Abstract
With the large-scale release of genetically modified (GM) crops, there are ecological concerns on transgene movement from GM crops to non-GM counterparts and wild relatives. In this research, we conducted greenhouse experiments to measure pollen-mediated gene flow (PGF) in the absence and presence of pollinators (Bombus ignitus, Apis mellifera and Pieris rapae) in one GM cotton (resistant to the insect Helicoverpa armigera and the herbicide glyphosate) and two non-GM lines (Shiyuan321 and Hai7124) during 2012 and 2013. Our results revealed that: (1) PGF varied depending on the pollinator species, and was highest with B. ignitus (10.83%) and lowest with P. rapae (2.71%); (2) PGF with B. ignitus depended on the distance between GM and non-GM cottons; (3) total PGF to Shiyuan321 (8.61%) was higher than to Hai7124 (4.10%). To confirm gene flow, we tested hybrids carrying transgenes for their resistance to glyphosate and H. armigera, and most hybrids showed strong resistance to the herbicide and insect. Our research confirmed that PGF depended on pollinator species, distance between plants and the receptor plant.
Genetic engineering has made it possible to transfer novel genes into plants, thus resulting in the production of genetically modified (GM) varieties conferring resistance to insects1, diseases2, herbicides3 and environmental stress4,5. The steady growth in global plantings of GM crops attests to their advantages for producers6. Despite these advantages, the production of GM crops raises concerns about their impacts on the environment1,5,9,10,11.
Critical environmental questions include whether introduced genes from GM crops will move to other plants, what would be the consequences of such movement, and whether the movement can be limited4,12. Pollen-mediated gene flow (PGF), defined as the dispersal of crop genes via pollen, has became a primary focus of risk assessment because of the potential for GM pollen to move to non-GM plants3,13,14,15. PGF is affected by many factors, including whether the crop is wind- or insect-pollinated, the distance of pollen dispersal, and the probability of fertilization13,16,17,18,19,20. Some studies have shown that PGF usually declines exponentially with increasing distance between the GM pollen donor and non-GM pollen recipient11,15,20,21,22, and the presence of pollinators, including the honeybee, Apis mellifera, which increased PGF18,19,23. Thus, the absence of efficient pollinators and long distance are important for attenuating the environmental risk. What has been less studied is the impact of pollinator species on PGF.
Cotton (Gossypium hirsutum L.) is primarily grown as an irrigated crop during summer in three major ecological regions of China: the Changjiang River Region, the Yellow River Region and the North-western Region. Cotton production is constrained by lepidopteran pests, especially Helicoverpa armigera18. Transgenic cotton, producing insecticidal proteins from Bacillus thuringiensis (Bt) has been used since 1997 in China to effectively suppress lepidopteran pests24,25. Cotton is primarily a self-pollinating plant, but a small amount of cross-pollination can occur in the presence of pollinators, and the large and sticky cotton pollen makes pollinators potentially important in cross-pollination18,19,26. Some studies have shown that PGF in cotton varied greatly depending on multiple factors, including location, environmental condition, the time period (different years) and the method used to measure the crossing rate16,19,20,27,28. However, the PGF due to different pollinator species has been less studied. Some researches confirmed that Bombus ignites and Apis mellifera are effective pollinators in the field and greenhouse29,30, and they are both used widely because of their commercial availability. Although Pieris rapae is not commercialized as a pollinator, its ability to pollinate plants has been reported31. In China where farmers primarily have small units of land on which they grow multiple crops, it is common to have crucifer crops, the host of P. rapae, near cotton. Thus, P. rapae serves as an example of potential pollinators for cotton and should be considered in a risk assessment for the spread of cotton transgenes.
In the present study, we used a GM cotton expressing Bt for insect resistance and resistance to a broad-spectrum herbicide (glyphosate) as the pollen donor. To measure PGF in GM cotton using the combination of molecular techniques and biological assay, we conducted greenhouse experiments with pollinators, including B. ignites, A. mellifera and P. rapae, and without pollinators, using one GM cotton and two non-GM counterparts. Our goals in the study were to: (1) measure PGF with different pollinators, (2) determine the influence of distance between GM pollen donor plants and non-GM pollen receptor plants on PGF, (3) analyze the impact of different non-GM cotton receptor plants on PGF, (4) confirm the insect and glyphosate resistance of hybrids.
Results
Greenhouse observation
The agronomic behavior of GM and non-GM cottons was similar to a standard cultivar in the field. Good cotton growth and flower production were achieved in three greenhouses during two years of this study. Time of flowering was synchronous between the GM and non-GM cotton throughout the flowering period, thus ensuring ample opportunity for pollen transfer. Insect proof nets prevented pollinators moving into the control groups. Wind velocities in the greenhouses were very low (<0.2 m/s) (Figure S1), minimizing any influences of wind on PGF.
Pollen-mediated gene flow in the absence and presence of pollinators
A total of 10,080 seeds (60 seeds/sample ×2 pollen receptors ×14 distances ×3 pollinators ×2 yr) were sampled to assess PGF by DNA analyses. PCR amplification products were consistently detected and clearly visible in the GM pollen donor and progeny from cross-pollination between GM and non-GM crops (Figure S2).
Hybridization was detected during the 2 yr of the study, and varied by pollinator species and pollen receptor. More specifically, the average PGF of all sampled distances and both pollen receptors during the two yr were 10.83%, 5.52% and 2.71% for the pollinators B. ignitus, A. mellifera and P. rapae, respectively (F2,285 = 27.971, P < 0.001). For both the two pollen receptors during the 2 yr of the study, the average PGF of all sampled distances and the three pollinators to the pollen receptor of Shiyuan321 (8.61%) was higher than that of Hai7124 (4.10%) (t = 4.768, df = 286, P < 0.001).
Generally, PGF declined with increasing distance between GM and non-GM cottons (Table 1, Fig. 1), especially in the treatment with B. ignitus (2012 Shiyuan321: F7,16 = 10.019, P < 0.001; Hai7124: F7,16 = 7.048, P = 0.001; 2013 Shiyuan321: F7,16 = 3.879, P = 0.012 ; Hai7124: F7,16 = 3.194, P = 0.026). PGF to Shiyuan321 with B. ignitus ranged from 30.00% at 1.6 m to 0.00% at 36 m in 2012 and from 21.67% at 1.6 m to 1.67% at 25.6 m in 2013. PGF to Hai7124 with B. ignitus ranged from 21.67% at 3.2 m to 0.00% at 36 m in 2012 and from 15.00% at 3.2 m to 0.00% at 25.6 m in 2013. In the treatment with A. mellifera, the PGF also significant decreased with increasing distance between GM and non-GM cottons in the pollen receptors plant Shiyuan321(2012: F7,16 = 8.258, P < 0.001; 2013: F7,16 = 3.109, P = 0.029), although there were not significant differences at all sampled distances in the pollen receptor cotton Hai7124 (2012: F7,16 = 0.825, P = 0.581; 2013: F7,16 = 0.762, P = 0.626). For the pollen receptor Shiyuan321, PGFs with P. rapae showed significant differences in 2012 (F7,16 = 3.918, P = 0.011), but not in 2013 (F7,16 = = 1.582, P = 0.211). For the receptor Hai 7124, PGFs with P. rapae in the both years were not related to distance (2012, F7,16 = 0.365, P = 0.910; 2013, F7,16 = 0.429, P = 0.870). Hybrids were not detected in the control group of three greenhouses during the 2 yr of the study.
Table 1. Pollen-mediated gene flow frequency with different pollinators from GM cotton to Shiyuan321 and Hai7124 during 2012 and 2013 (%).
| Distance(m) | Shiyuan321 |
Hai7124 |
||||
|---|---|---|---|---|---|---|
| B. ignitus | A. mellifer | P. rapae | B. ignitus | A. mellifer | P. rapae | |
| 2012 | ||||||
| 0.8 | 26.67 ± 6.67 | 20.00 ± 2.89 | 13.33 ± 3.33 | 20.00 ± 5.00 | 3.33 ± 3.33 | 1.67 ± 1.67 |
| 1.6 | 30.00 ± 5.77 | 18.33 ± 3.33 | 6.67 ± 1.67 | 13.33 ± 3.33 | 6.67 ± 4.41 | 0.00 ± 0.00 |
| 3.2 | 21.67 ± 4.41 | 11.67 ± 4.41 | 3.33 ± 3.33 | 21.67 ± 6.01 | 5.00 ± 5.00 | 1.67 ± 1.67 |
| 6.4 | 28.33 ± 6.01 | 16.67 ± 4.41 | 5.00 ± 2.89 | 15.00 ± 2.89 | 0.00 ± 0.00 | 1.67 ± 1.67 |
| 12.8 | 5.00 ± 2.89 | 3.33 ± 3.33 | 3.33 ± 1.67 | 1.67 ± 1.67 | 0.00 ± 0.00 | 3.33 ± 3.33 |
| 19.2 | 0.00 ± 0.00 | 1.67 ± 1.67 | 0.00 ± 0.00 | 5.00 ± 2.89 | 1.67 ± 1.67 | 1.67 ± 1.67 |
| 25.6 | 3.33 ± 1.67 | 0.00 ± 0.00 | 1.67 ± 1.67 | 0.00 ± 0.00 | 1.67 ± 1.67 | 0.00 ± 0.00 |
| 36 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 1.67 ± 1.67 |
| Average | 14.38 ± 2.91 | 8.96 ± 1.90 | 4.17 ± 1.07 | 9.58 ± 2.02 | 2.29 ± 0.95 | 1.46 ± 0.56 |
| 2013 | ||||||
| 0.8 | 18.33 ± 4.41 | 8.33 ± 3.33 | 5.00 ± 2.89 | 11.67 ± 4.41 | 1.67 ± 1.67 | 1.67 ± 1.67 |
| 1.6 | 21.67 ± 4.41 | 16.67 ± 4.41 | 11.67 ± 4.41 | 10.00 ± 2.89 | 5.00 ± 5.00 | 1.67 ± 1.67 |
| 3.2 | 15.00 ± 2.89 | 11.67 ± 3.33 | 6.67 ± 3.33 | 15.00 ± 2.89 | 5.00 ± 2.89 | 0.00 ± 0.00 |
| 6.4 | 5.00 ± 2.89 | 11.67 ± 3.33 | 0.00 ± 0.00 | 11.67 ± 1.67 | 5.00 ± 2.89 | 1.67 ± 1.67 |
| 12.8 | 11.67 ± 4.41 | 8.33 ± 1.67 | 1.67 ± 1.67 | 6.67 ± 4.41 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 19.2 | 11.67 ± 3.33 | 6.67 ± 3.33 | 1.67 ± 1.67 | 5.00 ± 2.89 | 1.67 ± 1.67 | 1.67 ± 1.67 |
| 25.6 | 1.67 ± 1.67 | 0.00 ± 0.00 | 5.00 ± 5.00 | 0.00 ± 0.00 | 1.67 ± 1.67 | 1.67 ± 1.67 |
| 36 | 8.33 ± 1.67 | 3.33 ± 1.67 | 1.67 ± 1.67 | 1.67 ± 1.67 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Average | 11.67 ± 1.64 | 8.33 ± 1.34 | 4.17 ± 1.15 | 7.71 ± 1.35 | 2.50 ± 0.85 | 1.04 ± 0.42 |
Means ± SE. Seeds from lower, middle, and upper flowers were set up as three replications.
Figure 1. Regression analysis for the pollen-mediated gene flow from GM cotton to Shiyuan321 (A) and Hai7124 (B) as a function of the distance.
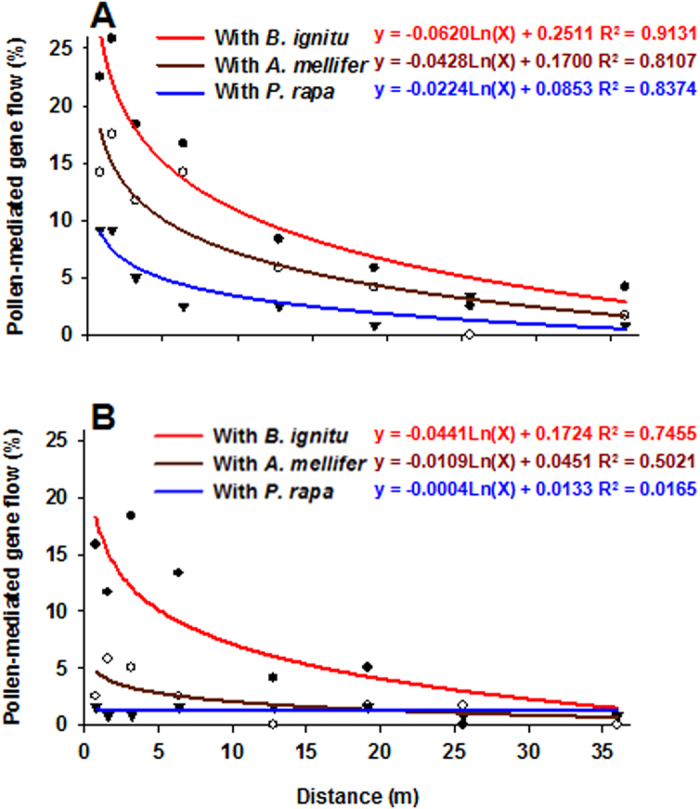
The correlation coefficient (R2) between PGF and distance was obtained using SigmaPlot 10.0 software. Regression analysis was based on the data of the two year study.
Hybrids expressing Cry I Ac were tested for Bt protein detection (Figure S3). Overall, there was low rate of false positives in Bt protein detection. During the 2 yr of the study, the false positives were 3.23% and 2.54% to the pollen receptors of Shiyuan321 and Hai7124, respectively.
Resistance test of hybrid
Insect resistance
The confirmed hybrids (progeny expressing Bt protein) were tested for insect resistance. The hybrid and GM cotton leaves significantly decreased the larval weight compared to non-GM cotton (Fig. 2A, F2,6 = 81.161, P < 0.001). The mortality of larvae fed on hybrids or GM cotton at the sixth day was higher than the control (F2,6 = 211.240, P < 0.001), although the mortality of insects fed on confirmed hybrids reached 75.00 ± 4.19%, and this was slightly lower than 91.67 ± 3.47% of the GM cotton (Fig. 2B).
Figure 2. Insect resistance detection of confirmed hybrid, Shiyuan321 and GM cotton.
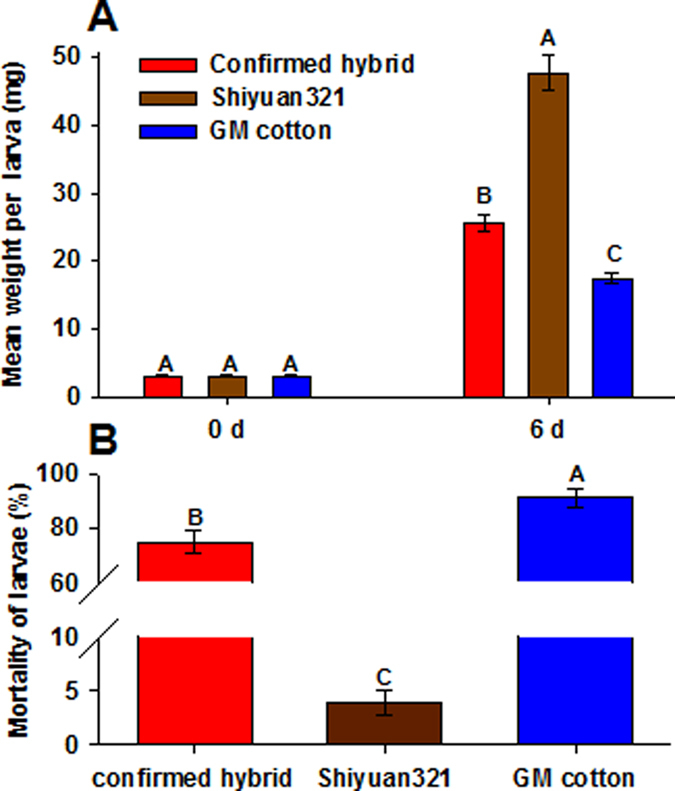
(A) Weight of H. armigera larvae fed on diet containing confirmed hybrid, Shiyuan321 and GM cotton. (B) Mortality effects of confirmed hybrid, Shiyuan321 and GM cotton on H. armigera larvae. Means ± SE within the same day after treatment and different diet followed by different letters are significantly different (Tukey’s multiple comparison test, P < 0.05).
Herbicide tolerance
Confirmed hybrids were screened for resistance to glyphosate. As shown in Table 2, non-GM cotton (Shiyuan321) grew slowly and was significantly shorter than GM cotton and the confirmed hybrids after glyphosate application, and none survived 42 d after glyphosate application. Although the height of confirmed hybrids was significantly shorter than that of GM cotton during 7 to 14 d, there was no significant difference with GM cotton until 28 d after glyphosate application. Most of confirmed hybrids showed resistance to glyphosate, and the survival rate reached 82.78% at 42 d after glyphosate application.
Table 2. Glyphosate resistance detection of confirmed hybrid, Shiyuan321, and GM cotton.
| Concentration (%) | Height of cotton (cm) |
survival rate (%) | ||||
|---|---|---|---|---|---|---|
| 0 d | 7 d | 14 d | 2814 d | |||
| GM cotton | 0.25 | 26.47 ± 0.83 A | 35.47 ± 0.64 A | 44.80 ± 1.10 A | 54.87 ± 1.36 AB | 100.00 ± 0.00 A |
| Confirmed hybrid | 0.25 | 26.83 ± 0.55 A | 31.77 ± 0.59 B | 39.27 ± 0.90 B | 50.97 ± 1.39 B | 82.78 ± 2.42 B |
| Shiyuan321 | 0.25 | 26.70 ± 1.13 A | 28.13 ± 0.90 C | 30.27 ± 1.25 C | 31.03 ± 1.84 C | 0.00 ± 0.00 C |
| GM cotton | 0 | 26.37 ± 0.88 A | 37.30 ± 0.55 A | 46.03 ± 0.87 A | 58.30 ± 1.21 A | 100.00 ± 0.00 A |
| Confirmed hybrid | 0 | 26.63 ± 0.73 A | 36.60 ± 0.64 A | 45.37 ± 0.79 A | 56.17 ± 0.79 AB | 100.00 ± 0.00 A |
| Shiyuan321 | 0 | 26.40 ± 0.60 A | 36.43 ± 0.39 A | 44.87 ± 0.65 A | 56.40 ± 0.62 AB | 100.00 ± 0.00 A |
| F | − | 0.053 | 31.909 | 42.022 | 65.275 | 1638.000 |
| P | − | 0.998 | <0.001 | <0.001 | <0.001 | <0.001 |
Means ± SE followed by different letters are significantly different (Tukey’s multiple comparison test, P < 0.05). Each treatment contained 50 seedlings, which was repeated 3 times.
Discussion
The risk of gene flow can occur within species, between species, between the higher plants and other organisms, but the most probable gene flow would occur between GM crops and their non-GM lines, or related wild species32. Molecular analysis offers a simple, effective and accurate method for measuring PGF3,15,23,33,34,35. In the present study, we used a combination of molecular methods and biological assays to measure PGF within species, the GM cotton with both Cry I Ac and CP4 EPSPS genes and non-GM cotton. We collected a total of 10,080 cotton seeds during the 2-yr study and then grew them to the 3-leaf stage to study PGF in the greenhouse with pollinators. Past studies on PGF were mainly conducted in fields11,33,34,36. Although these studies provided general patterns on PGF, it was difficult to evaluate the effects of an individual factor. Our study was conducted in greenhouses, which provided a relatively closed environment to avoid the effects of wind and other pollinators on PGF. We observed that PGF did occur at a higher frequency during our 2-yr study, compared to that in fields4,12,16,37. More importantly, our study was able to parse out the influences of various factors.
In our present study, we found that PGF varied with different pollinators, with B. ignitus and A. mellifera having higher PGF rates than P. rapae. Although the studies were conducted in the three greenhouses, the temperature and other environmental conditions were the same. We attribute the differences of PGF to be caused by different pollinators. B. ignitus and A. mellifera are widely used as agricultural pollinators29,38,39, especially B. ignitus. Bumblebees are effective pollinators because of the amount of pollen they collect, their working time and speed, frequency of visiting flowers and ability to work under a range of temperatures29,38,39,40,41,42. They often remove all available nectar from flowers in a single visit38, and deposit more pollen than honeybees39. The larger surface area of bumblebees allows more contact with stigmas and more pollen deposition compared to honeybees39,43,44. These merits of bumblebees provide better pollination, reduce geitonogamy, increase pollen flow distance, and thereby increase outcrossing rates40,45,46. On the other hand, honeybees are more sensitive to temperature, show more phototaxis, and need more time to adapt to the greenhouse condition compared to bumblebees30,47,48. Thus, these factors led to the higher PGF in the greenhouse with bumblebees, and potentially the higher environmental risk in the fields. Therefore, we suggested that B. ignites should not be used as a supplemental pollinator in GM crops. Although P. rapae adults provided pollination services in a botanical garden31, it was not an effective pollinator in the cotton field. Thus, it would provide almost no risk in the GM cotton field.
Umbeck et al.16 measured out-crossing from GM cotton to non-GM cotton in Mississippi using a seed germination assay and PCR analysis, and found that out-crossing went from 5.7% to < 1% within 7 m from the test plot, and <1% out-crossing continued to occur sporadically to a distance of 25 m. Van Deynze et al.19 found PGF from GM cotton declined exponentially with increasing distance from 7.65% at 0.3 m to < 1% beyond 9 m even with high pollinator activity by A. melifera. In the case of GE plums, the PGF ranged from 0.215% at 132 m from the center of the plot to 0.033-0.017% at longer distances (384–998 m)15. These results are similar to data gathered in Fujian province, China, where PGF from GM rice ranged from 0.28% at 0.2m to < 0.01% at 6.2 m12. Similar to these and other studies11,17,22,49, we found PGF with B. ignitus declined with increasing distance. PGF in the presence of pollinators would decrease significantly with increasing distance from GM plants not only because of decreased diffusion of the GM pollen and pollinator movement over the distance, but also because density of non-GM pollen would increase. One the other hand, the pollen grains may gradually sediment under the action of gravity after shedding during the dispersal. Furthermore, as GM pollen travels over longer distances and time, they may suffer slightly losses of competitiveness compared to non-GM pollen. These combined effects would lead to the sharp reduction of PGF with increased distance. Based on our results and the literature cited, it is apparent that increasing isolation distances is an effective method to minimize environmental risk for self-pollinating crops.
Biological confinement strategies (male sterility and gene insertion into chromosomes) might influence gene flow frequency. PGF from GM bentgrass, Agrostis stolonifera, to non-GM A. stolonifera (91 of 168) was higher than that to A. gigantea (13 of 39) as assessed by seedling progeny survival after spraying with glyphosate and PCR analysis50. Our experiment also showed that total PGF to the pollen receptor Shiyuan321 was higher than to Hai7124. The flowering time of cotton plant is longer than with bentgrass, and we chose cotton varieties with a similar growth period for our study. Therefore, we eliminated the effects of differences of flowering time on PGF. Differences in PGF of the two receptors were likely due to the genetic background because Shiyuan321 was the parent of the GM cotton and the relationship between the transgenic cotton and Shiyuan321 is closer than that to Hai7124. It is easier to hybridize with lines having a closer genetic relationship. The potential for out-crossing is highly dependent on sexual compatibility and relatedness between the parent species, and the opportunity for natural hybridization between two plants depends on many pre- and post-zygotic factors51.
PGF can be determined directly by progeny resistance detection because most of confirmed hybrids showed some resistance to glyphosate and targeted pests. For example, PGF was determined by seedling progeny survival after spraying with glyphosate4,19,37. In the current study, the confirmed hybrids showed strong resistance to both glyphosate and H. armigera. However, we think it is more reliable to determine PGF by PCR analysis. The reasons are (1) the resistance of confirmed hybrids was weaker than that of the GM cotton, (2) seedlings expressing resistance genes might not express functional proteins, although the level of false positive was very low in our present research.
In conclusion, our studies quantified the extent of PGF in cotton under greenhouse conditions in the presence of pollinators. Our results indicated that (1) PGF was dependent on pollinator species, and was highest with B. ignitus and lowest with P. rapae. (2) PGF with B. ignitus dropped exponentially with increasing distance. However, the level of PGF with P. rapae was very low and not obviously influenced by the distance. (3) Total PGF to the pollen receptor Shiyuan321 was higher than that for Hai7124. (4) Most confirmed hybrids using molecular methods showed strong resistance to glyphosate and H. armigera larvae. This study suggests that PGF was dependent on pollinator species, distances, and the receptor and suggests that these factors are important in a risk assessment.
Methods
Plant materials
In current study, the inbred line Shiyuan321 and island cotton Hai7124 were selected as the pollen receptors. The GM cotton as the pollen donor were obtained by transferring the insect resistance gene Cry I Ac and the glyphosate resistance gene CP4 EPSPS from Agrobacterium tumefaciens into the embryonic calli of Shiyuan321 via microprojectile bombardment. The promoters (mostly the 35S promoter of cauliflower mosaic virus) in GM cotton enhance the target gene expression in seeds and seedlings, so that even hemizygous progeny resulting from the cross-pollination with a non-GM cultivar can be easily identified by a variety of molecular analyses15,52,53,54. The GM and non-GM cottons have similar growth patterns and flower structure20. All plants were obtained from the Cotton Research Institute, Chinese Academy of Agricultural Sciences, and were purified by consecutive two-generations of self-crosses, to ensure that each line was homozygous.
Greenhouse layout and procedures
To evaluate the impacts of pollinators on PGF from a GM cotton to other non-GM cultivars, the trials were carried out in three greenhouses (25–28 °C, L60 m × W8 m × H4 m) in the Shangzhuang Experimental Station of China Agricultural University (40°14′N, 116°19′E), Beijing, China in 2012 and 2013. Each greenhouse contained one type of pollinator (Bombus ignitus, Apis mellifera, provided by the Bee Research Institute, Chinese Academy of Agricultural Sciences, or Pieris rapae reared in our lab). One small colony of honeybees (ca 4000 workers), one colony of bumblebees (ca 80 workers) and 80 butterflies were released in the greenhouses to evaluate gene flow. The appropriate quantity of each pollinator was determined by an expert in pollination, and appropriate for pollination in our greenhouse (60 m × 8 m). It has been reported some Lepidoptera act as pollinators of cotton55, and we found that there was high population density of P. rapae in cotton fields near the greenhouses in the current study. B. ignitus and A. mellifera, commonly used as agricultural pollinators, are effective pollinators in fields and greenhouses29,30,38,39,47. Thus, the three species were selected as pollinators in the present study. Wind speeds were monitored at 6:00, 14:00 and 22:00 using an anemograph (ZRQF-F303, Beijing Detector Instrument Co., Beijing, China) in the greenhouses20.
The GM cotton was planted in a 4 m × 8 m square in the middle of a 60 m × 8 m square area of non-GM cotton according to the scheme map presented in Fig. 3. Rows were spaced 0.8 m apart and plants within rows were spaced 0.5 m apart. Non-GM cotton in the west of greenhouse was set up as the control group and non-GM cotton in the east of greenhouse as the treatment group. Insect proof net (9 holes/cm2) was used to prevent pollinators moving into the control groups, and did not influence the pollen transfer. All plants were monitored at least twice weekly for florescence emergence and anthesis from early July through late August to ensure the flowering synchrony between GM and non-GM cotton37. Pollinators were released in three greenhouses in early August (25–28 °C, full-bloom stage for all plants). At harvest, seeds from pollen receptors were obtained by hand from lower, middle, and upper flowers at the distance of 0.8, 1.6, 3.2, 6.4, 12.8, 19.2, 25.6 and 36 m for the treatment group and 0.8, 1.6, 3.2, 6.4, 12.8 and 19.2 m for the control group from the donor plot when mature but before excessive shattering19. No significant differences in PGF were noted for flower positions on the pollen receptors16. Thus, seeds from lower, middle, and upper flowers were set up as three replications. Cotton was ginned separately, acid delinted, mixed and sub-sampled for the following detection.
Figure 3. Greenhouse trial design to measure gene flow from GM cotton to conventional lines during 2012 and 2013.
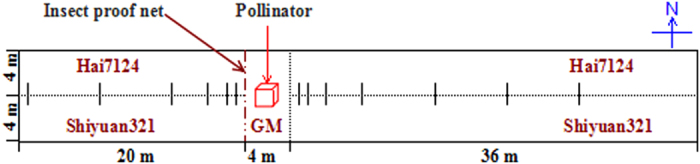
The GM cotton was planted in a 4 m × 8 m square in the middle of a 60 m × 8 m square planting of non-GM cotton. Non-GM cotton in the west of greenhouse was set up as the control group and non-GM cotton in the east of greenhouse as the treatment group. Bars indicate sampling points.
Hybrid confirmation
Seeds of the F1 generation, from the three greenhouses, were planted in flats containing moist potting mix composed of 75% vermiculite and 25% turf (by volume) in a greenhouse with a temperature of 22–35 °C and a 14 h photoperiod. Control flats were also seeded and evaluated, including the seeds of Shiyuan321 and Hai7124. Seedlings were allowed to grow to the 3-leaf stage, and one part of each seedling was used for DNA extraction and polymerase chain reaction (PCR) amplification, and the seedling grew continually. For each distance point, we screened 60 seedlings from each pollen receptor for hybrid confirmation.
The presence of transgenic DNA in putative hybrids was verified by PCR amplification of the fragment sequence of Cry I Ac. DNA was extracted from the leaf of a seedling using the cetyltrimethyl ammonium bromide (CTAB) protocol56 with the addition of 20 g/L polyvinylpyrrolidone (PVP) in the extraction buffer. A 1 μL sample of DNA was used to perform spectroscopic quantitation using a NanoDrop 2000 spectrophotometer (Thermo Fisher, USA). Primers of 5′-GAAGGATTGAGCAATCTCTAC-3′ and 5′-CAATCAGCCTAGTAAGGTCGT-3′ were used to amplify the Cry I Ac with 80 ng of DNA20. The PCR condition was: 95 °C for 4 min, followed by 30 cycles at 94 °C for 60 s, 56 °C for 60 s, 72 °C for 90 s, and a final extension step at 72 °C for 5 min20. Routine screening for PCR assay was performed by agarose gel electrophoresis (10 g agarose/L) with GM cotton as positive control and non-GM Shiyuan321 as negative control.
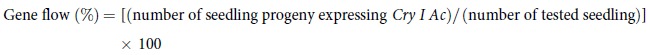 |
Total protein was extracted from 0.1 g of a leaf of seedling progeny expressing Cry I Ac, and was applied for Bt protein detection using Bt-Cry 1 Ab/Ac detection kits (Beijing Yintu Biological Technology Co., Beijing, China) in accordance with a previously described method20 (Figure S3). The appearance of a band indicated it was positive.
 |
Biological assay for detection of hybrid
The confirmed hybrid expressing Bt protein (cross-pollination with Shiyuan321), non-GM cultivar (Shiyuan321), and GM cotton were sprayed with a 0.25% (v/v) glyphosate solution at the 4-leaf stage, respectively. The height of a seedling that survived was measured at 0, 7, 14, 28 days after glyphosate application57, and the survival rate was recorded at 42 d after glyphosate application. Each treatment contained 50 seedlings, which was repeated 3 times.
Helicoverpa armigera larvae were used to determine the insect resistance of confirmed hybrids. H. armigera collected from a cotton field were reared in the IPM lab (China Agricultural University, 40°02′N, 116°28<mml:mspace width=”0.25em” depth=”0000”/>E) at 27 ± 1 °C, 75 ± 10% relative humidity (RH) and a 14: 10 light: dark photoperiod. This species has been cultured for more than 10 yr without exposure to Bt or other insecticides58,59. Larvae were reared on a synthetic diet60 and adults were supplied with a 10% honey solution. In the bioassay, larvae were reared on the artificial diet until the third instar. Then they were transferred into glass tubes (1 cm × 6 cm) containing leaves at the 5-leaf stage from the confirmed hybrid (cross-pollination with Shiyuan321), non-GM cotton (Shiyuan321 as control) or GM cotton according to the method described by Liu et al.58. Six d later, the weight and mortality of H. armigera larvae were recorded57. Each treatment contained 60 larvae, which was repeated 3 times.
Data analysis
Descriptive statistics were given as the mean values and standard errors of the mean. The data were analyzed by Tukey’s multiple comparison test at the P = 0.05 level of significance. Statistics were performed with SPSS 16.0 software (IBM, Armonk, NY). Regression analyses were performed using SigmaPlot 10.0 software (Systat Software Inc., San Jose). The correlation coefficient (R2) between PGF and distance was obtained from SigmaPlot 10.0 software.
Additional Information
How to cite this article: Yan, S. et al. Pollen-mediated gene flow from transgenic cotton under greenhouse conditions is dependent on different pollinators. Sci. Rep. 5, 15917; doi: 10.1038/srep15917 (2015).
Supplementary Material
Acknowledgments
This project was supported by the Special Research Projects of China for Developing Transgenic Plants (2014zx08011002–006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Conceived and designed the experiments: S.Y., J.L.Z., X.X.L. and Q.W.Z. Performed the experiments: S.Y., J.L.Z. and W.L.Z. Analyzed the data: S.Y. and Z.L. Contributed reagents/materials/analysis tools: S.Y., X.X.L., Q.W.Z., J.Y.L. and J.J.C. Wrote the paper: S.Y., A.M.S. and X.X.L.
References
- O’Callaghan M., Glare T. R., Burgess E. P. J. & Malone L. A. Effects of plants genetically modified for insect resistance on nontarget organisms. Annual Review of Entomology 50, 271–292 (2005). [DOI] [PubMed] [Google Scholar]
- Li C., Xiao P., Gray S. J., Weinberg M. S. & Samulski R. D. Combination therapy utilizing shRNA knockdown and an optimized resistant transgene for rescue of diseases caused by misfolded proteins. Proceedings of the National Academy of Sciences of the United States of America 108, 14258–14263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer J. et al. Field assessments of gene flow from transgenic to cultivated rice (Oryza sativa L.) using a herbicide resistance gene as tracer marker. Theoretical and Applied Genetics 103, 1151–1159 (2001). [Google Scholar]
- Lavigne C. et al. A pollen-dispersal experiment with transgenic oilseed rape. Estimation of the average pollen dispersal of an individual plant within a field. Theoretical and Applied Genetics 96, 886–896 (1998). [Google Scholar]
- Donegan K. K. et al. A field study with genetically engineered alfalfa inoculated with recombinant Sinorhizobium meliloti: effects on the soil ecosystem. Journal of Applied Ecology 36, 920–936 (1999). [Google Scholar]
- James C. Global status of commercialized biotech/GM crops: 2014. ISAAA Brief No. 49. ISAAA: Ithaca, NY (2014).
- Mannion A. M. & Morse S. Agronomic and environmental considerations and reflections based on 15 years of GM crops. Progress in Physical Geography 36, 747–763 (2012). [Google Scholar]
- Huang W. K. et al. Assessment of gene flow from glyphosate-resistant transgenic soybean to conventional soybean in China. Acta Physiologiae Plantarum 36, 1637–1647 (2014). [Google Scholar]
- Nap J. P., Metz P. L. J., Escaler M. & Conner A. J. The release of genetically modified crops into environment. Part I Overview of current status and regulations. The Plant Journal 33, 1–18 (2003). [DOI] [PubMed] [Google Scholar]
- Ammann K. Effects of biotechnology on biodiversity: herbicide-tolerant and insect-resistant GM crops. TRENDS in Biotechnology 23, 388–394 (2005). [DOI] [PubMed] [Google Scholar]
- Heuberger S., Ellers-Kirk C., Tabashnik B. E. & Carrière Y. Pollen- and seed-mediated transgene flow in commercial cotton seed production fields. PLoS ONE 5, e14128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J. et al. Dramatic reduction of crop-to-crop gene flow within a short distance from transgenic rice fields. New Phytologist 173, 346–353 (2007). [DOI] [PubMed] [Google Scholar]
- Chandler S. & Dunwell J. M. Gene flow, risk assessment and environmental release of transgenic plants. Critical Reviews in Plant Sciences 27, 25–49 (2008). [Google Scholar]
- Pasquet R. S. et al. Long-distance pollen flow assessment through evaluation of pollinator foraging range suggests transgene escape distances. Proceeding of the National Academy of Sciences of the United States of America 105, 13456–13461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorza R. et al. Spatial and temporal assessment of pollen- and seed-mediated gene flow from genetically engineered plum Prunus domestica. PLoS ONE 8, e75291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbeck P. F. et al. Degree of pollen dispersal by insects from a field-test of genetically engineered cotton. Journal of Economic Entomology 84, 1943–1950 (1991). [Google Scholar]
- Llewellyn D. & Fitt G. Pollen dispersal from two field trials of transgenic cotton in the Namoi valley, Australia. Molecular Breeding 2, 157–166 (1996). [Google Scholar]
- Llewellyn D. et al. Containment of regulated genetically modified cotton in the field. Agriculture Ecosystems and Environment 121, 419–429 (2007). [Google Scholar]
- Van Deynze A. E., Sundstrom F. J. & Bradford K. J. Pollen-mediated gene flow in California cotton depends on pollinator activity. Crop Science 45, 1565–1570 (2005). [Google Scholar]
- Zhu J. L., He J., Niu J. Q., Zhang Q. W. & Liu X. X. The influence of wind direction on pollen-mediated gene flow in transgenic insect-resistant cotton. Acta Ecologica Sinica 33, 6803–6812 (2013). [Google Scholar]
- Elfawal M. A., Bishr M. A. & Hassoub E. K. Natural cross pollination in Egyptian cotton (Gossypium barbadense L.). Journal of Agricultural Science 86, 205–209 (1976). [Google Scholar]
- Song Z. P., Lu B. R. & Chen J. K. Pollen flow of cultivated rice measured under experimental conditions. Biodiversity and Conservation 13, 579–590 (2004). [Google Scholar]
- Pu D. Q. et al. Flower-visiting insects and their potential impact on transgene flow in rice. Journal of Applied Ecology 51, 1357–1365 (2014). [Google Scholar]
- Wu K. M., Lu Y. H., Feng H. Q., Jiang Y. Y. & Zhao J. Z. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321, 1676 (2008). [DOI] [PubMed] [Google Scholar]
- Wu K., Li W., Feng H. & Guo Y. Seasonal abundance of the mirids, Lygus lucorum and Adelphocoris spp. (Hemiptera: Miridae) on Bt cotton in northern China. Crop Protection 21, 997–1002 (2002). [Google Scholar]
- Moffett J. O., Stith L. S., Burkhart C. C. & Shipman C. W. Honey bee visits to cotton flowers. Environmental Entomology 4, 203–206 (1975). [Google Scholar]
- Meredith W. R. & Bridge R. R. Natural crossing in cotton (Gossypium hirsutum L.) in the delta Mississippi. Crop Science 13, 551–552 (1973). [Google Scholar]
- Kareiva P., Morris W. & Jacobi C. M. Studying and managing the risk of cross-fertilization between transgenic crops and wild relatives. Molecular Ecology 3, 15–21 (1994). [Google Scholar]
- Dogterom M. H., Matteoni J. A. & Plowright R. C. Pollination of greenhouse tomatoes by the North American Bombus vosnesenskii (Hymenoptera: Apidae). Journal of Economic Entomology 91, 71–75 (1998). [Google Scholar]
- An J. D. et al. Foraging behavior and pollination ecology of Bombus lucorum L and Apis mellifera L in greenhouse peach garden. Chinese Journal of Applied Ecology 18, 1071–1076 (2007). [PubMed] [Google Scholar]
- Zhang H. F., Li L. Q., Liu Z. J. & Luo Y. B. The butterfly Pieris rapae resulting in the reproductive success of two transplanted orchids in a botanical garden. Biodiversity Science 18, 11–18 (2010). [Google Scholar]
- Kwon Y. W., Kim D. S. & Yim K. O. Herbicide-resistant genetically modified crop: assessment and management of gene flow. Weed Biology and Management 1, 96–107 (2001). [Google Scholar]
- Kavanagh V. B. et al. Molecular markers as a complementary tool in risk assessments: quantifying interspecific gene flow from triticale to spring wheat and durum wheat. Transgenic Research 22, 767–778 (2013). [DOI] [PubMed] [Google Scholar]
- Kim C. G. et al. Assessment of gene flow from genetically modified anthracnose-resistant chili pepper (Capsicum annuum L.) to a conventional crop. Journal of Plant Biology 52, 251–258 (2009). [Google Scholar]
- McCartney-Melstad E. et al. Population structure and gene flow of the yellow anaconda (Eunectes notaeus) in Northern Argentina. PLoS ONE 7, e37473 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieben S., Kalinina O., Schmid B. & Zeller S. L. Gene flow in genetically modified wheat. PLoS ONE 6, e29730 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. G. et al. Pollen-mediated gene flow from kentucky bluegrass under cultivated field conditions. Crop Science 46, 1990–1997 (2006). [Google Scholar]
- Irwin R. E. & Brody A. K. Nectar robbing in Ipomopsis aggregata: effects on pollinator behavior and plant fitness. Oecologia 116, 519–527 (1998). [DOI] [PubMed] [Google Scholar]
- Thomson J. D. & Goodell K. Pollen removal and deposition by honeybee and bumblebee visitors to apple and almond flowers. Journal of Applied Ecology 38, 1032–1044 (2001). [Google Scholar]
- Maloof J. E. The effects of a bumble bee nectar robber on plant reproductive success and pollinator behavior. American Journal of Botany 88, 1960–1965 (2001). [PubMed] [Google Scholar]
- Birmingham A. L. & Winston M. L. Orientation and drifting behaviour of bumblebees (Hymenoptera: Apidae) in commercial tomato greenhouses. Canadian Journal of Zoology 82, 52–59 (2004). [Google Scholar]
- Newman D. A. & Thomson J. D. Effects of nectar robbing on nectar dynamics and bumblebee foraging strategies in Linaria vulgaris (Scrophulariaceae). Oikos 110, 309–320 (2005). [Google Scholar]
- Thomson J. D. Pollen transport and deposition by bumblebees in Erythronium: influences of floral nectar and bee grooming. Journal of Ecology 74, 329–341 (1986). [Google Scholar]
- Snow A. A. & Roubik D. W. Pollen deposition and removal by bees visiting two tree species in Panama. Biotropica 19, 57–63 (1987). [Google Scholar]
- de Jong T. J., Waser N. M. & Klinkhamer P. G. L. Geitonogamy: the neglected side of selfing. Trends in Ecology and Evolution 8, 321–325 (1993). [DOI] [PubMed] [Google Scholar]
- Barrett S. C. H. & Harder L. D. Ecology and evolution of plant mating. Trends in Ecology and Evolution 11, 73–79 (1996). [DOI] [PubMed] [Google Scholar]
- Li J. L. et al. Strawberry pollination by Bombus lucorum and Apis mellifera in greenhouses. Acta Entomologica Sinica 49, 342–348 (2006). [Google Scholar]
- Stanghellini M. S., Ambrose J. T. & Schultheis J. R. The effects of honey bee and bumble bee pollination on fruit set and abortion of cucumber and watermelon. American Bee Journal 137, 386–391 (1997). [Google Scholar]
- Ma B. L., Subedi K. D. & Reid L. M. Extent of cross-fertilization in maize by pollen from neighbouring transgenic hybrids. Crop Science 44, 1273–1282 (2004). [Google Scholar]
- Watrud L. S. et al. Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker. Proceedings of the National Academy of Sciences of the United States of America 101, 14533–14538 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner A. J., Glare T. R. & Nap J. P. The release of genetically modified crops into the environment. Part II. Overview of ecological risk assessment. The Plant Journal 33, 19–46 (2003). [DOI] [PubMed] [Google Scholar]
- Pla M. et al. Assessment of real-time PCR based methods for quantification of pollen-mediated gene flow from GM to conventional maize in a field study. Transgenic Research 15, 219–228 (2006). [DOI] [PubMed] [Google Scholar]
- Michalski S. G. & Durka W. Assessment of provenance delineation by genetic differentiation patterns and estimates of gene flow in the common grassland plant Geranium pratense. Conservation Genetics 13, 581–592 (2012). [Google Scholar]
- de Jong T. J. & Rong J. Crop to wild gene flow: does more sophisticated research provide better risk assessment. Environmental Science and Policy 27, 135–140 (2013). [Google Scholar]
- Nanchong Normal School in Sichuan Province of China. The study of pollinators of cotton. China cotton 3, 23–27 (1978). [Google Scholar]
- Permingeat H. R., Romagnoli M. V. & Vallejos R. H. A simple method for isolating high yield and quality DNA from cotton (Gossypium hirsutum L.) leaves. Plant Molecular Biology Reporter 16, 1–6 (1998). [Google Scholar]
- Niu J. Q., Zhu J. L., Zhang Q. W. & Liu X. X. Glyphosate tolerance and effects of cotton bollworms on transgenic herbicide and insect resistant cotton. Chinese Journal of Applied Entomology 49, 889–894 (2013). [Google Scholar]
- Liu X. X., Sun C. G. & Zhang Q. W. Effects of transgenic Cry1A±CpTI cotton and Cry1Ac toxin on the parasitoid, Campoketis chlorideae (Hymenoptera: Ichneumonidae). Insect Science 12, 101–107 (2005). [Google Scholar]
- Liu X. X. et al. Effects of Bt transgenic cotton lines on the cotton bollworm parasitoid Microplitis mediator in the laboratory. Biological Control 35, 134–141 (2005). [Google Scholar]
- Wu K. J. & Gong P. Y. A new and practical artificial diet for the cotton bollworm. Entomologia Sinica 4, 277–282 (1997). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


