Abstract
Polyporus umbellatus, a species symbiotic with Armillaria mellea and it also exhibits substantial defence response to Armillaria mellea infection. There are no genomics resources databases for understanding the molecular mechanism underlying the infection stress of P. umbellatus. Therefore, we performed a large-scale transcriptome sequencing of this fungus with A. mellea infection using Illumina sequencing technology. The assembly of the clean reads resulted in 120,576 transcripts, including 38,444 unigenes. Additionally, we performed a gene expression profiling analysis upon infection treatment. The results indicated significant differences in the gene expression profiles between the control and the infection group. In total, 10933 genes were identified between the two groups. Based on the differentially expressed genes, a Gene Ontology annotation analysis showed many defence-relevant categories. Meanwhile, the Kyoto Encyclopedia of Genes and Genomes pathway analysis uncovered some important pathways. Furthermore, the expression patterns of 13 putative genes that are involved in defence response resulting from quantitative real-time PCR were consistent with their transcript abundance changes as identified by RNA-seq. The sequenced genes covered a considerable proportion of the P. umbellatus transcriptome, and the expression results may be useful to strengthen the knowledge on the defence response of this fungus defend against Armillaria mellea invasion.
In nature, organism resistance to exotic species infections is the rule rather than the exception. The defence mechanisms have been extensively studied in plants. For example, to understand the molecular mechanisms of the establishment of the infection of rice with blast fungus, extensive studies have characterized many rice and fungal genes that are involved in plant defence and pathogen attack1,2,3. Meanwhile, a lot of work has been done to explore the molecular mechanism of defence in animals such as Eriocheir sinensis and Portunus trituberculatus4,5. But there is little knowledge about the defence system of fungus in the published researches.
Polyporus umbellatus, belonging to Polyporaceae in the class of Basidiomycetes has been used as medicine fungus in China for more than 2500 years6. The P. umbellatus sclerotia can survive in soils for long periods of time, and produce new sclerotia directly from the old ones under appropriate conditions7. It is now well known that the growth of P. umbellatus sclerotia depends on a symbiotic relationship with the forest pathogenic fungus Armillaria mellea8, which is a serious root rot disease-causing fungus in the family of Physalacriaceae9. In the progress of symbiosis establishment, the rhizomorph of A. mellea adheres and invades sclerotia of P. umbellatus and the P. umbellatus sclerotia launch effective defence responses to fend off A. mellea invasion. For example, the cell wall of P. umbellatus sclerotia had become irregularly thickened to block the A. mellea further invasion and the P. umbellatus sclerotia can form a cavity structure in the medullar tissue and isolate A. mellea to prevent further intrusion of A. mellea hyphae under SEM or TEM observation10. Although in previous studies, the preliminary knowledge of interactions between P. umbellatus and A. mellea has been provided from the morphological characteristics, little information is available on mechanisms of defence response of P. umbellatus to A. mellea infection up to now.
In recent years, several differential screening techniques (e.g., differential display, subtraction libraries, and differential hybridization) have made it possible to characterize genes that are differentially expressed after certain treatments11. Recently, along with the rapid development and cost reduction of next generation sequencing (NGS), sequence-based assays of transcriptomes, namely RNA sequencing (RNA-seq), has been become a comprehensive and accurate tool for gene expression pattern analyses. Compared with the above screening techniques, RNA-Seq is known to have a wider dynamic range, higher technical reproducibility, and provide a better estimate of absolute expression levels12. Along with these advantages, RNA-seq has been used to elucidate the response of plants to various environmental stresses, such as cold13, salt14,15 and drought16,17. However, until now, no genes have been identified, and no molecular research of Polyporus umbellatus has been reported, despite the importance of the genus. Furthermore, seldom reports have been found on the response of P. umbellatus to A. mellea infection.
In this study, in order to identify defence-related genes, potential defence-responsive genes were first identified based on Illumina tag-sequencing, and then, the differentially expressed genes (DEGs) were screened and further validated by qPCR. To our knowledge, this is the first large-scale assessment of Polyporus genomic information. Our results will facilitate understanding of the response of P. umbellatus to A. mellea infection.
Results
Sequencing and de novo transcriptome assembly
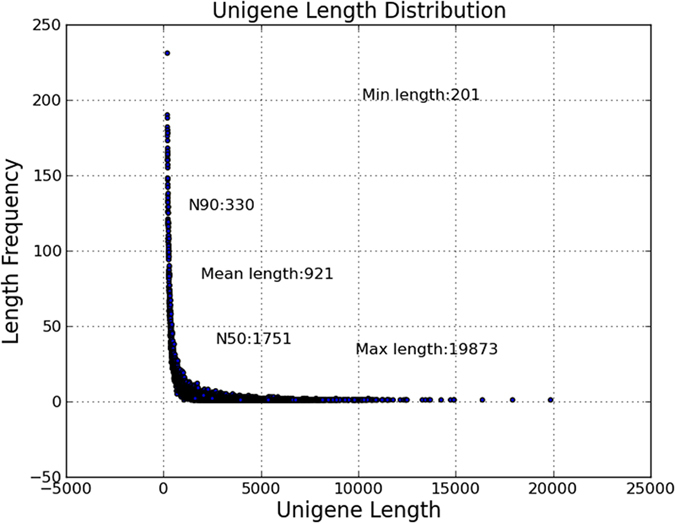
To obtain the P. umbellatus sclerotia transcriptome expression profile with A. mellea infection, one non-normalized library was constructed using sclerotia tissue with A. mellea infection and normal sclerotia tissue as control. Illumina sequencing data from P. umbellatus sclerotia was deposited in the NCBI SRA database under accession number SRP058382. In total, 186342740 Illumina PE raw reads were generated (Table 1). After removing adaptor sequences, ambiguous nucleotides and low-quality sequences, there were 177.117254 million clean reads remaining. Assembly of clean reads resulted in 38,444 unigenes in the range of 201–19,873 bp with a N50 length of 1751 bp (Fig. 1).
Table 1. Summary of sequences analysis.
| Sample | Raw Reads | Clean Reads | Clean Bases | Error (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| CT1_1 | 24750909 | 23407132 | 2.93G | 0.03 | 96.91 | 93.54 | 55.05 |
| CT1_2 | 24750909 | 23407132 | 2.93G | 0.04 | 93.22 | 87.58 | 54.98 |
| CT2_1 | 21572245 | 20537549 | 2.57G | 0.03 | 97.04 | 93.82 | 54.76 |
| CT2_2 | 21572245 | 20537549 | 2.57G | 0.04 | 92.54 | 86.61 | 54.67 |
| CK1_1 | 23683593 | 22616430 | 2.83G | 0.03 | 96.97 | 93.60 | 55.64 |
| CK1_2 | 23683593 | 22616430 | 2.83G | 0.04 | 93.22 | 87.57 | 55.53 |
| CK2_1 | 23164623 | 21997516 | 2.75G | 0.03 | 96.87 | 93.40 | 55.77 |
| CK2_2 | 23164623 | 21997516 | 2.75G | 0.05 | 92.41 | 86.21 | 55.66 |
| Total | 186342740 | 177117254 | 22.16G |
CT1, CT2: Controlled medullar tissue.
CK1, CK2: Treated medullar tissue.
Q20: The percentage of bases with a Phred value > 20.
Q30: The percentage of bases with a Phred value > 30.
Figure 1. Length distribution of assembled unigenes.
Sequence annotation
After eliminating repeated and short-length sequences, 38,444 non-redundant unigenes were subjected to the following 7 public databases (Table 2) for similarity searching. Analyses showed that 24,027 unigenes (62.49%) had significant matches in the Nr database, 2,977 (7.74%) in the Nt database. Our results also showed that 14,214 (36.97%) of non-redundant unigenes demonstrated similarity to the known genes in Swiss-Prot database. In total, there were 25,947 unigenes (67.49%) successfully annotated in at least one of the Nr, Nt, Swiss-Prot, KEGG, GO, COG and Pfam databases, with 1563 unigenes (4.06%) in all seven databases.
Table 2. BLAST analysis of non-redundant unigenes against public databases.
| Number of Unigenes | Percentage (%) | |
|---|---|---|
| Annotated in NR | 24027 | 62.49 |
| Annotated in NT | 2977 | 7.74 |
| Annotated in KO | 7418 | 19.29 |
| Annotated in SwissProt | 14214 | 36.97 |
| Annotated in Pfam | 16270 | 42.32 |
| Annotated in GO | 17668 | 45.95 |
| Annotated in KOG | 10466 | 27.22 |
| Annotated in all Databases | 1563 | 4.06 |
| Annotated in at least one Database | 25947 | 67.49 |
| Total Unigenes | 38444 | 100 |
According to Gene Ontology (GO), an international standardized gene functional classification system, 17,688 non-redundant unigenes were classified into three major functional ontologies (biological process, cellular component and molecular function) (Fig. 2). For biological process (B P), dominant subcategories were ‘cellular process’ (10,661) and ‘metabolic process’ (10,466). In the category of cellular component (C C), ‘cell’ (6,188), ‘cell part’ (6,187) and ‘organelle’ (4,329) were highly represented. Among molecular function (M F) terms, ‘binding’ (9,840) and ‘catalytic activity’ (9,126) were most represented, followed by ‘transporter activity’ (1,280). However, within each of the three categories, few genes were assigned to subcategories of ‘cell killing’, ‘extracellular matrix part’ and ‘antioxidant activity’.
Figure 2. GO categorization of non-redundant unigenes.
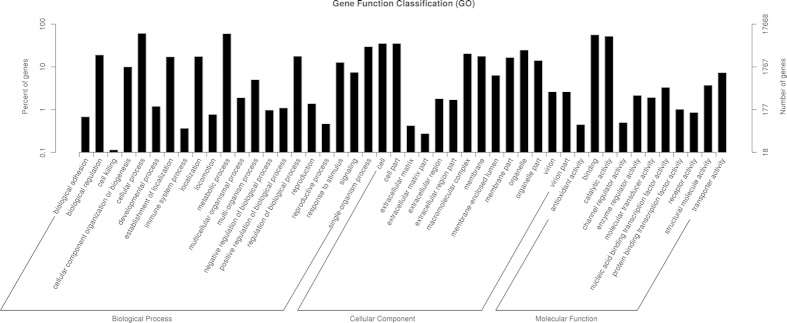
Each annotated sequence was assigned at least one GO term.
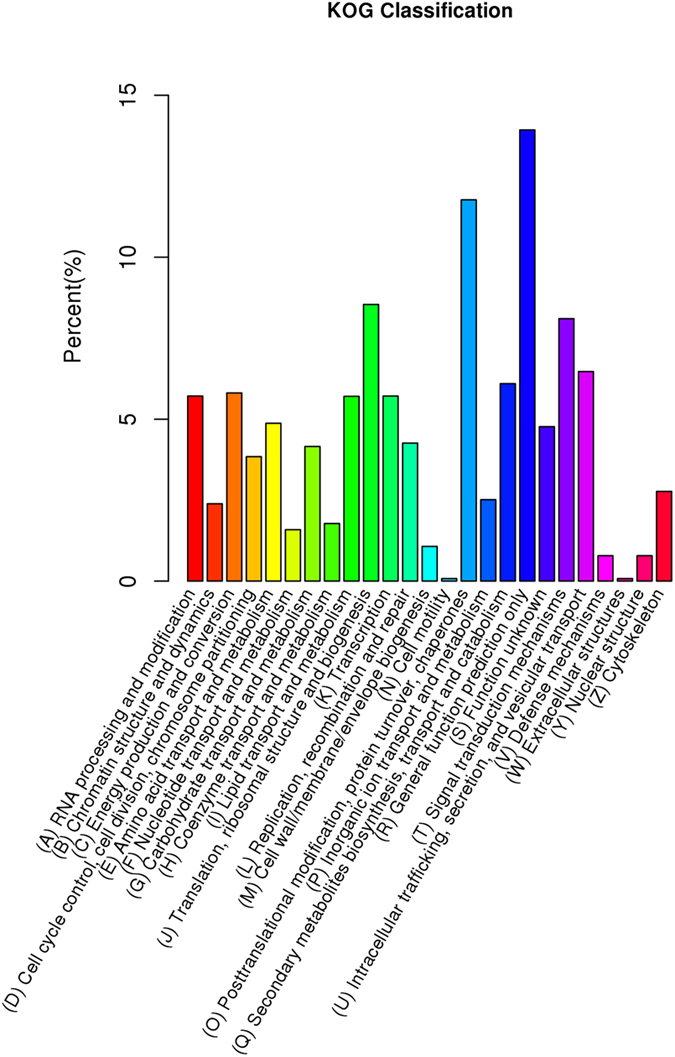
In addition, all unigenes were subjected to a search against the COG database for functional prediction and classification. 10,466 non-redundant unigenes (Table 2) were subdivided into 25 COG classifications (Fig. 3). Among them, the cluster of ‘general function prediction only’ (1,458) was the largest group, followed by ‘post-translational modification, protein turnover, chaperon’ (1,232), ‘translation’ (894), ‘signal transduction’ (848) and ‘Intracellular trafficking’ (677). Only a few unigenes were assigned to ‘extracellular structures’ (8) and ‘cell motility’ (8).
Figure 3. COG annotation of putative proteins.
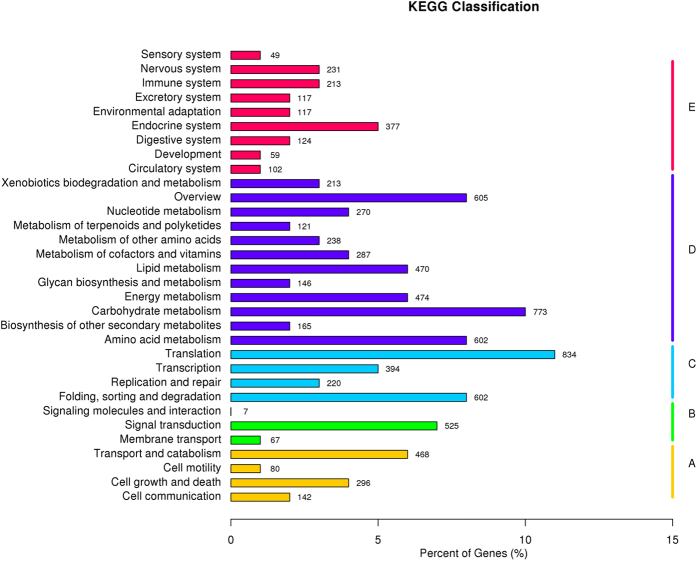
The unigene metabolic pathway analysis was also conducted using the KEGG annotation system. According to KEGG, 7418 unigenes (Table 2) were assigned to 266 pathways (Fig. 4). The pathways involving the largest number of unique transcripts were ‘translation’ (834), followed by ‘carbohydrate metabolism’ (773), whereas ‘signaling molecules and interaction’ (7) was the smallest group.
Figure 4. KEGG annotation of putative proteins.
Differential expression analysis of assembled P. umbellatus transcripts under A. mellea infection
To better survey the biological mechanism of defence, it is important to identify the DE genes between two different stages. To increase the accuracies of the measured expression levels for further analyses, data from two biological replicates were merged, and RPKM (Reads Per Kilo-base per Million reads) values were calculated based on the merged dataset. The resulting Pearson’s correlation coefficients (R) were significantly high between the replicates for CT (R = 0.83) and CK (R = 0.96) samples (Fig. 5). Differential expression analysis was firstly performed between the two medullar tissues. DEGs (q-value < 0.005 and |log2 (foldchange)| > 1) were defined as genes that were significantly enriched or depleted in one tissue relative to the other tissue.
Figure 5. Correlation of RPKM distribution between two biological replicates.
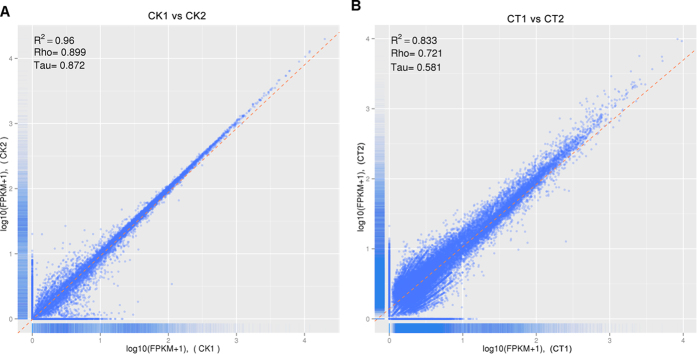
Comparisons of estimated RPKM distributions between biological replicates for (A) none-invading samples, (B) invading with A. mellea. Pearson’s correlation coefficients (R2) between replicates and statistical significance levels are presented.
Then, the DEGs between the CK and CT were analyzed. Of 38,444 unigenes, 10,933 were detected as significantly different by DEseq (Fig. 6). In this study, DEGs with higher expression levels in CT compared with CK were denoted as ‘up-regulated’, while those with lower expression levels in CT were ‘down-regulated’. The expression levels of 8780 of the 10993 genes were up-regulated in the CK while the other 2213 genes showed lower expression in the CK.
Figure 6. Volcano plot displaying differential expressed genes between CT and CK sample.
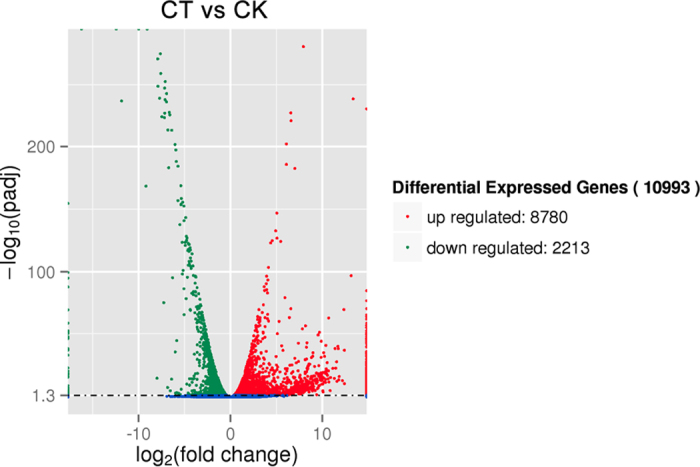
The y-axis corresponds to the mean expression value of log 10 (p-value), and the x-axis displays the log2 fold change value. The red dots represent the up regulated expressed transcripts (p < 0.05, false discovery rate (FDR) q < 0.05) between CT and CK; the green dots represent the transcripts whose expression down regulated (p < 0.05, FDR q < 0.05).
Functional distribution of differentially expressed genes
Differentially expressed genes were considered to be the important cause of defence response. To better survey the biological behavior of defence response, it is necessary to understand the functional distribution of these DE genes in P. umbellatus with A. mellea infection compared to the P. umbellatus without A. mellea infection.
GO analysis was conducted for the up-regulated DEGs in CT sample. The GO enrichment of up-regulated DEGs in CT was shown in supplementary Table S1. In the molecular function category, the top three enriched terms were threonine-type endopeptidase activity, threonine-type peptidase activity and sequence-specific DNA binding. In the cell components category, ‘proteasome complex’, ‘membrane part’ and ‘membrane’ were the three dominant enriched terms. For biological process, ‘protein folding’, ‘reductive tricarboxylic acid cycle’, and ‘chromatin assembly or disassembly’ was the mostly highly enriched. The iron transport was influenced, with ‘metal ion transmembrane transporter activity’ and ‘cation-transporting ATPase activity’ also enriched (P-value < 0.05). For the down-regulated DEGs in CK compared to CT sample, ‘response to organic substance’ and ‘cellular component biogenesis’ was the top-two biological process enriched by the down-regulated DEGs. The decline of organic substance and cellular component biogenesis might imply a great inhibition of biosynthetic process in the P. umbellatus. Consistent with this ‘biosynthesis’ was the top-three biological process enriched by down-regulated DEGs. The GO enrichment of down-regulated DEGs in CT was shown in supplementary Table S2. In the CK sample, ‘nucleoside monophosphate metabolic process’ and ‘pyrimidine-containing compound biosynthetic process’ were the third and fifth most enriched biological process, respectively. Taken together, proliferation of P. umbellatus cells was greatly limited with A. mellea infection, which may save much energy.
Biological behaviors of DEGs were complicated; the defence reaction is a coreaction result of many inner-cell metabolic pathways. KEGG pathway enrichment analysis for DEGs also revealed the complicated pathways related to defence response. Arginine and proline metabolism were the top-four enriched pathways (q ≤ 0.05) for the up-regulated DEGs in CT P. umbellatus cells, compared with CK, the KEGG pathway enrichment of up-regulated DEGs in the CT was shown in supplementary Table S3. There were 51 genes annotated as involved in this pathway. There were two DEGs annotated as Arginase (ARG), an important immune-related gene - comp31000_c0 and comp30397_c0 -with respectively 43.61- and 2.25-fold increased expression, which may lead to a defence reaction in the P. umbellatus cells. For the down-regulated DEGs in CT sample, ribosome were the top-one enriched pathways (q ≤ 0.05), the KEGG pathway enrichment of down-regulated DEGs in the CT was shown in supplementary Table S4. The ribosome is the place where mRNA is translated into protein. There were 11 DEGs annotated as ribosomal protein among the 49 genes annotated as involved in this ribosome pathway and all these 11 DEGs showed at least two fold lower expression in the CT sample. The decelebration of ribosome might imply a great inhibition of biosynthetic process, which is consistent with the biological process of GO analysis as discussed above leading to energy saving.
Validation of differentially expressed genes using qRT-PCR
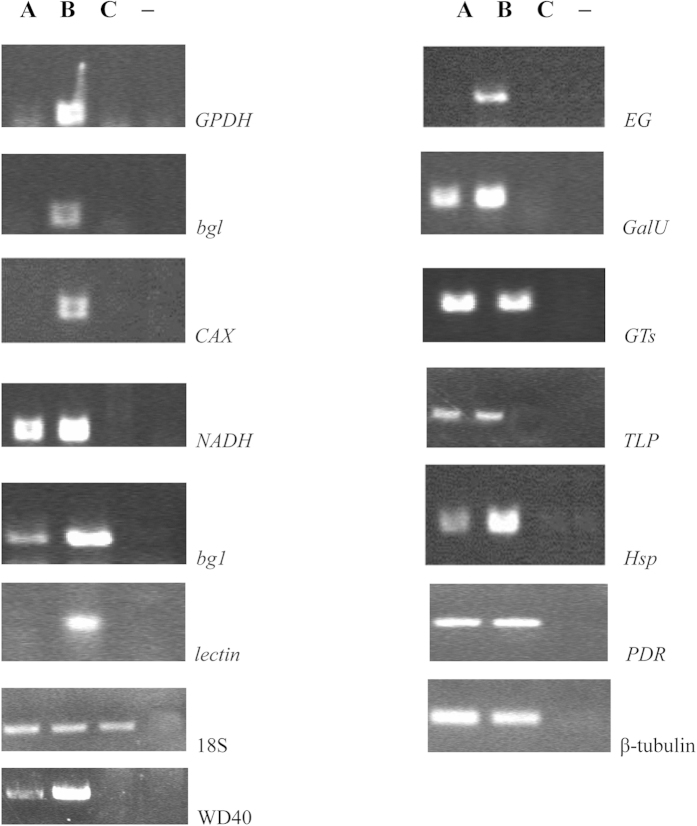
As the cDNA of CT sample was contaminated by the Armillaria mellea. Primers designed to the P. umbellatus beta-tubulin gene (EU442274) and ribosomal 18S (EU442272), as house-keeping genes, were amplified P. umbellatus transcripts in CK, CT cDNA and the genome DNA of A. mellea, which was isolated from the hyphae taken from the tissues of the P. umbellatus sclerotia18 respectively. However, only primers specific for beta-tubulin was not amplified a DNA fragment from the DNA of A. mellea, whereas amplicons were obtained for this sample with the ribosomal 18S specific primers which indicated that this beta-tubulin gene was specific in P. umbellatus (Fig. 7). For this reason, we only used beta-tubulin primers in the quantitative PCR experiments.
Figure 7. RT-PCR analyses of the P. umbellatus.
non-infected (A) and P. umbellatus, infected (B) tissues with A. mellea and A. mellea (C). −negative control.
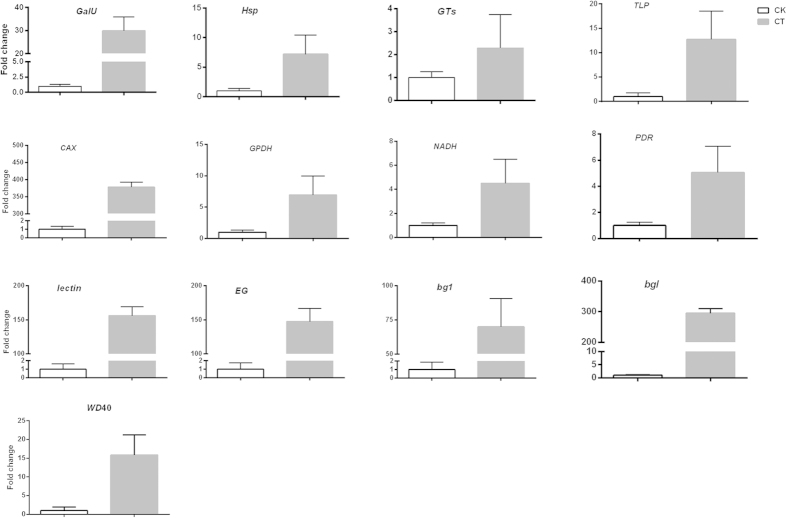
To confirm the reliability of the RNA-Seq data, the transcriptional level of 13 unigenes were examined by real-time quantitative PCR (Fig. 8). All the primers designed to the P. umbellatus were shown in supplementary Table S5. All the primers were PCR-amplified in the cDNA of CK, CT samples and genome DNA of A. mellea respectively. Only primers were not amplified for the sample of A. mellea DNA screened for further real-time quantitative PCR (qRT-PCR) analysis (Fig. 8). Since, new P. umbellatus sclerotia materials were used for the RNA extraction, the fold change did not exactly match the number revealed by the DEG analysis for these genes. All the 13 genes exhibited >2 fold higher expression in the P. umbellatus in response to A. mellea infection. Comp29904_c0 annotated as encoding lectin cannot be detected in the CK sample due to no/low expression, just as the result by the Illumina sequencing technology. Taken together, all the unigenes showed consistent expression patterns that were consistent with the RNA-Seq data, indicating that our experimental results were valid.
Figure 8. Quantitative real-time PCR (Q-PCR) validations of 12 genes that were differentially expressed between the CK (white bar) and CT (grey bar) samples.
For each Q-PCR validation, four technical replications were performed. beta-tubulin gene was used as internal control.
Discussion
In this subject, transcriptomes of P. umbellatus sclerotia were sequenced using the Illumina platform. In total, about 177 million high-quality reads with 22.16 Gb sequence coverage were obtained; there were 38,444 unigenes (≥200 bp) assembled and 100% were annotated. To the best of our knowledge, this is the first large-scale assessment of Polyporus genomic resources. Our results lay the foundation for development of molecular markers, construction of a genetic map and much other genomics research in Polyporus. To identify defence-responsive genes, 13 unigenes were selected. The expression of 13 unigenes was significantly up-regulated with A. mellea infection.
Molecular chaperones
The heat-shock proteins (Hsps) as one kind of molecular chaperones, are a ubiquitous feature of cells in which these proteins cope with stress-induced denaturation of other proteins19. The roles of Hsps in host-parasite interactions have received considerable attention. From the perspective of the host, Hsps expressed by invading parasites are potent antigens that elicit an immune response20,21,22. In this study, the expression of a small heat shock protein gene was induced by infection. Interestingly, our data agree with the observation that small heat shock protein gene was up-regulated by tobacco etch virus infection in Arabidopsis23, suggesting that the defence response may induce up-regulation of the small heat shock protein gene.
Reducing the oxidative damage
The induction of Reactive Oxygen Species (ROS)-related factors suggests that infection stress is accompanied by the production of ROS in organisms, and the induction of enzymes regenerating the reduced forms of antioxidants, and ROS interacting enzymes such as superoxide dismutase, peroxidases and catalases may be involved in the protection of tissues from oxidative damage under infection conditions24. The response and dynamics of antioxidant enzyme transcripts, including SOD, POD, PRX, APX and GR et al. are commonly used to study plant defence responses25. The expression pattern was also observed in this study, in which a GPDH was increased under infection by A. mellea in P. umbellatus. These results suggest that the antioxidative defence system was greatly activated in P. umbellatus. Consistent with the numerous studies that have shown a correlation between the ability to ameliorate ROS and survival under infection conditions, the high induction of ROS-related proteins in infected P. umbellatus showed that strong detoxification was critical for its survival.
Lectins as defence proteins
Lectins are a widespread class of proteins implicated in many essential cellular and molecular recognition processes such as cellular signaling, differentiation, host–pathogen interactions and tissue metastasis26. Fungal lectins take part in the defence of fungi since some present toxic activities such as insecticidal, vermicidal or antiviral27. For example, the β-trefoil lectins present in the sclerotia28 (compacted forms of mycelium) from S. sclerotiorum and R. solani are toxic towards the aphid Acyrthosiphon pisum29 and the cotton leafworm Spodoptera littoralis28, respectively. As expected and verified by qRT-PCR, similar to previous reports, a selected gene encoding lectin from P. umbellatus sclerotia was also induced by defence response in this study, suggesting that this P. umbellatus lectin have potential utility against A. mellea infection.
Pathogenesis-related proteins (PR-proteins)
Thaumatin-like proteins (TLPs) are a group of PR proteins (PR-5) that are induced in plants in response to infection by plant pathogens, elicitors, stress, and developmental signals30. The genes encoding PR-proteins exhibit strong in vitro antifungal activity. Several researches have demonstrated that the introduction of a single transgene encoding different antimicrobial proteins including PR-proteins in wheat resulted in enhanced resistance to a wide range of disease resistance31. Similarly, similar to the previous reports, a selected TLP from P. umbellatus was also induced by defence response in this study, which may suggest that the TLP can inhibit the growth of A. mellea for further invasion.
Secondary metabolism
Uridine triphosphate (UTP)-glucose-1-phosphate uridylyltransferase (GalU) is an enzyme that catalyzes the formation of uridine diphosphate (UDP)-glucose from UTP and glucose-1-phosphate. GalU is involved in virulence in a number of animal-pathogenic bacteria since its product, UDP-glucose, is indispensable for the biosynthesis of virulence factors such as lipopolysaccharide and exopolysaccharide32. The up-regulation pattern was also observed in this study, in which a GalU was increased under defence response in P. umbellatus. These results suggest that the GalU was a defence-induced gene in P. umbellatus and may enhance the virulence to prevent the further invasion of A. mellea.
Cell wall hydrolysis and fusion
The cell wall of orgnism represents an essential protection barrier against infection mechanisms and its integrity is important for the survival of the orgnism33. Fungal cell walls are complex structures but typically contain chitin, β-1, β-3- and β-1,6-glucan, mannan and cell wall proteins34. In our previous study, we found that P. umbellatus sclerotia has formed the cavity structure in the medullar tissue and isolate A. mellea to prevent further intrusion of A. mellea hyphae10. In order to form the cavity, the P. umbellatus sclerotia cell wall must first be hydrolyzed and then fused with other cell. The glucanolytic activities of several enzymes have been detected in the cell walls of some kind of fungi, such as Aspergillus fumigatus, Coccidioides posadasii and C. immitis. Some of these enzymes exhibit both glucanase and transglycosidase activities and, like the glucanases of yeasts, may play some roles in cell wall remodeling during morphogenesis35. β-1,3-Glucanases were also found up-regulated in the knob proteome of Monacrosporiu haptotylum, suggesting they may be important in fungal morphogenesis36. Interestingly, four genes annotated as encoding endoglucanase, beta-glucosidase, β-1,3-glucanase, glycosyltransferase were induced by defence in this research. The large-scale increase of mRNAs associated with glucanase and transglycosidase activities in P. umbellatus sclerotia indicated that defence response induces systemic inhibition of cell wall hydrolysis and fusion processes.
Regulating the fungus efflux
Many cellular processes, including plant responses to pathogens, are regulated by changes in cytosolic Ca2+ levels, where Ca2+ ions can serve to transduce a particular stimulus or stress that is imposed on a cell to target proteins that guide the cellular response37. It is well known that a Ca2+ elevation in the cytosol is an important event in plant cell perception of pathogen invasion and the calcium ion is a ubiquitous intracellular second messenger involved in plant defence responses38. In this study, a selected calcium/proton exchanger was induced in P. umbellatus by A. mellea infection (Fig. 8). This result may indicate that the role of calcium/proton exchanger in infection stress is to accumulate the calcium ion for mediating the signaling cascade.
Among the ABC transporters, the pleiotropic drug resistance (PDR) family is particular in that its members are found only in fungi and plants39. Several Saccharomyces cerevisiae PDR genes, such as PDR5 and SNQ2, have been functionally characterized in detail. They confer resistance to a large number of toxic compounds with no, or little, common structural or functional properties, such as fungicides, herbicides, pesticides, antibiotics, and detergents. PDR transporters are also involved in fungicide resistance of pathogenic fungi such as Candida albicans40 or Penicillium digitatum41. There was one unigene annotated as PDR in this study. Arabidopsis PDR12 was upregulated in response to oxylipins and the necrotrophic fungus Botrytis cinerea42,43. According to the above discussion, this may facilitate P. umbellatus defence response. Additionally, mRNA related to PDR (i.e. multidrug resistance-associated ABC transporter) was highly upregulated in the CT P. umbellatus. Thus, more attention needs to be paid to the relationship between PDR with P. umbellatus defence tolerance.
WD40 proteins
WD40 proteins were identified to play a crucial role in diverse protein-protein interactions by acting as scaffolding molecules and thus assisting the proper activity of the proteins44. It has been clarified that a subset of WD40 proteins involving in abiotic stress tolerance conducted in Arabidopsis and rice. HOS15, a WD40-repeat protein crucial for repression of genes associated with abiotic stress tolerance through histone deacetylation in Arabidopsis45. SRWD, a novel WD40 protein subfamily was regulated by salt stress in rice46. In this study, a unigene encoding for WD40 protein was significantly up-regulated by A. mellea infection, a kind of biotic stress, which suggest that the WD40 protein might involve in biotic stress tolerance of P. umbellatus. Thus, more attention needs to be paid to the biotic stress tolerance function studies of WD40 in other model plant.
In summary, the expression patterns of 13 putative genes that are involved in defence response were consistent with changes in their transcript abundance, mainly through molecular chaperones, reducing the oxidative damage, PR-proteins, and regulating secondary metabolism, cell wall hydrolysis and fusion. Meanwhile, regulating the fungus efflux plays an important role in defence response in P. umbellatus.
Conclusions
The combination of RNA-seq and DEGs analyses based on Illumina sequencing technology provided comprehensive information on gene expression. The substantially assembled sequences represented a considerable portion of the transcriptome of P. umbellatus. Based on the assembled de novo transcriptome, 10,933 DEGs were identified between the P. umbellatus sclerotial tissues with or without A. mellea infection. Kyoto Encyclopedia of Genes and Genomes pathway analysis uncovered that the differentially expressed genes were involved in important pathways, such as ‘metabolic pathways’. In addition, expression patterns of 13 selected DEGs were further validated with qRT-PCR, which reflected significant alteration in major signal pathway and metabolic pathways with the A. mellea infection. The information provided here can also further extend the knowledge regarding the molecular basis of the P. umbellatus defence reaction at the infection stage with A. mellea.
Methods
P. umbellatus sclerotia collection and preparation
As mentioned above, P. umbellatus sclerotia distributes in the nearer soil surface, such as 10–15 cm below the surface. In addition, it was hard to evaluate the infection stage by the infection time because of unlike the rice-pathogen interaction47, it took a long time such as 1–2 years for the rhizomorph of A. mellea adheres and invades P. umbellatus sclerotia48. Furthermore, it was difficult to determine the infection stage by the morphological characteristics of sclerotia medullar tissue. Therefore, in this study, we only set up two groups, P. umbellatus sclerotia without A. mellea infection (CK) and P. umbellatus sclerotia with A. mellea infection (CT). To increase the accuracies of the project, two biological replicates were included in each transcriptome sample.
P. umbellatus sclerotia (Fig. 9A) were collected from Guxian, Shanxi province in July 2014. The sclerotia were surface-disinfected by 75% ethanol for 30s, then cut into half and 100 mg of medullar tissue was removed. The tissue without invaded by the A. mellea was administered to the control group (CK), while the part invaded by the A. mellea rhizomorphs was treated as test group (CT) (Fig. 9B). Each sample was repeated twice, and all the four samples were frozen in liquid nitrogen immediately after collection and stored at −80 °C prior to RNA extraction.
Figure 9.
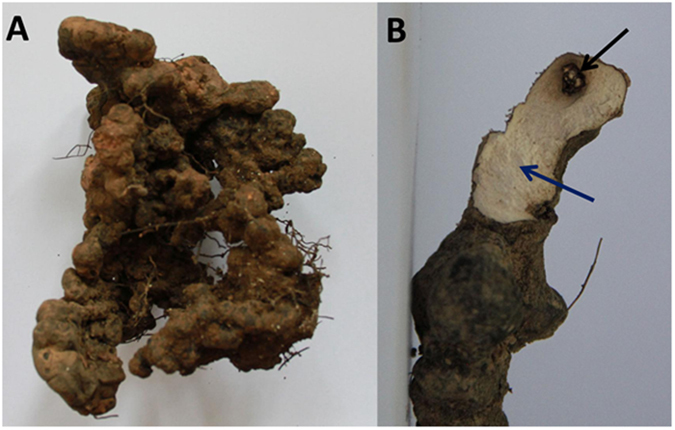
(A) The sclerotium of Polyporus umbellatus collected from Guxian, Shanxi. The surface of the sclerotium was attached to and penetrated by A. mellea rhizomorphs. (B) The internal part of the sclerotium. The black region (CT) surrounding the area invaded by rhizomorphs of A. mellea (black arrow) and and the none-invading region (CK) was white in color (blue arrow).
RNA extraction, library construction and sequencing
Total RNA was isolated from frozen medullar tissue by using the RNA plant mini kit with column DNAse digestion (Qiagen, Hilden, Germany) following the manufacturer’s instructions. RNA degradation and contamination was detected on 1.2% agarose gels. Then, we measured RNA concentration using Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA). Additionally, RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
A total amount of 3 μg RNA per sample was used as input material for the RNA preparations. Finally, four samples with RNA integrity number (RIN) values above 8 were used for libraries construction. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer. First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (NaseH−). Subsequently, second strand cDNA synthesis was performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments of preferential 150~200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and 125 bp paired-end reads were generated.
Sequence reads mapping, assembly and annotation
Raw data (raw reads) of fastq format were firstly processed through in-house Perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. At the same time, Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality.
The A, mellea reference genome and gene model annotation files were downloaded from genome website (http://www.icugi.org) directly. The Armillaria spp. reads derived from CT group were removed by the mapping of all reads against the A. mellea reference genome using Bowtie program (ver. 0.12.8)49 with the default parameters. Next, the remaining unmapped non-blast and non- Armillaria reads were used to examine the expression profiles of P. umbellatus. All the reads were then assembled using Trinity50 with min_kmer_cov and were set to 2 and all the other parameters were set to default.
Gene function was annotated based on the following seven databases: Nr (NCBInon-redundantproteinsequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG(Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), KO(KEGG Ortholog database) and GO (Gene Ontology), using BLAST with a cutoff E-value of 10−5.
Quantification and differential expression analysis of transcripts
Gene expression levels were estimated by RSEM51 for each sample. Clean data were mapped back onto the assembled transcriptome and then the readcount for each gene was obtained from the mapping results.
Differential expression analysis of two groups was performed using the DESeq R package (1.10.1). DESeq52 provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by DESeq were assigned as differentially expressed.
GO and KEGG enrichment analysis of differentially expressed transcripts
Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseqR packages based on Wallenius non-central hyper-geometric distribution53, which can be adjusted for gene length bias in DEGs.
KEGG54 is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from the molecular-level information, especially from large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). We used KOBAS software55 to test the statistical enrichment of differential expression genes in KEGG pathways.
Confirmation of the infection-responsive expression profiles by qRT-PCR
Differentially expressed genes identified by the above described method were validated using quantitative real-time PCR (qPCR). The real-time PCR was performed with the SYBR® Premix ExTaqTM (TaKaRa, Dalian, China) on the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The beta-tubulin gene was used as a reference control. The reaction was performed using the following conditions: denaturation at 95 °C for 30 s, followed by 40 cycles of amplification (95 °C for 5 s, 60 °C for 34 s). Each plate was repeated three times in independent runs for all reference and selected genes. Gene expression was evaluated by the 2−ΔΔCt method56.
Statistical analysis
For each sample, three technical replicates of the qRT-PCR assay were used with a minimum of three biological replicates. Results were expressed as means ± standard deviation (SD) of the number of experiments. A Student’s t-test for the values was performed at P < 0.05.
Additional Information
How to cite this article: Liu, M.-M. et al. Transcriptome analysis of genes involved in defence response in Polyporus umbellatus with Armillaria mellea infection. Sci. Rep. 5, 16075; doi: 10.1038/srep16075 (2015).
Supplementary Material
Acknowledgments
The research was financially supported by the National Natural Sciences Foundation of China (No. 30830117, No. 31201666), by the Program for Innovative Research Team in IMPLAD (PIRTI-IT1302).
Footnotes
Author Contributions M.M.L., Y.M.X. and S.X.G. conceived and designed the experiments. Y.M.X., D.W.Z. and M.M.L. performed the experiments. D.W.Z. analysed the data. M.M.L. wrote the paper. All authors read and approved the final manuscript.
References
- Chen X. & Ronald P. C. Innate immunity in rice. Trends Plant Sci 16, 451–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B. & Khang C. H. Recent advances in rice blast effector research. Curr Opin Plant Biol 13, 434–41 (2010). [DOI] [PubMed] [Google Scholar]
- Wilson R. A. & Talbot N. J. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7, 185–95 (2009). [DOI] [PubMed] [Google Scholar]
- Li X., Cui Z., Liu Y., Song C. & Shi G. Transcriptome analysis and discovery of genes involved in immune pathways from hepatopancreas of microbial challenged mitten crab Eriocheir sinensis. PLoS One 8, e68233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J. et al. Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS One 8, e82155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. M., Yin W. Q., Liu M. M., Wang C. L. & Guo S. X. Oxalic acid and sclerotial differentiation of Polyporus umbellatus. Sci Rep 5, 10759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. M. et al. Sclerotial formation of Polyporus umbellatus by low temperature treatment under artificial conditions. PLoS One 8, e56190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.-X. & Xu J.-T. Genesis and function of defense structure of sclerotia of Grifola umbellata after Armillaria mellea infection. Acta Mycol Sin 12, 283–288 (1993). (in Chinses) [Google Scholar]
- Hasegawa E., Ota Y., Hattori T. & Kikuchi T. Sequence-based identification of Japanese Armillaria species using the elongation factor-1 alpha gene. Mycologia 102, 898–910 (2010). [DOI] [PubMed] [Google Scholar]
- Guo S.-X. & Xu J.-T. Genic microsatellite markers in plants: features and applications. Acta Bot Sin 35, 44–50 (1993). (in Chinses) [Google Scholar]
- Yao Y. et al. Identification of differentially expressed genes in leaf and root between wheat hybrid and its parental inbreds using PCR-based cDNA subtraction. Plant Mol Biol 58, 367–84 (2005). [DOI] [PubMed] [Google Scholar]
- Marioni J. C., Mason C. E., Mane S. M., Stephens M. & Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18, 1509–17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T., Ye C. Y., Xia X. & Yin W. De novo sequencing and transcriptome analysis of the desert shrub, Ammopiptanthus mongolicus, during cold acclimation using Illumina/Solexa. BMC Genomics 14, 488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X. D. et al. Gene expression profiling of soybean leaves and roots under salt, saline-alkali and drought stress by high-throughput Illumina sequencing. Gene 512, 392–402 (2013). [DOI] [PubMed] [Google Scholar]
- Postnikova O. A., Shao J. & Nemchinov L. G. Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol 54, 1041–55 (2013). [DOI] [PubMed] [Google Scholar]
- Li H., Yao W., Fu Y., Li S. & Guo Q. De novo assembly and discovery of genes that are involved in drought tolerance in Tibetan Sophora moorcroftiana. PLoS One 10, e111054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. H. et al. Gene expression profiling of Sinapis alba leaves under drought stress and rewatering growth conditions with Illumina deep sequencing. Mol Biol Rep 39, 5851–7 (2012). [DOI] [PubMed] [Google Scholar]
- Fukuda M., Nakashima E., Hayashi K. & Nagasawa E. Identification of the biological species of Armillaria associated with Wynnea and Entoloma abortivum using PCR-RFLP analysis of the intergenic region (IGR) of ribosomal DNA. Mycol Res 107, 1435–41 (2003). [DOI] [PubMed] [Google Scholar]
- Feder M. E. & Hofmann G. E. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61, 243–82 (1999). [DOI] [PubMed] [Google Scholar]
- Maresca B. & Kobayashi G. S. Hsp70 in parasites: as an inducible protective protein and as an antigen. Experientia 50, 1067–74 (1994). [DOI] [PubMed] [Google Scholar]
- Kaufmann S. H. The cellular immune response to heat shock proteins. Experientia 48, 640–3 (1992). [DOI] [PubMed] [Google Scholar]
- Polla B. S. Heat shock proteins in host-parasite interactions. Immunol Today 12, A38–41 (1991). [DOI] [PubMed] [Google Scholar]
- Whitham S. A., Anderberg R. J., Chisholm S. T. & Carrington J. C. Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12, 569–82 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Czymmek K. J., Caplan J. L., Sweigard J. A. & Donofrio N. M. Suppression of plant-generated reactive oxygen species is required for successful infection by the rice blast fungus. Virulence 2, 559–62 (2011). [DOI] [PubMed] [Google Scholar]
- Mustroph A. et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152, 1484–500 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoving J. C., Wilson G. J. & Brown G. D. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol 16, 185–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrot A., Basheer S. M. & Imberty A. Fungal lectins: structure, function and potential applications. Curr Opin Struct Biol 23, 678–85 (2013). [DOI] [PubMed] [Google Scholar]
- Hamshou M. et al. High entomotoxicity and mechanism of the fungal GalNAc/Gal-specific Rhizoctonia solani lectin in pest insects. J Insect Physiol 59, 295–305 (2013). [DOI] [PubMed] [Google Scholar]
- Hamshou M. et al. Insecticidal properties of Sclerotinia sclerotiorum agglutinin and its interaction with insect tissues and cells. Insect Biochem Mol Biol 40, 883–90 (2010). [DOI] [PubMed] [Google Scholar]
- Anand A. et al. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot 54, 1101–11 (2003). [DOI] [PubMed] [Google Scholar]
- Oldach K. H., Becker D. & Lorz H. Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat. Mol Plant Microbe Interact 14, 832–8 (2001). [DOI] [PubMed] [Google Scholar]
- Liao C. T., Du S. C., Lo H. H. & Hsiao Y. M. The galU gene of Xanthomonas campestris pv. campestris is involved in bacterial attachment, cell motility, polysaccharide synthesis, virulence, and tolerance to various stresses. Arch Microbiol 196, 729–38 (2014). [DOI] [PubMed] [Google Scholar]
- Latge J. P. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol 66, 279–90 (2007). [DOI] [PubMed] [Google Scholar]
- Coronado J. E., Mneimneh S., Epstein S. L., Qiu W. G. & Lipke P. N. Conserved processes and lineage-specific proteins in fungal cell wall evolution. Eukaryot Cell 6, 2269–77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. J. Fungal cell wall chitinases and glucanases. Microbiology 150, 2029–35 (2004). [DOI] [PubMed] [Google Scholar]
- Andersson K. M. et al. Proteome of the nematode-trapping cells of the fungus Monacrosporium haptotylum. Appl Environ Microbiol 79, 4993–5004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D., Ranjeva R. & Pugin A. Calcium in plant defence-signalling pathways. New Phytol 171, 249–69 (2006). [DOI] [PubMed] [Google Scholar]
- Ma W. & Berkowitz G. A. Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol 190, 566–72 (2011). [DOI] [PubMed] [Google Scholar]
- Krattinger S. G. et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–3 (2009). [DOI] [PubMed] [Google Scholar]
- Prasad R. & Kapoor K. Multidrug resistance in yeast Candida. Int Rev Cytol 242, 215–48 (2005). [DOI] [PubMed] [Google Scholar]
- Nakaune R. et al. A novel ATP-binding cassette transporter involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Appl Environ Microbiol 64, 3983–8 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt N. A. Oxylipin signaling in plant stress responses. Plant Cell 20, 495–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham A. et al. Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS One 10, e0125666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A. K., Muthamilarasan M., Khan Y., Parida S. K. & Prasad M. Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS One 9, e86852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA 105, 4945–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. et al. SRWD: a novel WD40 protein subfamily regulated by salt stress in rice (OryzasativaL.). Gene 424, 71–9 (2008). [DOI] [PubMed] [Google Scholar]
- Kawahara Y. et al. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One 7, e49423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X., Ma X. & Guo S. Fungal species residing in the sclerotia of Polyporus umbellatus. Symbiosis 56, 19–24 (2012). [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K. & Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36, D480–4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J. G. & Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–93 (2005). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.