Abstract
A seed-borne fungus, Curvularia sp. EML-KWD01, was isolated from an indigenous wheat seed by standard blotter method. This fungus was characterized based on the morphological characteristics and molecular phylogenetic analysis. Phylogenetic status of the fungus was determined using sequences of three loci: rDNA internal transcribed spacer, large ribosomal subunit, and glyceraldehyde 3-phosphate dehydrogenase gene. Multi loci sequencing analysis revealed that this fungus was Curvularia spicifera within Curvularia group 2 of family Pleosporaceae.
Keywords: Curvularia group, Curvularia spicifera, Pleosporaceae, Seed-borne fungi, Wheat
The genus Curvularia (family: Pleosporaceae, order: Pleosporales) known as hyphomycete fungus was established by Boedijn [1] and typified by C. lunata (Wakker) Boedijn. Currently, 30 Curvularia species have been reported [2]. Curvularia and Bipolaris are closely related to each other with same teleomorph (Cochliobolus) [3,4]. Main differences among the genera of Bipolaris, Curvularia, Drechslera, and Exserohilum are their conidial shape, the presence and the shape of protruding hilum, and the conidial outline of the basal portion [5]. Curvularia species have dark mycelia and geniculate conidiophores with sympodial and distoseptate conidia [3,6]. Bipolaris contains about 45 species. They are mostly subtropical and tropical plant parasites as well as human pathogens. Bipolaris is characterized by fusiform to ellipsoidal shaped conidia, central cells not much darker but broader than the distal ones, hilum not protuberant, and bipolar germination. However, some Bipolaris species have short and straight conidia with intermediate conidial characteristics. These Bipolaris species may look different from the generic type B. maydis that has large and gently curving conidia.
In recent years, molecular biology has provided fundamental tools to study genera with complex taxonomy. rDNA sequencing has been used as a main tool to identify fungi and discriminate related species [7,8]. This has been especially useful for some Curvularia species. To assess the evolutionary relationships of Cochliobolus, PseudoCochliobolus, Curvularia, and Bipolaris, Berbee et al. [9] have conducted phylogenetic analyses of rDNA internal transcribed spacer (ITS) regions and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene sequences. Furthermore, molecular phylogenetic analysis based on multi loci has been increasingly used as a tool to determine new species [10,11]. Recently, Manamgoda et al. [12] have re-evaluated the taxonomy of the genera Bipolaris, Cochliobolus, and Curvularia based on combined genetic analysis of rDNA ITS, 28S, GAPDH, and translation elongation factor 1-α genes and confirmed that these genera were divided into two monophyletic groups: Bipolaris and Cochliobolus species clustered in group 1 together with their respective type species B. maydis (Y. Nisik. & C. Miyake) Shoemaker. In contrast, Curvularia including species that were first named as Bipolaris, Cochliobolus, PseudoCochliobolus but later re-classified as Curvularia were clustered in group 2 with its generic type C. lunata. Some Bipolaris species were re-classified into genus Curvularia, including C. australiensis, C. coisis, C. ellisii, C. graminicola, C. hawaiiensis, C. ovariicola, C. spicifera, C. ravenelli, and C. tripogonis. The objectives of this study were to describe an unrecorded species of C. spicifera EML-KWD01 based on morphological characteristics, phylogenetic analysis, and taxonomic re-evaluation of relationships within Curvularia and Bipolaris groups using multi loci sequences.
MATERIALS AND METHODS
Fungal isolates
Seeds harvested from an indigenous wheat in 2012 were sampled from farms and markets. Seeds were surface-sterilized with 2% sodium hypochlorite (NaOCl) for 30 sec, washed with sterilized distilled water, plated directly on a moist blotter, and incubated at 27℃ for 2~4 days to allow germination. After germination, seeds were stored in a freezer (-80℃) for 1 hr and then replaced in the same incubator. Recovered fungi were investigated with a stereo microscope, and fungal spores on the seed were inoculated to potato dextrose agar (PDA; Difco, Montreal, Canada) using a capillary tube. A seedborne fungus, EML-KWD01, was purely isolated from the wheat seed, subcultured on PDA, and then preserved in 20% glycerol stock in a deep freezer (-80℃) at Environmental Microbiology Laboratory (EML) culture collection, Chonnam National University, Gwangju, Korea. The strain was also deposited as ex-type (KOSPFGC0919) at culture collection of National Institute of Biological Resources (NIBR), Incheon, Korea.
DNA extraction and PCR
Isolates were cultured on PDA overlaid with cellophane at 27℃ for 3~5 days. Total genomic DNA was extracted with HiGene Genomic DNA Prep Kit for fungi (Biofact Co., Daejeon, Korea). Ribosomal DNA ITS and 28S regions were amplified with universal primer pairs ITS1/ITS4 and LROR/LR5F [14] in a 20 µL reaction mixture using AccuPower PCR Premix (Bioneer Co., Daejeon, Korea) containing Taq DNA polymerase, dNTPs, buffer, and tracking dye. PCR reaction was carried out with the following parameters: 2 min at 95℃ for initial denaturation, followed by 30 cycles of 1 min at 94℃ for denaturation, 30 sec at 54℃ for annealing, and 1min at 72℃ for extension, with 10 min at 72℃ for terminal extension. To amplify GAPDH gene, primers gpd1 and gpd2 [12] were used. PCR reaction was conducted using the following conditions; 2 min at 96℃ for initial denaturation, followed by 35 cycles of 1min at 96℃ for denaturation, 1 min at 52℃ for annealing, and 45 sec at 72℃ for extension, with 10 min at 72℃ for terminal extension [12]. PCR products were purified using AccuPrep PCR Purification Kit (Bioneer Co.), according to the manufacturer's instructions. Sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA).
Phylogenetic analysis
Sequences from this study and those from GenBank database (Table 1) were aligned to each other using ClustalX v. 1.83 [15] and edited with BioEdit v. 5.0.9.1 [16]. Phylogenetic analyses were performed using MEGA 6 [17], and neighbor-joining (NJ) tree was constructed using Kimura's two-parameter method. A maximum parsimony tree was constructed for combined datasets of rDNA ITS, 28S, and GAPDH gene sequences of EML-KWD01 isolate using the MEGA 6 program. Alternaria alternata was used as outgroup.
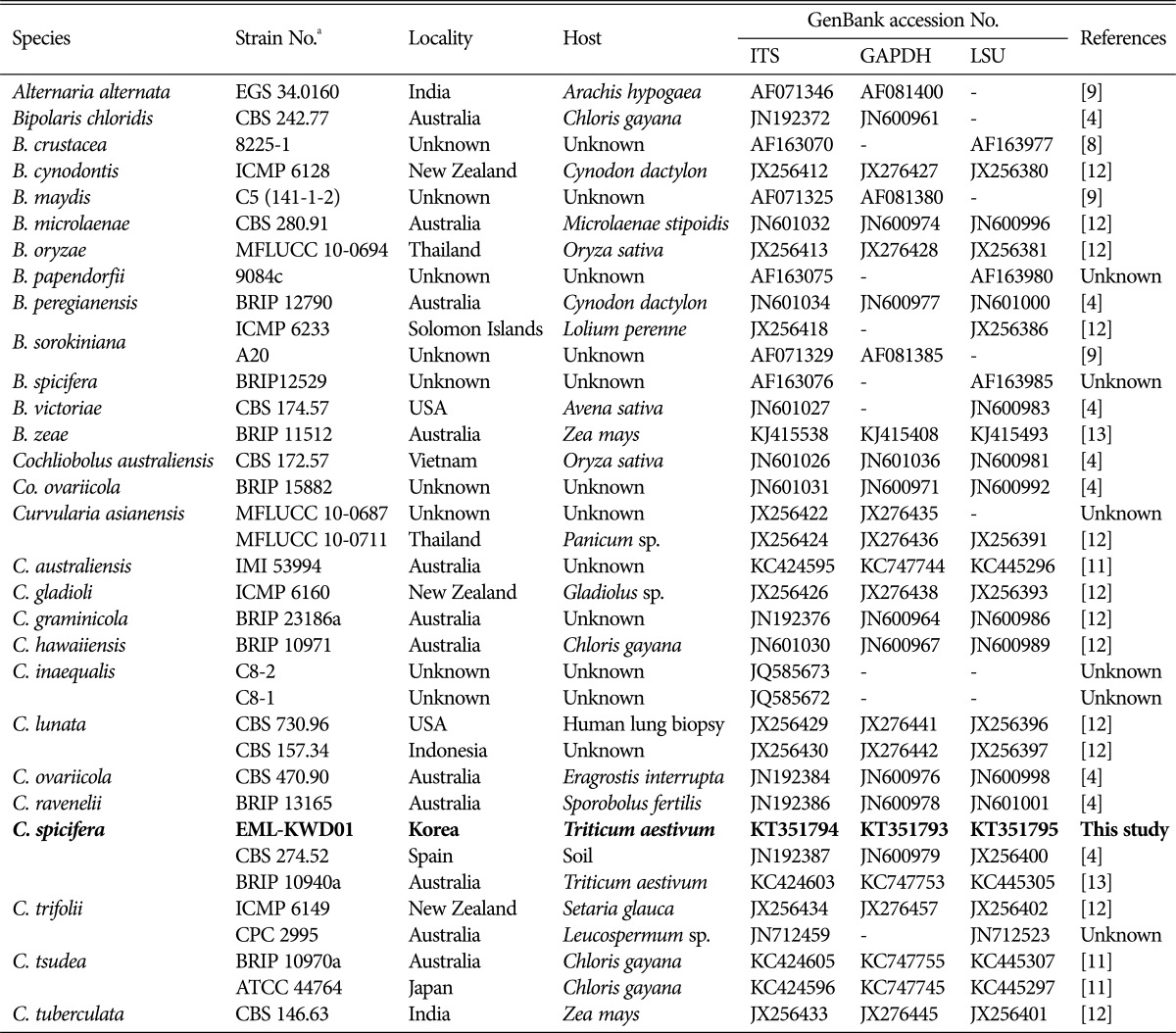
Table 1. Sequences of isolates used in this study with GenBank accession numbers.
ITS, internal transcribed spacer; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LSU, large subunit.
aCBS (Centraalbureau Voor Schimmelcultures, Utrecht, Netherlands), ATCC (American Type Culture Collection, Manassas, Virginia), BRIP (Plant Pathology Herbarium, Brisbane, Queensland, Australia), IMI (CABI Bioscience Genetic Resource Collection, CABI Bioscience UK Centre, Egham, UK), MFLUCC (Mae Fah Luang University Culture Collection, Chiang Rai, Thailand), EML (Environmental Microbiology Lab Fungal Herbarium, Chonnam National University, Gwangju, South Korea).
Morphological studies
For microscopic examination and determination of the growth rate, EML-KWD01 isolate was grown on PDA, oatmeal agar (OA; oatmeal 30 g/L, agar 15 g/L), malt extract agar (MEA; Difco), or V-8 juice agar (V-8; Campbell, Camden, NJ, USA) at 18℃, 25℃, 32℃, and 37℃ in the dark for 7 days. Morphological characteristics of fungal structures were determined under a light microscope (DFC290; Leica, Wetzlar, Germany) after preparing lactophenol slide mounts. Fine structures of the fungus were observed using scanning electron microscopy. Fungal samples were cultured on PDA medium in the dark at 27℃ for 7 days. Samples were fixed in 2.5% paraformaldehyde-glutaraldehyde buffer with 0.05 M phosphate (pH 7.2; Junsei, Tokyo, Japan) for 2 hr and washed in carcodylate buffer (Junsei). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (diluted in carcodylate buffer; Electron Microscopy Sciences, Hatfield, PA, USA) for 1 hr, washed again in carcodylate buffer, dehydrated in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei), and dried under a fume hood. Finally, these samples were covered with gold in a sputter coater and observed using a Hitachi S4700 field emission scanning electron microscope (Hitachi, Tokyo, Japan) at Korea Basic Science Institute, Gwangju, Korea.
Mycelial growth rate measurement
The diameter of growing colonies were measured every 2 days. Measurements of all colonies were made in two directions at right angles to each other for both controls. Temporal increase in the radius was plotted against time. Linear regression was calculated in order to estimate the growth rate in mm/day for each species under each set of environmental conditions [19]. All experiments were repeated three times.
RESULTS
Phylogenetic status
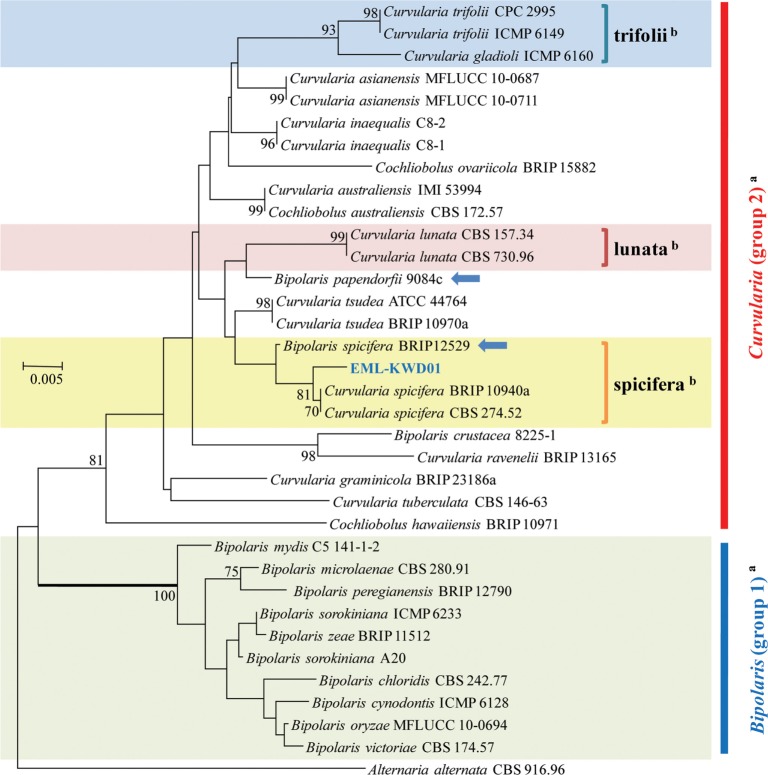
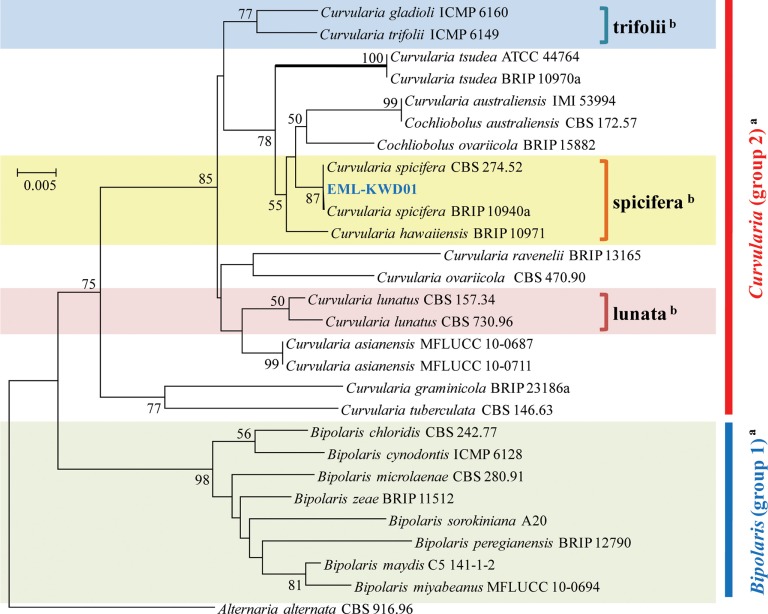
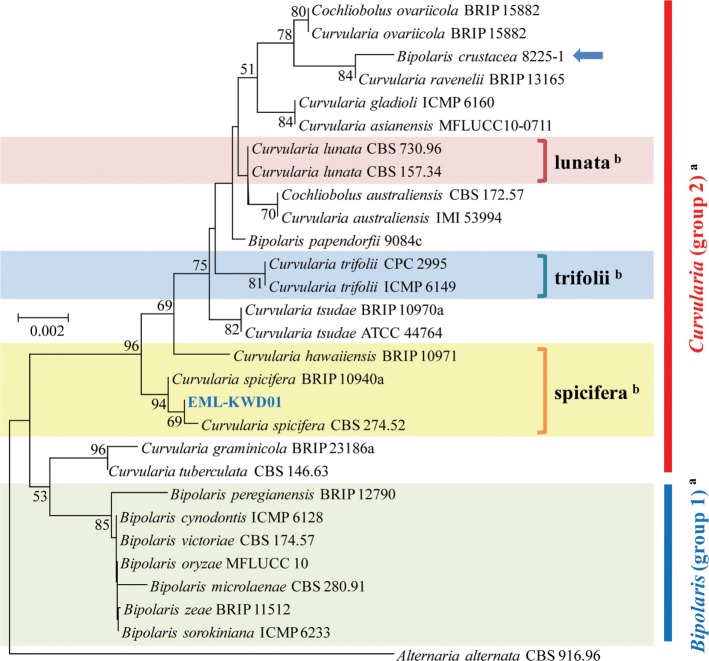
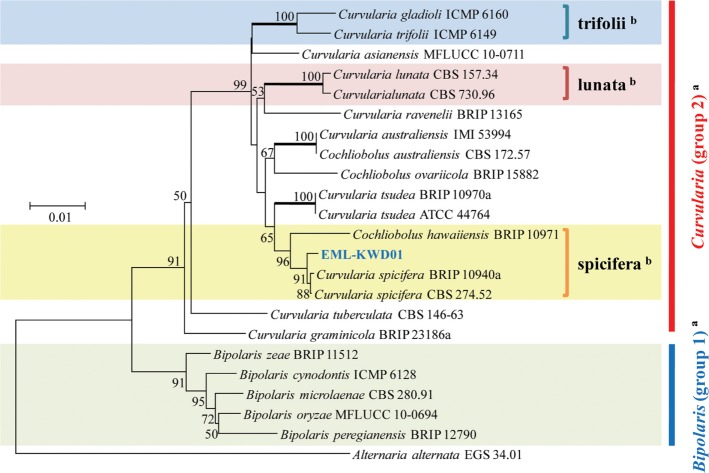
A phylogenetic tree was inferred using NJ method. Combined loci (ITS/28S/GAPDH) alignment was calculated for 36 strains (Table 1). Phylogenetic tree was constructed using the maximum parsimony method. A total of three most parsimonious trees were generated by a heuristic search using the combined dataset of 856 characters from the three loci. Basic Local Alignment Search Tool (BLAST) analysis of ITS sequences indicated that the EML-KWD01 isolate resembled C. spicifera R12 (accession No. KC315931) the most, displaying a 99.6% (523/525 bp) homology (Fig. 1). The GAPDH gene sequences of EML-KWD01 and that of C. spicifera CBS 274.52 (accession No. JN600979) shared 99.0% (514/519 bp) identities. The rDNA 28S sequence of EML-KWD01 shared 98.6% (820/832 bp) and 98.6% (821/832 bp) sequence identities with C. spicifera BRIP 10940a (accession No. KC445305) and C. spicifera CBS 274.52 (accession No. JX256400), respectively (Figs. 2 and 3). The tree generated from combined dataset of ITS, 28S, and GAPDH gene had a consistency index of 0.632 and a retention index of 0.814 (Fig. 4). Most nodes in the combined analysis showed increased clade support as measured by bootstrap analysis. The phylogenetic tree also showed that the unknown isolate EML-KWD01 belonged to family Pleosporaceae. Combining phylogenetic analyses of the ITS, 28S, and GAPDH gene sequences and morphological features, we were able to classify this species as C. spicifera belonging to Curvularia group 2 of family Pleosporaceae (Fig. 4).
Fig. 1. Neighbor-joining tree inferred from nuclear ribosomal rDNA internal transcribed spacer sequences of EML-KWD01 isolate and related taxa. Alternaria alternata was used as outgroup. Bootstrap support values ≥ 50% are indicated at nodes. Arrows represent that the genus name of Bipolaris should be changed to Curvularia. aClassification by Manamgoda et al. (2012) [12]. bClassification by Madrid et al. (2014) [18].
Fig. 2. Neighbor-joining tree inferred from glyceraldehyde 3-phosphate dehydrogenase gene sequences of EML-KWD01 isolate and related taxa. Alternaria alternata was used as outgroup. Bootstrap support values ≥ 50% are indicated at nodes. aClassification by Manamgoda et al. (2012) [12]. bClassification by Madrid et al. (2014) [18].
Fig. 3. Neighbor-joining tree inferred from large subunit rDNA sequences of EML-KWD01 isolate and related taxa. Alternaria alternata was used as outgroup. Bootstrap support values ≥ 50% are indicated at nodes. The arrow represents that the genus name of Bipolaris should be changed to Curvularia. aClassification by Manamgoda et al. (2012) [12]. bClassification by Madrid et al. (2014) [18].
Fig. 4. A phylogenetic tree showing the relationship of EML-KWD01 isolate with known species. The tree was generated from parsimony analysis based on the combination of rDNA internal transcribed spacer, 28S, and glyceraldehyde 3-phosphate dehydrogenase gene sequences using MEGA6. Alternaria alternata was used as outgroup. Bootstrap support values ≥ 50% are indicated at nodes. aClassification system by Manamgoda et al. (2012) [12]. bClassification system by Madrid et al. (2014) [18].
Taxonomy
Curvularia spicifera (Bainier) Boedijn, Bull. Soc. Mycol. Fr. 24: 81 (1909) [MB#278597] (Table 2, Figs. 5 and 6)
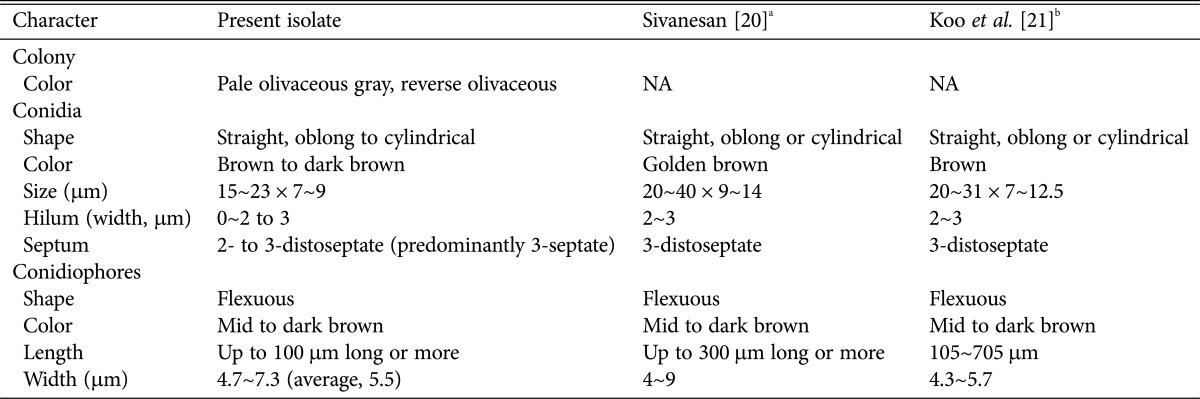
Table 2. Morphological characteristics comparison between isolate EML-KWD01 and references.
Fig. 5. Morphology of EML-KWD01 isolated from wheat seeds. A, Colonies grown on malt extract agar; B, Potato dextrose agar; C, V-8; and D, Oatmeal agar media at 25℃ for 7 days; E~J, Conidia with 2~3 transverse (predominantly 3-septate) septa on geniculate conidiophores; K, Oblong to cylindrical-shaped conidia without or with hilum (scale bars: F~K = 20 µm).
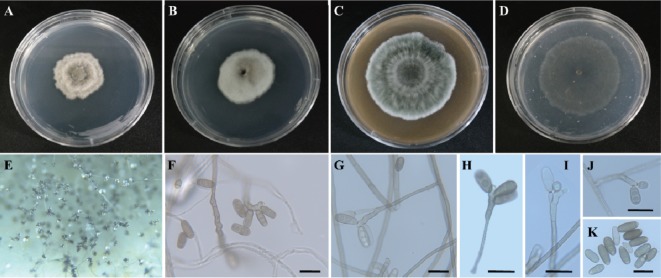
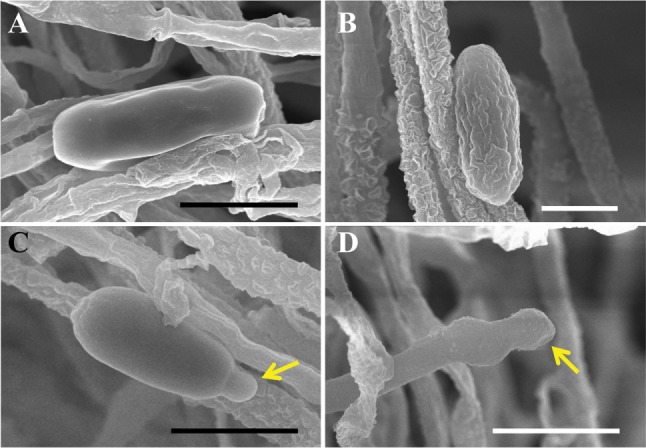
Fig. 6. Scanning electron micrograph of conidia and conidiophores of Curvularia spicifera EML-KWD01. A, B, Conidia with smooth or wrinkled surface; C, Germinating conidium (yellow arrow); D, Conidiophore with geniculate pore (yellow arrow) (scale bars: A, C, D = 10 µm, B = 5 µm).
≡ Drechslera spicifera (Bainier) Arx, The genera of fungi sporulating in pure culture: 222 (1970) [MB#313402]
≡ Bipolaris spicifera (Bainier) Subram, Hyphomycetes: an account of Indian species, except Cercosporae: 756 (1971) [MB#309557]
Etymology
This species was isolated from wheat seed collected from an agricultural farm in Korea.
Description
The colony diameter on PDA was approximately 4~6 mm at 25℃ 5 days after inoculation. The colony color was pale and olivaceous gray. The reverse of the colony was olivaceous. The conidiophores were simple, erect or ascendent, pigmented, geniculate from sympodial elongations, branched, variable length, with a width of 4.7~7.3 µm (average, 5.5 µm). Conidia were produced singly through pores, and frequently produced on several two side pores of tapering conidiophores. Conidia were not curved, oblong to cylindrical with 2~3 (predominantly 3-septate) transverse septa, hilum found (but not commonly), and measured 6.51~9.17 µm wide × 9.23~21.17 µm long.
Mycelial growth
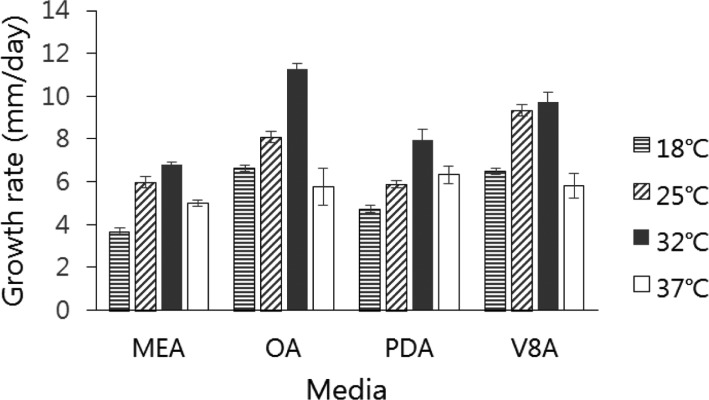
The isolate grew across a wide range of temperatures with various growth rates on MEA, OA, PDA, and V-8 media. The average colony sizes of EML-KWD01 isolate after 7 days of growth on MEA, OA, PDA, and V-8 were 6.8, 11.2, 7.9, and 9.7 mm, respectively. Optimal growth was observed on OA medium at 32℃. Slower growth was observed at 18℃ and 37℃ (Fig. 7).
Fig. 7. Effect of temperature and media on the growth of Curvularia spicifera EML-KWD01. Mycelial growth was evaluated on four kinds of media (malt extract agar [MEA], oatmeal agar [OA], potato dextrose agar [PDA], and V-8 agar [V8A]) after 7 days at 18℃, 25℃, 32℃, and 37℃.
DISCUSSION
Most species of Curvularia are found in subtropical to tropical regions, although a few are found in temperate zones. They are identified as major causal pathogens of sugarcane and grasses [6,20,21,22]. Some Curvularia species are also important to different industries owing to their ability of producing secondary metabolites, including substances with antimicrobial properties, enzymes, and precursors used for steroid production [13,23,24,25]. Curvularia spicifera (≡ Bipolaris spicifera) is known as a common allergen to humans. It can cause opportunistic infection in immunocompromised people such as acquired immune deficiency syndrome patients [18,26,27]. In Korea, this species has been detected from an imported grass seed without detailed descriptions [21]. However, it has not been detected from indigenous wheat seed.
Recently, ITS region has been used to identify fungi to discriminate species [28,29]. However, phylogenetic studies based on the ITS region have limited utility in identifying species, especially among members in the order of Pleosporales. In a recent phylogenetic study using multigene analysis, Manamgoda et al. [12] clarified the taxonomy of Curvularia. We used three loci to infer the phylogenetic relationships among species in the genus of Curvularia based on previous phylogenetic studies.
Although genus Curvularia can be easily distinguished from Bipolaris and Drechslera spp., there has been some difficulty to distinguish Curvularia and Bipolaris due to their conidial (wall) shape, size, and septation. Bipolaris and Cochliobolus species are not monophyletically clustered in Group 1 along with their type species. However, Curvularia species including species of Bipolaris, Cochliobolus, and Curvularia are clustered in Group 2 with its generic type [12]. Berbee et al. [9] have presented a phylogenetic analysis of Bipolaris and Curvularia species using combined sequences of ITS rDNA, large subunit (LSU) rDNA, and GAPDH gene.
In this study, rDNA ITS, and GAPDH genes provided sufficient phylogenetic information for the separation of Cochliobolus species into two groups. Our molecular phylogenetic data based on the sequence analyses of three loci of rDNA ITS, 28S, and GAPDH gene showed that the phylogenies were clustered into two main groups and that the isolate EML-KWD01 was an unrecorded Curvularia species, C. spicifera belonging to Curvularia group 2 within the Pleosporaceae family (Figs. 1, 2, 3, 4). Our results are in consistent with those obtained by Berbee et al. [9] and Manamgoda et al. [12] after using combined datasets of ITS, LSU (28S) rDNA, and GAPDH gene. As shown in Figs. 1 and 3, some Curvularia species were within the Bipolaris group. At the same time, some Bipolaris species were clustered in the Curvularia group 2. Our results revealed that the genus name of strains including Bipolaris crustacea 8225-1, Bipolaris papendorfii 9084c, and Bipolaris spicifera BRIP12529 in the phylogenetic trees should be changed to Curvularia species. Therefore, the status of some Bipolaris strains within the Curvularia group should be transferred to Curvularia group 2 based on the recent classification system. In Korea, Koo et al. [21] were the first ones who reported the species of Bipolaris spicifera. However, this old species name should be corrected to Curvularia spicifera based on recent classification system. However, studies with more Curvularia and Bipolaris isolates in comparison with type species based on multi loci sequence analyses are needed in the future.
ACKNOWLEDGEMENTS
This study was supported by the Project (201401205) on Survey and Exploration of Korean Indigenous Fungal Species and by the Graduate Program for the Undiscovered Taxa of Korea (201524202) of NIBR under the Ministry of Environment, Republic of Korea.
References
- 1.Boedijn KB. Über einige phragmosporen dematiazen. Bull Jard Bot Buitenzorg. 1933;13:120–134. [Google Scholar]
- 2.Ellis MB. Dematiaceous Hyphomycetes. Kew, Surrey: Commonwealth Mycological Institute; 1971. [Google Scholar]
- 3.Kirk P, Cannon PF, Minter DW, Stalpers JA. Ainsworth & Bisby's dictionary of the fungi. 10th ed. Wallingford: CAB International; 2008. [Google Scholar]
- 4.Manamgoda DS, Cai L, Bahkali AH, Chukeatirote E, Hyde KD. Cochliobolus: an overview and current status of species. Fungal Divers. 2011;51:3–42. [Google Scholar]
- 5.Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. London: Academic Press; 1980. [Google Scholar]
- 6.Amaradasa BS, Amundsen K. First report of Curvularia inaequalis and Bipolaris spicifera causing leaf blight of buffalo grass in Nebraska. Plant Dis. 2014;98:279. doi: 10.1094/PDIS-05-13-0487-PDN. [DOI] [PubMed] [Google Scholar]
- 7.Valente P, Ramos JP, Leoncini O. Sequencing as a tool in yeast molecular taxonomy. Can J Microbiol. 1999;45:949–958. doi: 10.1139/w99-094. [DOI] [PubMed] [Google Scholar]
- 8.Goh TK, Hyde KD, Lee DK. Generic distinction in the Helminthosporium-complex based on restriction analysis of the nuclear ribosomal RNA gene. Fungal Divers. 1998;1:85–107. [Google Scholar]
- 9.Berbee ML, Pirseyedi M, Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. [Google Scholar]
- 10.Tan YP, Madrid H, Crous PW, Shivas RG. Johnalcornia gen et comb. nov., and nine new combinations in Curvularia based on molecular phylogenetic analysis. Aust Plant Pathol. 2014;43:589–603. [Google Scholar]
- 11.Deng H, Tan YP, Shivas RG, Niu YC. Curvularia tsudae comb. nov. et nom. nov., formerly Pseudocochliobolus australiensis, and a revised synonymy for Curvularia australiensis. Mycoscience. 2015;56:24–28. [Google Scholar]
- 12.Manamgoda DS, Cai L, McKenzie EH, Crous PW, Madrid H, Chukeatirote E, Shivas RG, Tan YP, Hyde KD. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012;56:131–144. [Google Scholar]
- 13.Emami K, Hack E. Conservation of XYN11A and XYN11B xylanase genes in Bipolaris sorghicola, Cochliobolus sativus, Cochliobolus heterostrophus, and Cochliobolus spicifer. Curr Microbiol. 2002;45:303–306. doi: 10.1007/s00284-002-3618-8. [DOI] [PubMed] [Google Scholar]
- 14.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. pp. 315–322. [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madrid H, da Cunha KC, Gené J, Dijksterhuis J, Cano J, Sutton DA, Guarro J, Crous PW. Novel Curvularia species from clinical specimens. Persoonia. 2014;33:48–60. doi: 10.3767/003158514X683538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HB, Magan N. Environment factors influence in vitro interspecific interactions between A. ochraceus and other maize spoilage fungi, growth and ochratoxin production. Mycopathologia. 1999;146:43–47. doi: 10.1023/a:1007003316562. [DOI] [PubMed] [Google Scholar]
- 20.Sivanesan A. Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycol Pap. 1987;158:1–261. [Google Scholar]
- 21.Koo HM, Lee SH, Jung IM, Chun SC. A seedborne fungus Bipolaris spicifera detected from imported grass seeds. Plant Pathol J. 2003;19:133–137. [Google Scholar]
- 22.Lin SH, Huang CH, Deng ZY, Yan MX, Huang WH, Wei JJ, Qin ZQ. First report of leaf spot disease on sugarcane caused by Bipolaris spicifera in China. Aust Plant Dis Notes. 2012;7:51–53. [Google Scholar]
- 23.Hansen EH, Albertsen L, Schäfer T, Johansen C, Frisvad JC, Molin S, Gram L. Curvularia haloperoxidase: antimicrobial activity and potential application as a surface disinfectant. Appl Environ Microbiol. 2003;69:4611–4617. doi: 10.1128/AEM.69.8.4611-4617.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Schijndel JW, Vollenbroek EG, Wever R. The chloroperoxidase from the fungus Curvularia inaequalis: a novel vanadium enzyme. Biochim Biophys Acta. 1993;1161:249–256. doi: 10.1016/0167-4838(93)90221-c. [DOI] [PubMed] [Google Scholar]
- 25.Andrushina VA, Druzhinina AV, Yaderets VV, Stitsenko TS, Voishvillo NE. Hydroxylation of steroids by Curvularia lunata mycelium in the presence of methyl-β-cyclodextrine. Appl Biochem Microbiol. 2011;47:42–48. [PubMed] [Google Scholar]
- 26.Teran CG, Downes K, Medows M. Fatal Bipolaris spicifera infection in immunosuppressed child. BMJ Case Rep. 2014 Feb 03; doi: 10.1136/bcr-2013-009703. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buzina W, Braun H, Schimpl K, Stammberger H. Bipolaris spicifera causes fungus balls of the sinuses and triggers polypoid chronic rhinosinusitis in an immunocompetent patient. J Clin Microbiol. 2003;41:4885–4887. doi: 10.1128/JCM.41.10.4885-4887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwen PC, Hinrichs SH, Rupp ME. Untilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol. 2002;40:87–109. doi: 10.1080/mmy.40.1.87.109. [DOI] [PubMed] [Google Scholar]
- 29.Guarro J, Gené J, Stchigel AM. Developments in fungal taxonomy. Clin Microbiol Rev. 1999;12:454–500. doi: 10.1128/cmr.12.3.454. [DOI] [PMC free article] [PubMed] [Google Scholar]