 (A and B, per group); n = 5 to 6 (F, per group). ∗∗P < 0.01.
References
- 1.Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E.O., Schuppan D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 2.Shan L., Molberg Ø., Parrot I., Hausch F., Filiz F., Gray G.M., Sollid L.M., Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 3.Allegretti Y.L., Bondar C., Guzman L., Rua E.C., Chopita N., Fuertes M., Zwirner N.W., Chirdo F.G. Broad MICA/B expression in the small bowel mucosa: a link between cellular stress and celiac disease. PLoS One. 2013;8:e73658. doi: 10.1371/journal.pone.0073658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meresse B., Chen Z., Ciszewski C., Tretiakova M., Bhagat G., Krausz T.N., Raulet D.H., Lanier L.L., Groh V., Spies T., Ebert E.C., Green P.H., Jabri B. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 5.Rubio-Tapia A., Kyle R.A., Kaplan E.L., Johnson D.R., Page W., Erdtmann F., Brantner T.L., Kim W., Phelps T.K., Lahr B.D., Zinsmeister A.R., Melton L.J., 3rd, Murray J.A. Increased prevalence and mortality in undiagnosed celiac disease. Gastroenterology. 2009;137:88–93. doi: 10.1053/j.gastro.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sollid L.M., Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol. 2013;13:294–302. doi: 10.1038/nri3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geuking M.B., Cahenzli J., Lawson M.A., Ng D.C., Slack E., Hapfelmeier S., McCoy K.D., Macpherson A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 8.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw S.Y., Blanchard J.F., Bernstein C.N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol. 2010;105:2687–2692. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 10.Stefka A.T., Feehley T., Tripathi P., Qiu J., McCoy K., Mazmanian S.K., Tjota M.Y., Seo G.Y., Cao S., Theriault B.R., Antonopoulos D.A., Zhou L., Chang E.B., Fu Y.X., Nagler C.R. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candon S., Perez-Arroyo A., Marquet C., Valette F., Foray A.P., Pelletier B., Milani C., Ventura M., Bach J.F., Chatenoud L. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10:e0125448. doi: 10.1371/journal.pone.0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz Y., Palma G.D., Laparra M. Unraveling the ties between celiac disease and intestinal microbiota. Int Rev Immunol. 2011;30:207–218. doi: 10.3109/08830185.2011.599084. [DOI] [PubMed] [Google Scholar]
- 13.Mårild K., Stephansson O., Montgomery S., Murray J.A., Ludvigsson J.F. Pregnancy outcome and risk of celiac disease in offspring: a nationwide case-control study. Gastroenterology. 2012;142:39–45. doi: 10.1053/j.gastro.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canova C., Zabeo V., Pitter G., Romor P., Baldovin T., Zanotti R., Simonato L. Association of maternal education, early infections, and antibiotic use with celiac disease: a population-based birth cohort study in northeastern Italy. Am J Epidemiol. 2014;180:76–85. doi: 10.1093/aje/kwu101. [DOI] [PubMed] [Google Scholar]
- 15.Decker E., Engelmann G., Findeisen A., Gerner P., Laaβ M., Ney D., Posovszky C., Hoy L., Hornef M.W. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics. 2010;125:e1433–e1440. doi: 10.1542/peds.2009-2260. [DOI] [PubMed] [Google Scholar]
- 16.Aronsson C.A., Lee H.-S., Liu E., Uusitalo U., Hummel S., Yang J., Hummel M., Rewers M., She J.-X., Simell O., Toppari J., Ziegler A.G., Krischer J., Virtanen S.M., Norris J.M., Agardh D., Teddy Study Group Age at gluten introduction and risk of celiac disease. Pediatrics. 2015;135:239–245. doi: 10.1542/peds.2014-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vriezinga S.L., Auricchio R., Bravi E., Castillejo G., Chmielewska A., Crespo Escobar P., Kolaček S., Koletzko S., Korponay-Szabo I.R., Mummert E., Polanco I., Putter H., Ribes-Koninckx C., Shamir R., Szajewska H., Werkstetter K., Greco L., Gyimesi J., Hartman C., Hogen Esch C., Hopman E., Ivarsson A., Koltai T., Koning F., Martinez-Ojinaga E., te Marvelde C., Pavic A., Romanos J., Stoopman E., Villanacci V., Wijmenga C., Troncone R., Mearin M.L. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. 2014;371:1304–1315. doi: 10.1056/NEJMoa1404172. [DOI] [PubMed] [Google Scholar]
- 18.Verdu E.F., Galipeau H.J., Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2015;12:497–506. doi: 10.1038/nrgastro.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galipeau H.J., Rulli N.E., Jury J., Huang X., Araya R., Murray J.A., David C.S., Chirdo F.G., McCoy K.D., Verdu E.F. Sensitization to gliadin induces moderate enteropathy and insulitis in nonobese diabetic-DQ8 mice. J Immunol. 2011;187:4338–4346. doi: 10.4049/jimmunol.1100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marietta E., Black K., Camilleri M., Krause P., Rogers R.S., 3rd, David C., Pittelkow M.R., Murray J.A. A new model for dermatitis herpetiformis that uses HLA-DQ8 transgenic NOD mice. J Clin Invest. 2004;114:1090–1097. doi: 10.1172/JCI21055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slack E., Hapfelmeier S., Stecher B., Velykoredko Y., Stoel M., Lawson M.A., Geuking M.B., Beutler B., Tedder T.F., Hardt W.D., Bercik P., Verdu E.F., McCoy K.D., Macpherson A.J. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sánchez E., Nadal I., Donat E., Ribes-Koninckx C., Calabuig M., Sanz Y. Reduced diversity and increased virulence-gene carriage in intestinal enterobacteria of coeliac children. BMC Gastroenterol. 2008;8:50. doi: 10.1186/1471-230X-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whelan F.J., Verschoor C.P., Stearns J.C., Rossi L., Luinstra K., Loeb M., Smieja M., Johnstone J., Surette M.G., Bowdish D.M. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. An Am Thorac Soc. 2014;11:513–521. doi: 10.1513/AnnalsATS.201310-351OC. [DOI] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 25.Masella A.P., Bartram A.K., Truszkowski J.M., Brown D.G., Neufeld J.D. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye Y. Identification and quantification of abundant species from pyrosequences of 16s rRNA by consensus alignment. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2011;2010:153–157. doi: 10.1109/BIBM.2010.5706555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau N.M., Green P.H., Taylor A.K., Hellberg D., Ajamian M., Tan C.Z., Kosofsky B.E., Higgins J.J., Rajadhyaksha A.M., Alaedini A. Markers of celiac disease and gluten sensitivity in children with autism. PLoS One. 2013;8:e66155. doi: 10.1371/journal.pone.0066155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moeller S., Canetta P.A., Taylor A.K., Arguelles-Grande C., Snyder H., Green P.H., Kiryluk K., Alaedini A. Lack of serologic evidence to link IgA nephropathy with celiac disease or immune reactivity to gluten. PLoS One. 2014;9:e94677. doi: 10.1371/journal.pone.0094677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abadie V., Discepolo V., Jabri B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin Immunopathol. 2012;34:551–566. doi: 10.1007/s00281-012-0316-x. [DOI] [PubMed] [Google Scholar]
- 33.Bandeira A., Mota-Santos T., Itohara S., Degermann S., Heusser C., Tonegawa S., Coutinho A. Localization of gamma/delta T cells to the intestinal epithelium is independent of normal microbial colonization. J Exp Med. 1990;172:239–244. doi: 10.1084/jem.172.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhardt C., Bergentall M., Greiner T.U., Schaffner F., Östergren-Lundén G., Petersen L.C., Ruf W., Bäckhed F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez E., Donat E., Ribes-Koninckx C., Fernández-Murga M.L., Sanz Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl Environ Microbiol. 2013;79:5472–5479. doi: 10.1128/AEM.00869-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collado M.C., Donat E., Ribes-Koninckx C., Calabuig M., Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol. 2009;62:264–269. doi: 10.1136/jcp.2008.061366. [DOI] [PubMed] [Google Scholar]
- 37.Wacklin P., Laurikka P., Lindfors K., Collin P., Salmi T., Lähdeaho M.-L., Saavalainen P., Mäki M., Mättö J., Kurppa K., Kaukinen K. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am J Gastroenterol. 2014;109:1933–1941. doi: 10.1038/ajg.2014.355. [DOI] [PubMed] [Google Scholar]
- 38.Murray J.A., Dyke C.V., Plevak M.F., Dierkhising R.A., Zinsmeister A.R., Melton L.J. Trends in the identification and clinical features of celiac disease in a North American community, 1950–2001. Clin Gastroenterol Hepatol. 2003;1:19–27. doi: 10.1053/jcgh.2003.50004. [DOI] [PubMed] [Google Scholar]
- 39.Lionetti E., Castellaneta S., Francavilla R., Pulvirenti A., Tonutti E., Amarri S., Barbato M., Barbera C., Barera G., Bellantoni A., Castellano E., Guariso G., Limongelli M.G., Pellegrino S., Polloni C., Ughi C., Zuin G., Fasano A., Catassi C., SIGENP (Italian Society of Pediatric Gastroenterology, Hepatology, and Nutrition) Working Group on Weaning and CD Risk Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med. 2014;371:1295–1303. doi: 10.1056/NEJMoa1400697. [DOI] [PubMed] [Google Scholar]
- 40.Štepánková R., Tlaskalova-Hogenova H., Šinkora J., Jodl J., Fric P. Changes in jejunal mucosa after long-term feeding of germfree rats with gluten. Scand J Gastroenterol. 1996;31:551–557. doi: 10.3109/00365529609009127. [DOI] [PubMed] [Google Scholar]
- 41.Bhagat G., Naiyer A.J., Shah J.G., Harper J., Jabri B., Wang T.C., Green P.H., Manavalan J.S. Small intestinal CD8+ TCRγδ+ NKG2A+ intraepithelial lymphocytes have attributes of regulatory cells in patients with celiac disease. J Clin Invest. 2008;118:281–293. doi: 10.1172/JCI30989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer F., Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 43.Kverka M., Zakostelska Z., Klimesova K., Sokol D., Hudcovic T., Hrncir T., Rossmann P., Mrazek J., Kopecny J., Verdu E. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol. 2011;163:250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabst O., Mowat A. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa H., Tanaka K., Maeda Y., Aiba Y., Hata A., Tsuji N.M., Koga Y., Matsumoto T. Effect of intestinal microbiota on the induction of regulatory CD25+ CD4+ T cells. Clin Exp Immunol. 2008;153:127–135. doi: 10.1111/j.1365-2249.2008.03668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walton K., Galanko J., Balfour Sartor R., Fisher N. T cell-mediated oral tolerance is intact in germ-free mice. Clin Exp Immunol. 2006;143:503–512. doi: 10.1111/j.1365-2249.2006.03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hmida N.B., Ahmed M.B., Moussa A., Rejeb M.B., Said Y., Kourda N., Meresse B., Abdeladhim M., Louzir H., Cerf-Bensussan N. Impaired control of effector T cells by regulatory T cells: a clue to loss of oral tolerance and autoimmunity in celiac disease? Am J Gastroenterol. 2011;107:604–611. doi: 10.1038/ajg.2011.397. [DOI] [PubMed] [Google Scholar]
- 48.Korneychuk N., Ramiro-Puig E., Ettersperger J., Schulthess J., Montcuquet N., Kiyono H., Meresse B., Cerf-Bensussan N. Interleukin 15 and CD4+ T cells cooperate to promote small intestinal enteropathy in response to dietary antigen. Gastroenterology. 2014;146:1017–1027. doi: 10.1053/j.gastro.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 49.Ben Ahmed M., Belhadj Hmida N., Moes N., Buyse S., Abdeladhim M., Louzir H., Cerf-Bensussan N. IL-15 renders conventional lymphocytes resistant to suppressive functions of regulatory T cells through activation of the phosphatidylinositol 3-kinase pathway. J Immunol. 2009;182:6763–6770. doi: 10.4049/jimmunol.0801792. [DOI] [PubMed] [Google Scholar]
- 50.DePaolo R.W., Abadie V., Tang F., Fehlner-Peach H., Hall J.A., Wang W., Marietta E.V., Kasarda D.D., Waldmann T.A., Murray J.A., Semrad C., Kupfer S.S., Belkaid Y., Guandalini S., Jabri B. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mention J.J., Ben Ahmed M., Bègue B., Barbe U., Verkarre V., Asnafi V., Colombel J., Cugnenc P., Ruemmele F.M., McIntyre E., Brousse N., Cellier C., Cerf-Bensussan N. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. 2003;125:730–745. doi: 10.1016/s0016-5085(03)01047-3. [DOI] [PubMed] [Google Scholar]
- 52.Colpitts S.L., Stonier S.W., Stoklasek T.A., Root S.H., Aguila H.L., Schluns K.S., Lefrançois L. Transcriptional regulation of IL-15 expression during hematopoiesis. J Immunol. 2013;191:3017–3024. doi: 10.4049/jimmunol.1301389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suwanai H., Wilcox M.A., Mathis D., Benoist C. A defective Il15 allele underlies the deficiency in natural killer cell activity in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2010;107:9305–9310. doi: 10.1073/pnas.1004492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galipeau H.J., Wiepjes M., Motta J.P., Schulz J.D., Jury J., Natividad J.M., Pinto-Sanchez I., Sinclair D., Rousset P., Martin-Rosique R., Bermudez-Humaran L., Leroux J.C., Murray J.A., Smecuol E., Bai J.C., Vergnolle N., Langella P., Verdu E.F. Novel role of the serine protease inhibitor elafin in gluten-related disorders. Am J Gastroenterol. 2014;109:748–756. doi: 10.1038/ajg.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murphy E.F., Cotter P.D., Hogan A., O'Sullivan O., Joyce A., Fouhy F., Clarke S.F., Marques T.M., O'Toole P.W., Stanton C., Quigley E.M., Daly C., Ross P.R., O'Doherty R.M., Shanahan F. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut. 2013;62:220–226. doi: 10.1136/gutjnl-2011-300705. [DOI] [PubMed] [Google Scholar]
- 56.Olivares M., Neef A., Castillejo G., De Palma G., Varea V., Capilla A., Palau F., Nova E., Marcos A., Polanco I., Ribes-Koninckx C., Ortigosa L., Izquierdo L., Sanz Y. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2015;64:406–417. doi: 10.1136/gutjnl-2014-306931. [DOI] [PubMed] [Google Scholar]
- 57.Kutlu T., Brousse N., Rambaud C., Le Deist F., Schmitz J., Cerf-Bensussan N. Numbers of T cell receptor (TCR) alpha beta+ but not of TcR gamma delta+ intraepithelial lymphocytes correlate with the grade of villous atrophy in coeliac patients on a long term normal diet. Gut. 1993;34:208–214. doi: 10.1136/gut.34.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang W., Wang X., Zeng B., Liu L., Tardivel A., Wei H., Han J., MacDonald H.R., Tschopp J., Tian Z., Zhou R. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J Exp Med. 2013;210:2465–2476. doi: 10.1084/jem.20122490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Palma G., Cinova J., Stepankova R., Tuckova L., Sanz Y. Pivotal Advance: Bifidobacteria and Gram-negative bacteria differentially influence immune responses in the proinflammatory milieu of celiac disease. J Leukoc Biol. 2010;87:765–778. doi: 10.1189/jlb.0709471. [DOI] [PubMed] [Google Scholar]
- 60.De Palma G., Kamanova J., Cinova J., Olivares M., Drasarova H., Tuckova L., Sanz Y. Modulation of phenotypic and functional maturation of dendritic cells by intestinal bacteria and gliadin: relevance for celiac disease. J Leukoc Biol. 2012;92:1043–1054. doi: 10.1189/jlb.1111581. [DOI] [PubMed] [Google Scholar]
- 61.Cinova J., De Palma G., Stepankova R., Kofronova O., Kverka M., Sanz Y., Tuckova L. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS One. 2011;6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clean specific pathogen free (SPF) and germ-free C57BL/6 mice are protected from zein sensitization. Zein-treated clean SPF and germ-free C57BL/6 mice were orally sensitized to zein plus cholera toxin and challenged with zein three times a week for 2 weeks. Nonsensitized controls were treated with cholera toxin alone. A: Quantification of CD3+ cells in villi tips of jejunum sections, expressed as intraepithelial lymphocytes (IEL) per 100 enterocytes. B: Quantification of villus-to-crypt (V/C) ratios in jejunum sections. C: V/C ratios for clean SPF and germ-free control and zein-treated mice, expressed as a percentage of controls. Each dot represents an individual mouse. D: Representative hematoxylin and eosin–stained sections of jejunum. E: CD4+ T cells were isolated from mesenteric lymph nodes of zein-treated clean SPF and germ-free mice, labeled with carboxyfluorescein succinimidyl ester, and stimulated with pepsin-trypsin (PT)-gliadin, PT-zein, or medium. T-cell proliferation (% divided) was determined by flow cytometry. Data are presented as means ± SEM. n = 4 to 6 (per group). Original magnification, ×10 (D).
Gluten treatment does not induce changes in β T-cell receptor (TCR)+ or γδTCR+ intraepithelial lymphocyte (IEL) frequency or small-intestinal IL-15 levels in clean specific pathogen free (SPF) or germ-free NOD/DQ8 mice. A–C: IELs were isolated from the small intestine of nonsensitized controls and gluten-treated clean SPF and germ-free NOD/DQ8 mice, and the expression of βTCR and γδTCR was determined by flow cytometry. Quantification of γδTCR+ (A) and βTCR+ (B) cells gated on CD3+ lymphocytes. Each dot represents an individual mouse. Open circles represent clean SPF controls, closed circles represent clean SPF gluten-treated mice, open squares represent germ-free controls, closed squares represent germ-free gluten-treated mice. C: Representative flow cytometry plots for γδTCR+ and βTCR+ cells, gated on CD3+ IELs, are shown with the mean ± SEM indicated. D: IL-15 mRNA expression in the small intestine, normalized to GAPDH, and expressed as fold induction relative to controls. Data are presented as means ± SEM. n = 6 to 10 (per group).
Naive germ-free NOD/DQ8 mice have greater villus-to-crypt (V/C) ratios compared to naive clean specific pathogen free (SPF) NOD/DQ8 mice. A: Quantification of V/C ratios in jejunum sections from naive clean SPF and germ-free NOD/DQ8 mice. Each dot represents an individual mouse. B: Representative hematoxylin and eosin–stained jejunum sections from naive clean SPF and germ-free NOD/DQ8 mice. ∗P < 0.05. Original magnification, ×4 (B).
Gluten-treatment in germ-free NOD/DQ8 mice induces a proinflammatory gliadin-specific immune response, but is not associated with changes in small-intestinal regulator T cells (Tregs). A and B: CD4+ T cells were isolated from mesenteric lymph nodes of control and gluten-treated clean specific pathogen free (SPF) and germ-free mice and stimulated with pepsin-trypsin (PT)-gliadin, PT-zein, or medium for 3 days. Production of IL-6 (A) and IFN-γ (B) was measured in cell culture supernatant. Red line represents the limit of detection. C and D: Quantification of CD25+Foxp3+ cells gated on CD3+CD4+ lymphocytes in the small intestine of naïve mice (C) and control and gluten-treated clean SPF and germ-free mice (D). Each dot represents an individual mouse. E: Representative flow cytometry plots of small-intestinal CD25+Foxp3+ cells gated on CD3+CD4+ cells in naive nonsensitized control, and gluten-treated clean SPF and germ-free mice are shown with the mean ± SEM indicated. F: Quantification of IL-10 in small-intestinal tissue samples of nonsensitized control and gluten-treated clean SPF and germ-free mice. Data are presented as mean ± SEM. n = 3 (A and B, per group); n = 5 to 6 (F, per group). ∗∗P < 0.01.