Short abstract
A global analysis of sex-biased transcription in Drosophila shows extensive differential expression between the sexes. Most sex-differential expression is due to germ cells and nearly all genes with germline expression show sex-bias.
Abstract
Background
Sexual dimorphism results in the formation of two types of individuals with specialized reproductive roles and is most evident in the germ cells and gonads.
Results
We have undertaken a global analysis of transcription between the sexes using a 31,464 element FlyGEM microarray to determine what fraction of the genome shows sex-biased expression, what tissues express these genes, the predicted functions of these genes, and where these genes map onto the genome. Females and males (both with and without gonads), dissected testis and ovary, females and males with genetically ablated germlines, and sex-transformed flies were sampled.
Conclusions
Using any of a number of criteria, we find extensive sex-biased expression in adults. The majority of cases of sex differential gene expression are attributable to the germ cells. There is also a large class of genes with soma-biased expression. There is little germline-biased expression indicating that nearly all genes with germline expression also show sex-bias. Monte Carlo simulations show that some genes with sex-biased expression are non-randomly distributed in the genome.
Background
Sexual dimorphism varies enormously from the morphologically indistinct yeast mating types, to extreme differences that can lead to the misclassification of males and females as distinct species. While the Drosophila sexes show relatively modest sexually dimorphic somatic body plans there is striking sexual dimorphism in the germline [1]. Indeed, it is difficult to imagine cell types that differ more than eggs and sperm. During the last century a handful of genes involved in the regulation of somatic (for example, Sex-lethal [2], transformer [3], transformer-2 [4], doublesex [5], intersex [6]) and germline sexual identity (for example, ovo [7], sans fille [8], ovarian tumor [9], and stand still [10]) have been identified, but we know very little about the effector genes that actually result in a sexually dimorphic state. Major terminal genes in the current models of somatic and germline sex determination are a transcription factor (doublesex) and an RNA binding protein (Sex-lethal) [11,12]. These molecules must orchestrate a cascade of effector functions that result in sexually dimorphic gametes and the somatic support functions required for their union at fertilization.
Whilst we know relatively little about how germline sexual identity is determined in Drosophila, the downstream process of oogenesis has been well studied. Egg production occurs in the ovary [13]; each ovary consists of a cluster of 16-20 ovarioles where the assembly line-like production of germline and somatic cell units (egg chambers) progresses along the length of the ovariole. The germline component of each egg chamber contains the differentiated products of a single germline stem cell division. The differentiating stem cell daughter undergoes four rounds of incomplete cytokinesis to produce an interconnected 16-cell cyst surrounded by a somatic follicular epithelium.
One of these 16 germline cells becomes the oocyte and the remaining 15 cells develop into supporting nurse cells. The 16-germline cells within cysts are connected by an intercellular network facilitating the active transport of macromolecules from the nurse cells into the growing oocyte. These components include basic cellular machinery such as ribosomes, and a vast assortment of proteins and RNA species that support early embryonic development. For example, the dorsal/ventral and anterior/posterior axes of the future embryo are laid down during oogenesis [14]. The somatic follicular epithelium surrounding each egg chamber is an important source of structural proteins, such as yolk and egg shell proteins, that become incorporated into the oocyte, as well as patterning information. Most of the yolk proteins are produced distantly in non-gonadal fat body tissue [15]. Thus, while much of the egg is constructed by the cells of the egg chamber, there is significant contribution from distant organs. Finally, additional somatic functions in the female reproductive tract and female mating behaviors are required for productive gamete function [16]. A global analysis of gene expression in adult females therefore captures the genes required for all of the stages of oogenesis from stem cell to early embryo.
Analogously, the Drosophila testis contains developing gametes, from the stem cells at the apical tip of the testis to fully functional sperm [17]. As in the ovary, the germline stem cell division in the testis produces cysts of 16 primary spermatocytes. However, in males all these primary spermatocytes undergo meiosis resulting in a cyst of 64 spermatids. Each spermatid then follows an elaborate differentiation program of cytoskeletal and nuclear rearrangements to form a mature sperm cell. These changes are quite remarkable. For example, sperm chromatin is nearly crystalline and the nucleus changes from a round structure to a highly elongated and slightly hooked shape. Cytoskeletal rearrangement is equally dramatic. The round spermatid forms a flagellar axoneme that is nearly half the length of the adult. Mitochondrial differentiation in the axoneme is also striking. Individual mitochondria fuse into two large and interleaved structures extending along the length of the flagellum. Thus, while the structure of a sperm cell might suggest a simplistic developmental program for spermatogenesis (DNA and a motor), shedding the features that characterize virtually all other cells in the body (such as packing DNA into nucleosomes) is an enormous reengineering feat. As is the case in females, the male reproductive tract and male behavior are required for fertility [18]. Therefore a global analysis of gene expression in adult males captures the genes required for all of the stages of spermatogenesis from stem cell to the fertilized egg.
Not surprisingly, both genetic and classic molecular studies indicate that spermatogenesis and oogenesis are complex events requiring extensive and often sex-specifically deployed information [13,17]. More recently, global gene expression studies using printed cDNAs, expressed sequence tags (ESTs), and full transcriptome microarrays have revealed extensive overall sex differential expression [19-24], with gene expression in the germline and gonads being particularly striking [19,20,24]. Here we report gene expression profiles as a function of sexual dimorphism and sex determination in Drosophila as analyzed using a platform including 93% of predicted genes from version 1.0 of the Drosophila genome [25] and 75% of release 3.1. This article includes the dataset from Parisi et al. [24] on gene expression in adults, augmented with additional microarray experiments to further track the source of sexually dimorphic expression.
There are many stories embedded in the expression data reported here. We touch on only a few to illustrate the value of the dataset. The most significant aspect of the survey we report here is the creation of a dataset that can be mined by other researchers interested in gametogenesis and sexual dimorphism. To that end, genes showing differential expression have been organized into easy to browse tables that include internet links to FlyBase [26,27], the compendium of genome information for Drosophila. We have also deposited all the data at the Gene Expression Omnibus [28,29] so that those interested in large-scale reanalysis can easily download the entire dataset.
Results
Experimental design
We used a series of experiments to delimit germline and somatic sex-biased expression. By comparing flies with and without a germline we can identify genes with germline-dependent expression bias [21,23]. While most genes showing germline-dependent expression are expressed within the germline cells, this class also includes somatically-expressed genes induced or de-repressed by the presence of germ cells. Progeny of homozygous tudor1 mothers have no germline and are useful in differentiating somatic and germline-biased expression [30]. Another measure of germline contribution was obtained by dissection. The gonad samples provide a measure of gene expression in the germline and somatic components of the gonad. Whilst the gonads clearly contain both tissues, the vast majority of the mRNA is derived from the germline cells. Indeed, we were unable to collect sufficient mRNA (600 ng of polyA+) from dissected germline-less gonads to perform a single microarray experiment. Finally, wildtype Drosophila males are XY and females are XX. We reversed the sex of XX flies from female to male using mutations in tra2 and dsx [31]. These sex transformed flies have rudimentary testes with very few germline cells [32] and are thus phenotypically similar, although less extreme, than the male progeny of tudor1 mothers [1]. By performing replicate microarray experiments on these diverse samples we obtain a robust picture of sexually dimorphic gene expression in the germline and soma.
Pairs of mRNA samples (Figure 1) labeled using Cy3 or Cy5 labeled nonomers were applied to Incyte Fly Gene Expression Microarrays (FlyGEM) and the Cy3 and Cy5 channel intensity values were extracted for study as previously reported [33]. The microarray was designed against release 1 of the Drosophila genome and has been updated to release 3.1. There are many changes in the annotation and future changes are also likely. As a result some of the microarray elements no longer correspond to officially recognized genes (although some detect transcripts) and some transcripts are now recognized by multiple element species. Therefore we use the expression from the 14,611 DNA element species pairs corresponding to Drosophila transcripts (there are at least two of each printed on the array plus many control elements for a total of 31,464) as a metric for 'genes'. Using element species rather than gene names assures that the dataset can be updated as gene models change in the ongoing annotation effort.
Figure 1.

Microarray experimental design. Sex biased gene expression data was derived from 44 microarray hybridizations testing 15 conditions. All experimental conditions included at least one biological replicate and most also include dye-flip hybridizations for additional replicates. Abbreviated genotypes of the samples are shown in light ellipses with the total number of replicated hybridizations in dark circles between the two samples. Full genotypes of the flies used are as follows: X/Y;tra2- males are w67c/BsY;tra2B/Df(2R)trix, X/X;tra2- sex transformed males are w67c/+;tra2B/Df(2R)trix, X/X;dsxD/dsx- males are (+/+; dsxM+R45/dsxswe. 'tud' males and females are the progeny of homozygous tud1 bw1 sp1 females mated to tud1 bw1 sp1/CyO males and themselves are genotypically tud1 bw1 sp1 homozygotes. Females and males are whole adult y1 w67c flies. Ovary, testis and no gonad samples are also derived from y1 w67c flies.
In order to characterize a gene as showing biased expression in a particular tissue or sex, one is forced to use arbitrary statistical or magnitude cut-offs. In our replicated heterotypic hybridizations we used both a conservative measure to capture the highest confidence data and noisier, but more inclusive, clustering methods. We express sex-biased and tissue-biased gene expression in terms of percent of the genome for didactic purposes only, as these numbers depend greatly on the arbitrary cut-off. For gene lists, see the extensive set of tables and links to FlyBase in the Additional data files.
Expression analyzed by ratio
We have used conservative calls on competitive pairwise experiments at two-fold. An extensive set of homotypic hybridization experiments (mean r2 of 0.754, and a standard deviation of 0.097) has shown that the 99.5% tolerance interval for measurements on the FlyGEMs is <1.5-fold, and unlike the case with some array platforms the particular element sequence does not contribute significantly to variance [33]. Therefore, this global statistic can be applied to individual elements. Outliers are very effectively dealt with by replicates (Figure 2). In heterotypic hybridizations reported here, only 0.01% of the elements (representing 0-6 in each comparison) showed two-fold differences that fail to correlate with the experimental variables in question. Briefly, replication effectively eliminates false positives for tissue-biased expression in theory and practice.
Figure 2.
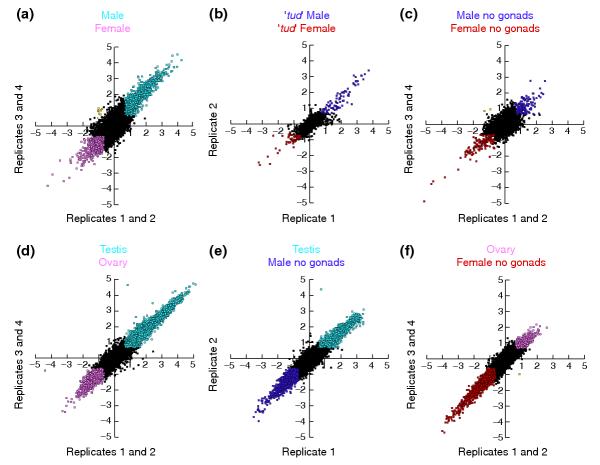
Sex-biased differential expression in Drosophila. Scatter plots show global expression in Drosophila testing different sex and tissue conditions. Data are pairwise comparisons of natural log (Ln) Cy3/Cy5 signal ratios averaged from dye flipped and biological replicate experiments. Black color indicates expression ratios that fall within a two-fold cutoff. Microarray element species greater than two-fold are color coded as indicated on each scatter plot. Yellow points indicate 2-fold differences that fail to correspond to the expression variables analyzed. The number of element species included in each pairwise comparison that met stringency conditions are indicated in parentheses. (a) y1 w67c males versus y1 w67c females (n = 10,688); (b) male versus female progeny of homozygous tud1 bw1 sp1 females (n = 12,836); (c)y1 w67c males, no gonads versus y1 w67 female, no gonads (n = 9,778); (d)y1 w67c testis versus y1 w67c ovary (n = 11,338); (e) y1 w67c testis versus y1 w67c y1 w67c male, no gonads (n = 12,461); (f) y1 w67c ovary versus y1 w67c female, no gonads (n = 11,223).
In comparisons of whole adult females and males, 17% of the element species reporting in all experiments showed a greater than two-fold sex-biased hybridization intensity (Figure 2a). Female-biased hybridization intensity was less extensive (6%) than male-biased hybridization intensity (11%). A series of experiments suggest that most sex-biased hybridization intensity is due to the germline (Figure 2; see also tables in the Additional data). We found that 1.5% of array element species show sex-biased hybridization in experiments using probes derived from flies with no germ cells (Figure 2b). This distribution was asymmetric, with greater numbers of element species showing high hybridization to male samples (0.9%) than to female samples (0.4%). Similarly, when male or female carcasses were compared, 3% of the element species showed sex-biased hybridization (Figure 2c). The greater degree of sex-biased hybridization observed in the carcass samples may reflect strain differences, which can be substantial [21,23], or germline-dependent expression in the soma. We are currently scrutinizing these possibilities with additional array experiments using flies of identical genotypes, with and without germlines. Briefly, these data indicate that relatively little of the sex-biased expression in whole flies is attributable to the soma.
While the contribution of the germline to sexually dimorphic gene expression can be approximated by examining the fraction of the sex-biased expression in whole wildtype flies minus the sex-biased expression in the soma, a more direct measurement is obtained by hybridizing mRNA from dissected gonads. As we show below, this substantially concentrates the mRNAs from germline cells and increases the depth of the observed expression profile. Ovary and testis samples exhibit the highest degree of sex-biased hybridization in our experiments (Figure 2d). A total of 29% of element species exhibit ovary-biased or testis-biased hybridization. This is in contrast to 14-16% of element species deduced to show ovary or testis-biased hybridization by subtracting the somatic sex-biased genes from the list of element species exhibiting sex-biased hybridization in whole adults. The expression profile is distinctly asymmetric with nearly twice as many element species showing greater than two-fold hybridization to testis samples (18%) versus ovary samples (11%). Additionally, there were far more element species showing a large magnitude of testis-biased hybridization. For example, 2% of element species showed >20-fold hybridization intensity in the testis channel, while <<1% of element species showed a similar magnitude in the ovary channel (n = 4). These data indicate that the cells of the testis deploy a larger battery of specific genes than those of the ovary. Additionally, these data confirm that the most sexually dimorphic tissues, the gonads, show the most distinct gene expression profiles.
A priori, one might expect that the soma would be quite different from the germline, but to what extent is this due to the expression of germline-specific or soma-specific gene batteries? To address this question, we have also examined germline versus somatic expression within a given sex. As expected we observed a high degree of testis-biased hybridization in competitive hybridizations with samples from either males with no or few germline cells (males from tudor1 mothers, and females transformed somatically into males; not shown) or from males with gonads removed (Figure 2e). A total of 14% of element species hybridize preferentially to labeled testis samples, supporting the idea that the gene batteries deployed in a testis-biased manner are extensive. Additionally, these experiments reveal a high degree of soma-biased hybridization - 10% of element species are preferentially hybridized to samples from males with the gonads removed (Figure 2e).
Somewhat surprisingly, comparing females with or without germlines did not show the same pattern of soma versus germline hybridization that was evident in the males. Ovary-biased hybridization was modest (4%) in experiments measuring hybridization of samples from ovary versus samples from females with the ovaries removed (Figure 2f). In contrast, soma-biased hybridization was extensive (12%) as was the case when examining male soma and germline (Figure 2e). Thus, unlike what is seen in males, the asymmetry in these ratio plots favors the soma. This suggests that most of the genes expressed in the female soma are also expressed in the ovary and that many genes are expressed specifically in the female soma. The soma is distinguished from the germline by a high degree of soma-biased gene expression.
Expression analyzed by intensity
An overview of all sex-biased gene expression is provided by examining a self-organized heat map of the cross-normalized intensities from each of the hybridizations (Figure 3). This also provides an independent method for building gene lists. As expected from the pairwise comparisons, the prominent clusters in this map highlight groups of element species exhibiting testis-biased (12%), ovary-biased (9%), and soma-biased (11%) hybridization. The clusters of element species showing testis-biased hybridization are most dramatic, both in terms of the number showing sex-bias and in terms of the degree of hybridization bias. This global overview shows very little overt sex-biased expression in the soma, and very little germline-biased expression. In agreement with the scatterplot analysis, the heat map indicates that the main batteries of differentially expressed genes distinguish the soma from the germline, and the female germline from the male germline. Again, the lack of an overt class of genes showing germline-biased expression indicates that the male and female germlines have very little in common, other than the absence of expression from a large class of genes that appear to have soma-biased function.
Figure 3.
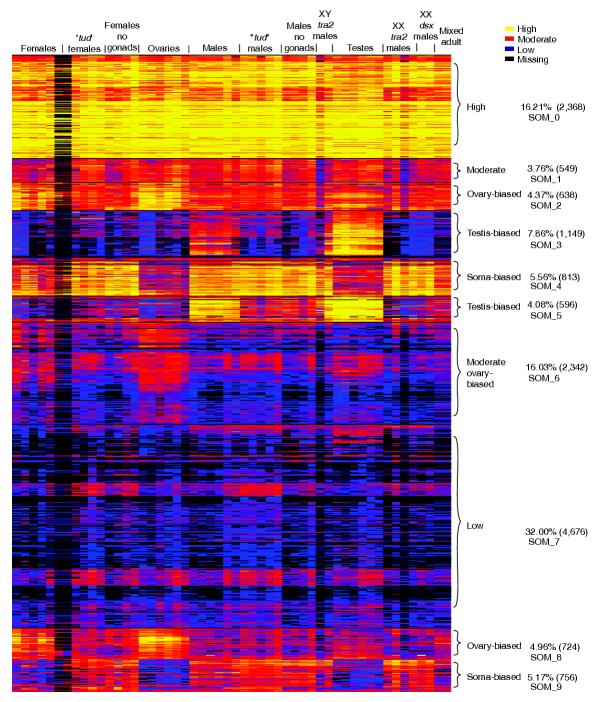
Heat diagram of intensities. Self organizing maps (SOMs) were used to generate an image of clustered intensity data from 26 pairwise experiments. The individual channels from these experiments are parsed out and arranged by tissue type as indicated in the top row text. The normalized intensities are indicated as high expression (yellow), moderate (red), low (blue) and missing value (below background; black). The diagram represents averaged data from the duplicated elements within each microarray. Brackets show ten SOM clusters with the tissue type and percentage of the total number of microarray elements in the right text column.
Cross-validation of expression by Northern blotting
It is standard practice to confirm the expression of genes of interest based on microarray data by RT-PCR or Northern blotting, although this is not practical when thousands of genes show biased expression. Additionally, this confirmation is usually restricted to genes showing differential expression, and therefore does not allow for a good estimate of sex-biased gene expression missed in the FlyGEM experiments. We selected 75 element species from the FlyGEM experiments, across the range of observed intensities and sex-biases, for Northern blotting experiments on adult males and females with and without germlines (Figure 4a). Signal intensities from each lane on the Northerns were measured for each experiment and then used to calculate differential expression ratios among the four sample lanes in a fashion analogous to pairwise microarray comparisons (absolute intensities correlated very poorly, indicating that the techniques have different signal and noise characteristics even though the same amplicons were used). Plotting the ratios from Northerns versus microarrays demonstrates consistent relative measurement of RNA between the two experimental systems (Figure 4b,c,d,e). The majority of the data points fall along the diagonal for wildtype males and females. In the other comparisons, both the microarray and Northern ratios tend to cluster near the origin (a ratio of 1 is 0 in log space) due to lower sex-biased expression in the germlineless flies of tudor1 mothers. Thus, ratios determined by Northern blotting and FlyGEM compare favorably.
Figure 4.

Northern and microarray comparison. A series of 75 Northern blots with total RNA from germline-less ('tud') male, wildtype (y1 w67c) male, germline-less ('tud') female and wildtype (y1 w67) female whole flies were probed with the PCR products identical to those printed as microarray elements. (a) 34 Northern blot images show a range of expression patterns among the input RNA samples. Phosphorimaging of the radioactive signal from Northern blots gave expression ratios between the RNA sample types for each Northern probe. (b-e) Corresponding (Ln) transformed Northern and microarray element ratios averaged from multiple experiments are shown as four scatterplots comparing expression between (b) y1 w67c males versus y1 w67c females; (c) 'tud' males versus 'tud' females; (d) y1 w67c males versus 'tud' males and (e) y1 w67c females versus 'tud' females. The y and x axes are expression ratios derived from Northern blot and microarray ratios respectively. Over two-, over four- and over tenfold differences in ratio values between the Northern and microarray experiments are indicated in yellow, red and blue.
Cross-validation of expression with the developmental time course
Comparing array results across platforms can cross-validate a larger fraction of the genome. Confidence is heightened for element species showing biased expression on two platforms [34]. A Drosophila developmental profile including males and females with and without a germline has also been performed on a cDNA array platform (GEO accession GPL218) [20]. This array is less complete than the FlyGEM and is biased against testis gene expression as no clones from testis cDNA libraries were included in the platform. Thus, overall estimates of sex-biased gene expression are expected to vary between the platforms. However, FlyGEM element species designed against the genes represented by cDNA inserts should correlate if both platforms are performing as expected. We extracted data from 2,536 GPL218 elements also present on the FlyGEM platform for comparative analysis.
The correlation between male/female hybridization ratios was quite good (Figure 5a,b). The trend lines clearly demonstrate that elements showing sex-biased expression on one platform show similar sex-biased expression on the other if one applies a greater than two-fold cut-off to these ratios. The only clearly artifactual result from this comparison is the presence of a substantial number of elements reporting sex-bias only in Arbeitman et al. [20]. This may be due to cross-hybridization as a result of low sequence complexity and repeats in the cDNA inserts - a common problem with cDNA arrays [35]. Nevertheless, this comparison clearly cross-validates the expression profiles for a substantial set of genes. Indeed, the r2 for male/female ratios across platforms is 0.447, which is similar to the correlation observed between replicate hybridizations within the time course dataset (r2 = 0.334 +/- 0.114).
Figure 5.

Meta-analysis of Arbeitman et al. [20,36] data. (a,b) Scatterplots of averaged (Ln) ratios for (a) male versus female and (b) 'tud' male versus 'tud' female comparing data from experiments performed with the FlyGEM and GPL218 platforms. Two-fold sex-biased expression in both datasets are indicated in blue (male-biased) and pink (female-biased). Those showing two-fold male-biased expression in one data set, but two-fold female-biased expression in the other are shown in yellow. (c,d) Comparison of Northern expression ratios for 16 probes versus averaged (Ln) ratios from Arbeitman et al. are shown in scatterplots for (c) male versus female and (d) 'tud' male versus 'tud' female. The 16 probes represent CG13263, CG8994, CG3972, CG10701, CG1088, CG7961, G10961, CG6206, CG4586, CG13095, CG5089, CG4847, CG6483, CG8549, CG7660, CG6457. Two-, four- and 10-fold deviations from 1:1 ratio are indicated by yellow, red and blue color (see Figure 4). (e,f) Clustering of FlyGEM normalized intensity data was performed for sex-biased somatic genes from Table 2 of Arbeitman et al. [36]. (e) 33 male and (f) 26 female genes are shown as intensity heat clustergrams. The normalized intensities are indicated as high expression (yellow), moderate (red), low (blue) and below background (black). (Intensities are represented by color as in Figure 4). Columns show 52 channels parsed from 26 pairwise FlyGEM microarray experiments. Clustering shows soma-bias corroborating the Table 2 lists. The array element species represented in rows from top are listed as follows: (e) CG12268, CG3359, CG3359, CG3359, CG5740, CG7050, CG7157, CG7178, CG7178, CG7748, CG8110, CG8552, CG9456, CG12558, CG14024, CG15097, CG16820, CG17843, CG18284, CG3359, CG5411, CG5455, CG6518, CG6716, CG6844, CG7178, CG7178, CG7342, CG8708, CG8708, CG8909, CG9519; (f) CG10281, CG10566, CG1090, CG11248, CG12269, CG1646, CG17012, CG7129, CG7702, CG7899, CG8327, CG8370, CG9547, CG10944, CG12740, CG14792, CG18525, CG3195, CG3751, CG4087, CG5821, CG7777, CG8453, CG8705, CG9696, CG9699.
We then tested whether expression ratios from our Northern analysis correlated to ratios derived from Arbeitman et al. [20]. We have Northern data in 16/2,536 elements common to both platforms. The male versus female and 'tud' male versus 'tud' female ratios were plotted (Figure 5c,d). The Arbeitman et al. [20] data to Northern (Figure 5c,d) and FlyGEM to Northern (Figure 4b,c) correlation was similar.
The Arbeitman et al. [20] data has been recently re-evaluated in a more statistically rigorous manner to extract the high confidence data from the moderately reproducible primary data [36]. The specific aim of this re-evaluation was to identify genes with sex-specific expression patterns. If the set of cDNA amplicons that most reliably hybridize to female or male samples can also be used to categorize female and male samples of differing genotype from another lab, then confidence in that list of genes is augmented. We therefore took a list of gene identifiers from this more recent study, extracted our corresponding data, and generated a cluster map of samples (Figure 5e,5f). This small subset of elements was sufficient for the unsupervised classification of whole females and males, despite disparate genetic backgrounds. Tissues were also appropriately juxtaposed on the sample tree. Collectively, the scatterplot and heat map comparisons indicate that FlyGEM and Arbeitman et al. [36] data are in broad agreement and suggests that a full meta-analysis of Drosophila expression data from multiple platforms will be fruitful.
Well-characterized genes are correctly scored
Genes known to have sex-biased expression or function should be present in the appropriate ovary-, testis-, or soma-biased lists. We have mapped the FlyGEM element species to the current release of the Drosophila genome and generated a large set of lists with links to Flybase gene report pages (see Additional data). Following these links leads to more links (for example, literature, additional array data, yeast two-hybrid maps) and following these trails lends further confidence in the array data. For example, on lists of element species showing ovary-bias (Table 21 in Additional data; see also Tables 5, 15, 17, 30, 24, 36, and 38), we find six that represent chorion proteins (Cp15, Cp16, Cp18, Cp19, Cp36, and Cp38). Chorion proteins are components of the egg shell, and are known to be highly over-expressed in the ovary [37,38]. Similarly, structural components of sperm such as beta-tubulin85D [39], tektin-A [40] and don juan [41] are found on testis-biased lists (Additional data, Table 22; see also Tables 6, 7, 9, 31, 33, and 39). A number of other genes showing biased hybridization are known to be required for female or male fertility. These include alpha-tubulin67C, which is represented by over 20 female sterile alleles [42] and deadhead, which is represented by four female sterile alleles and a single lethal allele [43]. Genes represented by male sterile alleles (for example, boule [34] and fuzzy onions [44]) are found on testis-biased lists. The boule gene is also required in mammalian spermatogenesis [34], highlighting the potential utility of this dataset for those interested in spermatogenesis in mammals. Genes known to encode products required maternally for germline formation in the resulting embryo are represented on lists of genes showing ovary-biased hybridization (for example, oskar [45], mago nashi [46], vasa [45], smaug [47], and cyclinB [48]) as are genes required for germline cell division within the ovary (for example, piwi and fs(1)Ya [49]). Even among the genes with little functional information, there is evidence to cross validate some of them. For example, BEST:GH14656 is represented by 13 testis ESTs from the NIH collection and just two ESTs from the BDGP head library [19,50].
In addition, there are a number of genes showing soma-biased expression in our experiments (Additional data, Table 23; see also Tables 9, 18, 37, and 40). Examples include actin88F [51], flightin [52], Tropomyosin1, Tropomyosin2, and upheld [53] that are known to be required for flight muscle function. Similarly, genes involved in phototransduction (arrestin-2 [54], ninaC [55] and five genes encoding rhodopsins [56]) exhibit soma-biased expression, as do genes expressed in the fat body which are immune response genes (Drosomycin [57]), or genes that may be required for energy storage (fat body protein 1 [58] and lipid storage droplet protein 1 [59]). Thus, many of the class of genes showing soma-biased expression are genes used for well-defined soma-specific functions.
Briefly, many of the genes expected to show biased expression do. Additionally, given the importance of these known genes, it seems likely that there will be interesting functions among the unstudied genes showing biased expression in our experiments.
Systematic analysis of gene function
Browsing offers some degree of confidence, but is not amenable to the analysis of large datasets. The Gene Ontology (GO) consortium [60] has provided a controlled vocabulary to describe gene function allowing us to determine rigorously which functions are over-represented in a particular expression profile. We queried our element species lists with the GO term analysis tool 'Database for Annotation, Visualization and Integrated Discovery' (DAVID) and 'Expression Analysis Systematic Explorer' (EASE) [61].
The results of GO term analysis are reassuring. Genes with oogenic functions [13] are highly over-represented among element species with ovary-biased expression (Figure 6a). The most highly over-represented categories are associated with metabolism (P < 1 × 10-20) and are probably associated with biosynthesis required for the manufacture of eggs. Similarly, ribosomes are highly over-represented cellular components (P < 2 × 10-42) as are ribosomal proteins in the protein GO category (P < 8 × 10-52). Genes encoding translational initiation complexes are also highly over-represented (P < 2 × 10-22). This is consistent with the high level of protein production during egg development and the high ribosomal content maternally loaded into the egg. Indeed, mutations in many ribosomal protein genes result in female sterility due to defective egg formation [62]. Also prominently represented are genes involved in DNA metabolism and cell cycle regulation (P < 1 × 10-12, P < 1 × 10-7). These functions may be required for the dramatic endoreplication of nurse cells during oogenesis. Similarly, the over-representation of transcripts from genes required for DNA replication (P < 1 × 10-9), chromatin architecture (P < 4 × 10-5), nuclear organization (P < 1 × 10-4), and DNA packaging (P < 5 × 10-5), may be required for nurse cell polyploidization [13] or for the rapidly dividing embryonic cells which rely on maternally deposited gene products. Finally, molecular genetic analysis of oogenesis has revealed that a substantial number of these gene products include RNA binding proteins, and this class is also over-represented in ovaries in our microarray data (P < 2 × 10-6).
Figure 6.

An analysis of Gene Ontology term categories. The results show categories over-represented in (a) ovary, (b) testis, (c) female soma and (d) male soma queried by the three GO ontologies of biological process (black bars), cellular component (dark gray bars) and molecular function (light gray bars). Histograms represent significant over-representation (P < 0.001) of element species for 116 GO categories. The y axis scale shows P-values of the modified F-statistic (EASE score). The GO terms for each column are: 1, behavior; 2, biosynthesis; 3, cell cycle; 4, cell growth and/or maintenance; 5, cell organization and biogenesis; 6, cell proliferation; 7, chromosome organization and biogenesis (sensu Eukarya); 8, cytoplasm organization and biogenesis; 9, cytoskeleton organization and biogenesis; 10, cytoskeleton-dependent intracellular transport; 11, DNA dependent DNA replication; 12, DNA metabolism; 13, DNA packaging; 14, DNA replication; 15, DNA replication and chromosome cycle; 16, eggshell formation; 17, eggshell formation (sensu Insecta); 18, establishment and/or maintenance of chromatin architecture; 19, G2/M transition of mitotic cell cycle; 20, gametogenesis; 21, insect chorion formation; 22, insemination; 23, intracellular transport; 24, M phase of mitotic cell cycle; 25, macromolecule biosynthesis; 26, mating behavior; 27, metabolism; 28, microtubule-based movement; 29, microtubule-based process; 30, mitotic cell cycle; 31, nuclear organization and biogenesis; 32, nucleobase, nucleoside, nucleotide and nucleic acid metabolism; 33, oogenesis; 34, oogenesis (sensu Insecta); 35, organelle organization and biogenesis; 36, ovarian follicle cell development (sensu Insecta); 37, oviposition; 38, physiological processes; 39, post-mating behavior; 40, protein biosynthesis; 41, protein metabolism; 42, protein modification; 43, proteolysis and peptidolysis; 44, regulation of cell cycle; 45, reproduction; 46, reproductive behavior; 47, S phase of mitotic cell cycle; 48, sexual reproduction; 49, sperm competition; 50, sperm displacement; 51, transport; 52, vitellogenesis; 53, cell; 54, chaperonin-containing T-complex; 55, cytoplasm; 56, cytoskeleton; 57, cytosol; 58, cytosolic large ribosomal subunit (sensu Eukarya); 59, cytosolic ribosome (sensu Eukarya); 60, cytosolic small ribosomal subunit (sensu Eukarya); 61, dynein complex; 62, eukaryotic 43S pre-initiation complex; 63, eukaryotic 48S initiation complex; 64, extracellular; 65, inner membrane; 66, intracellular; 67, large ribosomal subunit; 68, lysosome; 69, lytic vacuole; 70, membrane; 71, microtubule associated complex; 72, microtubule cytoskeleton; 73, mitochondrial inner membrane; 74, mitochondrial membrane; 75, mitochondrion; 76, pre-replicative complex; 77, replication fork; 78, ribonucleoprotein complex; 79, ribosome; 80, small ribosomal subunit; 81, alpha-mannosidase activity; 82, aminopeptidase activity; 83, ATP dependent helicase activity; 84, ATP dependent RNA helicase activity; 85, carbohydrate binding activity; 86, carrier activity; 87, catalytic activity; 88, chaperone activity; 89, chromatin binding; 90, chymotrypsin activity; 91, dynein ATPase activity; 92, endopeptidase activity; 93, enzyme activity; 94, exopeptidase activity; 95, galactose binding activity; 96, hormone activity; 97, hydrolase activity; 98, hydrolase activity, acting on acid anhydrides, involved in cellular and subcellular movement; 99, leucyl aminopeptidase activity; 100, metalloexopeptidase activity; 101, nucleic acid binding; 102, oxidoreductase activity; 103, peptidase activity; 104, RNA binding; 105, RNA dependent ATPase activity; 106, RNA helicase activity; 107, serine-type endopeptidase activity; 108, serine-type peptidase activity; 109, small protein conjugating enzyme activity; 110, structural constituent of chorion (sensu Insecta); 111, structural constituent of ribosome; 112, structural molecule activity; 113, thiolester hydrolase activity; 114, transporter activity; 115, trypsin activity; 116, ubiquitin conjugating enzyme activity.
The representation of GO terms in the testis-biased element species lists was similarly examined (Figure 6b). Again, the most significantly over-represented GO terms are consistent with what we know about spermatogenesis [17]. Genes encoding microtubule (P < 2 × 10-9) or movement (P < 3 × 10-8) function are among the element species with testis-biased expression. These are required for the extensive cytoskeletal reorganization during spermiogenesis [17]. Genes encoding transport proteins (P < 2 × 10-6), including those with dynein-associated ATPase activity (P < 1 × 10-9) are over-represented. Again, this is consistent with the motile nature of sperm. Chaperones (P < 3 × 10-4) are over-represented, which is in agreement with the known requirement for heat shock proteins during spermatogenesis [63]. In testis, cellular component analysis shows high over-representation of gene products associated with mitochondria (P < 9 × 10-14), membranes (P < 1 × 10-4) and the cytoskeleton (P < 3 × 10-7) which is almost certainly due to requirements for sperm axoneme structure.
Non-gonadal female somatic tissues show sex-biased expression of genes related to reproductive processes (Figure 6c). Vitellogenesis (P < 2 × 10-5) is required for egg development, as the vitellogenin or yolk proteins synthesized in the female fat body are the major component of a mature egg. The second category of proteolytic processing (P < 8 × 10-4) might reflect the process required for generating yolk proteins. Ribosomal protein encoding genes are also over-represented (P < 1 × 10-6) in the non-gonadal female soma transcription profile. Similarly, element species encoding lysosome components (P < 2 × 10-5) are over-represented. These functions probably reflect the dual anabolic and catabolic metabolism required to support egg development.
The male reproductive tract includes several additional somatic structures in addition to the testis. For example, the seminal fluid components of the ejaculate are produced in the accessory glands. These glandular secretions mediate sperm competition, reduce post copulatory mating by recipient females, and increase egg laying rates [18]. These element species with 'extracellular' function are over-represented in the male expression profile (P < 4 × 10-5) (Figure 6d).
Gene expression neighborhoods
A whole genome transcriptional profile enables us to investigate the genomic organization of tissue-specific gene batteries. Previous experiments have shown that there are significantly fewer genes with male-biased expression on the X chromosome [23,24]. The fourth chromosome is enriched in genes with soma-biased expression (not shown). On a more local level, Drosophila genes with tissue-, temporally-biased expression patterns have been shown to be concentrated in neighborhoods of contiguous genes [64]. We asked if genes with sex-biased expression show a similar sub-chromosomal level organization.
We first examined a heat map of cross-normalized intensities by position along the chromosome arms (not shown). While there were patterns of alternating bands of genes showing greater or lesser intensity along the chromosomes, similar but not as obvious as those reported for embryo and adult [64], these patterns were also seen in randomized datasets, albeit at a lower frequency. Additionally, these patterns were less striking than those seen when plotting normalized intensity versus process parameters such as position in the microarray. (The FlyGEM platform we used is printed randomly with respect to genome position, so these process artifacts do not affect our previously reported reduction in the density of genes with male-biased expression on the X chromosome [24], nor the neighborhoods reported below.)
Given that the concept of gene expression neighborhoods is relatively new, we were interested in identifying the most significant examples for detailed future study. To do so, we examined the distribution of element species with both highly ovary-, testis-, or soma-biased expression and high intensity to avoid low intensity noise. This resulted in 791 element species with testis-biased hybridization, 129 element species with ovary-biased hybridization, and 287 element species with soma-biased hybridization, which we mapped onto the genome (Figure 7). We also generated randomized datasets to determine if the distribution pattern of ovary-biased, testis-biased, or soma-biased expression is non-random. To do this, 100,000 randomizations were performed for each gene position over windows (consecutive genes along the chromosome) ranging between five to 200 genes. We used sliding windows of various sizes to determine how often an equal or greater number of elements reporting highly biased expression occurred in a random set versus the real data. A total of 20 gene expression neighborhoods were identified with a P-value < 10-3, 15 at P <10-4 and seven at the lowest possible P-value of <10-5 (0/100,000). The most clear-cut examples of gene expression neighborhoods include approximately 10 genes within a chromosomal region spanning between 20 to 40 genes (Figure 8). This is consistent with the report of 10- to 20-gene expression neighborhoods shown in microarray analysis of Drosophila embryonic versus adult transcription [64].
Figure 7.

Distribution of genes with high ovary-, testis-, and soma-biased expression in the genome. Gene positions are shown on the chromosome arms to scale with the positions of all genes on the five major chromosome arms (fourth and Y chromosomes not shown) presented as single tick marks for each in the upper column (All). The positions of genes represented by element species highly expressed in gonads and somatic tissues are presented in adjacent rows. The element species defined as highly expressed in the three tissue categories were up in multiple pairwise comparisons; ovary-biased (from microarrays versus both testis and female carcass), testis-biased (versus both ovary and male carcass) and somatic-biased (common among male carcass versus testis, and female carcass versus ovary). Asterisks indicate gene neighborhoods identified by Monte Carlo simulations with highly significant P values (*P < 10-3, **P < 10-4, ***P < 10-5).
Figure 8.

Gene neighborhoods. Monte Carlo simulations (see Materials and methods) were used to identify statistically significant clusters of testis- and soma-biased genes using a series of window sizes ranging from five to 200 consecutive genes. Examples are shown for six gene neighborhoods that were found zero times in 100,000 replicated randomization tests. GADFLY [26] annotation images of the chromosomal regions for each neighborhood are shown with the genes identified by element species showing expression-bias, indicated by red boxes. The normalized intensities are shown below each neighborhood indicating high expression (yellow), moderate (red), low (blue) and below background (black). (a) A testis-biased cluster at cytological position 2L:35D4-F435, (b) a testis-biased cluster at 2R:50B1-50C6, (c) a testis-biased cluster at 2R:56E1-F9, (d) a testis-biased cluster at 2R: 59C3-D6, (e) a soma-biased cluster at 2R:47E1-F5 and (f) a soma-biased cluster at 2R:55C6-C8.
There are several possible reasons for co-expression of neighboring genes. One possibility is that they are members of gene families or are part of the same transcription unit. Indeed, a recent article suggests that operons and duplicate genes account for many of the gene expression neighborhoods in Caenorhabditis elegans [65]. While there is no evidence for extensive operon structure in Drosophila, genome annotation errors might potentially introduce artifactual evidence for gene expression neighborhoods if genes thought to be distinct are in fact derived from a common transcript. Alternatively, gene expression neighborhoods might reflect the local transcriptional environment via a neighborhood-wide control structure [66,67]. In order to distinguish between these models, to a first approximation, we examined the seven P < 10-5 neighborhoods more closely.
All of the most significant gene expression neighborhoods contain genes expressed from either strand, indicating that neither polycistronic messages nor annotation errors are causal (Figure 8). Additionally, there are no overt gene families or groups of gene ontology terms associated with six of the neighborhoods, suggesting that they are unlikely to be derived by gene duplication, although one is a neighborhood of paralogous genes (Figure 8e). This latter soma-biased gene expression neighborhood consists of a group of distinct genes encoding trypsin-like proteases [68]. Such gene arrangements usually represent gene duplications from a common ancestor and are often co-expressed. The unusually high number of Drosophila genes encoding trypsins has been previously noted [69], it appears that repeated gene duplication may be the cause.
Discussion
A large fraction of the Drosophila genome shows sex-biased expression
We have carefully cataloged sex-biased expression in Drosophila adults using 88 samples of wild-type flies and tissues, germline-ablated flies, and flies with sex transformed genotypes with 14,611 pairs of elements representing exons from release 1 of the Drosophila genome. We have also carefully examined platform performance [33] and have extensively analyzed the dataset for internal consistency as well as cross-validation with other array datasets and Northern blotting. The lists of genes in the survey of sex-biased expression will be a valuable resource for ultimately understanding how sexual dimorphism is genetically encoded and executed. Additionally, this dataset has already proven valuable in the study of rapidly evolving genes with male-biased expression and in understanding X chromosome evolution [23]. Sex-biased expression may also be a convenient tool for understanding functional consequences and mechanistic underpinnings of gene expression neighborhoods.
In broad terms, we identify three large batteries of genes with ovary-biased, testis-biased and soma-biased expression. The germline and soma of Drosophila are the first lineages to separate in the developing embryo, so we tend to think of germline versus somatic tissue as being perhaps the most significant dichotomy. In addition to early divergence in the embryo, the germlines of females and males are functionally similar - they undergo meiosis and produce gametes. One might therefore expect that female and male germ cells would express many genes that are silent in somatic cells. There is a small core of genes showing germline-biased expression (~3%); our data, however, suggest that most of the genes showing germline-dependent expression also show sex-biased expression. Essentially, adult ovary and testis are as different from each other as they are from somatic cells. A major difference between the germline and soma is the absence of gene expression from loci encoding terminal differentiation proteins (for specific adult somatic structures such as eyes and muscles) in the germline.
The lack of a large battery of genes showing germline-bias could also be due to expression of sperm or egg differentiation products that overwhelm the signal from genes expressed in the stem cells, which show no overt sexual dimorphism. Early undifferentiated germ cells from males and females would be expected to have more similar gene expression profiles. However, preliminary global analysis of transcripts in undifferentiated germ cells are also revealing a modest set of genes with germline-biased expression. The germline transcription profile is defined by the transcripts from genes encoding somatic terminal differentiation products and by sex. Germline development is under the control of sex determination signals from the time the embryonic genome is activated [70]. Perhaps it is better to think of the male and female germlines as quite distinct entities, rather than lumping them together as 'germline'.
Sex-biased expression in ovaries and testis were by far the most dramatic in terms of both numbers of genes and the degree of sex-bias. While most of this sex-biased expression is clearly explained by expression within the germline, there also appears to be striking germline-dependent expression outside of the gonads. Flies with and without a germline are often used to distinguish between genes that are expressed in the germline from those that are expressed in the soma [30]. Technically, a gene expressed in wildtype adults but not in adults missing a germline is said to be germline-dependent, not germline expressed. In comparing flies without a germline to those whose gonads had been removed, we found that there was greater sex-biased expression in the latter. These data suggest that the germline acts at a distance to condition the soma. This does not appear to be due to strain differences, as preliminary analysis of gonadectomized flies of identical zygotic genotypes but which had or were missing germ cells, shows substantial gene expression differences. Thus, it appears that much of the sex-biased expression in the Drosophila soma is due to conditioning by the germline, rather than the somatic sex determination hierarchy.
Data mining
A catalog of gene expression data is of limited value if it cannot be easily accessed. In addition to depositing all the array data at GEO, we have run a number of queries and provided the resulting gene lists in the Additional data files. Any of the lists of element species in the Additional data associated with this manuscript may be browsed in order to confirm the performance of the platform with a gene of interest, or for finding a novel expression pattern for a known gene. We have highlighted just a few of the stories that are embedded in the array dataset reported here. To list just a few examples, the preliminary data reported here suggest that germ cells are dosage compensated [24]. The dot fourth chromosome is especially active in the soma. Odorant-binding protein-encoding genes with highly sex-biased expression might be quite interesting candidates in the pheromone response.
It is our hope that this and other datasets generated on this platform will be useful to those who are interested in data mining for candidate genes. Additionally, there was reasonably good agreement between the array datasets here and those of Arbeitman et al. [20,36], which suggests that Drosophila array data from different laboratories will be directly comparable. This further suggests that array data will be broadly useful to the wider Drosophila community. However, it is also clear that there will be artifacts in array datasets, and indeed in the underlying genome annotations. Strain differences and the presence of genetic markers are also likely to confound results; array data should be viewed with this firmly in mind.
Materials and methods
Animals and treatments
We used standard Drosophila techniques throughout. Flies were grown at 25 ± 0.5°C on GIF or PB media (KD Scientific, Columbia, MD, USA) and aged 5-7 days prior to use. Most alleles and transgenes have been previously described and can be found, with references, at FlyBase [26]. Flies wildtype for sex were of the genotype y1 w67c. To study the contribution of the body as a whole and the gonads, flies were dissected and snap frozen as described [19]. To study the germline-dependent gene expression profiles, samples were prepared from the progeny of homozygous tud1 bw1 sp1 females. Flies were dissected to confirm germline loss (tudor1 is not fully penetrant for germlineless-ness, only individuals with atrophic gonads were retained) as above. This sham dissection also facilitates comparisons to gonadectomized samples. The effects of somatic sex determination mutants on sex-biased gene expression included examining sex-transformed flies using doublesex (+/+; dsxM+R45/dsxswe) and transformer 2 (w67c/+;tra2B/Df(2R)trix) mutants. Sibling w67c/BsY;tra2B/Df(2R)trix were used in a direct comparison for one group of experiments.
Microarray hybridization
An extensive platform description is available [71] and at the Gene Expression Omnibus website [29] under accession number GPL20. RNA isolation, labeling, and hybridization have been previously described [71].
Microarray intensity quantification
Hybridized slides were read on a GenePix 4000 A to generate TIFF images which were imported into GemTools (Incyte Genomics, Palo Alto, CA, USA) for spot finding. Data was normalized and corrected in Array Qualifier (Novation Biosciences, Palo Alto, CA, USA). Intensities <2.5-fold above local background were discarded. Raw intensity data and corrected data from Array Qualifier were loaded into an Access database and analyzed in Excel (Microsoft, Redmond, WA, USA). The GEO data accessions for these hybridizations are: GSM2456, GSM2458, GSM2460, GSM2462, GSM2464, GSM2466, GSM16542, GSM16554, GSM16556, GSM16569, GSM16570, GSM16572, GSM16574, GSM16576, GSM16578, GSM16580, GSM2457, GSM2459, GSM2461, GSM2463, GSM2465, GSM2467, GSM16650, GSM16555, GSM16571, GSM16573, GSM16575, GSM16577, GSM16579, GSM16581, GSM16582, GSM16584, GSM15686, GSM16588, GSM16593, GSM16613, GSM16614, GSM16615, GSM16619, GSM16583, GSM16585, GSM16587, GSM16589, GSM16609.
Analysis of ratios
For analysis of ratios, the Cy3:Cy5 signal for each element was averaged over biological replicate experiments and dye flips and then averaged from duplicate elements. Elements failing to return a ratio value from duplicates were excluded. Boolean queries were run to generate gene lists of particular interest (for example, high in ovary and female, and low in all others). For analysis by intensity, array channels were cross-normalized in Array Qualifier.
For array to array comparisons, the GPL218 platform table and 40 sample datasets representing 20 biologically replicated adult stages were downloaded from the GEO website. For comparison to the GPL20 data, Log2 VALUE data (ratio of experiment to reference sample) from eight sample sets representing four biological replicates for five day adult Canton S or 'tud' males and females (GEO accessions GSM3551, GSM3552, GSM3518, GSM3519, GSM3531, GSM3532, GSM3537, GSM3538) were converted to Ln values. The ratios from each biological replicate pair were averaged and these experiment/reference sample ratios were then rearranged into male/female and 'tud' male/'tud' female ratios for parallel platform comparison. A total of 2,536 overlapping elements between GPL20 and GPL218 were selected based on Flybase IDs and plotted against the GPL20 counterpart (sample accession: GSM2456, GSM2458, GSM2457, GSM2459, GSM16582, GSM16583) or Northern ratios.
Clustering
To analyze sex-biased gene expression as normalized intensities among multiple microarray experiments, heat diagrams were generated by the Cluster 3.0 for Mac OSX program [72] formatted for OSX by Michiel deHoon [73]. A normalized set of signal intensity data was prepared for Cluster by averaging duplicate spots (the two duplicate features within an array) from 26 cross normalized microarray experiments. The sample columns were fixed, while genes were clustered. A self organizing map (SOM) was made using default parameters (10 clusters). The resulting SOM file was then used to perform k-means clustering. The output file was visualized with JavaTreeview (adapted by Alok Saldanha at Stanford University) [74].
A similar heat diagram was processed for analyzing the gene neighborhoods using this normalized dataset. A Treeview-generated image of the normalized intensities was made using an input data file that sorted the data for each gene by position along the chromosome. Data was sorted first by chromosome arm and then by distance along the chromosome using the first nucleotide of the microarray element amplimer as the positional reference relative to the annotation.
To produce the array to array comparison heat map, normalized intensity data from the FlyGEM platform was extracted for gene identifiers from Arbeitman et al., Table 2 [36]. Heat maps were performed by k-means clustering of 26 female and 33 male soma-biased element species. Both genes and samples were clustered.
GO term analysis
Analysis of gene function was done using the vocabulary of the Gene Ontology Consortium [75] and applying it to the pairwise expression ratio gene lists processed through the bioinformatics tools DAVID and EASE [76] (search performed November, 2003). The LocusLink identifiers for all microarray features showing greater than two-fold differential expression ratios were loaded into the DAVID website and search parameters set to include all available categories from the three GO ontologies (biological process, cellular component and molecular function). A statistical measure of the representation of all GO categories was done with the EASE bioinformatics tool. The statistical probability for representation of genes within a given category relative to the total number of genes with GO function terms (EASE score) was calculated for each category using the Drosophila-specific database as a background filter. Drosophila features with GO term categories with EASE scores of P < 0.001 were reported.
Monte Carlo
To test if genes with sex-biased expression are non-randomly distributed into neighborhoods, we performed 100,000 simulations per window. The input data are stringently selected lists of genes that are differentially expressed in ovaries (ovary versus testis or females, no gonads), testis (testis versus ovary or males, no gonads) and somatic tissue (female and male, no gonads versus their respective gonads). The microarray data elements include those shown to be greater than three standard deviations above the mean local background in one channel while the signal from the other channel was within background levels. These lists are binary. The chromosomal locations for all genes were mapped on the genomic scaffolds from version 3.1 of the Drosophila genome annotation using the first nucleotide position from the sequence of each microarray element. Redundant elements were then removed based on their primer sequences to minimize spurious clustering.
To control for non-random distribution of gene models on chromosomes, we made use of gene order rather than exact chromosomal position. This is addressed using a nonparametric permutation test approach [77]. Because there is a multiple testing issue [78], a large number of dependent tests (since the windows are allowed to overlap) are performed, one for every gene window, so the measures of significance have to be calculated to account for the number of tests performed.
We determined a significance level to associate with the test of clustering in a given window as follows. In the given window (w) we determine the number (n) of sex-biased genes. A series of 100,000 Monte Carlo iterations are carried out in which the sex-biased genes and the non-differentially expressed genes are shuffled among the gene positions and the relative frequency of at least n sex-biased genes occurring in some window of the same size as w on the chromosome is determined. Thus, our empirical P-value can be interpreted as the probability of such a degree of clustering occurring in at least some window of the size by chance alone.
Northern versus microarray
A subset of 75 element species were chosen as probes for Northern blotting on Hybond-N+ membranes (Amersham, Piscataway, NJ, USA) essentially as described in [79]. Probes were selected to cover the full range of absolute intensities and male/female differential expression revealed in microarray experiments. Blotted mRNAs were from flies wildtype with respect to sex and germline (y1 w67c) or from flies with no germline (progeny of homozygous tudor1 mothers). These same genotypes were used for labeling reactions in microarray experiments. Amplicon probes were made using the same primer pairs used in microarray construction and were labeled using Redi-prime II (Amersham). Northerns were hybridized at 42° in UltraHyb (Ambion, Houston, TX, USA) in 15 ml conical tubes in a bacterial shaker. Blots were images on a Storm 860 phosphorimager and quantified using ImageQuant (Molecular Dynamics, Sunnyvale, CA, USA). A total of 73 Northerns were successful (passing visual inspection and showing bands above background, see Figure 4a). While absolute intensities between microarrays and Northerns showed poor correlation (not shown), which is perhaps expected for different assays with different efficiencies, the relative measures showed very good correlation.
Additional data files
The following additional data files are available: a zip file containing 42 Excel tables (Additional data file 1) and an rtf file detailing the contents of the tables (Additional data file 2).
Supplementary Material
A zip file containing 42 Excel tables
An rtf file detailing the contents of the tables
Acknowledgments
Acknowledgments
We thank the Bloomington Stock Center for providing stocks. We also thank Dr. Margaret Cam, Dr. Thomas Brody, the Genome Biology referees and members of the Oliver lab for critical reading of the manuscript, and Bruce Baker for providing an advance copy of Arbeitman et al. [36].
References
- Oliver B. Genetic control of germline sexual dimorphism in Drosophila. Int Rev Cytol. 2002;219:1–60. doi: 10.1016/S0074-7696(02)19010-3. [DOI] [PubMed] [Google Scholar]
- Cline TW. Two closely linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics. 1978;90:683–698. doi: 10.1093/genetics/90.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown M, Belote JM, Boggs RT. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell. 1988;53:887–895. doi: 10.1016/s0092-8674(88)90369-8. [DOI] [PubMed] [Google Scholar]
- Belote JM, Baker BS. Sex determination in Drosophila melanogaster: analysis of transformer-2, a sex-transforming locus. Proc Natl Acad Sci USA. 1982;79:1568–1572. doi: 10.1073/pnas.79.5.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Chase BA, Baker BS. A genetic analysis of intersex, a gene regulating sexual differentiation in Drosophila melanogaster females. Genetics. 1995;139:1649–1661. doi: 10.1093/genetics/139.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B, Singer J, Laget V, Pennetta G, Pauli D. Function of Drosophila ovo+ in germ-line sex determination depends on X-chromosome number. Development. 1994;120:3185–3195. doi: 10.1242/dev.120.11.3185. [DOI] [PubMed] [Google Scholar]
- Oliver B, Kim YJ, Baker BS. Sex-lethal, master and slave: a hierarchy of germ-line sex determination in Drosophila. Development. 1993;119:897–908. doi: 10.1242/dev.119.3.897. [DOI] [PubMed] [Google Scholar]
- Pauli D, Oliver B, Mahowald AP. The role of the ovarian tumor locus in Drosophila melanogaster germ line sex determination. Development. 1993;119:123–134. doi: 10.1242/dev.119.1.123. [DOI] [PubMed] [Google Scholar]
- Pennetta G, Pauli D. stand still, a Drosophila gene involved in the female germline for proper survival, sex determination and differentiation. Genetics. 1997;145:975–987. doi: 10.1093/genetics/145.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 1988;2:477–489. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Sakamoto H, Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990;344:461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez-Arias A, editor. In The Development of Drosophila. Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Riechmann V, Ephrussi A. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev. 2001;11:374–383. doi: 10.1016/S0959-437X(00)00207-0. [DOI] [PubMed] [Google Scholar]
- Gelti-Douka H, Gingeras TR, Kambysellis MP. Yolk proteins in Drosophila: identification and site of synthesis. J Exp Zool. 1974;187:167–172. doi: 10.1002/jez.1401870120. [DOI] [PubMed] [Google Scholar]
- Bloch Qazi MC, Heifetz Y, Wolfner MF. The developments between gametogenesis and fertilization: ovulation and female sperm storage in Drosophila melanogaster. Dev Biol. 2003;256:195–211. doi: 10.1016/S0012-1606(02)00125-2. [DOI] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editor. In The Development of Drosophila. Cold Sping Harbor Laboratory Press; 1993. pp. 71–148. [Google Scholar]
- Wolfner MF. Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/S0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- Andrews J, Bouffard GG, Cheadle C, Lu J, Becker KG, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci USA. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Flybase, a Database of the Drosophila Genome http://flybase.bio.indiana.edu
- FlyBase The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI GEO, Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–448. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- Baker BS, Ridge KA. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthiger R, Jonglez M, Leuthold M, Meier-Gerschwiler P, Weber T. Sex determination in the germ line of Drosophila depends on genetic signals and inductive somatic factors. Development. 1989;107:505–518. doi: 10.1242/dev.107.3.505. [DOI] [PubMed] [Google Scholar]
- Johnston R, Wang B, Nuttall R, Doctolero M, Edwards P, Lu J, Vainer M, Yue H, Wang X, Minor J, et al. FlyGEM, a full transcriptome array platform for the Drosophila community. Genome Biol. 2004;5:R19. doi: 10.1186/gb-2004-5-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Lee DF, Klebes A, Turek PJ, Kornberg TB, Reijo Pera RA. Human BOULE gene rescues meiotic defects in infertile flies. Hum Mol Genet. 2003;12:169–175. doi: 10.1093/hmg/ddg017. [DOI] [PubMed] [Google Scholar]
- Yue H, Eastman PS, Wang BB, Minor J, Doctolero MH, Nuttall RL, Stack R, Becker JW, Montgomery JR, Vainer M, Johnston R. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 2001;29:E41–41. doi: 10.1093/nar/29.8.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–2021. doi: 10.1242/dev.01077. [DOI] [PubMed] [Google Scholar]
- DiBenedetto AJ, Lakich DM, Kruger WD, Belote JM, Baker BS, Wolfner MF. Sequences expressed sex-specifically in Drosophila melanogaster adults. Dev Biol. 1987;119:242–251. doi: 10.1016/0012-1606(87)90225-9. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL. Drosophila chorion genes: cracking the eggshell's secrets. Bioessays. 1991;13:97–105. doi: 10.1002/bies.950130302. [DOI] [PubMed] [Google Scholar]
- Raff EC. Genetics of microtubule systems. J Cell Biol. 1984;99:1–10. doi: 10.1083/jcb.99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano C, Bonaccorsi S, Gatti M. The kl-3 loop of the Y chromosome of Drosophila melanogaster binds a tektin-like protein. Genetics. 1993;133:569–579. doi: 10.1093/genetics/133.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Blumer N, Kampfer M, Renkawitz-Pohl R. Flagellar mitochondrial association of the male-specific Don Juan protein in Drosophila spermatozoa. J Cell Sci. 1998;111:3299–3309. doi: 10.1242/jcs.111.22.3299. [DOI] [PubMed] [Google Scholar]
- Matthies HJ, Messina LG, Namba R, Greer KJ, Walker MY, Hawley RS. Mutations in the alpha-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster. J Cell Biol. 1999;147:1137–1144. doi: 10.1083/jcb.147.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK, Flickinger TW, Mittendorf E, Pellicena-Palle A, Petschek JP, Albrecht EB. The Drosophila maternal effect locus deadhead encodes a thioredoxin homolog required for female meiosis and early embryonic development. Genetics. 1994;136:1075–1086. doi: 10.1093/genetics/136.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa JJ, Hiller MA, Fuller MT, Santel A. Differential expression of the Drosophila mitofusin genes fuzzy onions (fzo) and dmfn. Mech Dev. 2002;116:213–216. doi: 10.1016/S0925-4773(02)00141-7. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 2001;15:2886–2899. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B, Glover DM. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J. 1993;12:1219–1227. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. Asymmetric germ cell division and oocyte determination during Drosophila oogenesis. Int Rev Cytol. 2001;203:93–138. doi: 10.1016/S0074-7696(01)03005-4. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, Guarin H, Kronmiller B, Pacleb J, Park S, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3:research0080.1–0080.8. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U, Pasalodos-Sanchez S, Clark S, Clayton JD, Sparrow JC. Expression and function of the Drosophila ACT88F actin isoform is not restricted to the indirect flight muscles. J Muscle Res Cell Motil. 2001;22:111–119. doi: 10.1023/A:1010308326890. [DOI] [PubMed] [Google Scholar]
- Ayer G, Vigoreaux JO. Flightin is a myosin rod binding protein. Cell Biochem Biophys. 2003;38:41–54. doi: 10.1385/CBB:38:1:41. [DOI] [PubMed] [Google Scholar]
- Fyrberg E, Fyrberg CC, Beall C, Saville DL. Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J Mol Biol. 1990;216:657–675. doi: 10.1016/0022-2836(90)90390-8. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Lefkowitz RJ. Classical and new roles of beta-arrestins in the regulation of G-protein-coupled receptors. Nat Rev Neurosci. 2001;2:727–733. doi: 10.1038/35094577. [DOI] [PubMed] [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. Visual transduction in Drosophila. Annu Rev Cell Dev Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C, Laval M, Dahan A, Lepesant JA. The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol Cell Biol. 1994;14:4465–4474. doi: 10.1128/mcb.14.7.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, JR, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Saboe-Larssen S, Lyamouri M, Merriam J, Oksvold MP, Lambertsson A. Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics. 1998;148:1215–1224. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L, Karr TL, Nathan DF, Swift H, Srinivasan S, Lindquist S. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics. 1999;151:1065–1079. doi: 10.1093/genetics/151.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–243. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, Stuart J, Lund J, Kim S. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Oliver B, Parisi M, Clark D. Gene expression neighborhoods. J Biol. 2002;1:4. doi: 10.1186/1475-4924-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C, Riddell D, Higgins M, Holden J, White B. A gene family in Drosophila melanogaster coding for trypsin-like enzymes. Nucl Acids Res. 1985;13:6605–6619. doi: 10.1093/nar/13.18.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/S0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Williamson A, Lehmann R. Germ cell development in Drosophila. Annu Rev Cell Dev Biol. 1996;12:365–391. doi: 10.1146/annurev.cellbio.12.1.365. [DOI] [PubMed] [Google Scholar]
- Johnson R, Nuttall R, Doctolero M, Edwards P, Lü J, Wang B, Vainer M, Yue H, Wang X, Minor J, et al. FlyGEM, a full transcriptome array platform for the Drosophila community. Genome Biol. 2004;5:R19. doi: 10.1186/gb-2004-5-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Software, Cluster 3.0 (for Win, Mac and Linus) http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/software.htm
- Java Treeview http://genome-www.stanford.edu/~alok/TreeView
- Gene Ontology Consortium http://www.geneontology.org/
- DAVID, Database for Annotation, Visualization and Integrated Discovery http://apps1.niaid.nih.gov/david/upload.asp [PubMed]
- Good PI. Resampling Methods: A Practical Guide to Resampling Methods for Testing Hypotheses. New York: Springer-Verlag; 2000. [Google Scholar]
- Hsu J. Multiple Comparisons: Theory and Methods. Boca Raton: CRC Press; 1996. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A zip file containing 42 Excel tables
An rtf file detailing the contents of the tables


