Abstract
Arabidopsis LEAFY COTYLEDON1 (LEC1) transcription factor is a master regulator that shapes plant embryo development and post-embryonic seedling establishment. Loss-of-function of LEC1 alters the cotyledon identity, causing the formation of ectopic trichomes, which does not occur in wild-type seedlings, implying that LEC1 might regulate embryonic cell fate determination during post-embryonic development. To test this hypothesis, we compared the expression of trichome development-related genes between the wild-type and the lec1 mutant. We observed that transcripts of GLABROUS1 (GL1), GL2, and GL3, genes encoding the positive regulators in trichome development, were significantly upregulated, while the TRICHOMELESS1 (TCL2), ENHANCER OF TRY AND CPC1 (ETC1), and ETC2 genes, encoding the negative regulators in trichome development, were downregulated in the lec1 mutant. Furthermore, overexpression of LEC1 activated the expressions of TCL2, CAPPICE (CPC), and ETC1, resulting in production of cotyledonary leaves with no or fewer trichomes during vegetative development. In addition, we demonstrated that LEC1 interacts with TCL2 in yeast and in vitro. A genetic experiment showed that loss-of-function of GL2 rescued the ectopic trichome formation in the lec1 mutant. These findings strongly support that LEC1 regulates trichome development, providing direct evidence for the role of LEC1 in cell fate determination during post-embryonic development.
Keywords: Arabidopsis, LEAFY COTYLEDON1, trichome, cell fate determination, post-embryonic development
Introduction
In higher plants, embryogenesis generally terminates with a dormancy period for future sporophyte growth. Once conditions are favorable, seeds germinate, and undergo post-embryonic development, during which cells acquire specific fates. Extensive studies have shown that the specific cell fate determination during post-embryonic development is precisely controlled by multiple transcription factors (Peris et al., 2010; Perianez-Rodriguez et al., 2014). The plant Nuclear Factor Y (NF-Y) transcription factor LEAFY COTYLEDON1 (LEC1), a master regulator controlling embryogenesis (Meinke, 1992; West et al., 1994; Harada, 2001; Lee et al., 2003), was recently revealed to play a potential role in regulating post-embryonic development (Junker and Baumlein, 2012). LEC1 is expressed in both developing embryo and post-embryonic seedlings, but not in the vegetative true leaves (Kwong et al., 2003; Warpeha et al., 2007; Le et al., 2010). Loss-of-function of LEC1 causes a pleiotropic phenotype, including embryo desiccation intolerance and defects in the accumulation of seed storage compounds in developing seeds, as well as cotyledon with trichomes, premature development of vascular tissue and mesophyll cells, and short hypocotyls in early seedlings (Meinke, 1992; Meinke et al., 1994; West et al., 1994; Brocard-Gifford et al., 2003). By contrast, ectopic expression LEC1 allows seedlings to remain in the embryonic state with an unexpanded cotyledon after germination and is sufficient to induce conversion of true leaves into embryonic structures that lack trichomes (Lotan et al., 1998; Junker et al., 2012). These findings support the multifunctional roles of LEC1 in embryonic and post-embryonic development. Trichomes originate from the epidermal cell layer whose nuclei have undergone multiple rounds of endoreduplication, which then enlarge and expand away from the surface (Hulskamp et al., 1994; Pattanaik et al., 2014). The regulation of trichome formation has been well documented in many studies. The R2R3 MYB transcription factor GLABROUS1 (GL1) was the first identified positive regulator involved in trichome development (Marks and Feldmann, 1989; Oppenheimer et al., 1991). GL1 interacts with the bHLH transcription factor GLABRA3 (GL3), its close homolog ENHANCER OF GLABRA3 (EGL3), and the WD40-repeat factor TRANSPARENT TESTA GLABRA1 (TTG1), resulting in the formation of the MYB-bHLH-WD-repeat trichome-positive transcription complex (Zhao et al., 2008). This protein complex subsequently induces trichome formation by activating the expression of the downstream gene GLABRA2 (GL2), encoding a homeodomain-leucine zipper (HD-Zip) transcription factor (Rerie et al., 1994; Morohashi et al., 2007). Contrastingly, a small single-repeat R3 MYB protein family, including TRICHOMELESS1 (TCL1), TCL2, TRIPTYCHON (TRY), CAPPICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), and ETC2, plays redundant roles in the negative regulation of trichome development. These proteins compete with GL1 for binding to GL3/EGL3, resulting in the formation of an inactive protein complex that fails to activate GL2 expression and thus represses trichome formation (Wang and Chen, 2014; Zhou et al., 2014).
Trichome formation is considered a specific characteristic that distinguishes true leaves from cotyledons in Arabidopsis (Chandler, 2008). A hypothesis for the ectopic trichome formation on the cotyledons of the lec1 mutant is that the loss-of-function of LEC1 might result in the disturbance of embryonic cell fate determination, allowing the precocious post-germination events in which cotyledons are partially converted into true leaves (West et al., 1994; Chandler, 2008). However, the mechanism by which LEC1 functions in cell fate determination during post-embryonic development remains elusive.
Given that LEC1 mutation causes remarkable ectopic trichome growth on cotyledons, the trichome-related genes might serve as good candidates to study the function of LEC1 in the post-embryonic development phase. Here, we demonstrated that LEC1 represses trichome cell differentiation during post-embryonic development by regulating trichome-related genes. In addition, LEC1 was shown to interact with TCL2, a repressor of trichome development, which indicated that LEC1 might couple with other transcription factors to co-regulate trichome formation. A genetic experiment showed that loss-of-function of GL2 rescued the ectopic trichome development phenotype of the lec1 mutant. These results strongly supported the view that LEC1 functions in cell fate determination during the post-embryonic phase, providing new insights into LEC1’s role beyond the embryogenesis.
Materials and Methods
Plant Materials and Growth Conditions
All Arabidopsis plants used in this study are in the Col genetic background. The mutant gl2 was described previously (Wang et al., 2010). The mutant lec1-4 (Salk_095699) was obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). The estradiol-inducible pER10:LEC1-MYC transgenic line was obtained by kanamycin selection after transformation (Mu et al., 2008). The double mutant was generated by genetic crossing. After surface sterilization, seeds were sown on the half-strength Murashige and Skoog medium containing 0.8% agar and incubated at 4°C in the dark for 3 days. The seedlings were then transferred to the growth chamber at 22°C under long day conditions (16 h light/8 h dark). For estradiol treatment, 10 μM estradiol was added to the half-strength Murashige and Skoog medium plate and dimethyl sulfoxide, the solvent for estradiol, served as the mock treatment. After treatment, these seedlings were harvested for further analyses.
Microscopy
The formation of trichomes on the cotyledons and leaves was observed and photographed using a LEICA M165C stereoscope (Leica, Wetzlar, Germany).
Statistical Analysis
Five-day-old seedlings were observed to calculate frequency of the appearance of ectopic trichomes on cotyledons. At least 15 Col and 50 lec1-4 seedlings were used in each experiment. Data represent mean ± SD of three independent repeats. To calculate the frequency of different trichome types on lec1-4 cotyledons, at least 50 trichomes were analyzed in each experiment. Data represent the mean ± SD of seven independent repeats.
Gene Expression Analysis
Total RNA was extracted from 5-day-old Arabidopsis seedlings using the Plant RNA Kit (OMEGA, Atlanta, GA, USA), following the manufacturer’s instructions. One microgram of total RNA was used for the reverse transcription reaction. The first-strand cDNA was synthesized using the M-MLV reverse transcriptase (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Quantitative real-time PCR was performed using the LightCycler480 system (Roche, Basel, Switzerland) in a total volume of 10 μl, with 0.25 μl of each primer (10 μM), 1 μl of cDNA product and 5 μl of SYBR Premix ExTaq (Takara, Tokyo, Japan). The PCR program included an initial denaturation step at 94°C for 1 min, followed by 40 cycles of 10 s at 94°C and 1 min at 60°C. Each sample was quantified at least in triplicate and normalized using the TUB2 gene as the internal control. Primers used in this study are listed in Supplementary Table S1.
Yeast Two-Hybrid Assay
The full-length coding sequences of LEC1 and TCL2 were amplified and cloned into vectors pGBKT7 and pGADT7 (Clontech, Palo Alto, CA, USA), respectively. Primers are listed in Supplementary Table S1. Yeast two-hybrid assays were performed following the manufacturer’s instructions of the Yeastmaker Yeast Transformation System 2 (Clontech). In brief, a single colony of yeast AH109 was incubated at 30°C overnight (OD600 > 1.0). The cells were harvested by centrifugation and then resuspended in 25 mL ddH2O. After re-harvesting the cells by centrifugation, a 1.5-ml sterile 1× Tris-EDTA/LiAc solution was added to prepare yeast-competent cells. For the yeast two-hybrid assay, the bait (0.5 μg) and/or prey (0.5 μg) plasmids with 0.1 mg of carrier DNA were co-transformed into the yeast-competent cells using Polyethylene glycol/LiAc solution. After incubation at 30°C for 30 min, 70 μL dimethyl sulfoxide was added and incubation continued at 42°C for 15 min. The cells were centrifuged and washed using Tris-EDTA solution. After transformation, the yeast cells were grown on SD/-Trp/-Leu/-His/-Ade medium for the interaction test.
Pull-Down Assay
To produce glutathione-S-transferase (GST)-TCL1, GST-TCL2, GST-CPC, GST-ETC1, His-LEC1, and His-GFP recombinant proteins, the full-length coding sequences of the tested genes were cloned into vector pGEX-4T-1 (Pharmacia, Piscataway, NJ, USA) or pQE30 (QIAGEN, Dusseldorf, Germany), respectively. Primers are listed in Supplementary Table S1. These constructs were transformed into the Escherichia coli Rosetta strain DE3 (Novagen, Billerica, MA, USA). One hundred milliliters of (OD600 ≈ 0.5) Rosetta strains harboring various vectors were incubated at 16°C for 16 h with 0.1 mM isopropyl β-D-1-thiogalactopyranoside. The soluble GST fusion proteins were purified using glutathione sepharose beads (Amersham Biosciences, Piscataway, NJ, USA), while His fusion proteins were purified using Ni- nitrilotriacetic acid agarose beads (QIAGEN), according to the manufacturer’s instructions, respectively. For pull-down assays, 2 μg of His-LEC1 or His-GFP were incubated with immobilized GST or GST fusion proteins in the binding buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA) at 4°C for 4 h. After washing with binding buffer three times, proteins retained on the beads were subsequently resolved by sodium dodecyl sulfate polyacrylamide electrophoresis and detected with an anti-His antibody (GBI, Beijing, China).
Results
Ectopic Trichome Formation on the Cotyledons of the lec1 Mutant
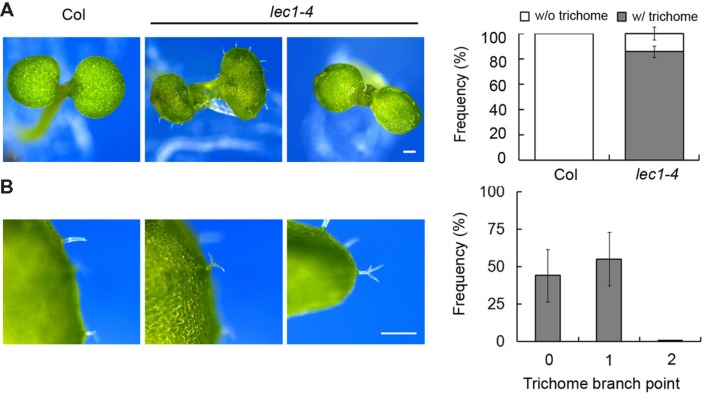
Extensive studies have shown that LEC1 serves as a key regulator in embryo development, which is consistent with its main expression in the developing seed (Kwong et al., 2003). However, recent observations suggested a potential role of LEC1 in post-embryonic cell differentiation, including hypocotyl elongation and the formation of vascular tissue, mesophyll cells and trichomes (Warpeha et al., 2007; Junker and Baumlein, 2012; Junker et al., 2012; Supplementary Figure S1A). Serving as a general model system in the study of cell fate determination in Arabidopsis (Yang and Ye, 2013), we focused on trichome formation to reveal the function of LEC1 in post-embryonic development. A desiccation-tolerant leaky allele lec1-4 whose mature seeds germinate normally (unpublished data) was used in this study. Consistent with previous reports, the wild-type cotyledons were glabrous, while ∼85% of lec1 cotyledons produced a varied number of ectopic trichomes (Figure 1A; Supplementary Table S2). Moreover, unlike the mostly three-branched trichomes developed on true leaves, the trichomes on lec1 cotyledons were mainly unbranched (∼44%) or two-branched (∼55%), and rarely three-branched (less than 1%; Figure 1B; Supplementary Table S3). These observations implied incomplete conversion of the lec1 cotyledons into true leaves. In addition, there was no significant difference observed in trichome numbers or the morphology of rosette leaves between the wild-type and lec1, which is consistent with the observation that no LEC1 transcripts accumulated in the vegetative tissues (Supplementary Figures S1A,B).
FIGURE 1.
Ectopic trichome formation on the lec1-4 mutant cotyledons. (A) Comparison of 5-day-old Col wild-type and lec1-4 mutant cotyledons. Bar = 0.5 mm. The right panel indicates the frequency of seedlings with (w/) or without (w/o) ectopic trichomes on their cotyledons. (B) Light micrographs of three types of trichome branch on lec1-4 mutant cotyledons. Bar = 0.5 mm. The right panel indicates the frequency of different types of trichomes. 0 indicates no branch on a trichome; 1 indicates that the trichome has two branches; 2 indicates a trichome with three branches.
Misregulation of Trichome-Related Genes in the lec1 Mutant
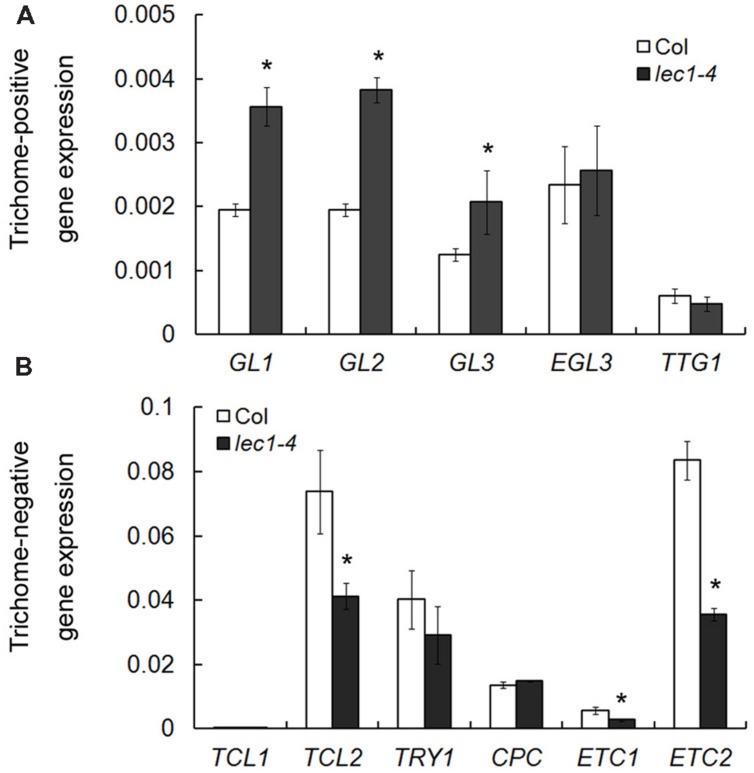
Trichome formation on leaves has been well studied during the past two decades and a number of trichome-related genes were identified, which could be divided into two groups: trichome-positive and -negative genes (Yang and Ye, 2013). To examine whether LEC1’s involvement in the inhibition of trichome formation on cotyledons occurs by mediating the regulation of trichome-related genes, five trichome-positive genes, and six trichome-negative genes were selected for quantitative real-time PCR analysis in wild-type and lec1 seedlings. Interestingly, among the trichome-positive genes, the expressions of GL1, GL2, and GL3 were significantly upregulated in the lec1 mutant compared with the wild-type, while those of EGL3 and TTG1 showed no change (Figure 2A). Moreover, the expressions of the trichome-negative genes TCL2, ETC1, and ETC2 were significantly downregulated in the lec1 mutant compared with the wild-type, while those of TRY and CPC showed little change. The TCL1 transcript was hardly detected in either the wild-type or lec1 seedlings (Figure 2B). These results indicated that the ectopic trichome formation might be caused by misregulation of the trichome-related genes in the lec1 mutant.
FIGURE 2.
Misregulation of trichome-related genes in the lec1-4 mutant seedlings. (A) The expression analysis of trichome-positive genes in 5-day-old Col wild-type and lec1-4 mutant seedlings. (B) The expression analysis of trichome-negative genes in 5-day-old Col wild-type and lec1-4 mutant seedlings. TUB2 was used as an internal control. Asterisks indicate significant differences between Col and lec1-4 mutant (p < 0.05, by Student’s t-test).
Overexpression of LEC1 Activates Several Trichome-Negative Transcription Factors
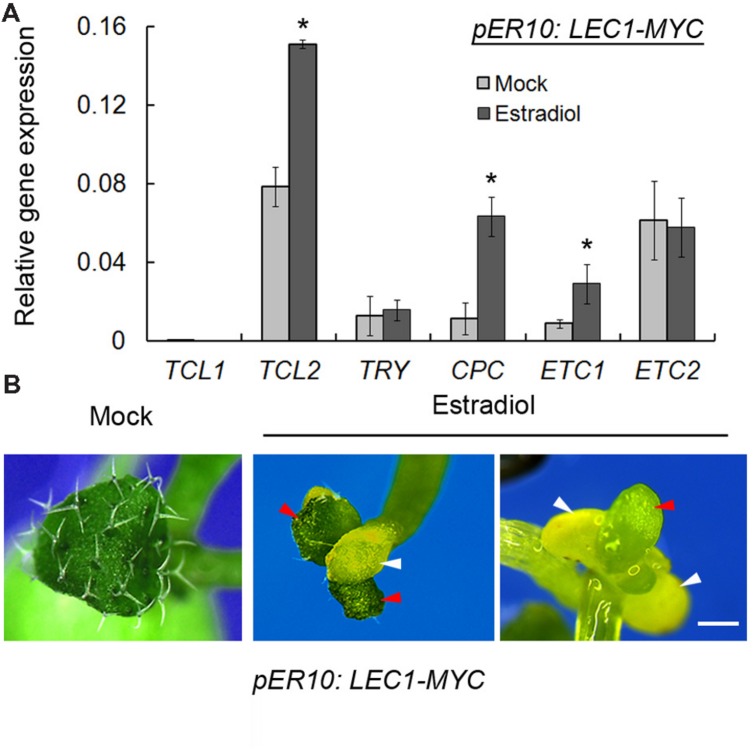
Given that loss-of-function of LEC1 resulted in misregulation of trichome-related genes, we wondered whether LEC1 overexpression could activate or repress these genes. LEC1 overexpression, driven by the 35S promoter, caused complex phenotypes and embryonic lethality (Lotan et al., 1998). Thus, instead, we employed an inducible transgenic plant harboring pER10:LEC1-MYC, in which LEC1 expression could be significantly induced by estradiol (Mu et al., 2008; Supplementary Figure S2). Unexpectedly, the trichome-positive genes GL1, GL2, GL3, EGL3, and TTG1 were not regulated by induced LEC1 (data not shown). Nevertheless, overexpression of LEC1 activated the expressions of TCL2, CPC, and ETC1, but had no effect on those of TCL1, TRY, and ETC2 (Figure 3A), which revealed the role of LEC1 in the regulation of trichome-negative genes. A previous study reported that TCL2, CPC, and ETC1 overexpression lines all presented a glabrous leaves phenotype (Wang and Chen, 2014); therefore, we asked whether ectopic LEC1 expression affects trichome formation on true leaves. The investigation demonstrated that cotyledons of pER10:LEC1-MYC under either mock or estradiol treatment displayed a similar glabrous phenotype to the wild-type (data not shown). Strikingly, under long-term induction by estradiol (14 days), the pER10:LEC1-MYC seedlings developed fewer trichomes and cotyledon-like rosette leaves (Figure 3B). Together with a previous report that the single-repeat R3 MYB quintuple mutant try cpc etc1 etc3 tcl1 produced ectopic trichomes on cotyledons similar to the lec1 mutant (Wang et al., 2008), our observations supported the hypothesis that LEC1 suppresses trichome formation via activation of TCL2, CPC, and ETC1 expression.
FIGURE 3.
Induced LEC1 overexpression suppresses trichome formation. (A) The expression analysis of trichome-negative genes in 5-day-old pER10:LEC1-MYC transgenic seedlings with 10 μM estradiol or mock treatment. TUB2 was used as an internal control. Asterisks indicate significant differences between estradiol and mock (p < 0.05, by Student’s t-test). (B) Induced LEC1 overexpression promotes conversion of true leaves into embryonic structures. Fourteen-day-old pER10:LEC1-MYC transgenic seedlings with 10 μM estradiol or mock treatment were used for phenotype observation. Red arrowheads indicate rosette leaves; white arrowheads indicate cotyledons. Bar = 1 mm.
LEC1 Physically Interacts with TCL2 In Vitro and in Yeast
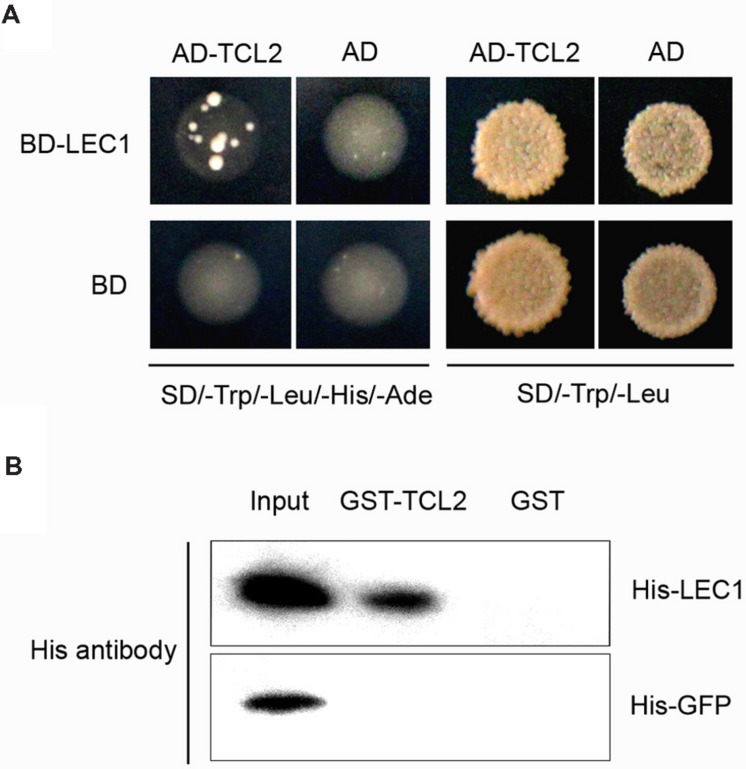
LEAFY COTYLEDON1 belongs to the NF-Y family, whose members interact with other transcription factors to co-regulate target gene expression (Yamamoto et al., 2009). To investigate whether LEC1 couples with cofactors to mediate trichome formation, we performed a yeast two-hybrid screening assay. Interestingly, the trichome transcriptional repressor TCL2 was observed to interact with LEC1 in yeast (Figure 4A). We then performed a pull-down assay to confirm the interaction between LEC1 and TCL2 using purified GST- and His-tagged proteins (Supplementary Figure S3). As expected, GST-TCL2 interacted with His-LEC1, but not with the His-GFP protein, while GST alone did not precipitate either of the His-tagged proteins (Figure 4B). TCL2 and its homologs TCL1, TRY, CPC, ETC1, and ETC2 exert a master role in repressing trichome formation during the whole life of plants; therefore, we further tested the binding between LEC1 and these homologous proteins. The pull-down results showed that LEC1 also interacted with TCL1, CPC, and ETC1 in vitro (Supplementary Figure S4); however, their bindings were not observed in the yeast two-hybrid assay, probably because of weak interactions in yeast (data not shown). These results indicated that, besides regulating the expression of the single-repeat R3 MYB genes, LEC1 might mediate trichome formation via direct protein interactions with these repressors, a result that requires further investigation.
FIGURE 4.
TCL2 physically interacts with LEC1 in yeast and in vitro. (A) Yeast two-hybrid assays showing that TCL2 interacts with LEC1. Transformed yeast cells were grown both on SD/-Trp/-Leu/-His/-Ade and SD/-Trp/-Leu medium. (B) A pull-down assay showing the direct interaction between His-LEC1 and GST-TCL2 fusion proteins in vitro. His-LEC1 or His-GFP protein was incubated with immobilized GST or GST-TCL2 proteins and the immunoprecipitated fractions were detected by an anti-His antibody.
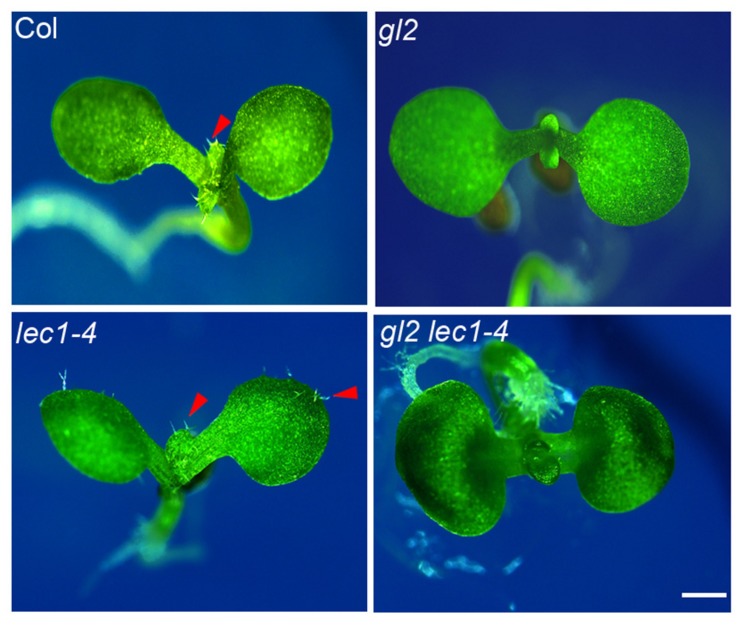
GL2 Genetically Acts Downstream of LEC1
GLABRA2 functions as the key transcription activator in trichome development, whose expression is activated by the trichome-positive MYB-bHLH-WD and repressed by the trichome-negative single-repeat R3 MYB-bHLH-WD transcription complexes (Zhao et al., 2008). Loss-of-function of GL2 completely abolished trichome formation on the first pair of true leaves in early seedlings (Figure 5). The elevated GL2 expression in the lec1 (Figure 2A) implied an epistatic effect of GL2 on LEC1 in trichome formation on cotyledons. To test this hypothesis, we created a gl2 lec1 double mutant and found that similar to gl2, the gl2 lec1 double mutant displayed glabrous true leaves (Figure 5). In contrast, the gl2 mutation repressed the ectopic trichome formation on cotyledons of lec1 seedlings remarkably (Figure 5), confirming the primary role of GL2 in trichome formation, regardless of the developmental stage. These results strongly supported the hypothesis that GL2 is required for LEC1-mediated repression of trichome formation on cotyledons.
FIGURE 5.
Loss-of-function of GL2 rescues the phenotype of ectopic trichome formation on lec1 cotyledons. Ten-day-old Col, gl2, lec1, and gl2 lec1 seedlings were used for the phenotype observation. A red arrowhead indicates the trichomes. Bar = 1 mm.
Discussion
Post-embryonic development is a vital transition state from embryonic status to vegetative growth, during which seedlings sense various environmental cues, such as light and temperature, to enable young sporophyte survival. At this stage, cell fate determination occurs to promote post-embryonic organogenesis that arises from the primary meristems, during which the visible features, such as cotyledons extension, hypocotyl elongation, and the emergence of true leaves with trichomes, can be observed in seedlings. Studies have shown that multiple endogenous factors, including phytohormones and transcription regulators, are involved in post-embryonic seedling development (Perianez-Rodriguez et al., 2014). LEC1, the master regulator in embryogenesis, was recently reported to be involved in light-mediated post-embryonic hypocotyl elongation (Junker and Baumlein, 2012). In this study, we provide molecular evidence that LEC1 and its genetic downstream partner GL2 are required for the repression of trichome formation on embryo-derived cotyledons, the characteristic that distinguishes them from true leaves. Mutation of LEC1 resulted in the misregulation of several trichome-related genes and thus triggered trichome formation on cotyledons. By contrast, ectopic LEC1 activates the expression of the single-repeat R3 MYB genes and induces embryonic leaves with few or no trichomes. Furthermore, the results indicated that LEC1 might regulate trichome formation by interacting with other transcription factors (e.g., TCL2 and its homologs). Taken together, our findings support the hypothesis that LEC1 plays an essential role in cell fate determination during the post-embryonic development.
The fact that LEC1 is expressed in both developing seeds and post-embryonic seedlings, combined with the pleiotropic phenotype of the lec1 mutant, supports the multifunctional roles of LEC1 in plant development (Junker and Baumlein, 2012; Supplementary Figure S1A); however, less is known about the downstream proteins of the LEC1-mediated regulatory process. A recent report indicated that LEC1 might regulate directly a number of genes involved in light, phytohormone signaling, and embryogenesis, including several bHLH and MYB transcription factors (Junker et al., 2012). However, using a chromatin immunoprecipitation assay, we observed that LEC1 does not target the selected trichome-related genes directly (data not shown), indicating that these genes might not act immediately downstream of LEC1. Instead, we observed that LEC1 interacts with the trichome development-related transcription repressors TCL1, TCL2, CPC, and ETC1 in vitro. TCL1 was reported to be highly expressed in developing seeds and its protein targets the GL1 promoter, while TCL2 is highly expressed in cotyledons, rather than in developing seeds, implying distinct roles of these two homologous genes in trichome regulation (Wang et al., 2007; Gan et al., 2011). The interactions between LEC1 and these repressors provide a possible mechanism by which LEC1 regulates embryonic cell fate determination spatio-temporally, via coupling with different transcription factors. Nevertheless, whether LEC1 and TCL2 together determine the post-embryonic trichome initiation requires further investigation.
Phytohormones serve as crucial endogenous integrators to mediate plant development by controlling precisely exact cell division and differentiation (Lumba et al., 2010; Vanstraelen and Benkova, 2012). Gibberellin (GA) has been reported to act as a positive endogenous factor to promote trichome formation, because the GA biosynthesis defective mutant ga1 displays fewer trichomes on its leaves (Perazza et al., 1998; Qi et al., 2014). LEC2 (Stone et al., 2001) and FUSCA3 (FUS3, Keith et al., 1994), two B3 domain-containing transcription factors, also defined as the LEC genes, promote GL1 expression via direct activation of the GA biosynthesis gene, ATGA3ox2. When crossed to ga1, the ectopic trichome formation on lec2 and fus3 cotyledons was completely suppressed (Curaba et al., 2004). However, the ectopic trichomes were not abolished in lec1 ga1 double mutant, which suggested that LEC1-mediated trichome formation is independent of GA (Curaba et al., 2004). Given that LEC2 and FUS3 act as potential downstream candidates of LEC1 (Junker et al., 2010), GA’s effect on the function of LEC factors might be under the control of a complicated regulatory network. Abscisic acid is also involved in LEC1-mediated trichome formation. A previous study reported that the conversion of leaves into cotyledon-like structures is enhanced in the LEC1 overexpression line when coupled with abscisic acid treatment (Junker et al., 2012). In addition, a number of abscisic acid-responsive genes were shown to be the direct targets of LEC1 (Junker et al., 2012). Nevertheless, the role of phytohormones in LEC1-mediated cell fate determination remains elusive and requires further investigation.
Accurate control of LEC1 expression is critical for plants to regulate cell fate determination during development. Epigenetic regulation plays key roles in the exclusion of LEC1 in the vegetative growth stage. PICKLE, a CHD3 chromatin-remodeling factor, represses LEC1 transcriptional activation in vegetative tissue by modifying histone methylation. In the pickle mutant, the seedling root produced an embryonic structure, termed the “PICKLE root”, caused by the ectopic expression of LEC1 (Ogas et al., 1997, 1999; Li et al., 2005). In addition, the polycomb group proteins CLF and SWN were demonstrated to couple with PICKLE to repress the conversion from vegetative tissue to an embryonic structure (Chanvivattana et al., 2004; Aichinger et al., 2009). Histone deacetylases (e.g., HDA6 and HDA19) also contribute to the repression of embryonic properties by regulating several embryogenesis regulators, including LEC1 (Tanaka et al., 2008). In addition to these epigenetic factors, some transcription factors, such as VP1/ABI3-LIKE and MYB115/118, were also reported as negative regulators of LEC1 (Suzuki et al., 2007; Wang et al., 2009; Jia et al., 2013). Embryonic development originates from the zygote after fertilization in higher plants (Lau et al., 2012). High expression during embryogenesis indicates that LEC1 is released from the suppressing environment of the vegetative tissue, thus allowing it to controls embryonic and post-embryonic cell fate determination.
Conclusion
In this study, we revealed that LEC1 interacts with other transcription factors and regulates trichome-related genes expression to repress the trichome formation on cotyledons. These findings provide evidence that supports the hypothesis that LEC1, probably by integrating phytohormone signals and epigenetic regulation, mediates cell fate determination during the post-embryonic development phase, providing new insights into the role of LEC1 beyond embryogenesis.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: LEC1 (AT1G21970), GL1 (AT3G27920), GL2 (AT1G79840), GL3 (AT5G41315), EGL3 (AT1G63650), TCL1 (AT2G30432), TCL2 (AT2G30424), TRY (AT5G53200), CPC (AT2G46410), ETC1 (AT1G01380), ETC2 (AT2G30420), and TUB2 (AT5G62690).
Author Contributions
MH and XH designed the research. MH, YH, XL, and YL performed experiments. MH, YH, and XH analyzed data. MH and XH wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shucai Wang for providing gl2 mutant seeds, the Arabidopsis Biological Resource Centre for lec1-4 seeds, and Jianru Zuo for pER10:LEC1-MYC plasmid.
Footnotes
Funding. This work was supported by grants from the National Science Foundation of China (No. 31370342, 31301055, and 31300239) and “Hundred Talents” program of the Chinese Academy of Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00955
References
- Aichinger E., Villar C. B. R., Farrona S., Reyes J. C., Hennig L., Kohler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5:605 10.1371/journal.pgen.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford I. M., Lynch T. J., Finkelstein R. R. (2003). Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 131 78–92. 10.1104/pp.011916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. W. (2008). Cotyledon organogenesis. J. Exp. Bot. 59 2917–2931. 10.1093/Jxb/Ern167 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y. H., Sung Z. R., et al. (2004). Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development 131 5263–5276. 10.1242/Dev.01400 [DOI] [PubMed] [Google Scholar]
- Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., et al. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136 3660–3669. 10.1104/pp.104.047266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L. J., Xia K., Chen J. G., Wang S. C. (2011). Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 11:176 10.1186/1471-2229-11-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada J. J. (2001). Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J. Plant Physiol. 158 405–409. 10.1078/0176-1617-00351 [DOI] [Google Scholar]
- Hulskamp M., Misera S., Jurgens G. (1994). Genetic dissection of trichome cell-development in Arabidopsis. Cell 76 555–566. 10.1016/0092-8674(94)90118-X [DOI] [PubMed] [Google Scholar]
- Jia H. Y., Mccarty D. R., Suzuki M. (2013). Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 163 1293–1305. 10.1104/pp.113.220988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A., Baumlein H. (2012). Multifunctionality of the LEC1 transcription factor during plant development. Plant Signal. Behav. 7 1718–1720. 10.4161/psb.22365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A., Hartmann A., Schreiber F., Baumlein H. (2010). An engineer’s view on regulation of seed development. Trends Plant Sci. 15 303–307. 10.1016/j.tplants.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Junker A., Monke G., Rutten T., Keilwagen J., Seifert M., Thi T. M., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71 427–442. 10.1111/j.1365-313X.2012.04999.x [DOI] [PubMed] [Google Scholar]
- Keith K., Kraml M., Dengler N. G., Mccourt P. (1994). fusca3 – a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6 589–600. 10.1105/Tpc.6.5.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong R. W., Bui A. Q., Lee H., Kwong L. W., Fischer R. L., Goldberg R. B., et al. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18. 10.1105/tpc.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Slane D., Herud O., Kong J. X., Jurgens G. (2012). Early embryogenesis in flowering plants: setting up the basic body pattern. Annu. Rev. Plant Biol. 63 483–506. 10.1146/annurev-arplant-042811-105507 [DOI] [PubMed] [Google Scholar]
- Le B. H., Cheng C., Bui A. Q., Wagmaister J. A., Henry K. F., Pelletier J., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107 8063–8070. 10.1073/pnas.1003530107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S., Fischer R. L., Goldberg R. B., Harada J. J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 100 2152–2156. 10.1073/pnas.0437909100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. C., Chuang K., Henderson J. T., Rider S. D., Bai Y., Zhang H., et al. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44 1010–1022. 10.1111/j.1365-313X.2005.02602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K. M., West M. A., Lo R., Kwong R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. 10.1016/S0092-8674(00)81463-4 [DOI] [PubMed] [Google Scholar]
- Lumba S., Cutler S., Mccourt P. (2010). Plant nuclear hormone receptors: a role for small molecules in protein-protein interactions. Annu. Rev. Cell Dev. Biol. 26 445–469. 10.1146/annurev-cellbio-100109-103956 [DOI] [PubMed] [Google Scholar]
- Marks M. D., Feldmann K. A. (1989). Trichome development in Arabidopsis-thaliana.1. T-DNA tagging of the Glabrous1 gene. Plant Cell 1 1043–1050. 10.1105/tpc.1.11.1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D. W. (1992). A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258 1647–1650. 10.1126/science.258.5088.1647 [DOI] [PubMed] [Google Scholar]
- Meinke D. W., Franzmann L. H., Nickle T. C., Yeung E. C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064. 10.1105/tpc.6.8.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Zhao M. Z., Yang M. L., Read B., Lloyd A., Lamb R., et al. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145 736–746. 10.1104/pp.107.104521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148 1042–1054. 10.1104/pp.108.126342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Cheng J. C., Sung Z. R., Somerville C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94. 10.1126/science.277.5322.91 [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 96 13839–13844. 10.1073/pnas.96.24.13839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer D. G., Herman P. L., Sivakumaran S., Esch J., Marks M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493. 10.1016/0092-8674(91)90523-2 [DOI] [PubMed] [Google Scholar]
- Pattanaik S., Patra B., Singh S. K., Yuan L. (2014). An overview of the gene regulatory network controlling trichorne development in the model plant, Arabidopsis. Front. Plant Sci. 5:259 10.3389/Fpls.2014.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D., Vachon G., Herzog M. (1998). Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 117 375–383. 10.1104/Pp.117.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perianez-Rodriguez J., Manzano C., Moreno-Risueno M. A. (2014). Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front. Plant Sci. 5:219 10.3389/Fpls.2014.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris C. I. L., Rademacher E. H., Weijers D. (2010). Green beginnings - pattern formation in the early plant embryo. Plant Dev. 91 1–27. 10.1016/S0070-2153(10)91001-6 [DOI] [PubMed] [Google Scholar]
- Qi T. C., Huang H., Wu D. W., Yan J. B., Qi Y. J., Song S. S., et al. (2014). Arabidopsis DELLA and JAZ proteins bind the WD-Repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26 1118–1133. 10.1105/tpc.113.121731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie W. G., Feldmann K. A., Marks M. D. (1994). The Glabra2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8 1388–1399. 10.1101/gad.8.12.1388 [DOI] [PubMed] [Google Scholar]
- Stone S. L., Kwong L. W., Yee K. M., Pelletier J., Lepiniec L., Fischer R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98 11806–11811. 10.1073/pnas.201413498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Wang H. H. Y., Mccarty D. R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143 902–911. 10.1104/pp.106.092320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146 149–161. 10.1104/pp.107.111674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M., Benkova E. (2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28 463–487. 10.1146/annurev-cellbio-101011-155741 [DOI] [PubMed] [Google Scholar]
- Wang S. C., Barron C., Schiefelbein J., Chen J. G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185 387–400. 10.1111/j.1469-8137.2009.03067.x [DOI] [PubMed] [Google Scholar]
- Wang S. C., Chen J. G. (2014). Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 5:133 10.3389/Fpls.2014.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. C., Hubbard L., Chang Y., Guo J. J., Schiefelbein J., Chen J. G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8:81 10.1186/1471-2229-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. C., Kwak S. H., Zeng Q. N., Ellis B. E., Chen X. Y., Schiefelbein J., et al. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134 3873–3882. 10.1242/Dev.009597 [DOI] [PubMed] [Google Scholar]
- Wang X. C., Niu Q. W., Teng C., Li C., Mu J. Y., Chua N. H., et al. (2009). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19 224–235. 10.1038/Cr.2008.276 [DOI] [PubMed] [Google Scholar]
- Warpeha K. M., Upadhyay S., Yeh J., Adamiak J., Hawkins S. I., Lapik Y. R., et al. (2007). The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143 1590–1600. 10.1104/pp.106.089904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Yee K. M., Danao J., Zimmerman J. L., Fischer R. L., Goldberg R. B., et al. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 1731–1745. 10.1105/tpc.6.12.1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58 843–856. 10.1111/j.1365-313X.2009.03817.x [DOI] [PubMed] [Google Scholar]
- Yang C. X., Ye Z. B. (2013). Trichomes as models for studying plant cell differentiation. Cell. Mol. Life Sci. 70 1937–1948. 10.1007/s00018-012-1147-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Morohashi K., Hatlestad G., Grotewold E., Lloyd A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135 1991–1999. 10.1242/Dev.016873 [DOI] [PubMed] [Google Scholar]
- Zhou L. M., Zheng K. J., Wang X. Y., Tian H. N., Wang X. L., Wang S. C. (2014). Control of trichome formation in Arabidopsis by poplar single-repeat R3 MYB transcription factors. Front. Plant Sci. 5:262 10.3389/Fpls.2014.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.