Abstract
Objective
Articular cartilage defects commonly result from traumatic injury and predispose to degenerative joint diseases. To test the hypothesis that aberrant healing responses and chronic inflammation lead to osteoarthritis, we examined spatiotemporal changes in joint tissues after cartilage injury in murine knees. Since intra-articular injection of hyaluronan (HA) can attenuate injury-induced osteoarthritis in wild-type (WT) mice, we investigated a role for HA in the response to cartilage injury in mice lacking HA synthase 1 (Has1−/−).
Design
Femoral groove cartilage of WT and Has1−/− mice was debrided to generate a non-bleeding wound. Macroscopic imaging, histology, and gene expression were used to evaluate naïve, sham-operated, and injured joints.
Results
Acute responses (1–2 weeks) in injured joints from WT mice included synovial hyperplasia with HA deposition and joint-wide increases in expression of genes associated with inflammation, fibrosis, and extracellular matrix (ECM) production. By 4 weeks, some resurfacing of damaged cartilage occurred, and early cell responses were normalized. Cartilage damage in Has1−/− mice also induced early responses; however, at 4 weeks, inflammation and fibrosis genes remained elevated with widespread cartilage degeneration and fibrotic scarring in the synovium and joint capsule.
Conclusions
We conclude that the ineffective repair of injured cartilage in Has1−/− joints can be at least partly explained by the markedly enhanced expression of particular genes in pathways linked to ECM turnover, IL-17/IL-6 cytokine signaling, and apoptosis. Notably, Has1 ablation does not alter gross HA content in the ECM, suggesting that HAS1 has a unique function in the metabolism of inflammatory HA matrices.
Keywords: cartilage, injury, synovium, inflammation, fibrotic scar, hyaluronan synthases
INTRODUCTION
Traumatic injuries to articular cartilage of the knee can result from excessive surface contact stresses after blunt impact or torsion, which occur frequently during sports and military training1. Resulting patellar dislocation2, joint incongruity, and instability can predispose to osteoarthritis (OA)3. Responses to cartilage injury share many features of wound healing, such as innate inflammation4 and activation of multipotent progenitor cells in the synovium or the articular surface5. However, in many cases, the repair response leads to fibrotic remodeling and scarring of the joint lining tissues, subchondral bone sclerosis, and chondrophyte or osteophyte development at the articular margins.
Whereas chronic inflammation is widely recognized as a driving factor in OA, the concept of a pathogenic role for fibrotic scarring is less well-studied6. Evidence for focal scarring, has been reported for synovial tissue and cartilages from both animal model and human OA7, each of which exhibit activation of multiple genes associated with collagen production and deposition (CRLF1, PLOD2, LOX, COL1A1, COL5A1, TIMP1). In addition, other laboratories8–10 also reported high expression of COL1A1, COL2A1, COL3A1, and COL5A1 in human OA cartilages. Moreover, mice deficient in genes that enhance collagen matrix formation and turnover in wound healing (Adamts5, Ddr2, Mmp13, Sdc4, and Tgm211–14) were variably protected from surgically induced OA.
In dermal wound healing, early inflammation is followed by formation of granulation tissue containing progenitor cells embedded in a provisional extracellular matrix (ECM) of collagens, fibronectin, hyaluronan (HA), and hyalectans15. These cells, following re-epithelialization, mature into fibroblasts to generate the functional collagenous repair tissue. Correspondingly, following cartilage injury, proliferation of cells and ECM deposition in synovial lining and adjacent adipose or joint capsule tissues occurs. This response can progress into fibrotic remodeling, often reported in inflammatory arthritis16. A similar response also develops in the meniscal destabilization mouse model of OA, where, at 2–4 weeks17, an inflammatory period is followed by elevated expression of profibrotic mediators such as type III collagen (Col3a1), fibromodulin (Fmod, a catalyst for TGF-β1 signaling), and prolargin (Prelp, an inhibitor of osteoclastic activity).
To examine repair responses specifically in the context of articular cartilage injury, we have adapted a murine model induced by surgical excision of cartilage from the patellar groove18. This allowed spatiotemporal macroscopic and microscopic evaluation of whole joint-responses and assay of gene expression in inflammatory, pro-fibrotic and ECM pathways, in both intact joints and separated tissue pools (meniscus and synovium (Men/Syn), cartilage and subchondral bone (C/SCB), and patellar tendon (PT)).
We have also examined such injury responses in mice deficient in HA synthase 1 (Has1), which have previously been reported to exhibit an aberrant dermal healing phenotype19. We show that Has1−/− mice, although not defective in overall HA production, are not able to control post-injury joint inflammation and develop extensive intra-articular scarring and severe OA-like symptoms.
METHODS
Murine Cartilage Injury Model
Wild-type (WT) and Has1−/− male C57Bl/6 mice (10–12 weeks old, ~30g) were used under approval of the Rush University Institutional Animal Care and Use Committee. Routinely, four C57Bl/6 males were caged with one C57Bl/6 female littermate, to minimize male aggression and prevent wounding in the pre-op and post-op maintenance periods. After anesthesia, an ~8-mm medial para-patellar incision was made on the right knee, medial parapatellar arthrotomy was performed, and the patella laterally luxated. Cartilage was debrided along the distal groove with a #15 scalpel without penetration of the subchondral bone. Joint surfaces were lavaged with sterile saline, and the patella repositioned, before the muscle layer and skin were closed with 6–0 Vicryl sutures. Supporting ligaments, menisci, and other cartilage surfaces were not damaged (Fig. S-1A), and no changes were detected in the contralateral joint post-injury (Fig. S1 -B). Sham surgery included all steps, except cartilage debridement. Mice were maintained at cage activity during the 4-week post-surgery period. The minimal number of mice needed for each outcome was determined based on previous studies20, and the numbers in each experimental group are given (Table S-1).
Macroscopic Joint Imaging, Histology, and HA Staining
Joint-wide pathology was assessed in operated and contralateral joints as previously described20. For histology, whole joints (after removal of skin and muscle) were fixed in 10% neutral-buffered formalin, decalcified with 5% EDTA in PBS, paraffin-embedded, and microtome-cut into 6-µm sections across the entire joint20. Sections 1–60, 61–120, and 121–190 spanned medial, central groove, and lateral compartments, respectively. Slides 1/2, 22/23, 42/43, 62/63, 82/83, 102/103, 122/123, 142/143 and 182/183 were stained with Safranin O (SafO), and adjacent sections with hematoxylin and eosin (H&E) or biotinylated HA binding protein (bHABP) to localize HA. It should be noted that the histological analysis was not used in this study to generate a numerical scale for cartilage grading (as per OARSI guidelines) but evaluated, in combination with the macro-images, to describe structural alterations in multiple tissue types adjacent to the injury and throughout the whole joint.
Quantitative PCR (qPCR)
For gene expression in whole joints from naïve, sham, and injured groups (n=3–4, detailed in Table S-1), hind legs were harvested immediately after sacrifice, the skin and muscle removed, and knee joints isolated by sharp dissection through the growth plates prior to storage at −20°C in RNAlater (Table S-1). To prepare separate tissue pools, twelve joints were used for Men/Syn or PT, and two for C/CSB. RNA purification, cDNA synthesis and qPCR (3 technical replicates) with Taqman®-primers (Table S-2) was done as previously described20. Transcript abundance was calculated as 1000×2−ΔCt, with ΔCt = [Ct (gene of interest) – Ct (Gapdh)] and Ct>35 considered 'non-detectable' (ND). RT2 Profiler PCR Arrays (Qiagen) were used for fibrosis (PAMM-120ZA) and NF-κB signaling target (PAMM-225ZA) genes. Injury-induced fold-change in expression was calculated as 2−ΔΔCt, where ΔΔCt = [ΔCt (post-injury time point) – ΔCt (naïve)]. Gene groupings indicated by Qiagen and Metacore™ software analysis of expression data were used to determine pathway associations.
Data and Statistical Analysis
For statistical comparisons across time points and between genotypes, qPCR assays were performed on whole joints (each joint an experimental unit), from naïve, sham, and injury groups, because the large number of mice (12 per experimental unit) required for generating multiple pools of separated tissue types was outside the scope of this study.
Gapdh Ct values from WT and Has1−/− samples were pooled to confirm normality of Ct values with the Shapiro-Wilk test, and analysis of variance (ANOVA) was used to compare Gapdh Ct values across groups to confirm selection of the housekeeping gene. For all combinations of WT/Has1−/− naïve/sham/injury joints (6 in total) were confirmed with the Shapiro-Wilk test to be normally distributed. For each gene, ANOVAs were performed on ΔCt values to compare expression in the following subgroups: 1-way ANOVA for WT naïve vs. 12 or 28 day post-sham; 2-way ANOVAs for WT vs. Has1−/−, naïve vs. 12 or 28 day post-injury (main effects: experimental end point, genotype). ANOVAs were followed by post hoc analysis of statistically significant effects using unpaired, two-tailed Student’s t tests with Bonferroni correction of the p value for multiple comparisons against naïve. Since only two end points at biologically distinct phases of sham and injury response were compared to naïve, Bonferroni correction was chosen as the most conservative option for post hoc comparisons after ANOVA.
For each gene from the arrays, unpaired two-tailed Student’s t tests were used to compare between WT and Has1−/− ΔCt values at naïve. Similar analysis was performed to compare array results of WT naïve to WT sham. ANOVA was used to compare genotypes for ΔCt values after injury (to determine the overall effect of genotype), followed by a post hoc unpaired, two-tailed Student’s t test with Bonferroni correction. The experiment-wide significance level was α = 0.05 for statistical analyses.
RESULTS
Macroscopic Imaging of the Murine Model of Cartilage Injury in WT Joints
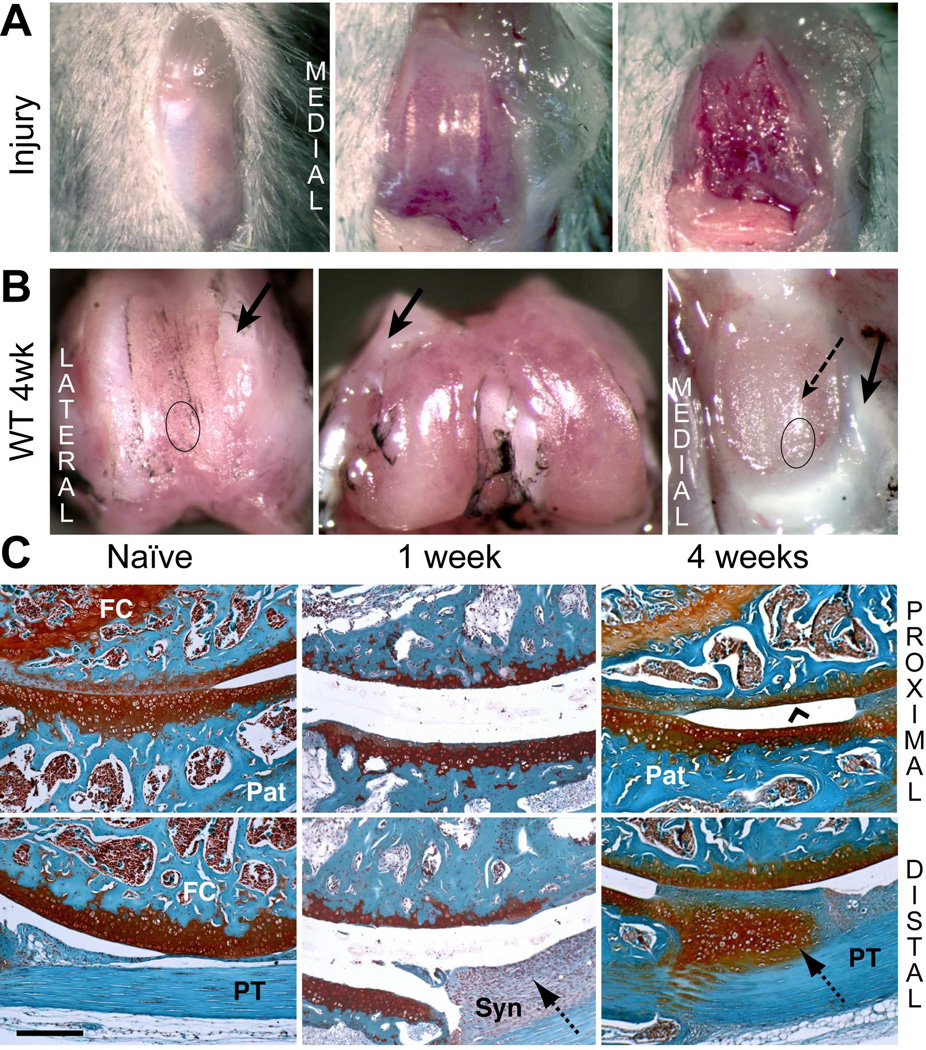
Cartilage was debrided along the full length of the patellar groove without damaging the trochlear ridges (Fig. 1A) or subchondral bone. Typical images of joints at 4 weeks post-surgery (Fig. 1B) illustrate that the trochlear ridges adjacent to the injury-site were covered with a whitish tissue (Fig. 1B, arrows) extending into the periosteum. SafO or H&E histology showed this to be dense fibrous tissue lacking chondroid staining and with few cells (data not shown). The damaged groove surface became covered with a thin layer of tissue, but the apposing patellar cartilage was markedly roughened, with evidence of tissue ingrowth from the margins (Fig. 1B). Contralateral joints remained macroscopically unaffected (Fig. S-1B), and sham operated joint surfaces also showed no damage after 4 weeks (Fig. S-1C).
Figure 1. Macroscopic and histological evaluation of joint tissue response to cartilage injury in wild type mice.
A) The joint capsule was opened via medial peri-patellar incision, and cartilage debride d along the patellar groove of the right knee. B) The macroscopic appearance of cartilage surfaces and adjacent soft tissues was examined in the injured joint at 4 weeks post-surgery. Dense fibrous tissue formation (solid arrows) at the margins of the injured area and cartilage wear on the patella (dashed arrow) are indicated. C) Safranin-O staining of tissue structures in the femoral-patellar compartment of naïve joints and at 1 and 4 week post-injury permitted histological evaluation. The approximate regions of the groove and the patella taken for histology are indicated by circles in panel B. Cartilage resurfacing (chevron arrowhead) is observed on the femoral groove. The hyperplastic tissue deposition at the synovium (dotted arrows) at week 1 developed into a chondrophytic deposit by week 4. (Pat = patella; FC = femoral condyle; PT = patellar tendon; Syn = synovium). Scale bar = 100 μm.
Histology of Joint Tissue Responses to Cartilage Injury in WT Mice
Typical images of SafO stained whole joint sections from naïve and 1- and 4-weeks post-injury WT mice (Fig. 1C) show the cartilage interfaces between the proximal groove and the center of the patella (top panels) and between the distal groove and the distal patellar tendon PT (bottom panels). In naïve mice, these showed full-depth patellar cartilage in apposition to the groove cartilage, transitioning into a SafO-poor fibrous tissue at the proximal end. At 1 week post-injury, a thin layer of calcified cartilage, remaining after the debridement covered the subchondral bone of the proximal groove (Fig. 1C, upper panel). The patellar cartilage apposing the injury site showed marked thinning relative to naïve, likely as a result of mechanical abrasion or degradative mediators released from the area of cartilage injury. Although subchondral bone was not penetrated, the cellularity in the marrow space of both bones was reduced at 1 week post-injury (Fig. 1C) and spaces filled with a disorganized ECM. A thick layer of hyperplastic synovium, extending between the patellar tendon and the distal patella was prominent at 1 week.
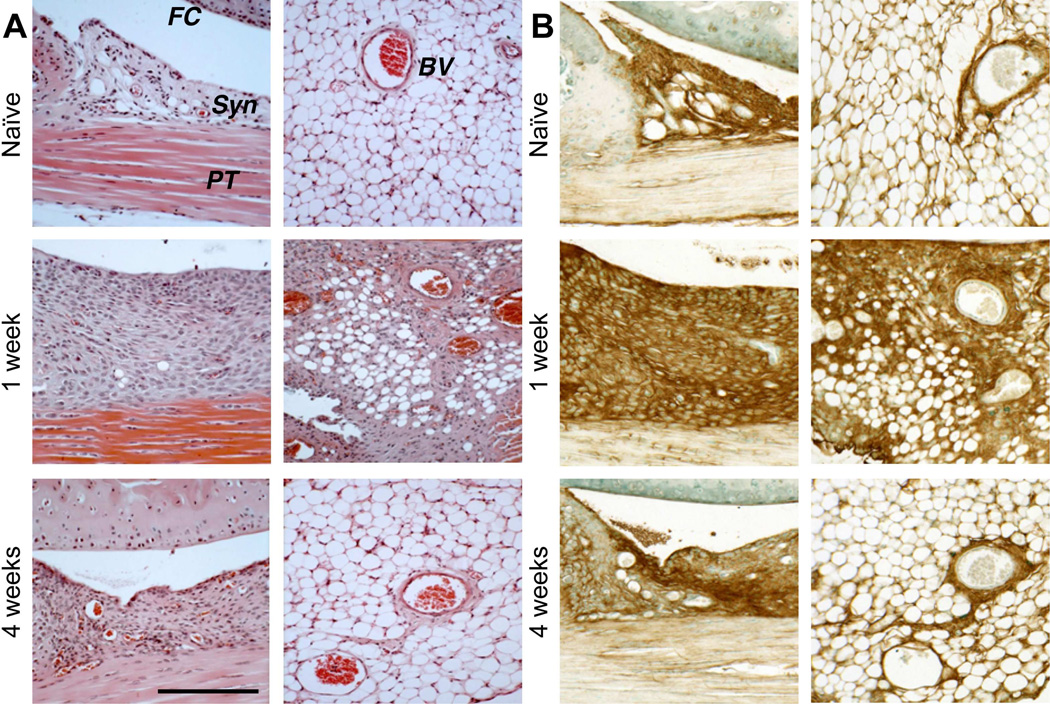
At 4 weeks, a continuous layer of SafO-stained “repair” cartilage covered the injured groove, and the cellularity of the marrow space had regenerated. The patellar cartilage surface was also restored, and all injured joints developed chondrophytic deposits with a fibrous covering (Fig. 1C, lower panel) derived from the hyperplastic synovium adjacent to the patella. Hyperplasia of both peri-patellar and perimeniscal synovium and the loss of adipocytes by 1 week was confirmed by H&E histology (Fig. 2A). By 4 weeks, normal cellularity was restored with minor thickening of the peri-vascular matrix. A transient influx of neutrophils to vascularized regions at the dermal incision site was evident at 3 days post-surgery (data not shown), but no infiltration of circulating inflammatory cells into the synovium or the joint space was detected, consistent with only innate inflammation.
Figure 2. Histological evaluation of hyperplasia in peripatellar and perimeniscal synovium and adipose tissue following cartilage injury in wild type mice.
A) H&E staining showed post-injury cellular hyperplasia and increased ECM deposition at the synovial lining, in the adipose stroma, and perivascular regions. B) Sections equivalent to those shown in (A) were stained for HA using bHABP. (FC = femoral condyle; PT = patellar tendon; Syn = synovium; BV = blood vessel). Scale bars = 100 μm.
When equivalent sections were examined for HA deposition, strong staining of synovium was seen at 1 week (Fig. 2B, left panel), but at 4 weeks HA staining essentially returned to naïve levels. The HA accumulation early post-injury is consistent with the increased expression of Has1 (p<0.01) and Has2 (p<0.05) at those times. Has1 expression remained significantly elevated (12d: p<0.01, 28d: p<0.05) at later times (Fig. S-3), suggesting that long-term ECM remodeling includes this prolonged expression response. Expression of Has1 and Has2 in contralateral joints was unaffected (Fig. S-3A). Furthermore, sham-operated joints at 2 weeks showed only mild peri-patellar synovial hyperplasia, which resolved at 4 weeks (data not shown). Minor fibrotic/chondroid remodeling at the patellar margins was observed (Fig S-1C), but femoral or tibial articular and growth plate cartilage appeared normal.
Post-injury Expression of HA-matrix Genes in WT Joint Tissues
Injury to skin21, intestine22, cartilage23, and other tissues is accompanied by inflammation and changes in HA accumulation, resulting from concurrent increased synthesis of HA itself and HA-associated proteins, such as TSG6 (encoded by Tnfaip6), the heavy chains of inter-alpha-trypsin inhibitor (encoded by Itih1–5 genes), and pentraxin.
Increased expression of some of these genes also occurred after cartilage injury (Table 1). In naïve joints, all transcripts except for Tnfaip6 were most abundant in the Men/Syn tissue pools, followed by PT and C/SCB pools, consistent with the staining intensity of HA (Fig. 2B). At 1 week, Has1 expression increased markedly in all tissues and remained elevated at 4 weeks. Has2 was also increased at 1 week in the Men/Syn and PT, remaining elevated in PT up to 4 weeks but normalizing in the Men/Syn (Table 1). Concurrent with the increased expression of Has1 and Has2, Tnfaip6 was activated at 1 week in all tissues, particularly in Men/Syn (43-fold) and PT (22-fold). Expression levels declined, but both time points by 4 weeks. Itih2 expression was minimally affected in all tissues at both time points except for a 7.7-fold increase in the PT at 4 weeks. Itih1 expression was virtually undetectable in any samples (data not shown).
Table 1.
Effect of cartilage injury on mRNA abundance relative to Gapdh of HA-network genes in isolated tissue pools from WT mice.
| Has1 | Has2 | Tnfaip6 | Itih2 | |
|---|---|---|---|---|
| Naive | ||||
| Cartilage and subchondral bone | 0.36 | 0.06 | 0.12 | 0.11 |
| Meniscus and synovium | 24.4 | 0.41 | 1.28 | 1.03 |
| Patellar Tendon | 2.58 | 0.27 | 2.32 | 0.51 |
| 1 week | ||||
| Cartilage and subchondral bone | 4.17 (12)† |
0.06 (1.1) |
1.75 (15) |
0.04 (0.4) |
| Meniscus and synovium | 78.5 (3.2) |
2.83 (7.0) |
55.6 (43) |
1.91 (1.9) |
| Patellar Tendon | 33.9 (13) |
2.68 (10) |
51.2 (22) |
0.65 (0.8) |
| 4 weeks | ||||
| Cartilage and subchondral bone | 3.73 (10) |
0.03 (0.5) |
1.06 (8.9) |
0.08 (0.8) |
| Meniscus and synovium | 54.6 (2.2) |
0.40 (1.0) |
10.4 (8.1) |
1.92 (1.9) |
| Patellar Tendon | 44.2 (17) |
1.19 (4.4) |
18.2 (7.9) |
3.90 (7.7) |
Italicized numbers in parentheses are fold change relative to naïve levels.
Post-injury Changes to Expression of ECM Genes in WT Tissues
We also assayed for changes in several ECM-related genes known to be associated with matrix remodeling in cartilaginous and fibrous tissue in individual tissue pools (Table 2). In naïve tissues, transcripts for Acan, Vcan V1 (formerly named the V0 isoform), and Col2a1 were highest in the C/SCB, and for Col3a1 in the Men/Syn. Notably, Col1a1 transcripts were very high relative to other genes in the three tissue pools, and Vcan V2 (formerly V1) was barely detectable.
Table 2.
Effect of cartilage injury on mRNA abundance relative to Gapdh of ECM genes in separate tissue pools from WT mice.
| Acan | Vcan V1 | Vcan V2 | Col1a1 | Col2a1 | Col3a1 | |
|---|---|---|---|---|---|---|
| Naive | ||||||
| Cartilage and subchondral bone | 1.87 | 26.9 | ND | 1720 | 1.22 | 1.6 |
| Meniscus and synovium | 0.85 | 5.21 | 0.16 | 1280 | 0.15 | 24.6 |
| Patellar Tendon | 0.04 | 6.33 | 0.05 | 962 | ND | 2.6 |
| 1 week | ||||||
| Cartilage and subchondral bone | 10.1 (5.4)† |
12.7 (0.5) |
0.47 (−) |
3550 (2.1) |
1.87 (1.5) |
62.9 (39 |
| Meniscus and synovium | 61.8 (72.3) |
15.2 (2.9) |
15.2 (92) |
10300 (8.0) |
0.38 (2.6) |
4010 (163) |
| Patellar Tendon | 130 (3386) |
11.1 (1.8) |
17.2 (366) |
10900 (11) |
0.42 (−) |
4451 (1734) |
| 4 weeks | ||||||
| Cartilage and subchondral bone | 7.97 (4.3) |
8.85 (0.3) |
0.18 (−) |
4880 (2.8) |
1.38 (1.1) |
54.8 (34) |
| Meniscus and synovium | 8.12 (9.5) |
2.80 (0.5) |
1.75 (10.6) |
2770 (2.2) |
0.80 (5.4) |
1080 (44) |
| Patellar Tendon | 83.7 (2180) |
6.87 (1.1) |
4.41 (94) |
7380 (7.7) |
17.7 (−) |
3200 (1245) |
Italicized numbers in parentheses are fold change relative to naïve levels. ND = not detected (Ct > 35), with no fold change calculation possible ( – ).
All genes were increased at 1 week post-injury, with the increases both tissue- and gene-specific. Increases in expression were highest for Acan, Vcan V2, and Col3a1 in the PT, possibly induced by manipulation at the time of surgery and through its proximity to the reactive synovium (Fig. 1 and 2). In contrast, expression of assayed genes was minimally affected in C/SCB. At 4 weeks post-injury, expression in Men/Syn and C/SCB decreased markedly to near naïve levels, but there was continued elevation of Acan, Vcan V2, and Col3a1 in the PT.
Gene Expression in Contralateral and Sham-operated Joints
A selected group of genes was assayed in contralateral and sham operated joints (Fig. 3-S). Expression was essentially unaffected in contralateral tissues, consistent with minimal systemic post-injury responses. In sham-operated joints, there was a significant increase in expression of Has1, Tnfaip6 and Col3a1 by 12 days, similar to that in fully injured joints. All genes returned to naïve levels in the sham at 28 days, except for Has1 and Col3a1, which both remained elevated, suggesting that these are specific indicators of joint damage itself.
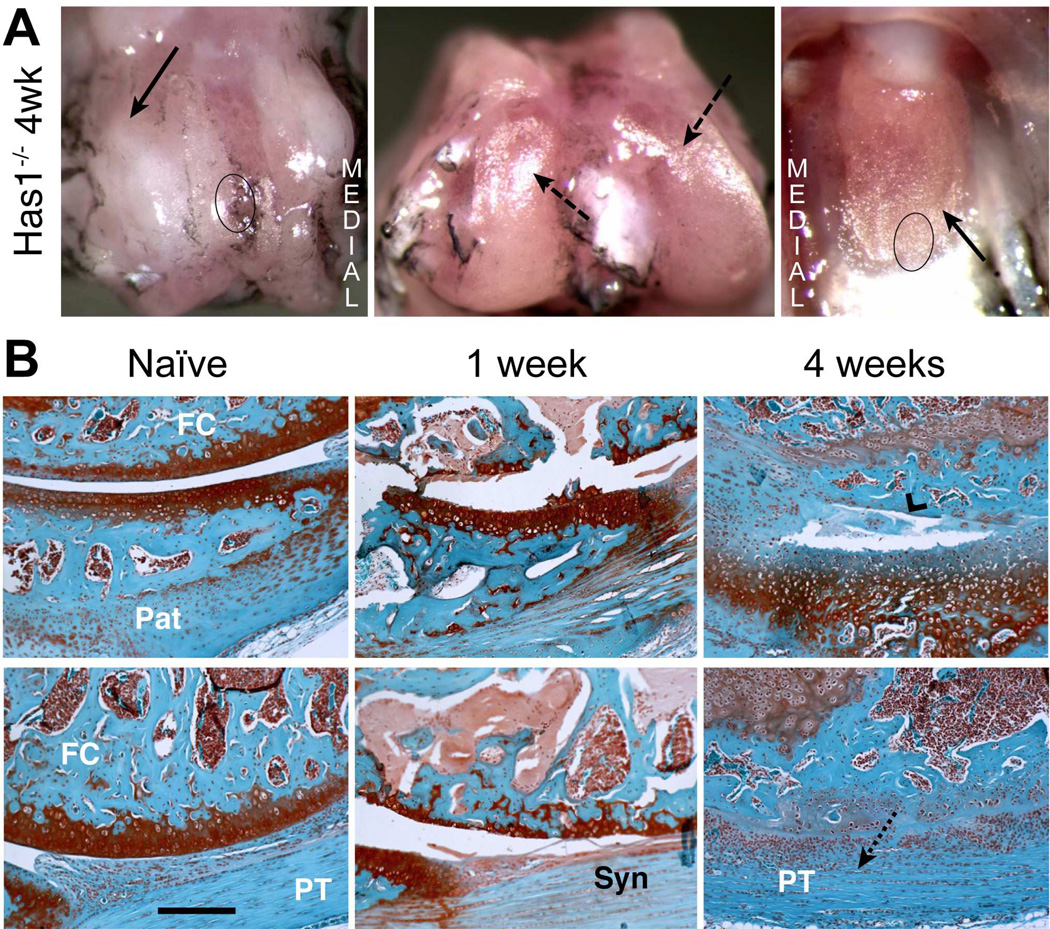
Figure 3. Macroscopic and histological evaluation of joint tissue response to cartilage injury in Has1−/− mice.
A) The macroscopic appearance of cartilage surfaces and adjacent soft tissues in the injured joint was examined at 4 weeks post-surgery. Extensive fibrotic deposits (solid arrow) were observed at the groove, and both the condylar and patellar cartilage showed signs of wear (dashed arrows). B) S ections from the femoral-patellar compartment of naïve joints and at 1 and 4 weeks post-injury were stained with Safranin-O. The approximate region of the groove and the patella taken for histology are indicated by circles in panel B. By 4 weeks post-injury, severe cartilage and bone loss (chevron arrowhead) combined with extensive fibrotic overgrowth (dotted arrow) results in a loss of joint space. (Pat = patella; FC = femoral condyle; PT = patellar tendon; Syn = synovium). Scale bar = 100 μm.
Has1 Ablation Modifies Joint Tissue Response to Cartilage Injury
Since Has1 was highly activated and sustained after cartilage injury (Table 1) and given the importance of HA in connective tissue healing, we examined cartilage injury responses in Has1−/− joints. Macroscopic examination of Has1−/− joints at 4 weeks post-injury (Fig. 3A) showed a dense collagenous tissue covering of the trochlear ridges and extending to the adjacent periosteum, whereas this was not seen in the WT (Fig. 1B). Instead of the partial regeneration of cartilage seen in WT joints, widespread damage to the cartilage and subchondral bone was seen (Fig. 3B) at 1 week, with extensive fibrotic overgrowth (devoid of any SafO staining) developing in these areas (Fig. 3B). Most notable was the finding that, at 4 weeks, the fibrotic tissue expanded from the injury site (Fig. 3B) into all soft tissues and also filled and thus eliminated the joint space.
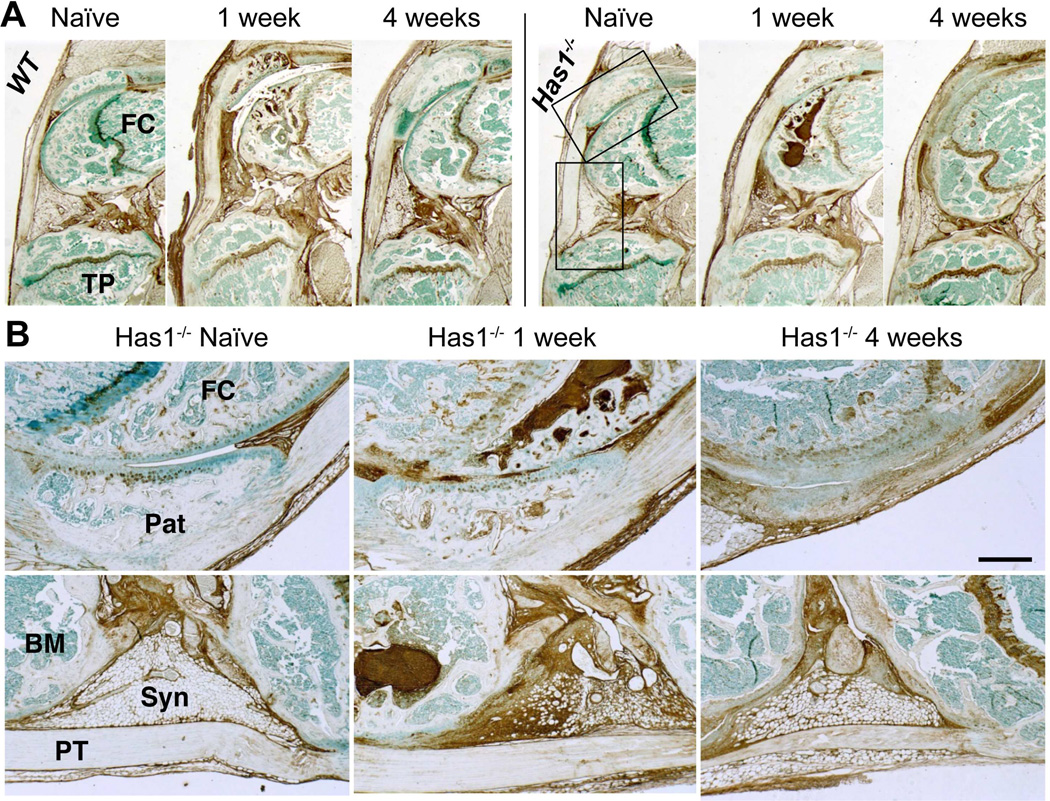
Has1 deficiency did not affect HA deposition at 1 week or its decrease at 4 weeks seen in the synovial lining, adherent adipose tissue, and SCB marrow (Fig. 4). This is consistent with the accepted notion that HAS2, but not HAS1 or HAS3, is the primary HA-producing synthase throughout all organs24, and therefore also responsible for the increased HA deposition seen in both the WT and Has1−/− joints.
Figure 4. Histological evaluation of HA distribution in naïve and post-injury joint tissues of WT and Has1−/− mice.
A) Sagittal sections stained with b HABP are shown at low magnification. B) High magnification images of patella /patellar groove/peri-patellar synovium (upper panel) and peri-meniscal adipose tissue (lower panel) from Has1−/− joints (see boxed areas in (A)) show the prominent increase in HA deposition in post-injury joints. (Pat = patella; FC = femoral condyle; TP = tibial plateau; PT = patellar tendon; Syn = synovium; BM = bone marrow) Scale bar = 100 μm.
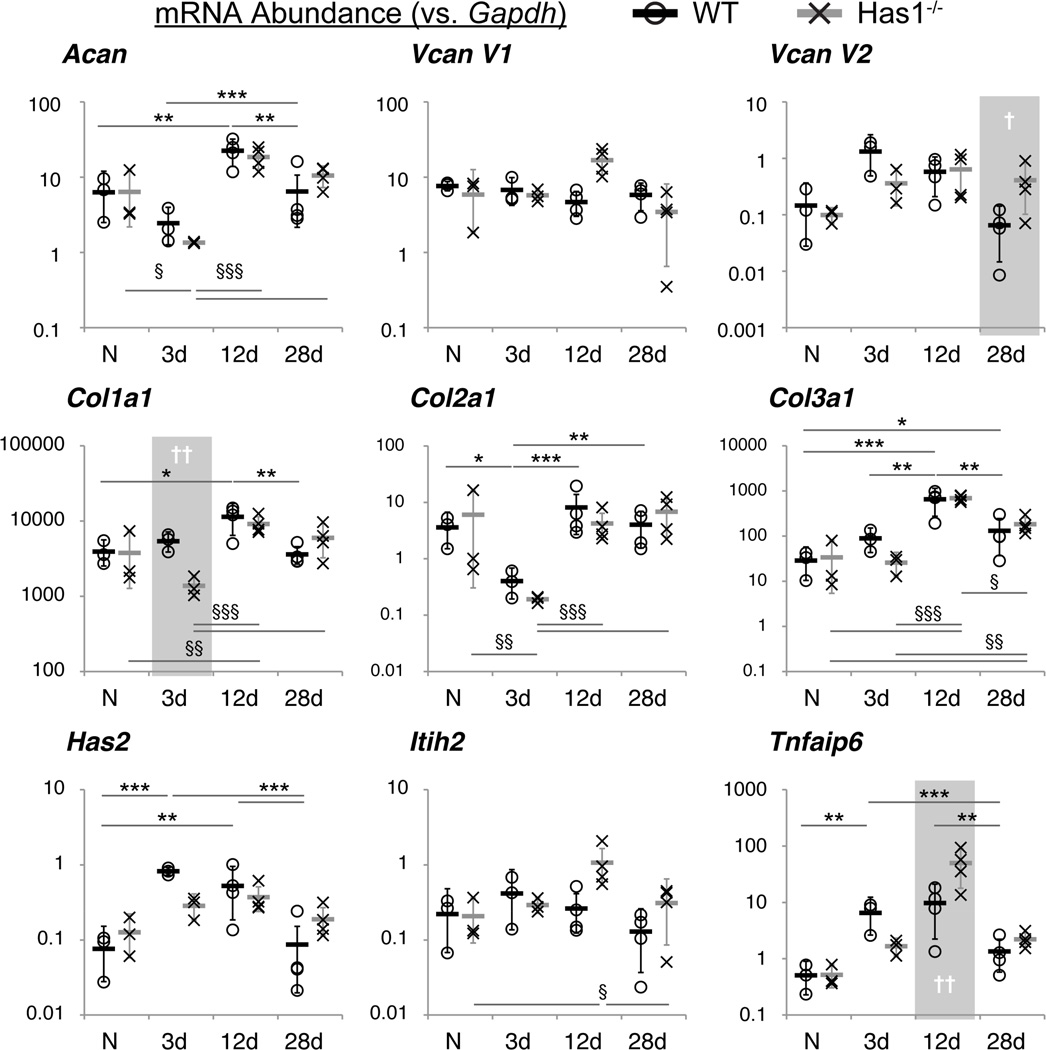
The tissue-specific and temporal post-injury gene expression trends (Table S-3) were similar for WT and Has1−/− mice, with transcripts for Has3 and Itih1 undetectable in both strains; however, there were significant differences between genotypes with time (Fig. 5). In WTs, Col2a1 (p<0.05) expression decreased at day 3 and returned to naïve levels by day 12, whereas expression of Has1 (p<0.01), Has2 (p<0.001), and Tnfaip6 (p<0.01) increased at 3 days and remained increased at 12 days (Fig. 5 and S-4). Expression of Col1a1 (p<0.05) and Col3a1 (p<0.001) peaked at day 12, and Col1a1, but not Col3a1 (p<0.05), returned to naïve levels by day 28 (Fig. 5). WT joints showed an increased expression of HA-matrix genes up to 12 days and a sustained increase of Col3a1 up to 28 days. In summary, whereas Has1−/− joints responded post-injury like WT for matrix genes, they did not show as clear a normalization of HA-matrix genes at later times.
Figure 5. Expression of selected ECM and HA network genes in WT and Has1−/− joints.
Gene expression, calculated as mRNA abundance relative to Gapdh (see Methods), of WT (circles) and Has1−/− (crosses) whole joints at each time point (naïve (N) & 3d: n = 3; 12d & 28d: n = 4) is shown for each biological replicate. Means within each group are shown as a thick black or gray bar, and the error bars represent the transformed upper and lower bounds of a 95% confidence interval for the two genotypes at each time point. Statistical significance between time points, after two-way ANOVA and correction for multiple comparisons (see Methods), at P < 0.05 (*/§), P < 0.01 (**/§§), and P < 0.001 (***/§§§), between time points are indicated for WT/Has1−/− joint gene expression. Statistically significant differences between genotypes are indicated for P < 0.05 (†) and P < 0.01 (††) and highlighted with a gray area.
Only in Has1−/− joints was Col1a1 (p<0.01) inhibited at 3 days and Vcan V2 (p<0.05) activated at 28 days (Fig. 5). Although temporal trends for HA-associated genes were mostly similar to WT, Has1−/− joints showed no significant changes with injury in Has2 and Tnfaip6 expression (Fig. 5). Thus, Has1 ablation was accompanied by a lower trend in activation of Has2 expression at 3 days, a much greater stimulation of Tnfaip6 at 12 days (p<0.01), and a significant activation of Itih2 expression (p<0.05) at 12 days, indicating it affects are not on HA production itself but on genes associated with forming the HA matrix.
Effect of Has1 Ablation on Gene Pathway Changes at Different Periods after Injury
The lack of cartilage regeneration, degenerative changes on adjacent cartilage surfaces, and extensive fibrotic remodeling of the synovium and joint capsule in Has1−/− mice are consistent with development of a chronic inflammatory environment in the post-injury joint. To establish a possible link between chronic inflammation and post-injury fibrotic scarring, we analyzed joints from both genotypes for NF-κB signaling target and fibrosis genes. Assays were done for naïve and 12 or 28 days post-injury in both genotypes, and data calculated as fold-change relative to genotype naïve (Tables S-6 and S-7). Notably, compared to WT, Has1−/− mice over-expressed a larger number (26 genes at >3 fold) of the NF-κB signaling target genes (Table S-4) and fibrosis genes (Table S-5).
Pathway associations were determined for genes that were modified at least 3-fold relative to naïve levels using Metacore™ software. For both genotypes, four functional gene groupings were identified: IL-17/IL-6 signaling, ECM remodeling, pro-apoptosis, and anti-apoptosis (Table 3). Moreover, within those groups, there were differences between WT and Has1−/− in expression of many genes, with 27 showing 6-fold or greater differences between WT and Has1−/− (Table 3): for IL-17/IL-6 signaling, Csf3, Map2k6 (12 days) and Cxcl1, .Il6, Mmp1a, Ptgs2 (28 days); for ECM remodeling, Col1a2 (p<0.01), Ctgf (p<0.05), Timp2 (12 days) and Plat, Serpine1, Tgfb3, Thbs2, Timp1 (28 days); for pro-apoptosis genes, Fasl, Ifng, Il4, Ins2 (p<0.01), Lta (12 days) and Il4, Ins2, Tnfsf10, Traf2 (28 days); and, for anti-apoptosis genes, Birc3, Cd40, Csf2, Fas, Il2, Rela (12 days) and Csf2 (28 days).
Table 3.
Effect of cartilage injury in multiple genes related to NF-κB signaling targets and fibrosis in whole joint preparations from WT and Has1−/− mice.
|
IL-17/IL-6 signaling |
WT (12d) |
Has1−/− (12d) |
WT (28d) |
Has1−/− (28d) |
ECM Remodeling |
WT (12d) |
Has1−/− (12d) |
WT (28d) |
Has1−/− (28d) |
|---|---|---|---|---|---|---|---|---|---|
| Akt1 | ↓ | + | ↓ | ↓ | Bmp7 | ↓↓↓ | ↓ | ↓↓ | ↓ |
| Ccl3 | ↓↓↓ | ↓↓ * | ↓ | + | Col1a2 | ↓↓ | +¶** | ↓ | ↔ |
| Ccl11 | + | ↔ | + | ++ | Col3a1 | +++ | ++++ | ++ | +++ |
| Ccr2 | ↓↓ | ↓↓↓ | ↓ | ↓ | Ctgf | ↓↓ | ++¶* | ↓ | ↔ |
| Cebpb | ↓↓↓ | ↓ | ↓↓↓ | ↓↓ | Egf | ↓ | + | ↓ | ↓ |
| Csf2rb | ++ | +++ | ++ | ++ | Egfr | ++ | +++ | ↓ | ↔ |
| Csf3 | ↓↓ | +++¶ | ↓ | ↓ | Grem1 | ↓↓↓↓ | ↓↓* | ↓↓ | ↓↓↓ |
| Cxcl1 | ++ | ++++ | ↔ | ++++¶ | Lox | + | + | ↓ | + |
| Icam1 | ↔ | ++ | ↓ | + | Mmp2 | ++ | +++ | + | ++ |
| II1a | ↓↓ | ↔ | ↓ | + | Mmp8 | ↓↓ | ↓↓ | ↓ | ↓ |
| II1b | ↓↓ | ↓ | ↓ | ↓ | Mmp13 | ↓↓ | ↓ | ↓ | ↓ |
| II6 | +++ | ++++ | + | ++++¶ | Mmp14 | + | ++ | ↓ | + |
| II10 | ↓↓ | ↓ | + | ↓ | Plat | ++ | ++ | ↓ | +++¶ |
| Jun | + | + | ↓ | + | Plau | + | ↓ | ↓ | ↓ |
| Map2k6 | ↓ | ++¶ | ↓ | + | Plg | ↓ | + | ↓ | + |
| Mmp1a | ↓↓↓↓ | ↓↓↓ | ↓↓↓ | +¶ | Serpina1a | ↓↓ | ↓↓ | ↓↓ | ↓↓ |
| Mmp3 | +++++ | +++++ | +++++ | +++++ | Serpine1 | ++ | +++ | + | +++¶ |
| Mmp9 | ↓ | + | ↓ | + | Serpinh1 | + | ++ | ↓ | + |
| Nfkb1 | ↓ | + | ↓ | + | Tgfb3 | +++ | ++++ | + | ++++¶ |
| Nfkbia | ↓ | + | + | ++ | Thbs2 | +++ | ++++ | + | ++++¶ |
| Ptgs2 | +++ | +++ | ↓ | +++¶ | Timp1 | + | ++ | + | +++++¶ |
| Rel | ↓ | ↓ | ↓ | + | Timp2 | ++ | ++++¶ | ↔ | ++ |
| Selp | +++ | +++ | +++ | ++++ | Timp3 | + | + | ↓ | + |
| Sp1 | ↓ | ↔ | ↓ | ↓ | Timp4 | ↓↓↓ | ↓ | ↓↓ | ↓ |
|
Pro Apoptosis |
WT (12d) |
Has1−/− (12d) |
WT (28d) |
Has1−/− (28d) |
Anti- Apoptosis |
WT (12d) |
Has1−/− (12d) |
WT (28d) |
Has1−/− (28d) |
| Agt | ↓↓ | ↓ | ↓↓ | ↓↓ | Adm | + | +++ | ↓ | + |
| Birc2 | ↓ | + | ↓ | ↓ | Bcl2 | ↓↓ | ↓ | ↓↓ | ↓ |
| Cd74 | ↓ | + | ↓ | ↓↓ | Bcl2a1a | ↓ | + | ↓ | ↓ |
| Fasl | ↓↓ | ++¶ | ↓ | + | Bcl2l1 | ↓↓ | ++ | ↓ | ↓ |
| Gadd45b | ++ | ++ | ↓ | + | Birc3 | ↓ | ++¶ | ↓ | ↓ |
| Ifnb1 | ↓↓↓↓ | + | ↓↓ | ↓ | Ccl12 | + | ++ | + | + |
| Ifng | ↓↓ | ++¶ | ↓↓ | ↓ | Cd40 | ↓ | ++¶ | ↓ | ↓ |
| II12b | ↔ | + | ↓ | ↓ | Cdkn1a | + | ++ | ↓ | ↔ |
| II2ra | ↓ | + | + | + | Csf2 | ↓↓↓ | ++¶ | ↓↓ | ↔¶ |
| II4 | ↓↓ | +¶ | ↓ | +++¶ | F3 | + | ++ | ↓ | + |
| Ins2 | ↓↓↓ | +¶* | ↓↓ | +¶ | Fas | ↓ | ++¶ | ↓ | + |
| Irf1 | ↓ | + | ↓ | ↓ | Il2 | ↓↓↓ | +¶ | ↓↓ | ↓ |
| Lta | ↓↓ | ++¶ | ↓ | ↔ | Myd88 | ↓ | + | ↓↓ | + |
| Mitf | ↔ | ++ | ↓ | + | Nfkb2 | + | ++ | ↓ | + |
| Myc | ↓ | + | + | + | Rela | ↓ | ++¶ | ↔ | + |
| Nqo1 | ↓ | + | ↔ | + | Relb | + | ++ | ↓ | ↔ |
| Nr4a2 | +++ | ++++ | + | +++ | Sod2 | ↔ | + | ↔ | + |
| Stat1 | ↓ | ↓ | ↓ | + | Stat5b | + | ++ | + | + |
| Tnfsf10 | ↓ | + | ↓ | ++¶ | Tnf | ↓ | ↓ | ↓ | + |
| Traf2 | ↓ | + | ↓ | ++¶ | Tnfrsf1b | ↓ | ↔ | ↓ | + |
| Trp53 | ↓ | + | ↔ | + | Xiap | ↓ | + | ↔ | + |
KEY: Increase/decrease by <2(+/↓), 2–4 (++/↓↓), 4–8 (+++/↓↓↓), 8–16 (++++/↓↓↓↓), or 16+ (+++++/↓↓↓↓↓) fold vs. genotype naïve.
No change (↔). >6-fold differential between genotypes (¶). Significant differences between genotypes
p<0.05,
p<0.01
DISCUSSION
Repair of articular cartilage defects in human joints remains problematic despite extensive research, due in part to insufficient information on self-healing mechanisms after injury. To mimic the focal cartilage defects commonly seen in highly active populations like athletes1 and the military25, we have used a non-bleeding, cartilage-only injury model to confine the repair response to factors from within the joint and minimize the role of cell infiltration from the circulation or the bone marrow.
Acute post-injury synovitis, peripatellar chondrophytes, and joint capsule fibrosis seen in this model have not been reported for other widely used models of murine OA26. This might be due to more severe responses to a cartilage injury or a reduced focus on the anterior joint compartment in other studies. In murine OA models, the focus is often on loss of cartilage of the tibiofemoral interface as observed on histology of coronal sections of the joint, while excluding examination of the anterior joint compartment, where severe changes in the synovium and joint capsule were seen in the present study, using sagittal sections. A unique effect of cartilage injury on joint response is suggested by the finding that, with the exception of Col3a1 and Mmp3 activation, structural and metabolic responses in the DMM model17 are distinct from those in the current work, suggesting pathogenesis of murine and likely human OA is dependent on type and severity of tissue injury, as well as mechanical perturbations.
A mechanistic link among inflammation, intra-articular scarring, and poor joint repair is also evident in this study, as we identified 27 genes related to IL-17/IL-6, ECM remodeling, and apoptosis (Table 3), with higher expression in Has1−/− joints. To determine whether these are linked to the pathology of Has1−/− joints, we researched whether they have previously been implicated in poor repair of connective tissue injury, using murine OA as reference. On the basis of this restricted search, we have concluded that the 27 genes identified are indeed good markers of ineffective joint repair.
With regard to genes related to IL-17/IL-6 pathways, IL-17 is highly linked to OA in a transcriptomic study of human hip cartilages10 and correlated with collagen expression in scleroderma and fibrosis27. The IL-17/IL-6 pathway genes found differentially activated here are increased in inflammation and fibrosis. Csf3 and Map2k6 promote inflammation in dermal repair28 and collagen-induced arthritis29, and Il-6 is central for development of keloid disease30,31. Mmp1a is upregulated in inflammatory ischemia32, Cxcl1 is a central inflammatory mediator in murine colitis33, and Ptgs2 is highly expressed in the inflammatory phase of murine experimental OA34.
Of the ECM-remodeling genes, the notably elevated Col1a1 and Col1a2 expression in injured Has1−/− joints supports the non-reparative fibrotic scarring response which appears to arise in progenitor-cell rich tissues such as synovium and periosteum, and the histological resemblance to hypertrophic scar tissue seen in keloid disease31. Ctgf expression is high in murine dermal sclerosis, but requires activation by IL-1735. The finding that Timp2 and Plat are upregulated in the Has1−/− joint appears to be inconsistent with the findings that they have shown protection in knockout studies36, 37. It is difficult to reconcile the Timp2 observations; however, the Plat difference could be explained by the simultaneous enhancement of Serpine1 (Table 3), an inhibitor of Plat. Enhanced expression of Tgfb3 occurs in experimental OA and is found in areas of osteophyte formation38. In a similar fashion, increased Thsp2 expression is linked to chondrophyte formation, since it stimulates chondrogenesis in a rabbit osteochondral defect model39. Finally, the increased expression of Timp1 expression in the Has1−/− mice is consistent with an increase in fibrous matrix deposition, since Timp1 activation also occurs in the anabolic phase of a murine model of OA when ECM genes, including Col1a1 and Col5a1, are also highly activated7.
The distinct elevated expression of apoptosis genes in Has1−/− joints (relative to WT) is consistent with an altered stress-induced apoptotic response in fibroblasts from Has1−/− and Has3−/− mice40. Moreover, Has1−/− joints showed elevated expression of Il4 (Table 3), a known stimulator of synovial fibrosis and fibroblast-to-myofibroblast transition41. Elevated Lta (TNFβ) expression results in an increased inflammatory response42, and increased Ins2 expression has been linked to enhanced collagen deposition in muscle repair after injury43. Lastly, increases in Ifng and Il2 expression are consistent with the findings that their joint fluid levels increase with OA severity44, and that there is cross-talk between IFN-γ and TGF-β in dermal wound healing45. Lastly, an increase in Rela would allow for a higher expression of Adamts5, which has been linked to fibrotic scarring in murine OA46.
The apparently normal content of HA in naïve and injured Has1−/− joints suggests that HAS1 is not responsible for the synthesis of the “bulk” HA. However, a more limited activation of Has2 expression in Has1−/− post-injury joints indicates that HAS1 may regulate HAS2 levels during the wound healing process. Moreover the absence of Has1 could abrogate the activation of anti-inflammatory phagocytic cells, as evidenced by the decrease in pro-inflammatory cells and improvement of collagen fiber orientation observed with lentiviral over-expression of HAS1 in dermal repair47, further, since HAS1 activity has been shown to be regulated by glucose concentration and several cytokines48, 49 during wound healing, it may synthesize a pericellular HA pool that controls mesenchymal cell fates in a post-injury environment19 (Figs. 1, 2, 4).
A link between inflammation and OA, particularly after injury, is well-documented50. However, mechanisms by which inflammation becomes chronic without the obvious presence of inflammatory cells in the joint remains to be established. The linkage between persistent activation of the NF-κB pathway, the excessive deposition of fibrotic scar tissue and the chronic cartilage damage in injured Has1−/− joints may provide a novel model in which to study the interplay of these pathways in the pathogenesis of injury-induced OA.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help of Dr. Zarema Arbieva of the University of Illinois at Chicago Genomics Core Facility for training and Katie Trella for assistance in the use of Metacore™-based gene array analyses. We also thank Dr. Guanghua Lei for assistance in murine surgeries and Dr. Katalin Mikecz for evaluating histological sections for inflammatory cell presence.
ROLE OF THE FUNDING SOURCE
Funding for authors include NIH (R01-AR057066; AP, WX, JL, JDS), the Arthritis Foundation (DDC), Seikagaku Corporation (DCC, JL) and the Katz-Rubschlager Endowment for OA Research (AP). These funding sources had no involvement in the study design, the data collection, analysis, and interpretation, the writing of this manuscript, and the decision to submit this work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONTRIBUTIONS
DDC – contributed to experimental design, carried out murine surgeries, gene expression analyses, data evaluation and manuscript preparation
JL – carried out murine surgeries, macroscopic imaging and histology
WFX – carried out murine surgeries, genotypic characterizing and maintenance of Has1−/− mouse colony
CdlM – consulted for experimental design, and provided assistance with interpretation of HA histology and gene array data
JDS – contributed to experimental design, data evaluation, and manuscript preparation
AP – directed experimental design, data interpretation, and manuscript preparation
AP (anna_plaas@rush.edu) takes responsibility for the integrity of the work as a whole, from inception to finished article.
COMPETING INTEREST STATEMENT
The authors have no conflicts of interest, perceived or actual, to declare.
Contributor Information
Deva D. Chan, Email: deva_chan@rush.edu.
Wenfeng Xiao, Email: wenfeng_xiao@163.com.
Jun Li, Email: jun_li@rush.edu.
Carol A. de la Motte, Email: delamoc@ccf.org.
John D. Sandy, Email: jsandy44@gmail.com.
Anna Plaas, Email: anna_plaas@rush.edu.
REFERENCES
- 1.Flanigan DC, Harris JD, Trinh TQ, Siston RA, Brophy RH. Prevalence of chondral defects in athletes’ knees: a systematic review. Med Sci Sports Exerc. 2010;42:1795–1801. doi: 10.1249/MSS.0b013e3181d9eea0. [DOI] [PubMed] [Google Scholar]
- 2.Mashoof AA, Scholl MD, Lahav A, Greis PE, Burks RT. Osteochondral injury to the mid-lateral weight-bearing portion of the lateral femoral condyle associated with patella dislocation. Arthroscopy. 2005;21:228–232. doi: 10.1016/j.arthro.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 3.Buckwalter JA, Anderson DD, Brown TD, Tochigi Y, Martin JA. The Roles of Mechanical Stresses in the Pathogenesis of Osteoarthritis: Implications for Treatment of Joint Injuries. Cartilage. 2013;4:286–294. doi: 10.1177/1947603513495889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu-Bryan R. Synovium and the innate inflammatory network in osteoarthritis progression. Curr Rheumatol Rep. 2013;15:323. doi: 10.1007/s11926-013-0323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candela ME, Yasuhara R, Iwamoto M, Enomoto-Iwamoto M. Resident mesenchymal progenitors of articular cartilage. Matrix Biol. 2014;39:44–49. doi: 10.1016/j.matbio.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell TM, Trudel G, Wong KK, Laneuville O. Genome-wide Gene Expression Analysis of the Posterior Capsule in Patients with Osteoarthritis and Knee Flexion Contracture. J Rheumatol. 2014 doi: 10.3899/jrheum.140079. [DOI] [PubMed] [Google Scholar]
- 7.Remst DF, Blom AB, Vitters EL, Bank RA, van den Berg WB, Blaney Davidson EN, et al. Gene expression analysis of murine and human osteoarthritis synovium reveals elevation of transforming growth factor beta-responsive genes in osteoarthritis-related fibrosis. Arthritis Rheumatol. 2014;66:647–656. doi: 10.1002/art.38266. [DOI] [PubMed] [Google Scholar]
- 8.Aigner T, Fundel K, Saas J, Gebhard PM, Haag J, Weiss T, et al. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–3544. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 9.Brew CJ, Clegg PD, Boot-Handford RP, Andrew JG, Hardingham T. Gene expression in human chondrocytes in late osteoarthritis is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Ann Rheum Dis. 2010;69:234–240. doi: 10.1136/ard.2008.097139. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Barter MJ, Swan DC, Rankin KS, Rowan AD, Santibanez-Koref M, et al. Identification of the pathogenic pathways in osteoarthritic hip cartilage: commonality and discord between hip and knee OA. Osteoarthritis Cartilage. 2012;20:1029–1038. doi: 10.1016/j.joca.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Echtermeyer F, Bertrand J, Dreier R, Meinecke I, Neugebauer K, Fuerst M, et al. Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat Med. 2009;15:1072–1076. doi: 10.1038/nm.1998. [DOI] [PubMed] [Google Scholar]
- 12.Little CB, Barai A, Burkhardt D, Smith SM, Fosang AJ, Werb Z, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60:3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlandi A, Oliva F, Taurisano G, Candi E, Di Lascio A, Melino G, et al. Transglutaminase-2 differently regulates cartilage destruction and osteophyte formation in a surgical model of osteoarthritis. Amino Acids. 2009;36:755–763. doi: 10.1007/s00726-008-0129-3. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Servais J, Polur I, Kim D, Lee PL, Chung K, et al. Attenuation of osteoarthritis progression by reduction of discoidin domain receptor 2 in mice. Arthritis Rheum. 2010;62:2736–2744. doi: 10.1002/art.27582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz GS, Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009;17:153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 16.Steenvoorden MM, Tolboom TC, van der Pluijm G, Lowik C, Visser CP, DeGroot J, et al. Transition of healthy to diseased synovial tissue in rheumatoid arthritis is associated with gain of mesenchymal/fibrotic characteristics. Arthritis Res Ther. 2006;8:165. doi: 10.1186/ar2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan M, Ferguson C, et al. Disease progression and phasic changes in gene expression in a mouse model of osteoarthritis. PLoS One. 2013;8:54633. doi: 10.1371/journal.pone.0054633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eltawil NM, De Bari C, Achan P, Pitzalis C, Dell’accio F. A novel in vivo murine model of cartilage regeneration Age and strain-dependent outcome after joint surface injury. Osteoarthritis Cartilage. 2009;17:695–704. doi: 10.1016/j.joca.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mack JA, Feldman RJ, Itano N, Kimata K, Lauer M, Hascall VC, et al. Enhanced inflammation and accelerated wound closure following tetraphorbol ester application or full-thickness wounding in mice lacking hyaluronan synthases Has1 and Has3. J Invest Dermatol. 2012;132:198–207. doi: 10.1038/jid.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Gorski DJ, Anemaet W, Velasco J, Takeuchi J, Sandy JD, et al. Hyaluronan injection in murine osteoarthritis prevents TGFbeta 1-induced synovial neovascularization and fibrosis and maintains articular cartilage integrity by a CD44-dependent mechanism. Arthritis Res Ther. 2012;14:151. doi: 10.1186/ar3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidgwick GP, Iqbal SA, Bayat A. Altered expression of hyaluronan synthase and hyaluronidase mRNA may affect hyaluronic acid distribution in keloid disease compared with normal skin. Exp Dermatol. 2013;22:377–379. doi: 10.1111/exd.12147. [DOI] [PubMed] [Google Scholar]
- 22.de la Motte CA, Hascall VC, Drazba J, Bandyopadhyay SK, Strong SA. Mononuclear leukocytes bind to specific hyaluronan structures on colon mucosal smooth muscle cells treated with polyinosinic acid:polycytidylic acid: inter-alpha-trypsin inhibitor is crucial to structure and function. Am J Pathol. 2003;163:121–133. doi: 10.1016/s0002-9440(10)63636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara Y, Plaas A, Osborn B, Margulis A, Nelson F, Stewart M, et al. Superficial zone chondrocytes in normal and osteoarthritic human articular cartilages synthesize novel truncated forms of inter-alpha-trypsin inhibitor heavy chains which are attached to a chondroitin sulfate proteoglycan other than bikunin. Osteoarthritis Cartilage. 2008;16:1343–1355. doi: 10.1016/j.joca.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Vigetti D, Karousou E, Viola M, Deleonibus S, De Luca G, Passi A. Hyaluronan: biosynthesis and signaling. Biochim Biophys Acta. 2014;1840:2452–2459. doi: 10.1016/j.bbagen.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Fitzpatrick K, Tokish JM. A military perspective to articular cartilage defects. J Knee Surg. 2011;24:159–166. doi: 10.1055/s-0031-1286052. [DOI] [PubMed] [Google Scholar]
- 26.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15:1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Fabro AT, da Silva PH, Zocolaro WS, de Almeida MS, Rangel MP, de Oliveira CC, et al. The Th17 pathway in the peripheral lung microenvironment interacts with expression of collagen V in the late state of experimental pulmonary fibrosis. Immunobiology. 2014 doi: 10.1016/j.imbio.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Chiang MF, Yang SY, Lin IY, Hong JB, Lin SJ, Ying HY, et al. Inducible deletion of the Blimp-1 gene in adult epidermis causes granulocyte-dominated chronic skin inflammation in mice. Proc Natl Acad Sci U S A. 2013;110:6476–6481. doi: 10.1073/pnas.1219462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammaker D, Topolewski K, Edgar M, Yoshizawa T, Fukushima A, Boyle DL, et al. Decreased collagen-induced arthritis severity and adaptive immunity in MKK-6-deficient mice. Arthritis Rheum. 2012;64:678–687. doi: 10.1002/art.33359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue H, McCauley RL, Zhang W, Martini DK. Altered interleukin-6 expression in fibroblasts from hypertrophic burn scars. J Burn Care Rehabil. 2000;21:142–146. doi: 10.1097/00004630-200021020-00010. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q, Yamaza T, Kelly AP, Shi S, Wang S, Brown J, et al. Tumor-like stem cells derived from human keloid are governed by the inflammatory niche driven by IL-17/IL-6 axis. PLoS One. 2009;4:7798. doi: 10.1371/journal.pone.0007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foley CJ, Kuliopulos A. Mouse matrix metalloprotease-1a (Mmp1a) gives new insight into MMP function. J Cell Physiol. 2014;229:1875–1880. doi: 10.1002/jcp.24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascaraque C, Aranda C, Ocon B, Monte MJ, Suarez MD, Zarzuelo A, et al. Rutin has intestinal antiinflammatory effects in the CD4+ CD62L+ T cell transfer model of colitis. Pharmacol Res. 2014;90C:48–57. doi: 10.1016/j.phrs.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Bateman JF, Rowley L, Belluoccio D, Chan B, Bell K, Fosang AJ, et al. Transcriptomics of wild-type mice and mice lacking ADAMTS-5 activity identifies genes involved in osteoarthritis initiation and cartilage destruction. Arthritis Rheum. 2013;65:1547–1560. doi: 10.1002/art.37900. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto Y, Hasegawa M, Matsushita T, Hamaguchi Y, Huu DL, Iwakura Y, et al. Potential roles of interleukin-17A in the development of skin fibrosis in mice. Arthritis Rheum. 2012;64:3726–3735. doi: 10.1002/art.34643. [DOI] [PubMed] [Google Scholar]
- 36.Mi M, Shi S, Li T, Holz J, Lee YJ, Sheu TJ, et al. TIMP2 deficient mice develop accelerated osteoarthritis via promotion of angiogenesis upon destabilization of the medial meniscus. Biochem Biophys Res Commun. 2012;423:366–372. doi: 10.1016/j.bbrc.2012.05.132. [DOI] [PubMed] [Google Scholar]
- 37.Cook AD, Braine EL, Campbell IK, Hamilton JA. Differing roles for urokinase and tissue-type plasminogen activator in collagen-induced arthritis. Am J Pathol. 2002;160:917–926. doi: 10.1016/S0002-9440(10)64914-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, Wang X, Kaplan DL. A 3D cartilage - inflammatory cell culture system for the modeling of human osteoarthritis. Biomaterials. 2011;32:5581–5589. doi: 10.1016/j.biomaterials.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong SY, Kim DH, Ha J, Jin HJ, Kwon SJ, Chang JW, et al. Thrombospondin-2 secreted by human umbilical cord blood-derived mesenchymal stem cells promotes chondrogenic differentiation. Stem Cells. 2013;31:2136–2148. doi: 10.1002/stem.1471. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Lauer ME, Anand S, Mack JA, Maytin EV. Hyaluronan Synthase 2 Protects Skin Fibroblasts against Apoptosis Induced by Environmental Stress. J Biol Chem. 2014;289:32253–32265. doi: 10.1074/jbc.M114.578377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattey DL, Dawes PT, Nixon NB, Slater H. Transforming growth factor beta 1 and interleukin 4 induced alpha smooth muscle actin expression and myofibroblast-like differentiation in human synovial fibroblasts in vitro: modulation by basic fibroblast growth factor. Ann Rheum Dis. 1997;56:426–431. doi: 10.1136/ard.56.7.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pantelidis P, Fanning GC, Wells AU, Welsh KI, Du Bois RM. Analysis of tumor necrosis factor-alpha, lymphotoxin-alpha, tumor necrosis factor receptor II, and interleukin-6 polymorphisms in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2001;163:1432–1436. doi: 10.1164/ajrccm.163.6.2006064. [DOI] [PubMed] [Google Scholar]
- 43.Krause MP, Moradi J, Nissar AA, Riddell MC, Hawke TJ. Inhibition of plasminogen activator inhibitor-1 restores skeletal muscle regeneration in untreated type 1 diabetic mice. Diabetes. 2011;60:1964–1972. doi: 10.2337/db11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teunis T, Beekhuizen M, Van Osch GV, Schuurman AH, Creemers LB, van Minnen LP. Soluble mediators in posttraumatic wrist and primary knee osteoarthritis. Arch Bone Jt Surg. 2014;2:146–150. [PMC free article] [PubMed] [Google Scholar]
- 45.Ishida Y, Kondo T, Takayasu T, Iwakura Y, Mukaida N. The essential involvement of cross-talk between IFN-gamma and TGF-beta in the skin wound-healing process. J Immunol. 2004;172:1848–1855. doi: 10.4049/jimmunol.172.3.1848. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Anemaet W, Diaz MA, Buchanan S, Tortorella M, Malfait AM, et al. Knockout of ADAMTS5 does not eliminate cartilage aggrecanase activity but abrogates joint fibrosis and promotes cartilage aggrecan deposition in murine osteoarthritis models. J Orthop Res. 2011;29:516–522. doi: 10.1002/jor.21215. [DOI] [PubMed] [Google Scholar]
- 47.Caskey RC, Allukian M, Lind RC, Herdrich BJ, Xu J, Radu A, et al. Lentiviral-mediated over-expression of hyaluronan synthase-1 (HAS-1) decreases the cellular inflammatory response and results in regenerative wound repair. Cell Tissue Res. 2012;351:117–125. doi: 10.1007/s00441-012-1504-7. [DOI] [PubMed] [Google Scholar]
- 48.Rilla K, Oikari S, Jokela TA, Hyttinen JM, Karna R, Tammi RH, et al. Hyaluronan synthase 1 (HAS1) requires higher cellular UDP-GlcNAc concentration than HAS2 and HAS3. J Biol Chem. 2013;288:5973–5983. doi: 10.1074/jbc.M112.443879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siiskonen H, Karna R, Hyttinen JM, Tammi RH, Tammi MI, Rilla K. Hyaluronan synthase 1 (HAS1) produces a cytokine-and glucose-inducible, CD44-dependent cell surface coat. Exp Cell Res. 2014;320:153–163. doi: 10.1016/j.yexcr.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 50.Olson SA, Horne P, Furman B, Huebner J, Al-Rashid M, Kraus VB, et al. The role of cytokines in posttraumatic arthritis. J Am Acad Orthop Surg. 2014;22:29–37. doi: 10.5435/JAAOS-22-01-29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.