Abstract
Objective
Clinical genetic testing is commercially available for rs61764370, an inherited variant residing in a KRAS 3′ UTR microRNA binding site, based on suggested associations with increased ovarian and breast cancer risk as well as with survival time. However, prior studies, emphasizing particular subgroups, were relatively small. Therefore, we comprehensively evaluated ovarian and breast cancer risks as well as clinical outcome associated with rs61764370.
Methods
Centralized genotyping and analysis were performed for 140,012 women enrolled in the Ovarian Cancer Association Consortium (15,357 ovarian cancer patients; 30,816 controls), the Breast Cancer Association Consortium (33,530 breast cancer patients; 37,640 controls), and the Consortium of Modifiers of BRCA1 and BRCA2 (14,765 BRCA1 and 7904 BRCA2 mutation carriers).
Results
We found no association with risk of ovarian cancer (OR= 0.99, 95% CI 0.94–1.04,p = 0.74) or breast cancer (OR = 0.98, 95% CI 0.94–1.01, p = 0.19) and results were consistent among mutation carriers (BRCA1, ovarian cancer HR = 1.09, 95% CI 0.97–1.23, p = 0.14, breast cancer HR = 1.04, 95% CI 0.97–1.12, p = 0.27; BRCA2, ovarian cancer HR = 0.89, 95% CI 0.71–1.13, p = 0.34, breast cancer HR = 1.06, 95% CI 0.94–1.19, p = 0.35). Null results were also obtained for associations with overall survival following ovarian cancer (HR = 0.94, 95% CI 0.83–1.07, p = 0.38), breast cancer (HR = 0.96, 95% CI 0.87–1.06, p = 0.38), and all other previously-reported associations.
Conclusions
rs61764370 is not associated with risk of ovarian or breast cancer nor with clinical outcome for patients with these cancers. Therefore, genotyping this variant has no clinical utility related to the prediction or management of these cancers.
Keywords: KRAS variant, Breast cancer, Ovarian cancer, Genetic association, Clinical outcome
1. Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that negatively regulate gene expression by binding partially complementary sites in the 3′ untranslated regions (UTRs) of their target mRNAs. In this way, miRNAs control many cancer-related biological pathways involved in cell proliferation, differentiation, and apoptosis [1]. To date, several inherited variants in microRNAs or miRNA target sites have been reported to confer increased cancer risks [2]. One such variant is located in the 3′ UTR of the KRAS gene (rs61764370 T > G) for which the rarer G allele has been reported to confer an increased risk of ovarian, breast, and lung cancer [3–7] as well as endometriosis [8], although not consistently [9–11].
For ovarian cancer, the rs61764370 G allele was also reported to be associated with increased risk (320 cases, 328 controls). Further increased risks were observed among 23 BRCA1 mutation carriers and 31 women with familial ovarian cancer, but without BRCA1 or BRCA2 mutations [3]. In contrast, no association with ovarian cancer risk was seen in another, much larger study, based on 8669 cases, 10,012 controls, and 2682 BRCA1 mutation carriers [9]. One criticism on the latter study was that some of the genotype data were for rs17388148, an imputed proxy for rs61764370; even though rs17388148 is highly correlated with rs61764370 (r2 = 0.97) and was imputed with high accuracy (r2 = 0.977) [12,13]. The minor allele of rs61764370 was also associated with shorter survival time in a study of 279 ovarian cancer patients diagnosed after age 52 years with platinum-resistant disease (28 resistant, 263 not resistant) and with sub-optimal debulking surgery after neoadjuvant chemotherapy (7 sub-optimal, 109 optimal) [14]. However, another study observed no association between rs61764370 and ovarian cancer outcome (329 cases) [15].
For breast cancer, a borderline significant increased frequency of the rs61764370 G allele was observed in 268 BRCA1 mutation carriers with breast cancer, but not in 127 estrogen receptor (ER)-negative familial non-BRCA1/BRCA2 breast cancer patients [5]. However, in a subsequent study, the variant was reported to be associated with increased risk of ER/PR negative disease (80 cases, 470 controls), as well as with triple negative breast cancer diagnosed before age 52 (111 cases, 250 controls), regardless of BRCA1 mutation status [6]. The validity of these findings has been questioned given the very small sample sizes and the number of subgroups tested [16,17]. Another report found no association with sporadic or familial breast cancer risk (695 combined cases, 270 controls), but found that the variant was associated with ERBB2-positive and high grade disease, based on 153 cases who used post-menopausal hormone replacement therapy [18].
It has also been reported, based on 232 women with both primary ovarian and breast cancer, that the frequency of the G allele at rs61764370 was increased for those who were screened negative for BRCA1 and BRCA2 (92 cases), particularly among those enrolled within two years of their ovarian cancer diagnosis (to minimize survival bias, 30 cases), those diagnosed with post-menopausal ovarian cancer (63 cases), those with a family history of ovarian or breast cancer (24 cases), and those with a third primary cancer (16 cases) [4].
This notable lack of consistency in findings between studies might be expected when appropriate levels of statistical significance are not used to declare positive findings from multiple small subgroup comparisons or post-hoc hypotheses [19]. In this respect, the dangers of subgroup analyses in the context of clinical trials are well-recognized [20]. These are important caveats, particularly since a genetic test for rs61764370 is currently marketed in the US for risk prediction testing to women who are at increased risk for developing ovarian and/or breast cancer or women who have been diagnosed with either ovarian or breast cancer themselves [21]. In general, much larger studies, with sufficient power to detect positive findings at much more stringent levels of statistical significance ought to be required to establish the clinical validity of a genetic test. Therefore, we conducted centralized genotyping of rs61764370 and other variants in the genomic region around the KRAS gene in 140,012 women to examine associations with risk and clinical outcome of ovarian and breast cancer.
2. Methods
2.1. Study participants
The following three consortia contributed to these analyses: the Ovarian Cancer Association Consortium (OCAC:41 studies, Supplementary Table S1) [22], the Breast Cancer Association Consortium (BCAC: 37 studies, Supplementary Table S2) [23], and the Consortium of Modifiers of BRCA1 and BRCA2 (CIMBA: 55 studies, Supplementary Table S3) [24,25]. OCAC and BCAC consisted of case–control studies of unrelated women, and CIMBA consisted of studies of women with germline deleterious BRCA1 or BRCA2 mutations primarily identified through clinical genetics centers. For the purpose of the current analyses, only participants of European ancestry were included. Following genotyping, quality control exclusions (described below), and analysis-specific exclusions, data from the following women were available for analysis: 46,173 OCAC participants (15,357 patients with invasive epithelial ovarian cancer and 30,816 controls), 71,170 BCAC participants (33,530 patients with invasive breast cancer and 37,640 controls), and 22,669 CIMBA participants (for ovarian cancer analyses: 2332 affected and 12,433 unaffected BRCA1 carriers, 599 affected and 7305 unaffected BRCA2 carriers; for breast cancer analyses: 7543 affected and 7222 unaffected BRCA1 carriers, 4138 affected and 3766 unaffected BRCA2 carriers). For OCAC, overall and progression-free survival data were available for 3096 patients from 13 studies. Overall survival data were available for 28,471 patients from 26 BCAC studies and for 2623 mutation carriers with breast cancer from 11 CIMBA studies (excluding studies with less than ten deaths) as described previously [26,27]. Each study was approved by its relevant governing research ethics committee, and all study participants provided written informed consent.
2.2. Genotyping and imputation
Genotyping for rs61764370 was performed using the custom iCOGS Illumina Infinium iSelect BeadChip, as previously described [22–25]. In total, DNA from 185,443 women of varying ethnic background was genotyped (47,630 OCAC participants, 114,255 BCAC participants, 23,558 CIMBA participants), along with HapMap2 DNAs for European, African, and Asian populations. Genotype data were also available for three OCAC genome-wide association studies (UK GWAS, US GWAS, Mayo GWAS) that had been genotyped using either the Illumina Human610-Quad Beadchip (12,607 participants) [28] or the Illumina HumanOmni2.5–8 Beadchip (883 participants). Raw intensity data files underwent centralized genotype calling and quality control [22–25]. HapMap2 samples were used to identify women with predicted European intercontinental ancestry; among these women, a set of over 37,000 unlinked markers was used to perform principal component (PC) analysis [29]. The first five and seven European PCs were found to control adequately for residual population stratification in OCAC and BCAC data, respectively. Samples with low conversion rate, extreme heterozygosity, non-female sex, or one of a first-degree relative pair (the latter for OCAC and BCAC only) were excluded. Variants were excluded if they were monomorphic or had a call rate <95% (minor allele frequency (MAF) >0.05) or <99% (MAF <0.05), deviation from Hardy–Weinberg equilibrium (p< 10−7),or >2%duplicate discordance.
In addition to rs61764370, 54 variants within 100 kb on either side of KRAS on chromosome 12 (25,258,179 to 25,503,854 bp in GRCh37.p12) were genotyped. Moreover, to provide a common set of variants across the region for analysis in all the data sets, we also used imputation to infer genotypes for another 1056 variants and for variants that failed genotyping. We performed imputation separately for OCAC samples, BCAC samples, BRCA1 mutation carriers, BRCA2 mutation carriers, and for each of the OCAC GWAS. We imputed variants from the 1000 Genomes Project data using the v3 April 2012 release as the reference panel [30]. To improve computation efficiency we initially used a two-step procedure, which involved pre-phasing using the SHAPEIT software [31] in the first step and imputation of the phased data in the second. We used the IMPUTE version 2 software [32]for the imputation for all studies with the exception of the US GWAS for which we used the MACH algorithm implemented in the minimac software version 2012.8.15 and MACH version 1.0.18 [33]. We excluded variants from association analyses if their imputation accuracy was r2 < 0.30 or their MAF was <0.005, resulting in 974 variants genotyped and imputed for OCAC, 989 variants genotyped and imputed for BCAC, and 1001 variants genotyped and imputed for CIMBA, including rs61764370 (Supplementary Tables S5, S6, and S7).
2.3. Analysis
Genotypes were coded for genotype dosage as 0,1, or 2, based on the number of copies of the minor allele. For ovarian cancer case–control analysis (i.e., OCAC studies), logistic regression provided estimated risks of invasive epithelial ovarian cancer with odds ratios (ORs) and 95% confidence intervals (CIs) adjusting for study, age, and the five European PCs. Subgroup analyses were conducted by histology, family ovarian and breast cancer history, menopausal status, time between ovarian cancer diagnosis and recruitment, and history of multiple primary cancers. For breast cancer case–control analysis (i.e., BCAC studies), the association between genotype and invasive breast cancer risk was evaluated by logistic regression, adjusting for study, age, and the seven European PCs, providing ORs and 95% CIs. Additional subgroup analyses were based on receptor status, first-degree family ovarian and breast cancer history, BRCA1 and BRCA2 mutation status, enrollment within two years of diagnosis, menopausal status (i.e. last menstruation longer than twelve months ago), age at diagnosis less than 52 years, and history of hormone replacement therapy use (i.e. longer than twelve months use). Risk analysis for BRCA1 and BRCA2 mutation carriers (i.e. CIMBA studies) was done using a Cox proportional hazard model to estimate hazard ratios (HRs) per copy of the minor allele, with age as follow-up time and stratified by country of residence; US and Canadian strata were further subdivided by self-reported Ashkenazi Jewish ancestry [24,25]. A weighted cohort approach was applied to correct for potential testing bias due to overrepresentation of cases in the study population [34]. We used robust variance estimation to allow for the non-independence of carriers within the same family [35]. To assess associations with ovarian cancer risk, mutation carriers were followed from birth until ovarian cancer diagnosis (event), a risk-reducing salpingo-oophorectomy (RRSO) or the age at enrollment, whichever occurred first. We also performed analyses restricted to women diagnosed or censored within two years before their enrollment. To assess associations with breast cancer risk, mutation carriers were followed from birth until a breast cancer diagnosis (i.e. either ductal carcinoma in situ or invasive breast cancer), ovarian cancer diagnosis, a risk-reducing bilateral prophylactic mastectomy or the age at enrollment, whichever occurred first.
Survival analysis of OCAC patients used Cox proportional hazards models estimating HRs and 95% CIs considering overall survival as well as progression-free survival following ovarian cancer diagnosis. Overall survival was adjusted for age at diagnosis, the five European PCs, histology, grade, FIGO stage, and residual disease after debulking surgery, and stratified by study, left truncating at the date of study entry and right censoring at five years to minimize events due to other causes. Progression-free survival was analyzed as for overall survival, but without adjustment for age and right censoring, and was defined as the time between the date of histologic diagnosis and the first confirmed sign of disease recurrence or progression, based on GCIG (Gynecological Cancer InterGroup) criteria [36]. We also performed subgroup analysis of patients suboptimally debulked after cytoreductive surgery (residual disease >1 cm) and of post-menopausal patients (age at diagnosis >52 years). Survival analysis of BCAC patients used Cox proportional hazard models estimating HRs and 95% CIs considering overall and breast cancer-specific survival following breast cancer diagnosis. Models were adjusted for age at diagnosis, tumor size, nodal status, grade, adjuvant hormonal and/or chemotherapy, and stratified by study, left-truncating at the date of study entry and right censoring at ten years. In addition, we performed subgroup analysis on ER-positive and ER-negative patients. For CIMBA breast cancer patients associations between genotype and overall survival were evaluated using Cox proportional hazard models estimating HRs and 95% CIs. Models were adjusted for age at diagnosis, tumor size, nodal status, grade, adjuvant hormonal and/or chemotherapy, and preventive bilateral oophorectomy and stratified by study, left-truncating at the date of study entry and right censoring at twenty years. Analyses were performed using STATA version 12.0 (StataCorp, Texas, USA).
3. Results
The results of the overall analysis as well as the subgroup analyses investigating the association between the minor allele at rs61764370 and ovarian cancer risk, breast cancer risk, and ovarian and breast cancer risks in BRCA1 and BRCA2 mutation carriers are shown in Table 1. Associations with clinical outcomes in and ovarian and breast cancer patients including BRCA1 and BRCA2 mutation carriers are shown in Table 2 and Supplementary Table S4.
Table 1. Associations between KRAS rs61764370 and risk of ovarian and breast cancer.
For BRCA1 and BRCA2 mutation carrier analyses, cases are affected BRCA1/BRCA2 mutation carriers and controls are unaffected BRCA1/BRCA2 mutation carriers, and relative risks are estimated by hazard ratios; for other analyses, relative risks are estimated by odds ratios; ovarian cancer analyses used OCAC data adjusted for study, age, and the five European principal components; breast cancer analyses used BCAC data adjusted for study, age, and the seven European principal components; BRCA1 and BRCA2 mutation carrier analyses used CIMBA data with age as follow-up time and stratified for country; 95% CI, 95% confidence interval.
| Number |
Minor allele frequency |
Relative risk (95% CI) | ||||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | p-Value | ||
| Ovarian cancer | ||||||
| All invasive | 15,357 | 30,816 | 0.0914 | 0.0949 | 0.99 (0.94–1.04) | 0.74 |
| Histology | ||||||
| High-grade serous | 6938 | 30,816 | 0.0946 | 0.0949 | 1.04 (0.97–1.11) | 0.26 |
| Endometrioid | 2151 | 30,816 | 0.0834 | 0.0949 | 0.90 (0.80–1.00) | 0.06 |
| Clear cell | 1015 | 30,816 | 0.0994 | 0.0949 | 1.09 (0.94–1.27) | 0.27 |
| Mucinous | 1000 | 30,816 | 0.0902 | 0.0949 | 0.99 (0.85–1.16) | 0.91 |
| Low-grade serous | 485 | 30,816 | 0.0705 | 0.0949 | 0.76 (0.59–0.97) | 0.03 |
| First-degree family history | ||||||
| Ovarian cancer | 483 | 342 | 0.0803 | 0.0849 | 0.87 (0.60–1.27) | 0.47 |
| Breast or ovarian cancer | 477 | 18,442 | 0.0977 | 0.0915 | 1.09 (0.93–1.28) | 0.28 |
| BRCA1/2 mutation negative | 346 | 15,492 | 0.1050 | 0.0997 | 1.09 (0.85–1.41) | 0.49 |
| BRCA1 mutation carriers | 2332 | 12,433 | 0.0954 | 0.0922 | 1.09 (0.97–1.23) | 0.14 |
| BRCA2 mutation carriers | 599 | 7305 | 0.0952 | 0.0966 | 0.89 (0.71–1.13) | 0.34 |
| Enrolled within two years of diagnosis | ||||||
| All invasive | 10,121 | 30,815 | 0.0942 | 0.0949 | 0.99 (0.95–1.04) | 0.68 |
| BRCA1 mutation carriers | 1095 | 10,802 | 0.0950 | 0.0940 | 1.05 (0.90–1.23) | 0.52 |
| BRCA2 mutation carriers | 270 | 6509 | 0.0907 | 0.0979 | 0.85 (0.60–1.20) | 0.36 |
| Menopausal status | ||||||
| Pre- or peri-menopausal | 4264 | 8789 | 0.0915 | 0.0927 | 1.02 (0.92–1.13) | 0.68 |
| Post-menopausal | 11,058 | 15,903 | 0.0916 | 0.0951 | 0.99 (0.93–1.06) | 0.81 |
| Prior breast cancer | ||||||
| Enrolled within two years of diagnosis | 426 | 30,815 | 0.0943 | 0.0949 | 0.91 (0.71–1.17) | 0.46 |
| Post-menopausal ovarian cancer | 341 | 15,903 | 0.0810 | 0.0951 | 0.90 (0.68–1.21) | 0.49 |
| First degree breast or ovarian cancer family history | 202 | 30,815 | 0.0916 | 0.0949 | 0.99 (0.70–1.40) | 0.95 |
| Breast cancer | ||||||
| All invasive | 33,530 | 37,640 | 0.0904 | 0.0930 | 0.98 (0.94–1.01) | 0.19 |
| Receptor status | ||||||
| ER—/PR— | 4009 | 37,043 | 0.0940 | 0.0932 | 1.04 (0.96–1.13) | 0.36 |
| ER—/PR—/ERBB2— | 1673 | 28,480 | 0.0885 | 0.0947 | 0.97 (0.85–1.10) | 0.62 |
| First-degree family history | ||||||
| Breast cancer | 4357 | 1943 | 0.0942 | 0.0954 | 0.96 (0.84–1.10) | 0.59 |
| Ovarian or breast cancer | 4593 | 2265 | 0.0933 | 0.0949 | 0.96 (0.85–1.09) | 0.52 |
| Age diagnosis <52 | ||||||
| ER—/PR— | 1530 | 37,043 | 0.0980 | 0.0932 | 1.07 (0.95–1.22) | 0.28 |
| ER—/PR—/ERBB2— | 546 | 27,690 | 0.0908 | 0.0948 | 0.99 (0.81–1.20) | 0.90 |
| BRCA1/2 mutation negative | 1431 | 1097 | 0.0853 | 0.0925 | 0.91 (0.75–1.11) | 0.35 |
| BRCA1 mutation carriers | 7543 | 7222 | 0.0935 | 0.0919 | 1.04 (0.97–1.12) | 0.27 |
| BRCA2 mutation carriers | 4138 | 3766 | 0.1005 | 0.0921 | 1.06 (0.94–1.19) | 0.35 |
| Enrolled within two years of diagnosis | ||||||
| All invasive | 20,444 | 34,349 | 0.0924 | 0.0934 | 0.99 (0.95–1.04) | 0.73 |
| BRCA1 mutation carriers | 2595 | 5976 | 0.0896 | 0.0924 | 0.95 (0.85–1.05) | 0.30 |
| BRCA2 mutation carriers | 1359 | 3365 | 0.0960 | 0.0926 | 1.05 (0.90–1.23) | 0.52 |
| Menopausal status | ||||||
| Pre- or peri-menopausal | 7086 | 8642 | 0.0934 | 0.0933 | 0.98 (0.91–1.07) | 0.70 |
| Post-menopausal | 16,346 | 18,605 | 0.0904 | 0.0943 | 0.98 (0.93–1.03) | 0.36 |
Table 2. Associations between KRAS rs61764370 and outcome in ovarian and breast cancer.
Ovarian cancer analyses used OCAC data adjusted for age at diagnosis (overall survival only), the five European principal components, histology (serous, mucinous, endometrioid, clear cell, and other epithelial), grade (low versus high), FIGO stage (I–IV), residual disease after debulking surgery (nil versus any), and stratified by study; breast cancer analyses used BCAC data adjusted for age at diagnosis, tumor size, nodal status, grade, adjuvant hormonal and/or chemotherapy and was stratified by study; analyses for BRCA1 and BRCA2 mutation carriers used CIMBA data adjusted for age at diagnosis, tumor size, nodal status, grade, adjuvant hormonal and/or chemotherapy, and preventive bilateral oophorectomy and was stratified by study; 95% CI, 95% confidence interval.
| No. of patients |
No. of events |
Hazard ratio (95% CI) |
p-Value |
|
|---|---|---|---|---|
| Ovarian cancer | ||||
| Overall survival | ||||
| All patients | 3096 | 1421 | 0.94 (0.83–1.07) | 0.38 |
| Patients who were suboptimally debulked after cytoreductive surgery |
1114 | 784 | 0.94 (0.78–1.13) | 0.50 |
| Post-menopausal patients > 52 years |
2226 | 1276 | 0.97 (0.84–1.12) | 0.70 |
| Progression-free survival | ||||
| All patients | 3096 | 2144 | 1.01 (0.90–1.13) | 0.84 |
| Patients who were suboptimally debulked after cytoreductive surgery |
1114 | 961 | 1.03 (0.87–1.21) | 0.74 |
| Post-menopausal patients >52 years |
2226 | 1603 | 1.02 (0.90–1.16) | 0.76 |
| Breast cancer | ||||
| Overall survival | ||||
| All patients | 28,471 | 3013 | 0.96 (0.87–1.06) | 0.38 |
| ER-positive patients | 20,071 | 1754 | 0.96 (0.85–1.10) | 0.58 |
| ER-negative patients | 4778 | 771 | 0.97 (0.81–1.18) | 0.78 |
| Breast cancer-specific survival | ||||
| All patients | 28,471 | 1693 | 0.95 (0.83–1.08) | 0.40 |
| Overall survival | ||||
| BRCA1 mutation carriers | 1706 | 241 | 0.72 (0.48–1.08) | 0.11 |
| BRCA2 mutation carriers | 917 | 162 | 0.98 (0.65–1.46) | 0.90 |
We found no evidence for association between the rs61764370 Gallele and ovarian or breast cancer risk. The most statistically significant association was observed for risk of low-grade serous ovarian cancer (n = 485; OR 0.76, 95% CI 0.59–0.97, p = 0.031), but this finding was not significant after Bonferroni correction for multiple testing. We also evaluated the association for additional specific subgroups in which an association with rs61764370 had been reported previously [3–6]. Ovarian cancer subgroups considered BRCA1 mutation carriers as well as BRCA1 and BRCA2 screened-negative patients with first degree family histories of breast or ovarian cancer and patients who had been diagnosed with breast cancer before their ovarian cancer diagnoses. For breast cancer these included, among others, BRCA1 mutation carriers, patients diagnosed with ER- and PR-negative tumors, and patients diagnosed with triple negative tumors before age 52 years. Importantly, we observed no evidence for association of rs61764370 with any of these subgroups (detailed in Table 1), with all ORs close to unity and very narrow CIs including unity.
Similarly, case-only analyses did not reveal any associations between rs61764370 genotype and ovarian and breast cancer clinical features or outcome (Table 2 and Supplementary Table S4). For example, the previously reported association between rs61764370 and risk of ERBB2-positive and high grade breast cancer in hormone replacement therapy users [18] was not replicated (Supplementary Table S4), and in ovarian cancer analyses we found no evidence of reduced survival among patients diagnosed after age 52 years or patients with suboptimal debulking after cytoreductive surgery (Table 2) [14]. The G allele of rs61764370 was also not associated with survival of breast cancer patients (Table 2).
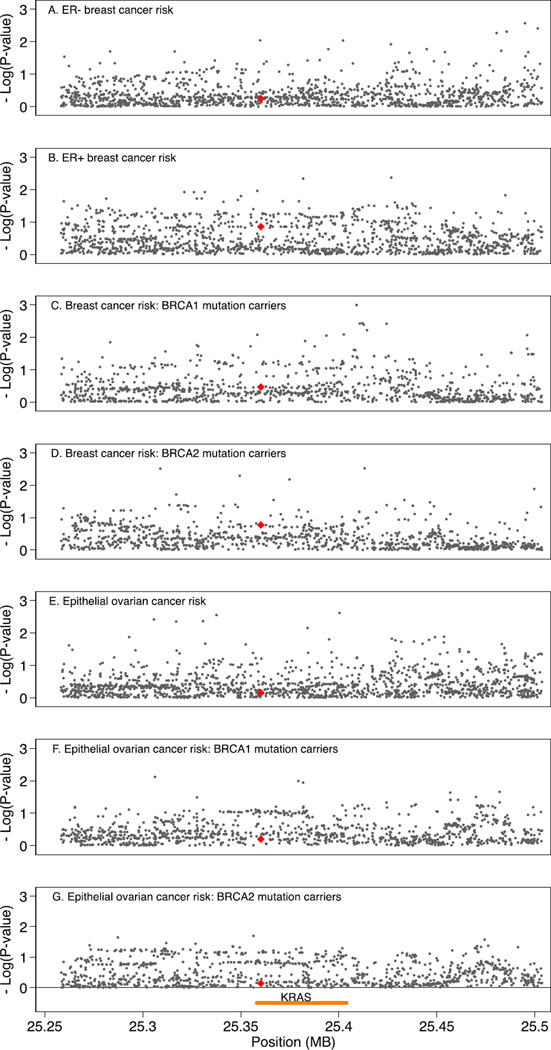
Finally, we evaluated the association between the primary phenotypes of interest and common genetic variation (MAF > 0.02) in the genomic region of KRAS (i.e., within 100 kb on either side of the gene), using imputed and genotyped data on 974 variants for OCAC, 989 variants for BCAC, and 1001 variants genotyped and imputed for CIMBA (Supplementary Tables S5, S6, and S7). We found no evidence of association for any of these variants, including rs61764370 and rs17388148, with these phenotypes that would withstand Bonferroni correction for multiple testing, as detailed in Supplementary Tables S5, S6, and S7 and shown in regional association plots (Fig. 1).
Fig. 1.
Regional association plots for variants within the genomic region 100 kb either side of KRAS and risk of ovarian and breast cancer. X-axis position is referent to position (bp) on chromosome 12, build GRCh37.p12; yellow line indicates position of KRAS; red triangle indicates rs61764370. Y-axis is — log10(p-values) from association tests for risk of A) ER-negative breast cancer, B) ER-positive breast cancer, C) breast cancer in BRCA1 mutation carriers, D) breast cancer in BRCA2 mutation carriers, E) epithelial ovarian cancer, F) epithelial ovarian cancer in BRCA1 mutation carriers, and G) epithelial ovarian cancer in BRCA2 mutation carriers. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Our analysis of 140,012 women genotyped for inherited variants in the KRAS region provides definitive clarification of the role of these variants in ovarian and breast cancer susceptibility and outcome. We have found no evidence to support an association between rs61764370 and ovarian or breast cancer risk, or clinical outcomes in patients with ovarian or breast cancer. In the absence of any association and with ORs close to unity we would not typically consider sub-group analyses, particularly sub-groups for which differential associations would not be expected to occur. However, given the previous positive associations reported for a myriad of different subgroups, we tested for association among each of these subgroups and found no evidence to support the previously reported associations.
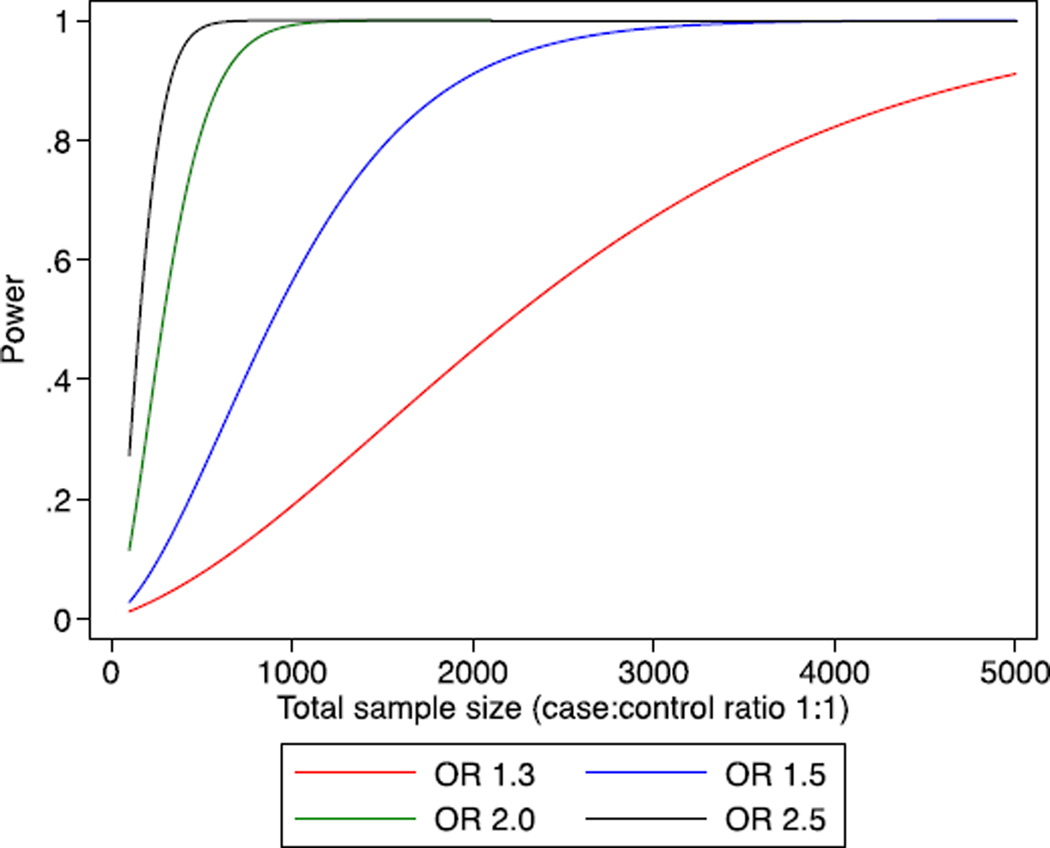
Our study has notable strengths. The vast majority (i.e. >95%) of the samples were genotyped using the same genotyping platform and employing a common approach to genotype calling and quality control; additional samples used denser arrays and nearly identical procedures. The very large sample sizes for all the major phenotypes of interest provide substantial statistical power to exclude any clinically relevant associated risks for the major phenotypes of interest (Fig. 2). The null results found here are thus not due to lack of statistical power, and this analysis also had greater than 80% power to detect association for most of the subgroups, although for some subgroups it was not possible to exclude modest risks. In contrast to the current findings, other genetic association analyses using the same genotyping platform and the same studies as included here have identified more than 90 common germline variants associated with ovarian or breast cancer risk at p < 5 × 10−8 [22,23,37]. While critiques on a previous null KRAS report have suggested that inclusion of male controls, use of “prevalent” cases, and reliance on a surrogate genetic variant may have led to falsely negative conclusions, these are not issues in the present data set. Rather, we demonstrate the importance of international collaboration to identify true associations as well as to refute false associations, an equally important objective.
Fig. 2.
Power curve for modest risk variants according to the total sample size. X-axis is total sample size for which case–control ratio is 1:1. Y-axis is the statistical power (range 0 · 5–1 · 0) for variants given a range of risks, assuming alpha = 0 · 01 and minor allele frequency 0 · 09.
The rise of individualized medicine including the use of panels of common variants to predict cancer risk more accurately than using family history alone holds great promise [38]. For example, the 31 prostate cancer susceptibility alleles confirmed as of 2011 (at p < 5 × 10−8) can be combined to identify men in the top one percent of the risk distribution having a 3.2-fold increased risk [39]. Prediction has since then improved with now over 70 prostate cancer susceptibility alleles [40] and the utility of these genetic tests is currently under clinical evaluation. A similar clinical examination in ovarian and breast cancer is not far behind, with now over 18 and 77 confirmed susceptibility alleles, respectively, for these cancers [22,23]. The genotype at rs61764370, however, does not predict ovarian or breast cancer risk, even among particular subgroups of women or for particular subtypes of disease, nor is it a marker of differential outcome following diagnosis with these cancers. Therefore, genetic test results for rs61764370 should not be used to counsel women about their ovarian or breast cancer risks or outcome. Our results highlight the dangers of developing clinical tests without appropriate data from carefully conducted, large-scale studies to establish clinical validity.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in this study and all the researchers, clinicians and administrative staff who have made possible the many studies contributing to this work.
The COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 -HEALTH-F2-2009-223175). The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/ RPCI.07). The scientific development and funding for this project were in part supported by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112). This study made use of data generated by the Wellcome Trust Case Control consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/. Funding for the project was provided by the Wellcome Trust under award 076113.
G.C.-T. and P.M.W. are supported by the National Health and Medical Research Council; P.A.F. is supported by the Deutsche Krebshilfe; B.K. holds an American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN); K.-A.P. is an Australian National Breast Cancer Foundation Fellow; and A.B. holds the Barbara Thomason Ovarian Cancer Research Professorship from the American Cancer Society (SIOP-06-090-06). R. Balleine was a Cancer Institute NSW Clinical Research Fellow.
OCAC, in particular, acknowledges D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); U. Eilber and T. Koehler (GER); L. Gacucova (HMO); P. Schürmann, F. Kramer, W. Zheng, T.-W. Park-Simon, K. Beer-Grondke and D. Schmidt (HJO); Sharon Windebank, Christopher Hilker and Jason Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY (NHS); L. Paddock, M. King, U. Chandran, A. Samoila, and Y. Bensman (NJO); L. Brinton, M. Sherman, A. Hutchinson, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini, P. Harrington, the SEARCH team and ECRIC (SEA); the Scottish Gynaecological Clinical Trails group and SCOTROC1 investigators (SRO); W-H. Chow and Y-T. Gao (SWH); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan and J. Ford (UKO); and Carole Pye (UKR). Funding of the constituent OCAC studies was provided by the American Cancer Society (CRTG-00-196-01-CCE); the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); the Canadian Institutes for Health Research; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/ A10124, C536/A13086, C536/A6689); the Celma Mastry Ovarian Cancer Foundation the Danish Cancer Society (94-222-52); ELAN Funds of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Imperial Experimental Cancer Research Centre (C1312/A15589); the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA15083, P50-CA105009, P50-CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063678, R01-CA063682, R01-CA064277, R01-CA067262, R01-CA071766, R01-CA076016, R01-CA080978, R01-CA087538, R01-CA092044, R01-095023, R01-CA106414, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA149429, R01CA83918, R03-CA113148, R03-CA115195, R37-CA070867, R37-CA70867, U01-CA069417, U01-CA071966 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17-98-1-8659, DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-07-0449); the National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01 GB 9401); the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Women's Health Theme” and the Royal Marsden Hospital; and WorkSafeBC.
CIMBA studies also acknowledge the following. BCFR: This work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names and commercial products, or organizations imply endorsement by the US Government or the BCFR. BCFR-AU: Maggie Angelakos, Judi Maskiell, Gillian Dite, Helen Tsimiklis. BCFR-NY: We wish to thank members and participants in the New York site of the Breast Cancer Family Registry for their contributions to the study. BCFR-ON: We wish to thank members and participants in the Ontario Familial Breast Cancer Registry for their contributions to the study. BFBOCC: BFBOCC is partly supported by: Lithuania (BFBOCC-LT): Research Council of Lithuania grant LIG-07/2012; Latvia (BFBOCC-LV) is partly supported by LSC grant 10.0010.08 and in part by a grant from the ESF Nr.2009/0220/1DP/1.1.1.2.0/09/APIA/VIAA/016 and Liepaja's municipal council. BFBOCC-LT: we acknowledge Vilius Rudaitis, Laimonas Griškevičius, Ramūnas Janavičius (if not in the authorship). BFBOCC-LV acknowledges Drs Janis Eglitis, Anna Krilova and Aivars Stengrevics. BIDMC: BIDMC is supported by the Breast Cancer Research Foundation. BMBSA: BRCA-gene mutations and breast cancer in South African women (BMBSA) was supported by grants from the Cancer Association of South Africa (CANSA) to Elizabeth J. van Rensburg. BMBSA: We wish to thank the families who contribute to the BMBSA study. BRICOH: SLN was partially supported by the Morris and Horowitz Familes Endowed Professorship. We wish to thank Yuan Chun Ding and Linda Steele for their work in participant enrollment and biospecimen and data management. CBCS: This work was supported by the NEYE Foundation. CNIO: This work was partially supported by Spanish Association against Cancer (AECC08), RTICC 06/0020/1060, FISPI08/1120, Mutua Madrileña Foundation (FMMA) and SAF2010-20493. We thank Alicia Barroso, Rosario Alonso and Guillermo Pita for their assistance. COH-CCGCRN: City of Hope Clinical Cancer Genetics Community Network and the Hereditary Cancer Research Registry, supported in part by Award Number RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. CONSIT TEAM: Italian Association for Cancer Research (AIRC) and funds from Italian citizens who allocated the 5 × 1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1000’). CORE: The CIMBA data management and data analysis were supported by Cancer Research — UK grants C12292/A11174 and C1287/A10118. SH is supported by an NHMRC Program Grant ot GCT. ACA is a Cancer Research — UK Senior Cancer Research Fellow. GCT is an NHMRC Senior Principal Research Fellow. DEMOKRITOS: This research has been co-financed by the European Union (European Social Fund — ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) — Research Funding Program of the General Secretariat for Research & Technology: ARISTEIA. Investing in knowledge society through the European Social Fund. DKFZ: The DKFZ study was supported by the DKFZ. EMBRACE: EMBRACE is supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles and Elizabeth Bancroft are supported by Cancer Research UK Grant C5047/A8385. Epidemiological study of BRCA1 & BRCA2 mutation carriers (EMBRACE): Douglas F. Easton is the PI of the study. EMBRACE Collaborating Centres are: Coordinating Centre, Cambridge: Debra Frost, Steve Ellis, Elena Fineberg, Radka Platte. North of Scotland Regional Genetics Service, Aberdeen: Zosia Miedzybrodzka, Helen Gregory. Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison, Lisa Jeffers. West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Kairen Ong, Jonathan Hoffman. South West Regional Genetics Service, Bristol: Alan Donaldson, Margaret James. East Anglian Regional Genetics Service, Cambridge: Marc Tischkowitz, Joan Paterson, Amy Taylor. Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark T. Rogers, Emma McCann. St James's Hospital, Dublin & National Centre for Medical Genetics, Dublin: M.John Kennedy, David Barton. South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous, Sarah Drummond. Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman, Kathryn Hill. West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson, Eshika Haque, Ed Tobias, Alexis Duncan. South East Thames Regional Genetics Service, Guy's Hospital London: Louise Izatt, Chris Jacobs, Caroline Langman. North West Thames Regional Genetics Service, Harrow: Huw Dorkins. Leicestershire Clinical Genetics Service, Leicester: Julian Barwell. Yorkshire Regional Genetics Service, Leeds: Julian Adlard, Gemma Serra-Feliu. Cheshire & Merseyside Clinical Genetics Service, Liverpool: Ian Ellis, Catherine Houghton. Manchester Regional Genetics Service, Manchester: D Gareth Evans, Fiona Lalloo, Jane Taylor. North East Thames Regional Genetics Service, NE Thames, London: Lucy Side, Alison Male, Cheryl Berlin. Nottingham Centre for Medical Genetics, Nottingham: Jacqueline Eason, Rebecca Collier. Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber, Irene Jobson. Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durell, Barbara Stayner. The Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins, Elena Castro, Emma Killick, Sue Martin, Gillian Rea, Anjana Kulkarni. North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley. South West Thames Regional Genetics Service, London: Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester, Charlotte Eddy, Vishakha Tripathi, Virginia Attard, Anna Lehmann. Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Donna McBride, Sarah Smalley. FCCC: The authors acknowledge support from The University of Kansas Cancer Center (P30 CA168524) and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. was funded by 5U01CA113916, R01CA140323, and by the Chancellors Distinguished Chair in Biomedical Sciences Professorship. We thank Ms. JoEllen Weaver and Dr. Betsy Bove for their technical support. GC-HBOC: The German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC) is supported by the German Cancer Aid (grant no 109076, Rita K. Schmutzler) and by the Center for Molecular Medicine Cologne (CMMC). GEMO: The study was supported by the Ligue National Contre le Cancer; the Association “Le cancer du sein, parlons-en!” Award; and the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program. Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) study: National Cancer Genetics Network “UNICANCER Genetic Group”, France. We wish to thank all the GEMO collaborating groups for their contribution to this study. GEMO Collaborating Centers are: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Hospices Civils de Lyon — Centre Léon Bérard, & Equipe “Génétique du cancer du sein”, Centre de Recherche en Cancérologie de Lyon: Olga Sinilnikova, Sylvie Mazoyer, Francesca Damiola, Laure Barjhoux, Carole Verny-Pierre, Alain Calender, Sophie Giraud, Mélanie Léone; and Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Carole Tirapo, Antoine de Pauw. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Olivier Caron. Centre Jean Perrin, Clermont–Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona, Sandrine Handallou. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, Audrey Remenieras, François Eisinger. CHU Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis. Hôpital René Huguenin/Institut Curie, St Cloud: Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Nicolas Sevenet, Michel Longy. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung, Magalie Peysselon. CHU Dijon: Fanny Coron, Laurence Faivre. CHU St-Etienne: Fabienne Prieur, Marine Lebrun, Caroline Kientz. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU Limoges: Laurence Vénat-Bouvet. CHU Nantes: Capucine Delnatte. CHU Bretonneau, Tours: Isabelle Mortemousque. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier. CHU Vandoeuvre-les-Nancy: Johanna Sokolowska, Myriam Bronner. CHU Besançon: Marie-Agnès Collonge-Rame, Alexandre Damette. Creighton University, Omaha, USA: Henry T. Lynch, Carrie L. Snyder. G-FAST: Bruce Poppe is a senior clinical investigator for the Fund for Scientific Research Flanders (FWO). We wish to thank the technical support of Ilse Coene en Brecht Crombez. GOG: This study was supported by National Cancer Institute grants to the Gynecologic Oncology Group (GOG) Administrative Office and Tissue Bank (CA 27469), the GOG Statistical and Data Center (CA 37517), and GOG's Cancer Prevention and Control Committee (CA 101165). Drs. Greene, Mai and Savage were supported by funding from the Intramural Research Program, NCI. HCSC: Was supported by a grant RD12/00369/0006 and 12/00539 from ISCIII (Spain), partially supported by European Regional Development FEDER funds. We acknowledge Alicia Tosar for her technical assistance. HEBCS: The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society and the Sigrid Juselius Foundation. HEBCS would like to thank Karl von Smitten, Tuomas Heikkinen, Dario Greco, and Irja Erkkilä. HEBON: The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the NWO grant 91109024, the Pink Ribbon grant 110005 and the BBMRI grant CP46/NWO. HEBON stands for The Hereditary Breast and Ovarian Cancer Research Group Netherlands and consists of the following Collaborating Centers: Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: M.A. Rookus, F.B.L. Hogervorst, F.E. van Leeuwen, S. Verhoef, M.K. Schmidt, J.L. de Lange; Erasmus Medical Center, Rotterdam, NL: J.M. Collée, A.M.W. van den Ouweland, M.J. Hooning, C. Seynaeve, C.H.M. van Deurzen; Leiden University Medical Center, NL: C.J. van Asperen, J.T. Wijnen, R.A. Tollenaar, P. Devilee, T.C.T.E.F. van Cronenburg; Radboud University Nijmegen Medical Center, NL: C.M. Kets, A.R. Mensenkamp; University Medical Center Utrecht, NL: M.G.E.M. Ausems, R.B. van der Luijt; Amsterdam Medical Center, NL: C.M. Aalfs, T.A.M. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, NL: E.B. Gómez-Garcia, M.J. Blok; University Medical Center Groningen, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits, G.H. de Bock. The Netherlands Foundation for the detection of hereditary tumors, Leiden, NL: H.F. Vasen. HUNBOCS: Hungarian Breast and Ovarian Cancer Study was supported by Hungarian Research Grant KTIA-OTKA CK-80745. We wish to thank to Hungarian Breast and Ovarian Cancer Study Group members (Janos Papp, Tibor Vaszko, Aniko Bozsik, Judit Franko, Maria Balogh, Gabriella Domokos, Judit Ferenczi, Department of Molecular Genetics, National Institute of Oncology, Budapest, Hungary) and the clinicians and patients for their contributions to this study. ICO: Contract grant sponsor: Asociación Española Contra el Cáncer, Spanish Health Research Fund; Carlos III Health Institute; Catalan Health Institute and Autonomous Government of Catalonia. Contract grant numbers: ISCIIIRETIC RD06/0020/1051, RD12/0036/008, PI10/ 01422, PI10/00748, PI13/00285 and 2009SGR290. We wish to thank the ICO Hereditary Cancer Program team led by Dr. Gabriel Capella and all ICO study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. IHCC: Katarzyna Jaworska is a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University, supported by the Polish Foundation of Science. ILUH: The ILUH group was supported by the Icelandic Association “Walking for Breast Cancer Research” and by the Landspitali University Hospital Research Fund. INHERIT: This work was supported by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program, the Canadian Breast Cancer Research Alliance-grant #019511 and the Ministry of Economic Development, Innovation and Export Trade — grant # PSR-SIIRI-701. We would like to thank Dr Martine Dumont, Martine Tranchant for sample management and skillful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. IOVHBOCS: The study was supported by Ministero della Salute and “5 × 1000” Istituto Oncologico Veneto grant. KCONFAB: kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. GCT and ABS is an NHMRC Senior Research Fellow. We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (funded 2001–2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia #628333) for their contributions to this resource, and the many families who contribute to kConFab. MAYO: MAYO is supported by NIH grant CA128978, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a U.S. Department of Defence Ovarian Cancer Idea award (W81XWH-10-1-0341) and a grant from the Breast Cancer Research Foundation. MCGILL: Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade. MODSQUAD: The work was supported by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/ 2.1.00/03.0101) and MH CZ — DRO (MMCI, 00209805). MSKCC: MSKCC is supported by Breast Cancer Research Foundation, the Niehaus Family Genetics Research Fund and the STARR Cancer Consortium Grants. NAROD: 1R01 CA149429-01. NCI: The research of Drs. MH Greene, PL Mai and SA Savage was supported by the Intramural Research Program of the US National Cancer Institute, NIH, and by support services contracts NO2-CP-11019-50 and N02-CP-65504 with Westat, Inc., Rockville, MD. NICCC: NICCC is supported by Clalit Health Services in Israel. Some of its activities are supported by the Israel Cancer Association and the Breast Cancer Research Foundation (BCRF), NY. We wish to thank the NICCC National Familial Cancer Consultation Service team led by Sara Dishon, the lab team led by Dr. Flavio Lejbkowicz, and the research field operations team led by Dr. Mila Pinchev. NNPIO: This work has been supported by the Russian Federation for Basic Research (grants 11-04-00227,12-04-00928 and 12-04-01490) and the Federal Agency for Science and Innovations, Russia (contract 02.740.11.0780). OSU CCG: OSUCCG is supported by the Ohio State University Comprehensive Cancer Center. Leigha Senter, Kevin Sweet, Caroline Craven and Michelle O'Connor were instrumental in accrual of study participants, ascertainment of medical records and database management. Samples were processed by the OSU Human Genetics Sample Bank. PBCS: This work was supported by the ITT (Istituto Toscano Tumori) grants 2011–2013. SMC: This project was partially funded through a grant by the Israel cancer association and the funding for the Israeli Inherited breast cancer consortium. SMC team wishes to acknowledge the assistance of the Meirav Comprehensive breast cancer center team at the Sheba Medical Center for assistance in this study. SWE-BRCA: SWE-BRCA collaborators are supported by the Swedish Cancer Society. Swedish scientists participating as SWE-BRCA collaborators are: from Lund University and University Hospital: Åke Borg, Håkan Olsson, Helena Jernström, Karin Henriksson, Katja Harbst, Maria Soller, Niklas Loman, Ulf Kristoffersson; from Gothenburg Sahlgrenska University Hospital: Anna Öfverholm, Margareta Nordling, Per Karlsson, Zakaria Einbeigi; from Stockholm and Karolinska University Hospital: Anna von Wachenfeldt, Annelie Liljegren, Annika Lindblom, Brita Arver, Gisela Barbany Bustinza, Johanna Rantala; from Umeå University Hospital: Beatrice Melin, Christina Edwinsdotter Ardnor, Monica Emanuelsson; from Uppsala University: Hans Ehrencrona, Maritta Hellström Pigg, Richard Rosenquist; and from Linköping University Hospital: Marie Stenmark-Askmalm, Sigrun Liedgren. UCHICAGO: UCHICAGO is supported by NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, U01 CA161032 and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women's Cancer Research Alliance and the Breast Cancer research Foundation. OIO is an ACS Clinical Research Professor. We wish to thank Cecilia Zvocec, Qun Niu, physicians, genetic counselors, research nurses and staff of the Cancer Risk Clinic for their contributions to this resource, and the many families who contribute to our program. UCLA: Patricia Ganz and the Jonsson Comprehensive Cancer Center Foundation; Breast Cancer Research Foundation. We thank Joyce Seldon MSGC and Lorna Kwan, MPH for assembling the data for this study. UCSF: UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. We would like to thank Dr. Robert Nussbaum and the following genetic counselors for participant recruitment: Beth Crawford, Kate Loranger, Julie Mak, Nicola Stewart, Robin Lee, Amie Blanco and Peggy Conrad. And thanks to Ms. Salina Chan for her data management. UKFOCR: UKFOCR was supported by a project grant from CRUK to Paul Pharoah. We thank Carole Pye, Patricia Harrington and Eva Wozniak for their contributions towards the UKFOCR. UPENN: National Institutes of Health (NIH) (R01-CA102776 and R01-CA083855); Breast Cancer Research Foundation; Rooney Family Foundation; Susan G. Komen Foundation for the cure, Basser Research Center for BRCA. VFCTG: Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. Geoffrey Lindeman, Marion Harris, and Martin Delatycki of the Victorian Familial Cancer Trials Group. We thank Sarah Sawyer and Rebecca Driessen for assembling this data and Ella Thompson for performing all DNA amplification. WCP: The Women's Cancer Program (WCP) at the Samuel Oschin Comprehensive Cancer Institute is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN).
BCAC studies also acknowledge the following. We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. Part of this work was supported by the European Community's Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). This work was partly supported by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program (J.S. & D.E.), and the Ministry of Economic Development, Innovation and Export Trade of Quebec — grant # PSR-SIIRI-701 (J.S. & D.E., P. Hall). The BCAC is funded by CR-UK (C1287/A10118 and C1287/ A12014). Meetings of the BCAC have been funded by the European Union COST program (BM0606). D.F.E. is a Principal Research Fellow of CR-UK. J.S. is chair holder of the Canada Research Chair in Oncogenetics. ABCFS: Maggie Angelakos, Judi Maskiell, and Gillian Dite. The ABCFS, NC-BCFR and OFBCR work was supported by the United States National Cancer Institute, National Institutes of Health (NIH) under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Northern California Cancer Center (U01 CA69417), and University of Melbourne (U01 CA69638). Samples from the NC-BCFR were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names and commercial products, or organizations imply endorsement by the US Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia Fellow and a Victorian Breast Cancer Research Consortium Group Leader. M.C.S. is a NHMRC Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. The ABCS study was supported by the Dutch Cancer Society [grants NKI 2007-3839; 2009 4363]; BBMRI-NL, which is a Research Infrastructure financed by the Dutch government (NWO 184.021.007); and the Dutch National Genomics Initiative. BBCC: The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. BBCS: Eileen Williams, Elaine Ryder-Mills, Kara Sargus. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). BIGGS: ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London, United Kingdom. IT is supported by the Oxford Biomedical Research Centre. Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones. BSUCH: The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). Peter Bugert, Medical Faculty Mannheim. CECILE: The CECILE study was funded by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Ligue contre le Cancer Grand Ouest, Agence Nationale de Sécurité Sanitaire (ANSES), and Agence Nationale de la Recherche (ANR). CNIO-BCS: The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI081120). The Human Genotyping-CEGEN Unit (CNIO) is supported by the Instituto de Salud Carlos III. Guillermo Pita, Charo Alonso, Daniel Herrero, Nuria Álvarez, Pilar Zamora, Primitiva Menendez, the Human Genotyping-CEGEN Unit (CNIO). CTS: The CTS was supported by the California Breast Cancer Act of 1993; National Institutes of Health (grants R01 CA77398 and the Lon V Smith Foundation [LVS39420]); and the California Breast Cancer Research Fund (contract 97-10500). Collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. ESTHER: The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). Hartwig Ziegler, Sonja Wolf, Volker Hermann. GENICA: The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The GENICA Network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany; [H.B., Wing-Yee Lo, Christina Justenhoven], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch], Institute of Pathology, University of Bonn, Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [U.H.], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Germany [T.B., Beate Pesch, Sylvia Rabstein, Anne Spickenheuer], Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth]. HEBCS: The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation. Karl von Smitten, Tuomas Heikkinen, Dario Greco, Irja Erkkilä. HMBCS: The HMBCS was supported by a grant from the Friends of Hannover Medical School and by the Rudolf Bartling Foundation. Peter Hillemanns, Hans Christiansen and Johann H. Karstens. HUBCS: The HUBCS was supported by a grant from the German Federal Ministry of Research and Education (RUS08/ 017). KARBAC: The KARBAC study was supported by the Swedish Cancer Society, the Gustav V Jubilee Foundation and the Bert von Kantzow foundation. KBCP: The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland and by the strategic funding of the University of Eastern Finland. Eija Myöhänen, Helena Kemiläinen. kConFab/AOCS: kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684, 288704, 454508]. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], Cancer Council Victoria, Queensland Cancer Fund, Cancer Council New South Wales, Cancer Council South Australia, The Cancer Foundation of Western Australia, Cancer Council Tasmania and the National Health and Medical Research Council of Australia [NHMRC; 400413, 400281,199600]. G.C.T. and P.W. are supported by the NHMRC. Heather Thorne, Eveline Niedermayr, the AOCS Management Group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green, P Webb), the ACS Management Group (A Green, P Parsons, N Hayward, P Webb, D Whiteman). LMBC: LMBC is supported by the ‘Stichting tegen Kanker’ (232-2008 and 196-2010). Diether Lambrechts is supported by the FWO and the KULPFV/10/016-SymBioSysII. Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts. MARIE: The MARIE study was supported by the Deutsche Krebshilfe e.V [70-2892-BR I], the Hamburg Cancer Society, the German Cancer Research Center and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. Tracy Slanger, Elke Mutschelknauss, Ramona Salazar, S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese, K. Smit. MBCSG: MBCSG was funded by grants from the Italian Association for Cancer Research (AIRC) and thanks Siranoush Manoukian of the Istituto Nazionale dei Tumori, Milano, Italy; Monica Barile and Irene Feroce of the Istituto Europeo di Oncologia, Milan, Italy; Giuseppe Giannini of the Sapienza University, Rome, Italy; Loris Bernard end per personnel of the Cogentech Cancer Genetic Test Laboratory, Milan, Italy. MCBCS: The MCBCS was supported by the NIH grants [CA122340, CA128978], an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201], the Breast Cancer Research Foundation, and the Komen Race for the Cure. MCCS: MCCS cohort recruitment in the study was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. MEC: The MEC was support by NIH grants CA63464, CA54281, CA098758 and CA132839. MTLGEBCS: The authors gratefully acknowledge Martine Tranchant for DNA extraction, sample management and skillful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. The work of MTLGEBCS was supported by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program — grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade — grant # PSR-SIIRI-701. NBCS: The NBCS was supported by grants from the Norwegian Research council, 155218/V40, 175240/S10 to ALBD, FUGE-NFR 181600/V11 to VNK and a Swizz Bridge Award to ALBD. NBHS: The NBHS was supported by NIH grant R01CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. We thank study participants and research staff for their contributions and commitment to this study. NHS: The NHS was funded by NIH grant CA87969. OBCS: The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland, the University of Oulu, and the Oulu University Hospital. Meeri Otsukka, Kari Mononen. OFBCR: Teresa Selander, Nayana Weerasooriya. ORIGO: The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). We thank E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. The LUMC survival data were retrieved from the Leiden hospital-based cancer registry system (ONCDOC) with the help of Dr. J. Molenaar. PBCS: The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. Louise Brinton, Mark Sherman, Stephen Chanock, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner. pKARMA: The pKARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The Swedish Medical Research Counsel. RBCS: The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk, the Erasmus MC Family Cancer Clinic. SASBAC: The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The Swedish Medical Research Counsel. SBCS: The SBCS was supported by Yorkshire Cancer Research S295, S299, S305PA Sue Higham, Helen Cramp, and Dan Connley. SEARCH: SEARCH is funded by program grants from Cancer Research UK [C490/A11021 and C490/ A10124]. The SEARCH and EPIC teams. SKKDKFZS: SKKDKFZS is supported by the DKFZ. We thank all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. SZBCS: The SZBCS was supported by Grant PBZ_KBN_122/P05/2004; Katarzyna Jaworska is a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University, supported by the Polish Foundation of Science. UKBGS: The UKBGS is funded by Breakthrough Breast Cancer and the Institute of Cancer Research (ICR). ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. We thank Breakthrough Breast Cancer and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. Genome Quebec: The authors would like to acknowledge the contribution of the staff of the genotyping unit under the supervision of Dr. Sylvie LaBoissière as well as Frédérick Robidoux from the McGill University and Génome Québec Innovation Centre.
Footnotes
Conflict of interest statement
There are no conflicts of interest to disclose.
Antoinette Hollestelle and Ellen L Goode had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygyno.2015.04.034.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006 Apr;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Salzman DW, Weidhaas JB. SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther. 2013 Jan;137(1):55–63. doi: 10.1016/j.pharmthera.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70(16):6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilarski R, Patel DA, Weitzel J, McVeigh T, Dorairaj JJ, Heneghan HM, et al. The KRAS variant is associated with risk of developing double primary breast and ovarian cancer. PLoS ONE. 2012;7(5):e37891. doi: 10.1371/journal.pone.0037891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollestelle A, Pelletier C, Hooning M, Crepin E, Schutte M, Look M, et al. Prevalence of the variant allele rs61764370 T > G in the 3′UTR of KRAS among Dutch BRCA1, BRCA2 and non-BRCA1/BRCA2 breast cancer families. Breast Cancer Res Treat. 2011 Jul;128(1):79–84. doi: 10.1007/s10549-010-1080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case–control and genetic analysis. Lancet Oncol. 2011 Apr;12(4):377–386. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68(20):8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E, et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med. 2012 Mar;4(3):206–217. doi: 10.1002/emmm.201100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pharoah PD, Palmieri RT, Ramus SJ, Gayther SA, Andrulis IL, Anton-Culver H, et al. The role of KRAS rs61764370 in invasive epithelial ovarian cancer: implications for clinical testing. Clin Cancer Res. 2011;17(11):3742–3750. doi: 10.1158/1078-0432.CCR-10-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson HH, Christensen BC, Plaza SL, Wiencke JK, Marsit CJ, Kelsey KT. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010 Jul;69(1):51–53. doi: 10.1016/j.lungcan.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luong HT, Nyholt DR, Painter JN, Chapman B, Kennedy S, Treloar SA, et al. No evidence for genetic association with the let-7 microRNA-binding site or other common KRAS variants in risk of endometriosis. Hum Reprod. 2012 Dec;27(12):3616–3621. doi: 10.1093/humrep/des329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidhaas JB, Slack FJ. KRAS rs61764370 in epithelial ovarian cancer-Letter. Clin Cancer Res. 2011 Oct 15;17(20):6600. doi: 10.1158/1078-0432.CCR-11-1195. [DOI] [PubMed] [Google Scholar]
- 13.Risch HA, Berchuck A, Paul DP. KRAS rs61764370 in epithelial ovarian cancer-Response. Clin Cancer Res. 2011 Oct 15;17(20):6601. doi: 10.1158/1078-0432.CCR-11-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratner ES, Keane FK, Lindner R, Tassi RA, Paranjape T, Glasgow M, et al. A KRAS variant is a biomarker of parker of poor outcome, platinum chemotherapy resistance and a potential target for therapy in ovarian cancer. Oncogene. 2011;31(42):4559–4566. doi: 10.1038/onc.2011.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caiola E, Rulli E, Fruscio R, Buda A, Broggini M, Marabese M. KRas-LCS6 polymorphism does not impact on outcomes in ovarian cancer. Am J Cancer Res. 2012;2(3):298–308. [PMC free article] [PubMed] [Google Scholar]
- 16.Pharoah P, Antoniou A, Berchuck A, Chenevix-Trench G, Gayther S, Goode E, et al. Association between KRAS rs61764370 and triple-negative breast cancer - a false positive? Lancet Oncol. 2011;12(8):723–724. doi: 10.1016/S1470-2045(11)70156-5. [DOI] [PubMed] [Google Scholar]
- 17.Weidhaas J, Slack F, Miller N, Harris L, Tuck D, Zhu Y, et al. Association between KRAS rs61764370 and triple-negative breast cancer — a false positive? Authors' reply. Lancet Oncol. 2011;12(8):724. doi: 10.1016/S1470-2045(11)70156-5. [DOI] [PubMed] [Google Scholar]
- 18.Cerne JZ, Stegel V, Gersak K, Novakovic S. KRAS rs61764370 is associated with HER2-overexpressed and poorly-differentiated breast cancer in hormone replacement therapy users: a case control study. BMC Cancer. 2012;12(105) doi: 10.1186/1471-2407-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kivimaki M, Batty GD, Kawachi I, Virtanen M, Singh-Manoux A, Brunner EJ. Don'T let the truth get in the way of a good story: an illustration of citation bias in epidemiologic research. Am J Epidemiol. 2014;180(4):446–448. doi: 10.1093/aje/kwu164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peto R. Current misconception 3: that subgroup-specific trial mortality results often provide a good basis for individualising patient care. Br J Cancer. 2011;104(7):1057–1058. doi: 10.1038/bjc.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. http://www.miradx.com.
- 22.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013 Apr;45(4):362–370. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9(3):e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudet MM, Kuchenbaecker KB, Vijai J, Klein RJ, Kirchhoff T, McGuffog L, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9(3):e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White KL, Vierkant RA, Fogarty ZC, Charbonneau B, Block MS, Pharoah PD, et al. Analysis of over 10,000 cases finds no association between previously reported candidate polymorphisms and ovarian cancer outcome. Cancer Epidemiol Biomark Prev. 2013 May;22(5):987–992. doi: 10.1158/1055-9965.EPI-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weischer M, Nordestgaard BG, Pharoah P, Bolla MK, Nevanlinna H, Van't Veer LJ, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30(35):4308–4316. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010 Oct;42(10):874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 30.Project G. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012 Feb;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 32.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009 Jun;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012 Aug;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antoniou AC, Goldgar DE, Andrieu N, Chang-Claude J, Brohet R, Rookus MA, et al. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol. 2005 Jul;29(1):1–11. doi: 10.1002/gepi.20074. [DOI] [PubMed] [Google Scholar]
- 35.Boos DD. On generalised score tests. Am Stat. 1992;46:327–333. [Google Scholar]
- 36.Rustin GJ, Vergote I, Eisenhauer E, Pujade-Lauraine E, Quinn M, Thigpen T, et al. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG) Int J Gynecol Cancer. 2011 Feb;21(2):419–423. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 37.Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015 Feb;47(2):164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowdhury S, Dent T, Pashayan N, Hall A, Lyratzopoulos G, Hallowell N, et al. Incorporating genomics into breast and prostate cancer screening: assessing the implications. Genet Med. 2013 Jun;15(6):423–432. doi: 10.1038/gim.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pashayan N, Duffy SW, Chowdhury S, Dent T, Burton H, Neal DE, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104(10):1656–1663. doi: 10.1038/bjc.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013 Apr;45(4):385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.