Abstract
Staphylococcus aureus is a common nosocomial pathogen with property to develop resistance to antimicrobial agents. But in the modern era, drug resistance had been developed by microbes due to its continuous usage of antibiotics. This study was carried out to evaluate antibiotic resistant pattern of methicillin resistant Staphylococcus aureus (MRSA) using molecular genotyping. In view of the present problem, the study has been conducted to detect the molecular genotyping of mecA gene from MRSA and confirmation of its restriction sites using EcoRI and BamHI. The pus samples were swabbed out, and clinical strains were isolated using standard microbiological procedures. Then the strains were subjected to in vitro antibiotic susceptibility assay and identified MRSA. Further molecular genotyping of mecA gene was determined by polymerase chain reaction technique. The percentage analysis was done. The clinical strains were isolated from the wound infected patients. A total of 60 samples were collected, of 60 samples, 40 (66.7%) were showed positive to strains of S. aureus. The in vitro antibiotic susceptibility assay was carried to find the drug sensitive and resistant patterns. Further methicillin resistant strains (35%) of S. aureus were screened and subjected to molecular genotyping of mecA gene and was confirmed by restriction digestion. Overall, 70% of plasmids show positive for the presence of mecA gene, although all strains have restriction sites. Hence, the present study revealed that the early detection of antibiotic resistant character using molecular genotyping will help the infected patient to cure short period and will reduce the development of multidrug resistance.
Keywords: Antibiogram, in vitro, mecA gene, methicillin resistant Staphylococcus aureus, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus caused a wide range of major and minor infection in man and animals and is characterized by its ability to clot blood plasma by the action of the enzyme coagulase. The emerging drug resistant strain of S. aureus associated with risk factors such as drug resistant in Intensive Care Unit, changes in blood cells etc., in children.[1]
Methicillin resistant Staphylococcus aureus (MRSA) is present account for 3–5% of all isolates of S. aureus. The recent research carried out by Abbas et al.[2] was reported that hospital acquired-MRAS and community acquired-MRSA were more prevalent at a tertiary care hospital. The detailed molecular mechanism with respect to MRSA resistant genes and their regulation was propounded by Sangappa and Thiagarajan.[3]
The present study is undertaken to investigate the antibiotic resistant pattern of MRSA and determination of the presence of mecA gene and amplification of mecA gene by polymerase chain reaction (PCR) and analysis of its restriction sites.
MATERIALS AND METHODS
Collection of specimens
The wound swabs were collected from patients admitted in Sundaram Medical Foundation, Chennai, Tamil Nadu. The samples were collected aseptically with ice box and transported to the laboratory immediately, stored in refrigerator, and the analysis was carried out within 2 h of times.
Wound swabs
A sterile dry cotton swab was pressed and rolled over the infected tissue, and the swab was inserted in the alkaline peptone water. The swab was pushed down one-third into the alkaline peptone water, and the cap was screwed.
Isolation and biochemical screening of Staphylococcus aureus
The specimen was inoculated on nutrient agar and incubated at 37°C overnight. The biochemical tests such as catalase, oxidase, coagulase, and sugar fermentation were done for further confirmation.[4]
In vitro antibiotic susceptibility test by disc diffusion assay
The pure clinical isolates of S. aureus were Gram-stained before the test. The S. aureus cultures were inoculated in peptone water and incubated at 37°C to get turbidity equal. A sterile cotton swab was dipped into the inoculation, and the excess was removed by pressing the swab to the sides of the tube. The entire agar surface was swabbed. The inoculation was allowed to dry for 15 min. Methicillin and other antibiotic discs such as ampicillin, chloramphenicol, gentamycin, tetracycline, and erythromycin were applied on the medium. The plates were incubated at 37°C and examined after 18–24 h.
Plasmid isolation by alkaline lysis method
The pure culture of S. aureus was inoculated into LB broth with ampicillin and incubated overnight at 37°C. To this, 10 ml of well-growth culture was transferred into a centrifuge tube. The cells were pelleted at 10,000 rpm for 5 min in a refrigerated centrifuge, and the supernatant was discarded. The pellet was resuspended in 150 μl of solution A and was incubated in ice for 30–40 min. Added 250 μl of solution B into the centrifuge tube and vortex it and then incubated on ice for 10 min. Similarly, 150 μl of solution C was added and incubated in ice for 60 min. Centrifugation was done at 6000 rpm for 10 min. The supernatant was transferred into a fresh microfuge tube. Added equal amount of ice cold 95% ethanol and kept it in −20°C for overnight. The solution D was added after 24 h and kept for 10 min. Then centrifugation was done at 6000 rpm for 10 min. The supernatant was removed, and the pellet was dried. The dried plasmid DNA was dissolved in 50 μl of Tris buffer. Loading dye was mixed with 20 μl of sample and run in agarose gel electrophoresis.
Agarose gel electrophoresis
Tris-acetate-ethylenediaminetetraacetic acid (TAE) is most commonly used buffer, but it gets easily exhausted during extended or high voltage electrophoresis. Agarose gel electrophoresis is carried out in a horizontal submarine electrophoresis unit. Prepare 1 L of tank buffer using ×100 TAE buffer (take 10 ml of × 1 TAE buffer and then add 990 ml of double distilled water). For the preparation of gel electrophoresis, dissolve by heating 1.0 g of agarose in 100 ml of × 1 TAE buffer, before pouring into the gel cast add ethidium bromide. Load the gel with 15 μl of sample and 5 μl of gel loading dye. Electrophoresis is carried for 1 h depending upon the length of the gel, until the gel loading dye migrates more than half the length of gel. At the end of the electrophoresis, visualize the plasmid DNA under UV transilluminator (Biorad, USA).
RESULTS
The pus samples were collected from 60 wound patients for the isolation of MRSA. Of the 60 pus samples, 40 (66.7%) samples showed positive result for S. aureus. Of 40 isolated strains, 14 are MRSA.
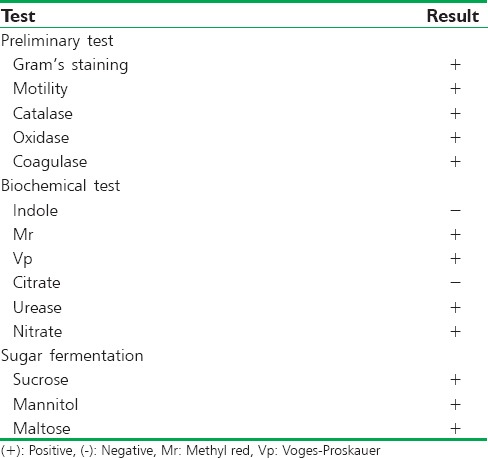
The organisms were identified from the morphological, cultural, and biochemical testing. The S. aureus was isolated in nutrient agar and blood agar. In nutrient agar, the colony shows circular, convex, shiny, opaque, smooth colonies with slightly golden yellow pigment. In blood agar, beta-hemolytic white colonies were observed. S. aureus was confirmed using selective media like Mannitol Salt Agar. S. aureus strains are detected by the sugar fermentation test. Further, S. aureus was confirmed from catalase, oxidase, coagulase, and various biochemical characterization test like indole production test, methyl red test, Voges–Proskauer test, citrate utilization test, urease production test, and Nitrate reduction test. The results are shown in Table 1.
Table 1.
The biochemical test for the identification of isolated Staphylococcus aureus
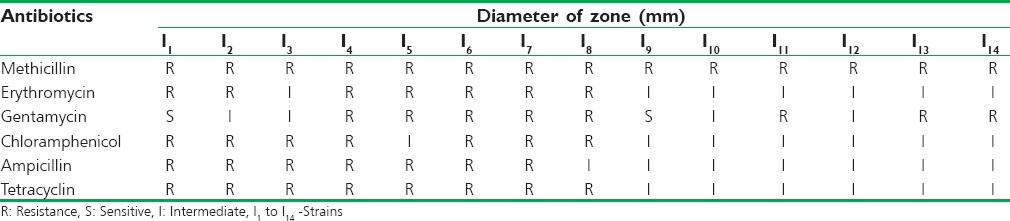
The in vitro antibiotic sensitivity patterns of the S. aureus toward the antibiotics like ampicillin, penicillin, chloramphenicol, tetracycline, erythromycin, and methicillin. The organisms showed resistant toward methicillin are called as MRSA [Figure 1]. The sensitivity was determined from comparing the results with zone size interpretative chart provided by Hi-Media Laboratories Ltd. The results are shown in Table 2.
Figure 1.

Staphylococcus aureus showing methicillin resistant
Table 2.
In vitro antibiotic sensitivity pattern of Staphylococcus aureus
The plasmid profile was done by alkaline lysis method [Figure 2] and was carried out to study the level of sensitivity pattern between the antibiotic resistant strains of MRSA and determining the presence of mecA gene by PCR [Figure 3] and analyzing of its restriction sites using EcoRI and BamHI [Figures 4 and 5].
Figure 2.
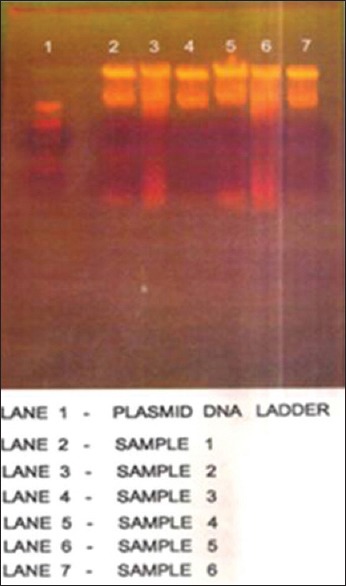
Extraction of plasmid DNA from methicillin resistant Staphylococcus aureus
Figure 3.
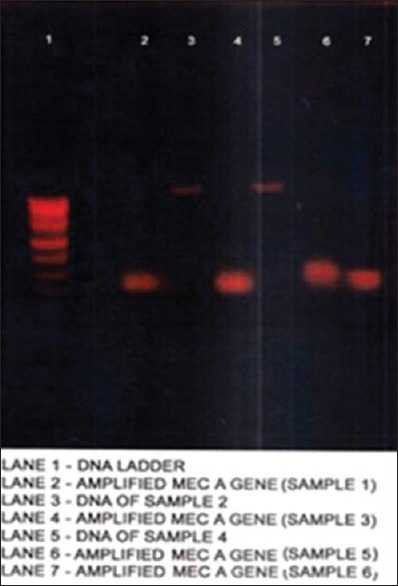
Polymerase chain reaction amplification of mecA gene
Figure 4.
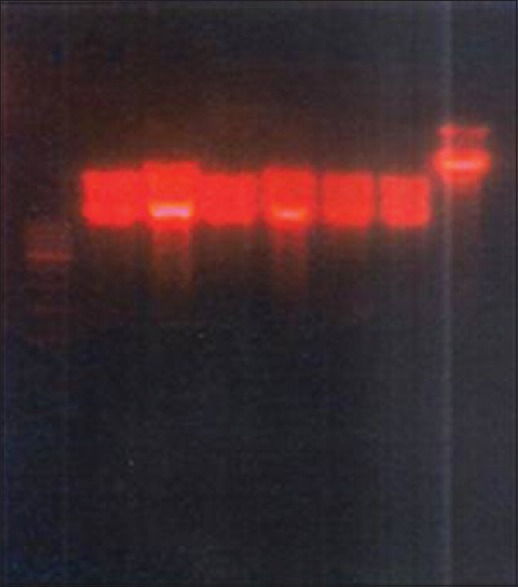
Restriction analysis of plasmid DNA by EcoRI
Figure 5.
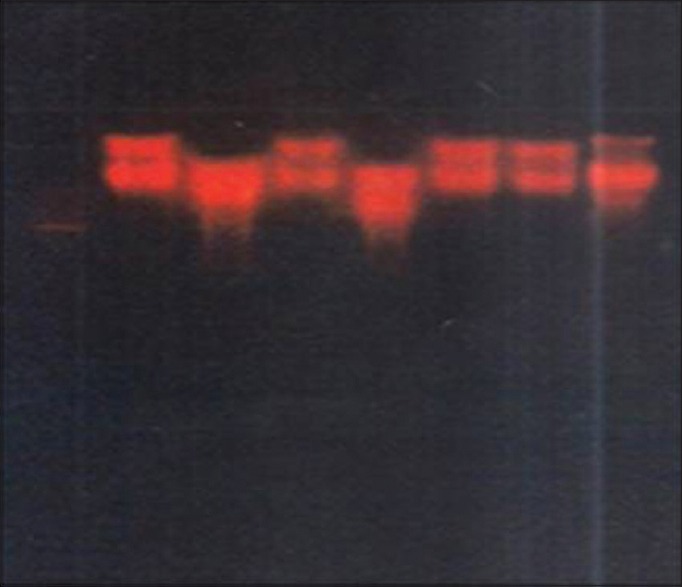
Restriction analysis of plasmid DNA by BamHI
DISCUSSION
In the recent years, there has been more report on the prevalence and drug resistant of S. aureus. As these bacteria are increasingly gaining resistance to commonly used antibacterial agents, there is immediate need for finding authentic detection of drug resistant strains for controlling risk factors due to infection. Therefore, the present study has made an attempt in exploring molecular detection of mecA gene from MRSA from wound infected patients and confirmation of its restriction sites using EcoRI and BamHI.
Qureshi et al.[5] had screened the clinical isolates of S. aureus using a set of primers specific for mecA gene. The researchers reported that 76.44% were identified as MRSA and 19.6% as MRSA. The mecA gene was found positive in 89 (MRSA and MSSA) isolates (87.22%), 20 of 98 isolates that were categorized as MSSA by disc diffusion method, 11 showed amplification of mecA gene by PCR. Sasirekha et al.[6] had evaluated different conventional phenotypic methods in detecting MRSA and determined the prevalence of bioflim producer among MRSA. A total of 153 clinical isolates of S. aureus were subjected to antibiotic susceptibility test by disc diffusion. Of 153 isolates, 42 (57.7%) were found to be methicillin resistant, and drug resistance patterns of MRSA isolates were performed. The another study conducted by Kaur et al.[7] had estimated the frequency of Panton-Valentine leukocidin (PVL) positive S. aureus in Belgaum, South India. The clinical isolates were identified, phenotypically characterized as methicillin sensitive and methicillin resistant strains of S. aureus by disc diffusion method by employing oxacillin disc (1 μg) and genetically by multiplex PCR for mecA and femB genes. The researchers were confirmed 38.6% as MRSA by PCR for mecA and also showed high percentage of PVL positive MRSA and MSSA, respectively.
Hussain et al.[8] had screened the antimicrobial susceptibility pattern of S. aureus. They were isolated 80 strains, in which 53 (66.25%) strains were found to be methicillin resistant while the rest of the 27 (33.75%) strains were methicillin-susceptible. In contrast to the present study, we had isolated 66.7% of S. aureus, of which 35% of clinical strains were showed resistance to methicillin.
The plasmid DNA was extracted by alkaline lysis method and bands observed on agarose gel electrophoresis revealed that there is no remarkable change in the molecular level between MRSA strains. The isolated plasmid DNA was used as template in the PCR study. The protocol developed by Merlino et al.[9] was followed in the present study for amplification of plasmid DNA. Amplification was performed using specific primers mecA gene forward primer (5‘-AAAATCGATGGTAAAGGTTGGC-3’) and mecA reverse primer (5‘-AGTTCTGCAGTACCGGATTTGC-3’).
Irayyif et al.[10] had identified MRSA by a rapid PCR technique. The researchers were collected blood sample from the wound infected patients. The bacterial genomic DNA was isolated and amplified using the forward primer, 5’CTGCTATCCACCCTCAAACAG3’ and the reverse primer, 5’CACCTTGTCCGTAACCTGAATC3’, respectively. Further, the plasmid isolation was done by alkaline lysis method, and confirmation of the clones was subjected to restriction digestion using BamH1 and EcoR1. Another study conducted by Khan et al.[11] had amplified mecA and nuc genes in multidrug resistant S. aureus strains from hospital personnel. The mecA gene was amplified using primer 5‘AGTTGTAGTTGTCGGGTTT-3’ and 5‘AGTGGAACGAAGGTATCATC-3’. The result revealed that of 17 clinical isolates, 7 (41.18%) strains were showed the present of mecA gene.
For molecular genotyping, the gene of interest is mecA gene that is responsible for the resistance to the drug called methicillin and codes for penicillin-binding protein 2A.[12] But in the recent year, apart from studying mecA gene, the researchers[13,14] were found a new mechanism of MRSA for screening mecC gene for genotypic confirmation of MRSA.
The PCR was performed in thermal cycler (Eppendorf Company, USA) using PCR conditions. The final PCR products were resolved in 1% agarose using TE buffer.
The results clearly indicated that 4 strains among 6 carried mecA gene. The resistance for the remaining 2 strains may have been encoded by different gene, such as femA gene.[15]
The difference in strains may also be analyzed by screening for restriction sites using restriction enzymes. Hence, screening for restriction sites were carried out by employing EcoRI and BamHI restriction enzymes. The results in our study showed that the presence of restriction sites for both the enzymes in all the positive strains.
CONCLUSIONS
S. aureus is a common nosocominal pathogen with property to develop a resistant to antimicrobial agents. In the present study, isolation of clinical strains from wound sample, microbiological characterization of S. aureus, Plasmid isolation, mecA gene amplification by PCR and restriction digestion pattern of the MRSA were carried out. Among 60 samples from the infected patients, 40 (66.7%) samples were showed positive for S. aureus. From this, 14 (35%) were resistant to the antibiotic-methicillin, so called MRSA. All the MRSA isolates have the same molecular weight plasmid. The plasmid DNA on restriction digestion with EcoRI and BamHI showed banding pattern that confirms the presence of restriction sites. Dialyzed plasmid DNA was amplified for the confirmation of the presence of mecA gene. Seventy percentage of plasmids show positive for the presence of mecA gene.
Several research works[16,17,18,19,20,21,22,23] had been conducted in India with reference to the molecular detection of mecA gene from MRSA. But no studies have been reported in India by detecting mecA gene using mecA gene forward primer (5‘-AAAATCGATGGTAAAGGTTGGC-3’) and mecA gene reverse primer (5 ‘-AGTTCTGCAGTACCGGATTTGC-3’), which was prescribed and developed by Merlino et al.[9] To the best of our knowledge, this constitutes the first report in India on the molecular detection of mecA gene from S. aureus. In addition, 70% of MRSA plasmids confirm the presence of mecA gene although all MRSA strains have restriction sites. Henceforth, early detection of MRSA could reduce morbidity and mortality and also reduce the development of multidrug antibiotic resistance.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Kapil A. Risk factors associated with MRSA infection in children. Indian Pediatr. 2015;52:22–4. [PubMed] [Google Scholar]
- 2.Abbas A, Nirwan PS, Srivastava P. Prevalence and antibiogram of hospital acquired-methicillin resistant Staphylococcus aureus and community acquired-methicillin resistant Staphylococcus aureus at a tertiary care hospital National Institute of Medical Sciences. Community Acquir Infect. 2015;2:13–5. [Google Scholar]
- 3.Sangappa M, Thiagarajan P. Methicillin resistant Staphylococcus aureus: Resistance genes and their regulation. Int J Pharm Pharm Sci. 2012;4:658–67. [Google Scholar]
- 4.Versalovic J, Caroll KC, Funke G, Jogensen JH, Landry ML. 10th ed. Washington: ASM Press; 2007. Manual of Clinical Microbiology. [Google Scholar]
- 5.Qureshi A, Ingle R, Musaddiq M, Ali Y, Khan Z. Status and distribution of mecA gene in hospitalized patient's MRSA isolates. Biosci Discov. 2012;3:52–7. [Google Scholar]
- 6.Sasirekha B, Usha MS, Amrutha AJ, Ankit S, Brinda N, Divya R. Evaluation and comparison of different phenotypic tests to detect methicillin resistant Staphylococcus aureus and their bioflim production. Int J PharmTech Res. 2012;4:532–41. [Google Scholar]
- 7.Kaur H, Purwar S, Saini A, Kaur H, Karadesai SG, Kholkute SD, et al. Status of Methicillin resistant Staphylococcus aureus infections and evaluation of PVL producing strains in Belgaum, South India. J Krishna Inst Med Sci Univ. 2012;1:43–51. [Google Scholar]
- 8.Hussain JH, Thakur A, Mishra B, Dogra V, Jaggi T. Antimicrobial susceptibility pattern of methicillin-resistant strains of Staphylococcus aureus in a super specialty hospital. Int J Health Allied Sci. 2015;4:69–72. [Google Scholar]
- 9.Merlino J, Watson J, Rose B, Beard-Pegler M, Gottlieb T, Bradbury R, et al. Detection and expression of methicillin/oxacillin resistance in multidrug-resistant and non-multidrug-resistant Staphylococcus aureus in Central Sydney, Australia. J Antimicrob Chemother. 2002;49:793–801. doi: 10.1093/jac/dkf021. [DOI] [PubMed] [Google Scholar]
- 10.Irayyif SM, Senthil Kumar R, Malla S. Identification of methicillin resistant Staphylococcus aureus by a rapid polymerase chain reaction technique. Asian J Pharm Clin Res. 2014;7:16–9. [Google Scholar]
- 11.Khan AU, Sultan A, Tyagi A, Zahoor S, Akram M, Kaur S. Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel. J Infect Dev Ctries. 2007;1:289–95. [PubMed] [Google Scholar]
- 12.Wielders CL, Fluit AC, Brisse S, Verhoef J, Schmitz FJ. mecA gene is widely disseminated in Staphylococcus aureus population. J Clin Microbiol. 2002;40:3970–5. doi: 10.1128/JCM.40.11.3970-3975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walther B, Wieler LH, Vincze S, Antão EM, Brandenburg A, Stamm I, et al. MRSA variant in companion animals. Emerg Infect Dis. 2012;18:2017–20. doi: 10.3201/eid1812.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paterson GK, Morgan FJ, Harrison EM, Peacock SJ, Parkhill J, Zadoks RN, et al. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother. 2014;69:598–602. doi: 10.1093/jac/dkt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM, Qian Q. Methicillin-resistant Staphylococcus aureus: Comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol. 2001;39:3946–51. doi: 10.1128/JCM.39.11.3946-3951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadig S, Namburi P, Raghunath D, Arakere G. Genotyping of methicillin-resistant Staphylococcus aureus Isolates from Indian hospitals. Curr Sci. 2006;91:1364–9. [Google Scholar]
- 17.Khan AU, Sultan A, Tyagi A, Zahoor S, Akram M, Kaur S, et al. Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel. J Infect Dev Ctries. 2007;1:289–95. [PubMed] [Google Scholar]
- 18.Mohanasoundaram KM, Lalitha MK. Comparison of phenotypic versus genotypic methods in the detection of methicillin resistance in Staphylococcus aureus. Indian J Med Res. 2008;127:78–84. [PubMed] [Google Scholar]
- 19.Bennimath VD, Gavimath CC, Kalburgi PB, Kelmani C. Amplification and sequencing of mecA gene from methicillin resistance Staphylococcus aureus. Int J Adv Biotechnol Res. 2011;2:310–4. [Google Scholar]
- 20.Sasikala S, Ramganesh S, Sundararaj T. Drug resistance of Staphylococcus aureus in sinusitis patients. Int J Biosci. 2011;1:63–71. [Google Scholar]
- 21.Pramodhini S, Thenmozhivalli PR, Selvi R, Dillirani V, Vasumathi A, Agatha D. Comparison of various phenotypic methods and mecA based PCR for the detection of MRSA. J Clin Diagn Res. 2011;5:1359–62. [Google Scholar]
- 22.Sajith Khan AK, Shetty PJ, Lakshmi Sarayu Y, Chidambaram A, Anganathan R. Detection of mecA genes of methicillin-resistant Staphylococcus aureus by polymerase chain reaction. Int J Health Rehabil Sci. 2012;1:64–8. [Google Scholar]
- 23.Pillai MM, Latha R, Sarkar G. Detection of methicillin resistance in Staphylococcus aureus by polymerase chain reaction and conventional methods: A comparative study. J Lab Physicians. 2012;4:83–8. doi: 10.4103/0974-2727.105587. [DOI] [PMC free article] [PubMed] [Google Scholar]


