Abstract
A total of 4 aquatic plants, Eleocharis kuroguwai Ohwi, Hydrocharis dubia Backer, Salvinia natans All., and Zizania latifolia Turcz., were sampled from representative two wetlands of South Korea. A total of 38 endophytic fungal strains were isolated from aquatic plants native to the Daepyeong wetland, and 27 strains were isolated from the Jilnal wetland. The internal transcribed spacer regions of fungal isolates were sequenced and a phylogenetic analysis was performed. In addition, endophytic fungal diversity from each wetland and host plant species was deduced. A total of 25 fungal genera were purely isolated, and 16 fungal genera were isolated from each of the two wetlands. Commonly isolated genera from both wetlands were Aspergillus, Cladosporium, Clonostachys, Fusarium, Leptosphaeria, Penicillium, and Talaromyces. This study revealed that fungal diversity varied with environmental conditions and by host plant in representative two wetlands.
Keywords: Aquatic plant, Diversity index, Endophytic fungi, Fresh water, Wetland
Wetlands are now known to be global controllers of their surrounding environments, due to their roles in natural purification, remediation, material cycling, and buffering [1]. Freshwater marsh always undergoes biological or microbial succession, which aids in the maintenance of biodiversity [2]. Furthermore, diverse aquatic plants have colonized areas surrounding well-developed wetlands, which were formed long before the agricultural stage in Korea [3].
Aquatic plants in wetlands carry out photosynthesis as primary producers in the ecosystem, with blooms at the surfaces of freshwater. Thus, they affect the penetration of sunlight into the freshwater bed, the concentration of dissolved oxygen and CO2 concentration, and the structure of the aquatic ecosystem. Therefore, aquatic plants are considered useful resources because of their ability to purify water [4,5,6].
Meanwhile, endophytic fungi distributed in the leaves or roots of plants exhibit symbiotic relationships with their host plant. The plant growth promoting activity and the induction of systemic resistance (ISR) by these fungi in their host plants has been widely researched [7,8,9,10,11]. Despite the potential of these microbial resources, research regarding the endophytic fungi of aquatic plants native to the wetlands of Korea has not been conducted [11].
The aim of this study was to identify the distribution and diversity of fungi from 4 representative aquatic plant species native to the Daepyeong and Jilnal wetlands, which were designated as natural monuments for the purpose of wetland preservation. Fungal colonies from the roots of each aquatic plant were isolated and identified by the amplification of the internal transcribed spacer (ITS) region of genomic DNA. Fungal strains were then categorized into several groups based on the phylogeny. Furthermore, the fungal diversity of each plant was assessed and comparatively analyzed. This study is the first research that provides basic data on the relationship between aquatic plants and their endophytic fungi. Promising microbial resources with benefits for aquatic plants that purify water environments can be identified by this study.
MATERIALS AND METHODS
Sampling and isolation of endophytic fungi
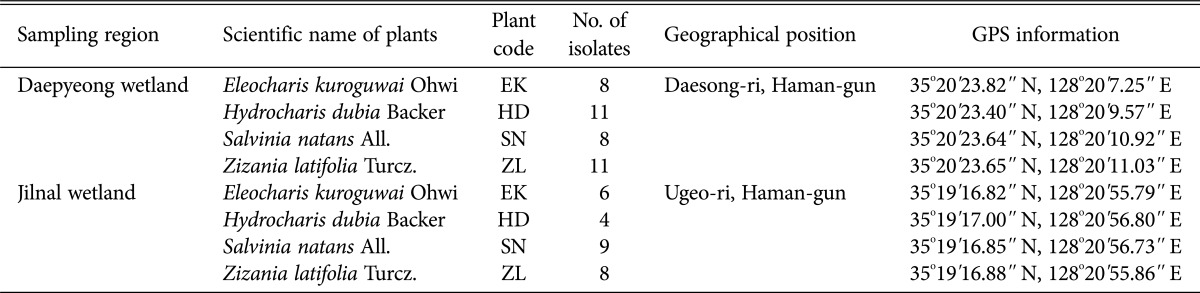
A total of 4 representative aquatic plants, Eleocharis kuroguwai Ohwi (belonging to the order Cyperales), Hydrocharis dubia Backer (belonging to the order Psammophytes), Salvinia natans All. (a pteridophyte belonging to the order Polypodiales), and Zizania latifolia Turcz. (a gramineous plant, belonging to the order Braminales) are native to the Daepyeong and Jilnal wetlands. These species commonly live at the edge of water or float at the surface of freshwater (Table 1). Sampled plants (16 individuals per each species) were harvested along with freshwater from their habitats to minimize physiological changes. Sterile distilled water (SDW) and sterilized 0.1% Tween 80 solution (Sigma-Aldrich, St. Louis, MO, USA) was sprayed on the surface of samples to eliminate suspended solids or normal microflora on the plant surfaces. The plants were submerged in 1.0% perchloric acid (HClO4) 2 times for 10 min each, and were subsequently washed with SDW 3~4 times. Residual water was eliminated with dried, sterile gauze and 50 pieces of root from a plant sample was cut to a length of 3~4 cm. Pre-treated samples were loaded into Hagem minimal medium containing 80 ppm of streptomycin (Sigma-Aldrich) to exclude root bacteria, and incubated at 25℃ for 15 days [11]. Sub-culturing of endophytic fungi for pure isolation was performed with the same media and conditions. Finally, pure isolates were incubated on potato dextrose agar (Difco, Detroit, MI, USA) and selected based on morphological differences.
Table 1. Information of aquatic plants collected from the wetlands.
EK, Eleocharis kuroguwai Ohwi; HD, Hydrocharis dubia Backer; SN, Salvinia natans All.; ZL, Zizania latifolia Turcz.
Extraction of genomic DNA and polymerase chain reaction (PCR)
All endophytic fungi from the 4 aquatic host plants were inoculated into potato dextrose broth (Difco) media and incubated at 25℃ for 7 days with 120 rpm using a rotary shaker. Filtered mycobionts were lyophilized for 2 days. The DNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) was used for the extraction of genomic DNA from lyophilized mycobionts and primers targeting the ITS regions, ITS1 and ITS4 were used for amplification [12].
The PCR conditions were pre-denaturation (94℃, 4min), denaturation (94℃, 1 min), annealing (55~58℃, 1 min), and extension (72℃, 2min) for a total of 35 cycles, followed by a final extension (72℃, 2 min).
The PCR products were confirmed by electrophoresis (1.5% agarose gel, stained with ethidium bromide) and observation of the resulting band pattern under a UV transilluminator. The AccuPrep PCR & Gel Extraction Kit (Bioneer, Daejeon, Korea) was used for the purification of PCR products, and an ABI 3730XL DNA analyzer (Applied Biosystems, Carlsbad, CA, USA) was used for the sequencing of ITS regions [10].
Phylogenetic analysis and examination of the diversity of endophytic fungi
The ITS region sequences of endophytic fungi were compared with sequences of other fungal species showing similarity over 99%, as determined by analyzing data from the GenBank databases of National Center for Biotechnology Information (NCBI). Phylogenetic relationships were analyzed using the MEGA program ver. 6.0 with an alignment of sequences that was prepared using ClustalW software [13]. The phylogenetic trees were inferred with the neighbor-joining algorithm with the Kimura 2-parameter. The stability of relationships was evaluated by a bootstrap analysis with a resampling of 1,000 times [13]. The diversity of the endophytic fungi from each sampling site was analyzed and compared. Diversity at the genus level was revealed using the Margalef's richness index (Dmg) [14] and Mehinick's index (Dmn) [15].
RESULTS AND DISCUSSION
Isolation of endophytic fungi and phylogenetic analysis
Plant community of E. kuroguwai and H. dubia, around the Daepyeong and Jilnal wetlands, had floral axes of about 70~80 cm. S. natans had a floral axis of about 70 cm in length, and Z. latifolia has an axis of about 80~90 cm with a leaf width of 3~4 cm. Sampling information of aquatic plants is presented in Table 1.
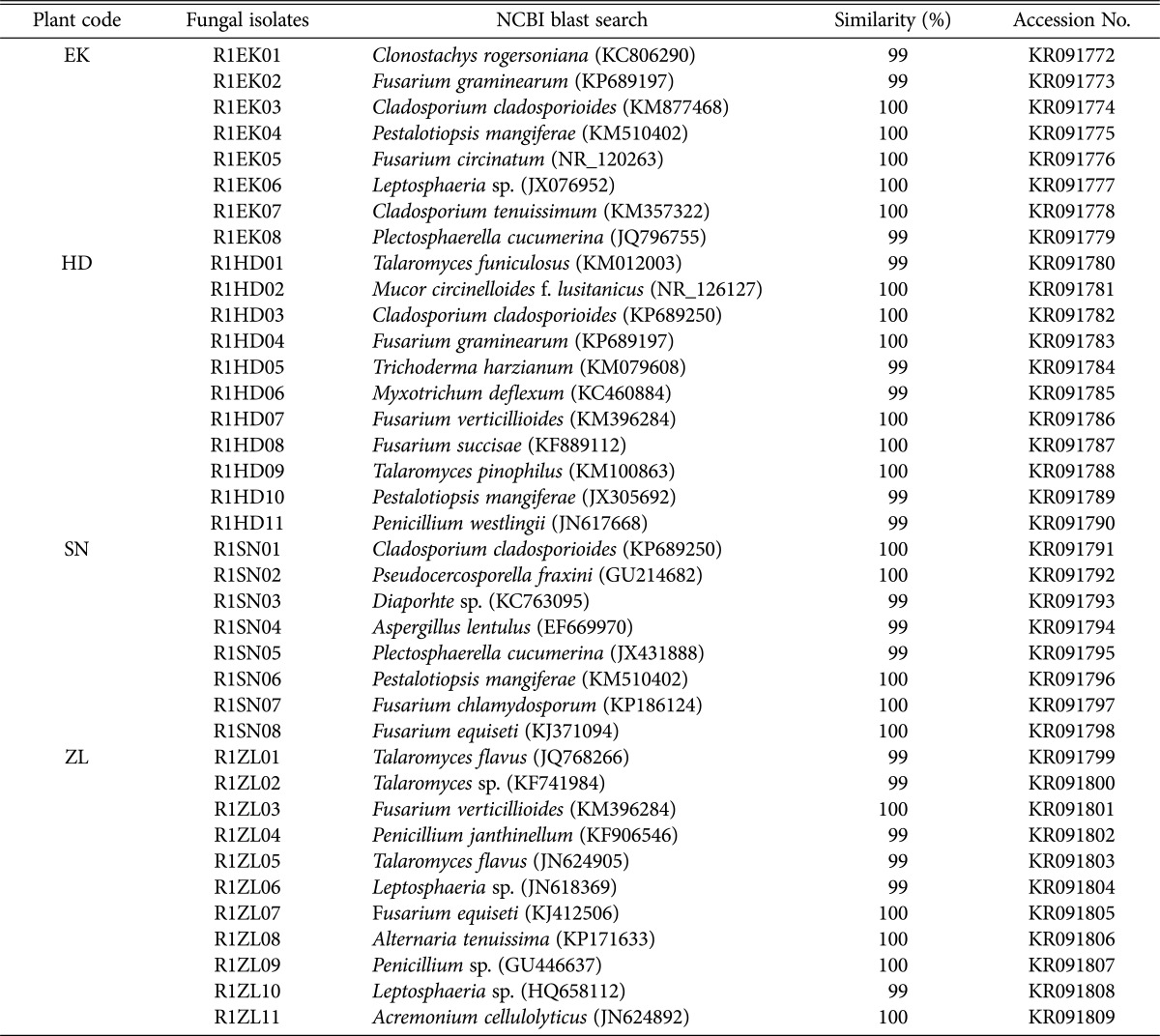
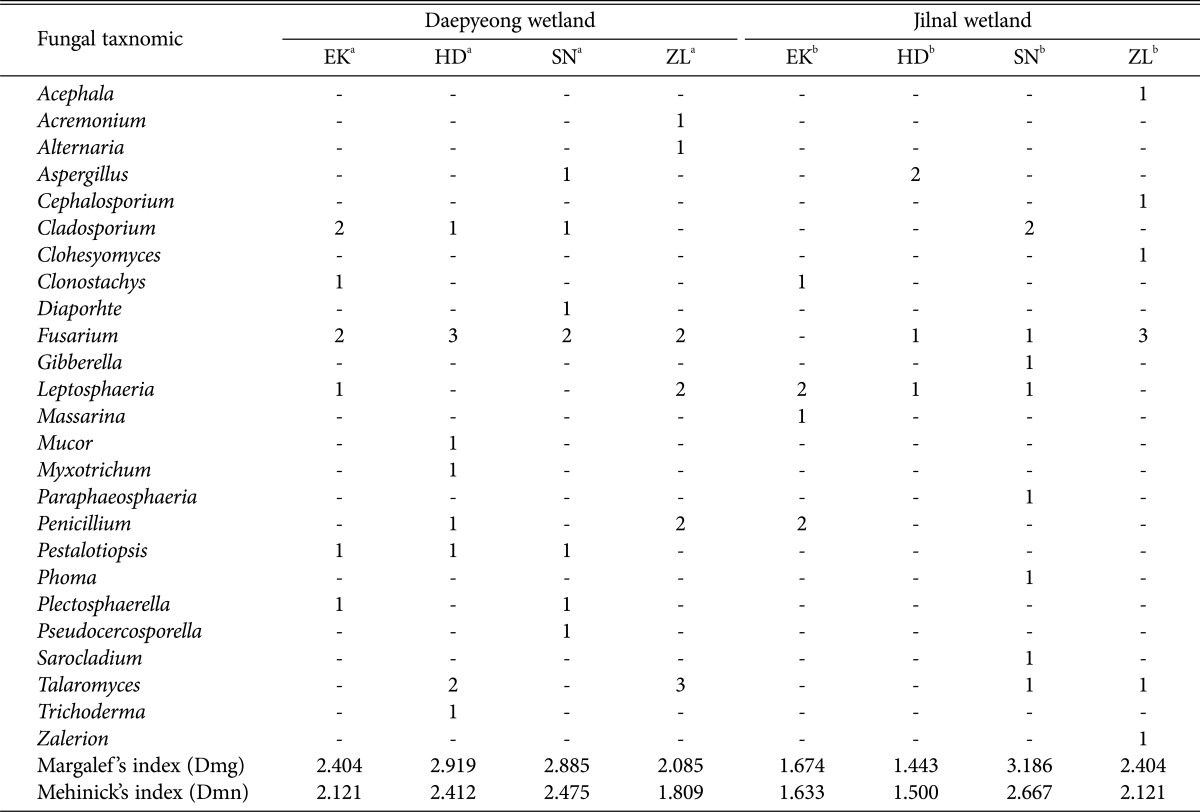
Based on the phylogenetic analysis, we found the following fungal genera represented in the Daepyeong wetland. A total of 8 strains isolated from E. kuroguwai belonged to the genera Cladosporium, Clonostachys, Fusarium, Leptosphaeria, Pestalotiopsis, and Plectosphaerella. A total of 11 strains from H. dubia belonged to genera Cladosporium, Fusarium, Mucor, Myxotrichum, Penicillium, Pestalotiopsis, Talaromyces, and Trichoderma. A total of 8 strains from S. natans belonged to 7 genera, including Aspergillus, Cladosporium, Diaporhte, Fusarium, Pestalotiopsis, Plectosphaerella, and Pseudocercosporella. Finally, 11 strains from Z. latifolia belonged to 6 genera, including Acremonium, Alternaria, Fusarium, Leptosphaeria, Penicillium, and Talaromyces (Table 2).
Table 2. Endophytic fungi isolated from aquatic plants in the Daepyeong wetland.
EK, Eleocharis kuroguwai Ohwi; HD, Hydrocharis dubia Backer; SN, Salvinia natans All.; ZL, Zizania latifolia Turcz.
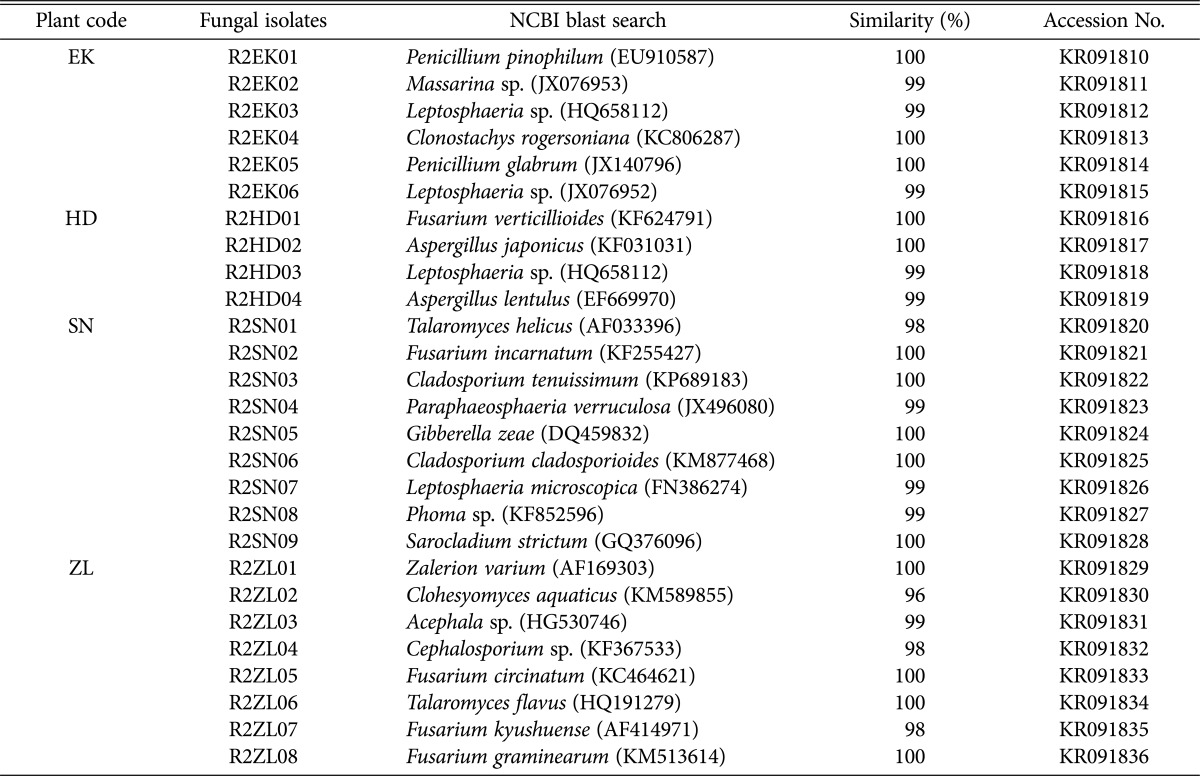
Similarly, we found the following fungal genera represented in the Jilnal wetland. A total of 6 strains isolated from E. kuroguwai belonged to 4 genera, including Clonostachys, Leptosphaeria, Massarina, and Penicillium, and 4 strains from H. dubia belonged to 3 genera, including Aspergillus, Fusarium, and Leptosphaeria. A total of 9 strains from S. natans belonged to 8 genera, including Cladosporium, Fusarium, Gibberella, Leptosphaeria, Paraphaeosphaeria, Phoma, Sarocladium, and Talaromyces, and 8 strains from Z. latifolia belonged to 6 genera, including Acephala, Cephalosporium, Clohesyomyces, Fusarium, Talaromyces, and Zalerion (Table 3).
Table 3. Endophytic fungi isolated from aquatic plants in Jilnal wetland.
EK, Eleocharis kuroguwai Ohwi; HD, Hydrocharis dubia Backer; SN, Salvinia natans All.; ZL, Zizania latifolia Turcz.
The fungi sampled varied by wetland. A total 38 fungal strains from 16 genera were isolated from the Daepyeong wetland, while 27 strains from 16 genera were isolated from the Jilnal wetland. The 65 total strains from the two wetlands belonged to 25 genera. Common isolates from both the Daepyeong and the Jilnal wetlands were identified as belonging to the species Aspergillus, Cladosporium, Clonostachys, Fusarium, Leptosphaeria, Penicillium, and Talaromyces. On the other hand, there were 9 unique genera from the Daepyeong wetland, including Acremonium, Alternaria, Diaporthe, Mucor, Myxotrichum, Pestalotiopsis, Plectosphaerella, Pseudocercosporella, and Trichoderma. In contrast, there were 8 unique genera from the Jilnal wetland, including Acephala, Cephalosporium, Clohesyomyces, Gibberella, Massarina, Paraphaeosphaeria, Phoma, Sarocladium, and Zalerion (Tables 2 and 3).
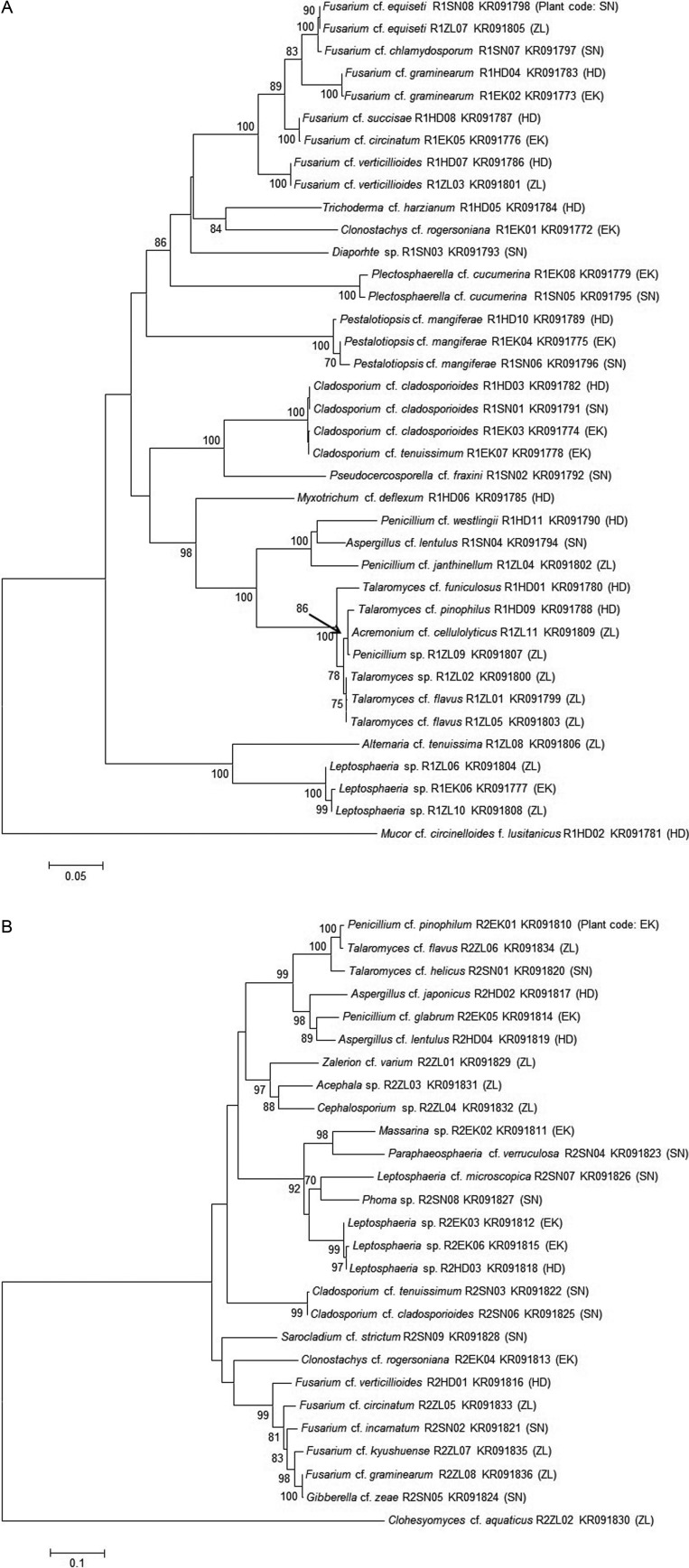
The ITS sequences of endophytic fungal strains from each wetland were registered into the GenBank database of NCBI, including isolates of E. kuroguwai (KR091772~KR091779), H. dubia (KR091780~KR091790), S. natans (KR091791~KR091798), and Z. latifolia (KR091799~KR091809) from the Daepyeong wetland, and isolates of E. kuroguwai (KR091810~KR091815), H. dubia (KR091816~KR091819), S. natans (KR091820~KR091828), and Z. latifolia (KR091829~KR091836) to the Jilnal wetland. Phylogenetic trees of endophytic fungi isolated from the roots of aquatic plants native to the each wetland were constructed (Fig. 1A and 1B).
Fig. 1. Phylogenetic analysis of endophytic fungi isolated from the aquatic plants in Daepyeong and Jilnal wetlands. This phylogenetic tree was constructed by using the neighbor-joining method (1,000 bootstrap replications). Bootstrap values (70%) are indicated at relevant nodes. Dendrogram of endophytic fungi isolated from the aquatic plants in Daepyeong (A) and Jilnal wetlands (B).
Diversity of endophytic fungi
The richness of fungal isolates from the Daepyeong and Jilnal wetlands was analyzed at the genus level using Mehinick's index (Dmn) and Margalef's richness index (Dmg). In terms of generic richness calculated by Margalef's index, the fungal biota from each aquatic plant was as follows: E. kuroguwai (2.404, 1.674), H. dubia (2.919, 1.443), S. natans (2.885, 3.186), and Z. latifolia (2.085, 2.404). Using Mehinick's index, the generic richness was calculated as follows: E. kuroguwai (2.121, 1.633), H. dubia (2.412, 1.500), S. natans (2.475, 2.667), and Z. latifolia (1.809, 2.121) (Table 4).
Table 4. Diversity index of endophytic fungi isolated from the aquatic plants in wetlands.
EK, Eleocharis kuroguwai Ohwi; HD, Hydrocharis dubia Backer; SN, Salvinia natans All.; ZL, Zizania latifolia Turcz.
aAquatic plants code collected from the Daepyeong wetland.
bAquatic plants code collected from the Jilnal wetland.
E. kuroguwai and H. dubia from the Daepyeong wetland showed higher diversity values than those from the Jilnal wetland. In contrast, S. natans and Z. latifolia from the Jilnal wetland showed higher values than those from the Daepyeong wetland. The high values of fungal diversity from E. kuroguwai and H. dubia in the Daepyeong wetland may be due to the greater number of isolates or variety of confirmed genus than from the Jilnal wetland. Similarly, the high values of fungal diversity from S. natans and Z. latifolia in the Jilnal wetland may be due to the greater number of confirmed genera than from the Daepyeong wetland. Mehinick's index is similar, conceptually, to Margalef 's richness index for analyzing species richness. The deduced diversity values from each of the two indices showed similar patterns in this study. Because of the endophyte sample size, Shannon's diversity index (H') [16] and Simpson's diversity index (D) [17] were not used.
Phylogenetic and diversity analyses of the endophytic fungi were conducted. Isolated fungi were found to belonging to 7 genera, and Aspergillus, Cladosporium, Fusarium, and Penicillium were commonly isolated. Some fungal species belonging to the genus Fusarium or Leptosphaeria have been revealed as plant pathogens, but the genus Clonostachys has not been studied well. Other genera have been isolated from diverse environments. Each strain belonging to the 9 genera that can be differentiated by the source wetland is thought that result from the unique environment features of the two habitats. This result indicates that fungal biota of host plants can be differentiated by habitat location, even if they are from the same plant species, and that this could be a result of adaptation to their unique environments. Hydroecological endophytes are well known for producing effective metabolites in their host plants [2,18]. These positive roles might be applied to aquatic plants to the purpose of effective water purifying [1,7]. First, endophyte diversity from water purifying aquatic plants native to freshwater marshes must be secured prior to this tactic [9]. However, these researches have not been vigorously conducted. This study provides endophytic diversity as basic data from these wetland environments and on aquatic plant-endophyte interactions that will be valuable for further study.
Inland wetlands suffer from water stress that is very unfavorable to mesophytic growth [18]. For this reason, aquatic plants adapted to unique environmental features dominate. Differences in the thicknesses of a root epidermis or root cap between each aquatic plants species can lead to distinctive types of fungal biota. Furthermore, these plants have evolved with independent morphological characteristics, even if the species diversity of aquatic plants is less than that of mesophytic plants [18]. Therefore, microbial distribution and diversity may differ from one another [11]. Up to now, endosymbiotic microorganisms of aquatic plants have not been studied well, so this study can serve as a starting point for further research.
The 4 aquatic plants used in this study can be categorized by taxonomical criteria. All plants are tracheophytes. E. kuroguwai, H. dubia, and Z. latifolia form flowers and fruit and are perennial plants. In contrast, S. natans, a pteridophyte, is an annual plant that does not form flowers and fruit and that thrives by sporulation [2,18]. Generally, pteridophytes are more primeval type of vascular plants than others, and they hold a key position in the evolution of vascular plants [19]. Pteridophytes adapted to the drastic environmental changes of primitive ages, and sporulation allows for an explosion of the population under favorable conditions [2,18]. S. natans can eliminate nitrogen and phosphorus from eutrophied water, and a very effective due to their strong propagation ability. Because of this, S. natans is a highly valued bio-resource that can remediate contaminated natural environments. Therefore, research on how endophytes interact with S. natans has become an important field [19]. However, S. natans can also cover the surface of a wetland with blooms and decrease the dissolved oxygen concentration in the water [6,18]. In conclusion, endophytes may play a major role in growth modulation of S. natans, an outstanding eutrophication controller. However, the interaction between the water purifying S. natans and their endophytes has not been studied well. In this study, S. natans showed the highest fungal diversity value of the plants surveyed (Table 4). This may have resulted from a prolonged period of endosymbiosis with pteridophytes that appeared at an early stage of tracheophyte evolution. Researches that screen the biological activity (ISR, plang growth promoting) from endophytes secured in this study and application of promising endophytes to S. natans to promote (control) their growth or increase the efficiency of water purification activity in eutrophied water, have to be done. This study compared the distribution of endophytic fungi from aquatic plants native to two representative wetlands in Korea for the purpose of discovering diverse and effective microorganisms. This study provides basic information about microbial resources of wetland aquatic plants.
ACKNOWLEDGEMENTS
This work was carried out with the support of "Research Program for Agriculture Science & Technology Development (Project title: Diagnosis of horticultural and herbal crops diseases and insect pests, Project No. PJ01136802)" Rural Development Administration, Republic of Korea.
References
- 1.Whitaker V, Matvienko B. The denitrification potential and hydrological conditions in the wetlands of the lobo resevoir. Verh Int Ver Theor Angew Limnol. 1998;26:1377–1380. [Google Scholar]
- 2.Denny P. Biodiversity and wetlands. Wetl Ecol Manag. 1994;3:55–61. [Google Scholar]
- 3.Kuczyńska-Kippen N. Habitat choice in rotifer communities of three shallow lakes: impact of macrophyte substratum and season. Hydrobiologia. 2007;593:27–37. [Google Scholar]
- 4.Carpenter SR, Lodge DM. Effects of submersed macrophytes on ecosystem processes. Aquat Bot. 1986;26:341–370. [Google Scholar]
- 5.Desmet NJ, Van Belleghem S, Seuntjens P, Bouma TJ, Buis K, Meire P. Quantification of the impact of macrophytes on oxygen dynamics and nitrogen retention in a vegetated lowland river. Phys Chem Earth. 2011;36:479–489. [Google Scholar]
- 6.Yeh TY, Ke TY, Lin YL. Algal growth control within natural water purification systems: macrophyte light shading effects. Water Air Soil Pollut. 2011;214:575–586. [Google Scholar]
- 7.Redman RS, Sheehan KB, Stout RG, Rodriguez RJ, Henson JM. Thermotolerance generated by plant/fungal symbiosis. Science. 2002;298:1581. doi: 10.1126/science.1072191. [DOI] [PubMed] [Google Scholar]
- 8.Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Hückelhoven R, Neumann C, von Wettstein D, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci U S A. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You YH, Kwak TW, Kang SM, Lee MC, Kim JG. Aspergillus clavatus Y2H0002 as a new endophytic fungal strain producing gibberellins isolated from Nymphoides peltata in fresh water. Mycobiology. 2015;43:87–91. doi: 10.5941/MYCO.2015.43.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You YH, Yoon H, Kang SM, Woo JR, Choo YS, Lee IJ, Shin JH, Kim JG. Cadophora malorum Cs-8-1 as a new fungal strain producing gibberellins isolated from Calystegia soldanella. J Basic Microbiol. 2013;53:630–634. doi: 10.1002/jobm.201200002. [DOI] [PubMed] [Google Scholar]
- 11.You YH, Yoon H, Kang SM, Shin JH, Choo YS, Lee IJ, Lee JM, Kim JG. Fungal diversity and plant growth promotion of endophytic fungi from six halophytes in Suncheon Bay. J Microbiol Biotechnol. 2012;22:1549–1556. doi: 10.4014/jmb.1205.05010. [DOI] [PubMed] [Google Scholar]
- 12.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Inis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. pp. 315–322. [Google Scholar]
- 13.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margalef DR. Information theory in ecology. Gen Syst. 1958;3:36–71. [Google Scholar]
- 15.Whittaker RH. Evolution of species diversity in land communities. Evol Biol. 1977;10:1–67. [Google Scholar]
- 16.Pielou EC. Ecological diversity. New York: John Wiley & Sons; 1975. [Google Scholar]
- 17.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 18.Arnold G, Van der V. The biology of freshwater wetlands. Oxford: Oxford University Press; 2012. [Google Scholar]
- 19.Fernández NV, Messuti MI, Fontenla SB. Occurrence of arbuscular mycorrhizas and dark septate endophytes in pteridophytes from a Patagonian rainforest, Argentina. J Basic Microbiol. 2013;53:498–508. doi: 10.1002/jobm.201100613. [DOI] [PubMed] [Google Scholar]