Abstract
In this study, transcriptome analysis of twelve laccase genes in Pleurotus ostreatus revealed that their expression was differentially regulated at different developmental stages. Lacc5 and Lacc12 were specifically expressed in fruiting bodies and primordia, respectively, whereas Lacc6 was expressed at all developmental stages. Lacc1 and Lacc3 were specific to the mycelial stage in solid medium. In order to investigate their biochemical characteristics, these laccases were heterologously expressed in Pichia pastoris using the pPICHOLI-2 expression vector. Expression of the laccases was facilitated by intermittent addition of methanol as an inducer and sole carbon source, in order to reduce the toxic effects associated with high methanol concentration. The highest expression was observed when the recombinant yeast cells were grown for 5 days at 15℃ with intermittent addition of 1% methanol at a 12-hr interval. Investigation of enzyme kinetics using 2,2-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid (ABTS) as a substrate revealed that the primordium-specific laccase Lacc12 was 5.4-fold less active than Lacc6 at low substrate concentration with respect to ABTS oxidation activity. The optimal pH and temperature of Lacc12 were 0.5 pH units and 5℃ higher than those of Lacc6. Lacc12 showed maximal activity at pH 3.5 and 50℃, which may reflect the physiological conditions at the primordiation stage.
Keywords: Heterologous expression, Laccase, Mushroom, Primordia
Laccases (EC 1.10.3.2) are multi-copper polyphenol oxidases produced by fungi, bacteria, and plants. These enzymes are involved in the formation of plant cell wall by catalyzing the polymerization of monolignols to lignin through the generation of free radical via Cu2+-mediated single-electron transfer [1]. Contrary to plants, fungi and bacteria use laccases to mineralize recalcitrant lignin in dead wood making the microbial laccases key players in recycling of carbon-based materials in nature. In addition, laccases have attracted a great deal of attention in biotechnological applications due to their high catalytic activity toward a broad range of aromatic substrates. Laccases, which are potentially useful for bioremediation of aqueous environments polluted by endocrine disruptors [2], polycyclic aromatic hydrocarbons [3,4], and organic dyes [5], have also been used for denim bleaching in the textile industry [6] and pulp delignification in paper pulp manufacturing processes [7]. Recent studies have also demonstrated the potential utility of laccases in organic synthesis [8] and biotechnological applications, particularly in the fabrication of biosensors and biofuel cells [9,10].
In fungi, laccase is involved in a variety of cellular physiological events. This enzyme catalyzes the production of pigments, such as melanin in Lentinula edodes [11] and Cryptococcus neoformans [12], and cinnabarin in Pycnoporus mushrooms [13]. Melanins play important roles in the pathogenesis of C. neoformans in human [12], as well as in primordiation of L. edodes [11]. Cinnabarin, synthesized by Pycnoporus coccineus, exhibits antimicrobial activity [13]. Moreover, white rot fungi, such as Trametes versicolor and Pleurotus ostreatus, appear to produce laccases as a defense mechanism against environmental microorganisms [14]. An example of this defense mechanism is the degradation of aflatoxin by laccase from P. ostreatus [15].
Genomes of filamentous fungi, particularly basidiomycetes, contain multiple copies of laccase genes [16,17]. Laccase genes are induced by substrate analogs [18,19,20], copper ions [4,20,21], and nutritional factors [22]; however, the presence of an inducer does not result in all the copies of the laccase genes being expressed. P. coccineus expresses a single laccase gene, encoding a major extracellular protein, upon induction by copper ions, despite the presence of 7~9 isogenes in the genome [4]. Similarly, P. ostreatus has 12 homologous laccase genes but produces only two major laccase proteins, LCC1 and LCC2, in the culture supernatant [23]. Recent studies using chemically defined medium have demonstrated that the expression of laccase genes in P. ostreatus is rigorously regulated in response to different environmental conditions, in a strain-dependent manner, at the transcription level [24]. In particular, two laccases, Lacc2 and Lacc10, which were highly overexpressed in the inducible medium, were mainly responsible for the laccase activity observed in the culture supernatant [24]. Lacc2, characterized under the name of POXA3, was found to form heterodimer with a 16-kDa small subunit protein [25,26]. Lacc9 and Lacc10 were also characterized under the names POX1 [27] and POX2 or POXC [28,29], respectively.
This study was conducted to extend our knowledge of the physiological and biochemical characteristics of P. ostreatus laccases. In order to accomplish this, we investigated the expression of 12 laccase genes at the transcription level during different developmental stages. We next constructed a heterologous expression system in Pichia pastoris and optimized the culture conditions for maximal laccase production. Finally, the biochemical characteristics of two laccase enzymes were examined.
MATERIALS AND METHODS
Strains and culture media
A commercial strain of Pleurotus ostreatus Chunchu was obtained from Culture Collection of Wild Mushroom, Incheon University, Korea. P. ostreatus was grown in a polypropylene bottle with a solid substrate consisting of poplar tree sawdust (50%), beet pulp (30%), and cottonseed hull (20%). The water content of the solid substrate was adjusted to 75%. Pichia pastoris GS115 (Life Technology, San Diego, CA, USA) was used as the host strain for laccase expression. The yeast cells harboring the expression vector were grown in buffered methanol-complex medium (BMMY) consisting of yeast extract (10 g/L), tryptone (10 g/L), yeast nitrogen base (13.4 g/L), biotin (0.4 mg/L), histidine (40mg/L), 0.1M potassium phosphate buffer (pH 6.0), and zeocin (100 mg/mL).
Preparation of mRNA from mycelia, primordia, and fruiting bodies of P. ostreatus for transcriptome analysis
In order to obtain mycelia, primordia, and fruiting bodies of P. ostreatus, mycelia were grown in the substrate bottle by incubating at 25℃ for 30 days in the dark. Fruiting was induced by shifting the temperature to 18℃ after physical shock. For mRNA preparation, mycelia, primordia, and fruiting bodies were collected from the culture bottle and frozen in liquid nitrogen. The frozen samples were ground with a pestle and mortar. Total RNA was extracted from the ground samples using TRIzol reagent (Solgent Co., Daejeon, Korea). RNA-Seq was conducted using a commercial sequencing service (Macrogen, Seoul, Korea). The mRNA level determined by RNA-Seq was expressed as Reads Per Kilobase per Million mapped reads (RPKM).
Construction of expression vectors and transformation
The DNA sequences for P. ostreatus laccase genes were obtained from the P. ostreatus genome sequence in the Comparative Fungal Genomics Platform site (http://cfgp.snu.ac.kr/ [30]) by BLAST search with known fungal laccase gene sequences. The laccase genes were synthesized, with codon optimization, for Pichia expression using a commercial gene synthesis service (Bioneer Co., Daejeon, Korea). The synthesized genes were amplified using a forward primer (5'-GGCTTAGTCGACATGGCCGTCTCTGTC-3') and reverse primer (5'-CATAATGCGGCCGCTGCGGGCAGACGATG-3'). The PCR products were inserted between the SalI and NotI sites of a Pichia expression vector, pPICHOLI-2 (MoBiTec GmbH, Göttingen, Germany). Transformation of P. pastoris was performed by electroporation using competent yeast cells grown to OD600 = 1. Laccase gene expression in pPICHOLI-2 was placed under the control of the methanol-inducible alcohol oxidase I (AOX1) promoter.
Laccase activity assay
Laccase activity was measured using 2mM 2,2-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS; Sigma-Aldrich, St. Louis, MO, USA) in 50 mM sodium acetate buffer (pH 4.0). The reaction mixture, containing 0.99 mL substrate solution and 0.01 mL enzyme, was incubated for 3 min at 25℃. The reaction was tracked using a UV-Vis spectrophotometer (Model Ultraspec 2100 Pro; Amersham Biosciences, Piscataway, NJ, USA) at 420 nm. The absorbance at 420 nm was converted into the amount of product using the molecular extinction coefficient of the oxidized ABTS (ε420 nm = 36,000/M/cm). One unit of enzyme activity was defined as the amount of laccase required to produce 1 µmole of the product per minute.
Purification of the laccase protein
The culture supernatant of P. pastoris harboring pPICHOLI-Lacc was collected by centrifugation at 2,500 ×g for 10 min. The supernatant was precipitated by ammonium sulfate fractionation. The precipitant was dissolved in phosphate buffered saliine buffer and then loaded onto a DEAE-Sephadex column (Amersham Biosciences). The bound protein was eluted by NaCl gradient in 30 mM sodium phosphate buffer (pH 7.0). The flow rate was 1 mL/min.
RESULTS
Laccase genes in P. ostreatus and their differential expression
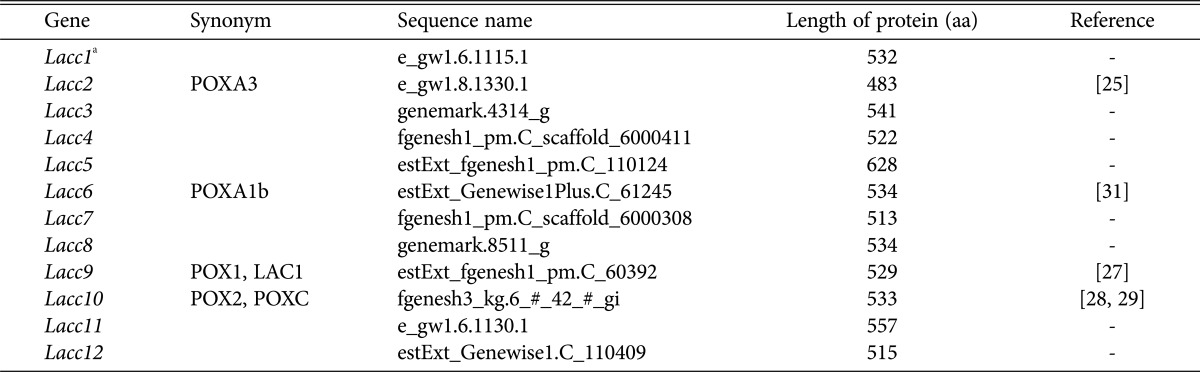
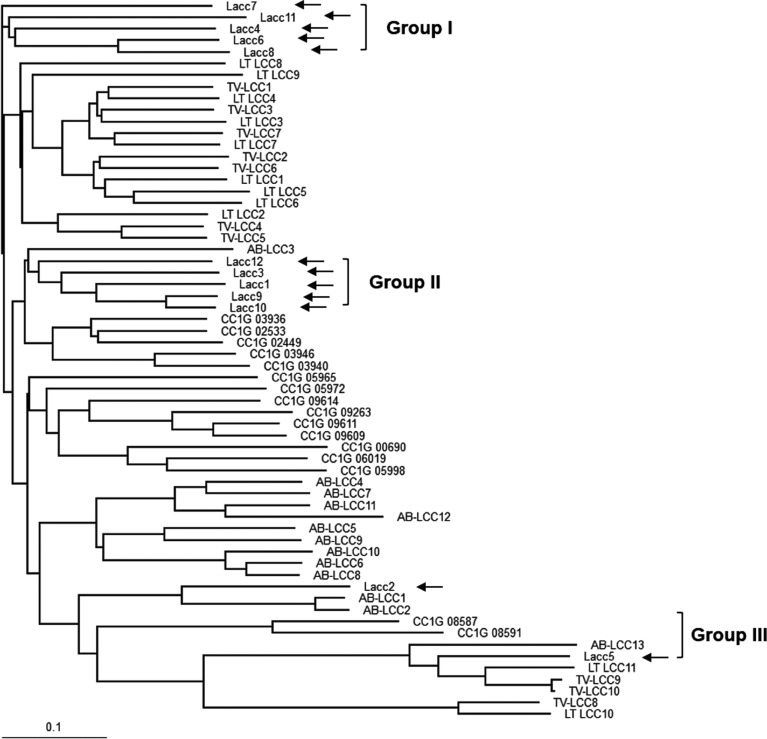
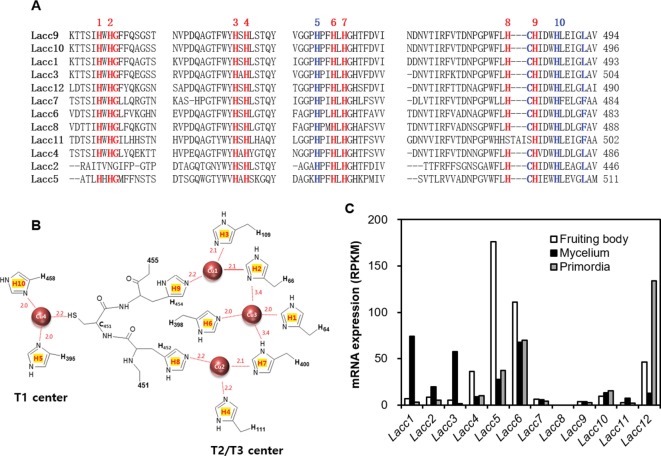
The P. ostreatus genome contains 12 homologous laccase genes (Table 1). Comparison of the translated protein sequences with known laccase sequences from four mushroom species revealed that the P. ostreatus laccases are clustered into three distinct groups (Fig. 1). P. ostreatus laccase proteins commonly contain a highly conserved mononuclear copper center, T1, and a trinuclear copper center, T2/T3, with conserved sequence signatures, including HXHG, HXH, HXXHXH, and HCHXXXHXXXXL/F (Fig. 2A). Ten histidine residues in the conserved sequence signatures form coordinate bonds with four copper ions, thereby constituting the enzyme active site, which makes them essential for laccase activity (Fig. 2B). Interestingly, the conserved HXHG sequence was not found in Lacc2, and Lacc11 did not contain the copper binding cysteine residue in the T1 center (Fig. 2A).
Table 1. Laccase genes in Pleurotus ostreatus.
aLaccase gene names were taken from Castanera et al. [24].
Fig. 1. Phylogenetic analysis of laccases from Pleurotus ostreatus. Laccase protein sequences were obtained from the Comparative Fungal Genomics Platform. AB, Agaricus bisporus; CC, Coprinopsis cinerea; LT, Lentinula tigrinus.
Fig. 2. Conserved sequence regions and laccase expression in Pleurotus ostreatus. A, Conserved sequence regions; the numbers above the sequence indicate conserved histidine residues; B, Arrangement of histidine residues in the catalytically important T1 and T2/T3 centers; coordinate bonds between the amino acid residues and copper ions in the laccase structure were redrawn using the laccase structure of Trametes versicolor (PDB code: 1GYC); C, RNA-Seq analysis of laccase genes in the mycelia, primordia, and fruiting bodies of P. ostreatus. RPKM, Reads Per Kilobase per Million mapped reads.
Transcriptional regulation of the laccase genes during different developmental stages was investigated by transcriptome analysis of the mycelium, primordium, and fruiting body (Fig. 2C). Among the twelve genes investigated, Lacc5 was highly expressed in the fruiting body, whereas Lacc1 and Lacc3 were expressed in the mycelium. Lacc12 was specifically expressed in the primordium. None of these genes have been extensively studied to date. Notably, Lacc6 was highly expressed at all stages. Lacc6 encodes a laccase protein similar to the blue laccase (GeneBank No. CAA06291) from P. ostreatus [31]. Lacc2 and Lacc7~Lacc11 were not significantly expressed under these conditions.
Heterologous expression of laccase genes in Pichia pastoris
Data from the transcriptome analysis indicated that Lacc5 and Lacc12 were the main laccases active during fruiting body maturation and primordiation stages, respectively, whereas Lacc2, Lacc8, and Lacc11 were not expressed at any stage (Fig. 2C); in addition, Lacc2 and Lacc11 contained unusual active site arrangements (Fig. 2A). Lacc6, which was ubiquitously expressed, is the most studied laccase in P. ostreatus [31]. Moreover, these laccases were scattered among the three phylogenetic groups: Lacc4, Lacc6, and Lacc11 in group I; Lacc12 in group II; and Lacc2 and Lacc5 in group III. Therefore, theses laccases were selected for further characterization. We constructed laccase expression vectors by cloning the six synthesized laccase genes adjacent to the AOX1 promoter in the Pichia expression vector pPICHOLI-2. Next, culture conditions for the production of laccase protein in P. pastoris GS115, using Lacc6 as a model gene, were investigated. In this system, methanol serves as the carbon source for yeast cell growth, as well as the inducer for gene expression. However, the methanol concentration needed to be optimized, as methanol exerts toxic effects on the cells on reaching a critical concentration in the culture medium.
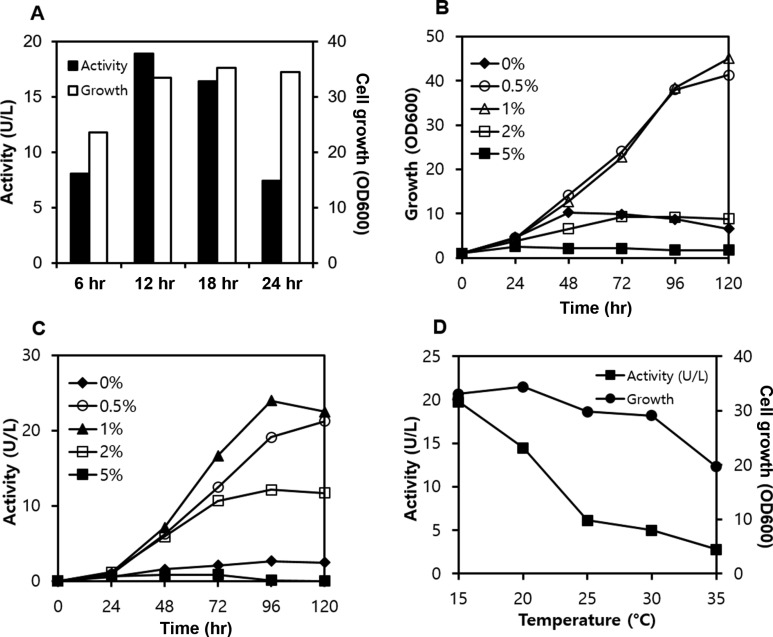
In order to overproduce mushroom laccase in P. pastoris harboring pPICHOLI-Lacc6, the effects of methanol feeding frequency, in terms of cell growth and extracellular laccase activity, were investigated in batch culture. When 1% methanol was fed every 6 hr, both cell growth and production of the laccase protein decreased due to methanol toxicity (Fig. 3A). Cell growth was maintained at high levels at a feeding interval of 12~24 hr, whereas extracellular laccase production decreased significantly when methanol was fed every 24 hr. Extracellular laccase activity and cell growth reached maximal levels when methanol was fed at 12-hr intervals. Next, the effects of methanol concentration were examined by feeding methanol at different concentrations every 12 hr. Cell growth and laccase activity approached maximum levels after 120 hr of incubation (Fig. 3B and 3C). Methanol was found to inhibit both cell growth and laccase production at concentration of ≥ 2%. The optimal concentration ranged from 0.5% to 1% (Fig. 3B and 3C).
Fig. 3. Optimization of culture conditions for expression of laccase genes in Pichia pastoris. A, Optimal methanol feeding interval; methanol (1%) was fed intermittently at the time intervals indicated; B, C, Optimal methanol concentration for cell growth and laccase production; D, Effect of culture temperature on cell growth and production of extracellular laccase protein; yeast cells were grown for 72 hr at specific temperatures with intermittent addition of 1% methanol every 12 hr.
Heterologous proteins are generally expressed at 30℃ in P. pastoris while numerous studies have shown that temperature is one of the important variables in the Pichia expression system [32,33,34]. Accordingly, the effects of culture temperature on enzyme production were examined in the present study. Yeast cells grew to optical densities of 30~35 at 600 nm (OD600) at temperatures ranging from 15℃ to 30℃ (Fig. 3D), with longer lag times at 15℃ (data not shown). Cell growth was significantly inhibited at temperatures higher than 35℃. Extracellular laccase activity in the culture broth was also temperature-dependent; laccase activity decreased with increasing culture temperature (Fig. 3D). Cells grown at 20℃ produced more laccase during earlier stages of the culture; however, the laccase productivity was overtaken by cells grown at 15℃ when the culture time exceeded 48 hr (data not shown). Laccase activity remained low at temperatures higher than 25℃ (Fig. 3D). The requirement for low temperatures may be attributed to the instability of laccase protein at elevated temperatures.
Biochemical characterization of P. ostreatus laccase enzymes
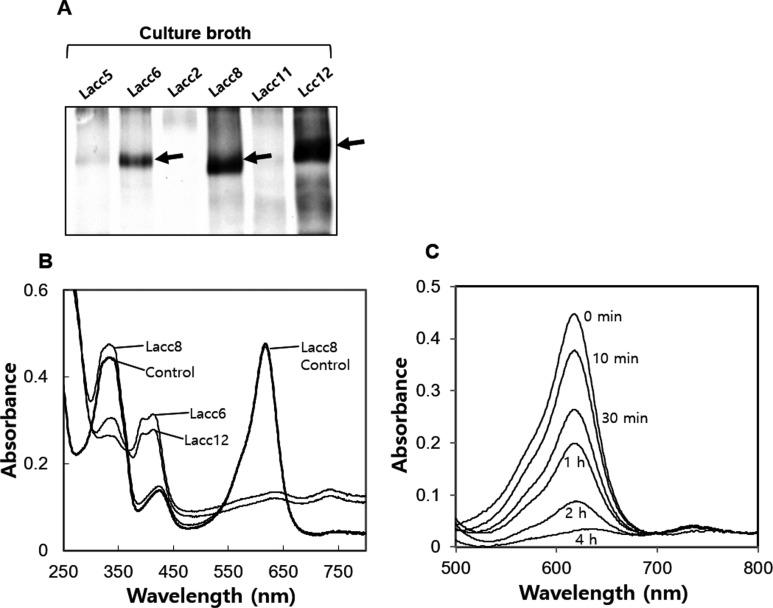
P. ostreatus laccase enzymes were produced using optimized culture conditions (15℃, and 1% methanol fed at 12-hr intervals) in P. pastoris. Lacc6, Lacc8, and Lacc12 were observed as major protein bands, whereas Lacc2, Lacc5, and Lacc11 were not detected or detected as smaller and minor protein bands (Fig. 4A). Notably, Lacc2 and Lacc11 encode defective proteins that lack HXHG and contain incomplete HCH signatures, respectively (Fig. 2A). Laccase activity was assessed by decolorization of malachite green, as described in our previous report [4]. As shown in Fig. 4B, Lacc6 and Lacc12 induced complete decolorization of malachite green, as indicated by the disappearance of the maximum absorbance peak at 622 nm, in a timedependent manner (Fig. 4C). Lacc2, Lacc5, Lacc8, and Lacc11 did not decolorize malachite green (Fig. 4B). Despite its high concentration, Lacc8 was non-functional (Fig. 4B). Additionally, the Lacc8 gene was not expressed during any stage of mushroom development (Fig. 2C), suggesting that Lacc8, which is closely related to Lacc6 (Fig. 1), appears to be in the degenerate state at the level of both transcription and translation.
Fig. 4. Extracellular production of laccase protein by Pichia pastoris harboring laccase genes. A, SDS-PAGE analysis of culture supernatant; arrows indicate the electrophoretic position corresponding to laccase protein; B, Decolorization of malachite green by culture supernatant containing laccase proteins; C, Time-dependent decolorization of malachite green by Lacc12.
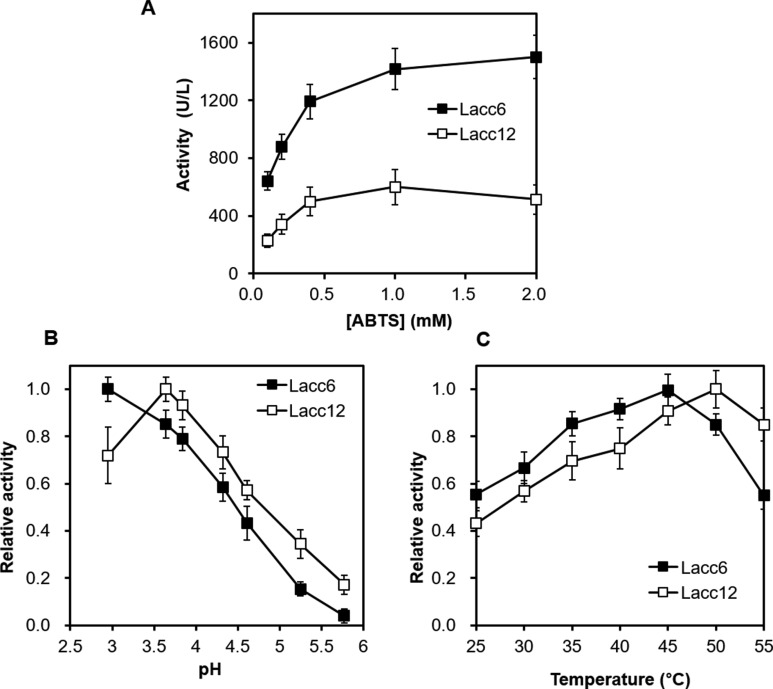
We next investigated the biochemical characteristics of two functional laccase enzymes, Lacc6 and Lacc12. The oxidation activity of laccase was measured using various concentrations of ABTS. The activity increased proportionally to ABTS concentration, and became saturated at above 2 mM, showing maximum activities of 1,560 units/L and 600 units/L for Lacc6 and Lacc12, respectively, and Km values of 0.152 mM and 0.316 mM for Lacc6 and Lacc12, respectively (Fig. 5A). Vmax/Km, which represents the enzyme catalytic activity at low substrate concentration, was 10,263 units/L/mM and 1,898 units/L/mM for Lacc6 and Lacc12, respectively. These findings indicate that Lacc6 is 5.4-fold more active than Lacc12 at low substrate concentration. Both enzymes were highly active at acidic pH. Notably, Lacc6 showed lower pH optimum than Lacc12 with optimum pH values of < 3.0 and 3.5 for Lacc6 and Lacc12, respectively (Fig. 5B). The effects of temperature on laccase activity were also slightly different. Specifically, Lacc12 showed a 5℃ higher temperature optimum than Lacc6; however, both enzymes were essentially thermophilic, with optimum temperatures of 45℃ and 50℃ for Lacc6 and Lacc12, respectively (Fig. 5C).
Fig. 5. Biochemical characteristics of Lacc6 and Lacc12. A, Activity vs. substrate concentration profile; B, Effect of pH on laccase activity; C, Effect of temperature on laccase activity. ABTS, 2,2-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid).
DISCUSSION
The genome of P. ostreatus contains 12 laccase genes clustered into three major groups (Fig. 1). Lacc2 and Lacc10 have been shown to be overexpressed in the mycelia of P. ostreatus in submerged culture when induced by wheat straw extract in a chemically defined medium [24]. However, the present study showed that expression of the two laccases was negligible in sawdust medium at all developmental stages (Fig. 2C), indicating that P. ostreatus expresses different laccase enzymes depending on the medium conditions. Lacc1 and Lacc3 were specific to the mycelial stage in solid medium (Fig. 2C). Lacc6 was highly expressed in all developmental stages whereas Lacc5 and Lacc12 were specific for fruiting bodies and primordia, respectively (Fig. 2C). Lacc6 was characterized as a blue laccase under the name POXA1b [31].
A comparative study of the enzymatic characteristics of the primordium-specific laccase Lacc12 and the constitutive laccase Lacc6 showed that the ABTS oxidation activity of Lacc12 was 40% that of Lacc6. Moreover, the optimal pH and temperature for Lacc12 activity were 0.5 pH units and 5℃ higher than those for Lacc6 (Fig. 5B and 5C). These findings may reflect the physiological conditions during the primordiation stage. In L. edodes, laccase activity in the sawdust substrate decreased dramatically at the primordial stage, along with a sharp increase in cellulase activity [35]. These findings suggest that laccase activity is necessary for the vegetative growth of mycelia in the sawdust substrate and potentially for lignin degradation. Degradation of lignin or production of lignin-derived compounds may induce primordiation. Upon primordiation, laccase activity in the substrate is no longer required. Instead, activity within primordia is necessary for the completion of primordiation, with cellulases of the mycelia beneath the primordia digesting the cellulosic substrate to provide a source of carbon and energy. Volvariella volvacea shows a high level of expression of a laccase gene, lac1, as well as high laccase activity, particularly during the primordial stage [36]. Taken together, these findings indicate that different laccases function at various stages of development as well as in lignin degradation.
ACKNOWLEDGEMENTS
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ00999304)", Rural Development Administration, Republic of Korea. MP, MSK, SIK, and BSH were supported by a scholarship from the BK 21 Plus Program, the Ministry of Education, Korea.
References
- 1.Wang Y, Chantreau M, Sibout R, Hawkins S. Plant cell wall lignification and monolignol metabolism. Front Plant Sci. 2013;4:220. doi: 10.3389/fpls.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soares A, Jonasson K, Terrazas E, Guieysse B, Mattiasson B. The ability of white-rot fungi to degrade the endocrinedisrupting compound nonylphenol. Appl Microbiol Biotechnol. 2005;66:719–725. doi: 10.1007/s00253-004-1747-7. [DOI] [PubMed] [Google Scholar]
- 3.Majcherczyk A, Johannes C, Hüttermann A. Oxidation of polycyclic aromatic hydrocarbons (PAH) by laccase of Trametes versicolor. Enzyme Microb Technol. 1998;22:335–341. [Google Scholar]
- 4.Park JW, Kang HW, Ha BS, Kim SI, Kim S, Ro HS. Strain-dependent response to Cu2+ in the expression of laccase in Pycnoporus coccineus. Arch Microbiol. 2015;197:589–596. doi: 10.1007/s00203-015-1090-7. [DOI] [PubMed] [Google Scholar]
- 5.Kang HW, Yang YH, Kim SW, Kim S, Ro HS. Decolorization of triphenylmethane dyes by wild mushrooms. Biotechnol Bioprocess Eng. 2014;19:519–525. [Google Scholar]
- 6.Reid ID. Biological pulping in paper manufacture. Trends Biotechnol. 1991;9:262–265. [Google Scholar]
- 7.Muller M, Shi C. Laccase for denim processing. AATCC Rev. 2001;1:4–5. [Google Scholar]
- 8.Riva S. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 2006;24:219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Chawla S, Rawal R, Shabnam, Kuhad RC, Pundir CS. An amperometric polyphenol biosensor based on laccase immobilized on epoxy resin membrane. Anal Methods. 2011;3:709–714. doi: 10.1039/c0ay00679c. [DOI] [PubMed] [Google Scholar]
- 10.Minson M, Meredith MT, Shrier A, Giroud F, Hickey D, Glatzhofer DT, Minteer SD. High performance glucose/O2 biofuel cell: effect of utilizing purified laccase with anthracenemodified multi-walled carbon nanotubes. J Electrochem Soc. 2012;159:G166–G170. [Google Scholar]
- 11.Nagai M, Kawata M, Watanabe H, Ogawa M, Saito K, Takesawa T, Kanda K, Sato T. Important role of fungal intracellular laccase for melanin synthesis: purification and characterization of an intracellular laccase from Lentinula edodes fruit bodies. Microbiology. 2003;149(Pt 9):2455–2462. doi: 10.1099/mic.0.26414-0. [DOI] [PubMed] [Google Scholar]
- 12.Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol. 2003;38:143–158. doi: 10.1016/s1087-1845(02)00526-1. [DOI] [PubMed] [Google Scholar]
- 13.Eggert C, Temp U, Dean JF, Eriksson KE. Laccase-mediated formation of the phenoxazinone derivative, cinnabarinic acid. FEBS Lett. 1995;376:202–206. doi: 10.1016/0014-5793(95)01274-9. [DOI] [PubMed] [Google Scholar]
- 14.Baldrian P. Increase of laccase activity during interspecific interactions of white-rot fungi. FEMS Microbiol Ecol. 2004;50:245–253. doi: 10.1016/j.femsec.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Alberts JF, Gelderblom WC, Botha A, Van Zyl WH. Degradation of aflatoxin B1 by fungal laccase enzymes. Int J Food Microbiol. 2009;135:47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Kües U, Rühl M. Multiple multi-copper oxidase gene families in basidiomycetes: what for? Curr Genomics. 2011;12:72–94. doi: 10.2174/138920211795564377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 18.Dekker RF, Barbosa AM, Giese EC, Godoy SD, Covizzi LG. Influence of nutrients on enhancing laccase production by Botryosphaeria rhodina MAMB-05. Int Microbiol. 2007;10:177–185. [PubMed] [Google Scholar]
- 19.Garcia TA, Santiago MF, Ulhoa CJ. Properties of laccases produced by Pycnoporus sanguineus induced by 2,5-xylidine. Biotechnol Lett. 2006;28:633–636. doi: 10.1007/s10529-006-0026-3. [DOI] [PubMed] [Google Scholar]
- 20.Tong P, Hong Y, Xiao Y, Zhang M, Tu X, Cui T. High production of laccase by a new basidiomycete, Trametes sp. Biotechnol Lett. 2007;29:295–301. doi: 10.1007/s10529-006-9241-1. [DOI] [PubMed] [Google Scholar]
- 21.Saparrat M, Balatti PA, Martinez MJ, Jurado M. Differential regulation of laccase gene expression in Coriolopsis rigida LPSC No. 232. Fungal Biol. 2010;114:999–1006. doi: 10.1016/j.funbio.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Eugenio ME, Carbajo JM, Martín JA, González AE, Villar JC. Laccase production by Pycnoporus sanguineus under different culture conditions. J Basic Microbiol. 2009;49:433–440. doi: 10.1002/jobm.200800347. [DOI] [PubMed] [Google Scholar]
- 23.Mansur M, Arias ME, Copa-Patiño JL, Flärdh M, González AE. The white-rot fungus Pleurotus ostreatus secretes laccase isozymes with different substrate specificities. Mycologia. 2003;95:1013–1020. doi: 10.1080/15572536.2004.11833017. [DOI] [PubMed] [Google Scholar]
- 24.Castanera R, Pérez G, Omarini A, Alfaro M, Pisabarro AG, Faraco V, Amore A, Ramírez L. Transcriptional and enzymatic profiling of Pleurotus ostreatus laccase genes in submerged and solid-state fermentation cultures. Appl Environ Microbiol. 2012;78:4037–4045. doi: 10.1128/AEM.07880-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmieri G, Cennamo G, Faraco V, Amoresano A, Sannia G, Giardina P. Atypical laccase isoenzymes from copper supplemented Pleurotus ostreatus cultures. Enzyme Microb Technol. 2003;33:220–230. [Google Scholar]
- 26.Giardina P, Autore F, Faraco V, Festa G, Palmieri G, Piscitelli A, Sannia G. Structural characterization of heterodimeric laccases from Pleurotus ostreatus. Appl Microbiol Biotechnol. 2007;75:1293–1300. doi: 10.1007/s00253-007-0954-4. [DOI] [PubMed] [Google Scholar]
- 27.Palmieri G, Giardina P, Marzullo L, Desiderio B, Nitti G, Cannio R, Sannia G. Stability and activity of a phenol oxidase from the ligninolytic fungus Pleurotus ostreatus. Appl Microbiol Biotechnol. 1993;39:632–636. doi: 10.1007/BF00205066. [DOI] [PubMed] [Google Scholar]
- 28.Giardina P, Aurilia V, Cannio R, Marzullo L, Amoresano A, Siciliano R, Pucci P, Sannia G. The gene, protein and glycan structures of laccase from Pleurotus ostreatus. Eur J Biochem. 1996;235:508–515. doi: 10.1111/j.1432-1033.1996.00508.x. [DOI] [PubMed] [Google Scholar]
- 29.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi J, Cheong K, Jung K, Jeon J, Lee GW, Kang S, Kim S, Lee YW, Lee YH. CFGP 2.0: a versatile web-based platform for supporting comparative and evolutionary genomics of fungi and Oomycetes. Nucleic Acids Res. 2013;41:D714–D719. doi: 10.1093/nar/gks1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giardina P, Palmieri G, Scaloni A, Fontanella B, Faraco V, Cennamo G, Sannia G. Protein and gene structure of a blue laccase from Pleurotus ostreatus. Biochem J. 1999;341(Pt 3):655–663. [PMC free article] [PubMed] [Google Scholar]
- 32.Hong F, Meinander NQ, Jönsson LJ. Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol Bioeng. 2002;79:438–449. doi: 10.1002/bit.10297. [DOI] [PubMed] [Google Scholar]
- 33.Gasser B, Maurer M, Rautio J, Sauer M, Bhattacharyya A, Saloheimo M, Penttilä M, Mattanovich D. Monitoring of transcriptional regulation in Pichia pastoris under protein production conditions. BMC Genomics. 2007;8:179. doi: 10.1186/1471-2164-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P, Anumanthan A, Gao XG, Ilangovan K, Suzara VV, Düzgüneş N, Renugopalakrishnan V. Expression of recombinant proteins in Pichia pastoris. Appl Biochem Biotechnol. 2007;142:105–124. doi: 10.1007/s12010-007-0003-x. [DOI] [PubMed] [Google Scholar]
- 35.Ohga S, Royse DJ. Transcriptional regulation of laccase and cellulase genes during growth and fruiting of Lentinula edodes on supplemented sawdust. FEMS Microbiol Lett. 2001;201:111–115. doi: 10.1111/j.1574-6968.2001.tb10741.x. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, Ge W, Buswell JA. Biochemical and molecular characterization of a laccase from the edible straw mushroom, Volvariella volvacea. Eur J Biochem. 2004;271:318–328. doi: 10.1046/j.1432-1033.2003.03930.x. [DOI] [PubMed] [Google Scholar]