Abstract
Background
The spread of carbapenemase-producing Enterobacteriaceae (CPE) is a great problem of healthcare worldwide. Study of the spread for blaOXA-48-like genes coding epidemically significant carbapenemases among hospital pathogens is important for the regional and global epidemiology of antimicrobial resistance.
Methods
Antibacterial resistant isolates of Klebsiella pneumoniae (n = 95) from 54 patients, P.mirabilis (n = 32) from 20 patients, Enterobacter aerogenes (n = 6) from four patients, and Enterobacter cloacae (n = 4) from four patients were collected from January, 2013 to October, 2014 in neurosurgical intensive care unit (ICU) of the Burdenko Neurosurgery Institute, Moscow. Characteristics of the isolates were done using susceptibility tests, PCR detection of the resistance genes, genotyping, conjugation, DNA sequencing, and bioinformatic analysis.
Results
Major strains under study were multi drug resistant (MDR), resistant to three or more functional classes of drugs simultaneously—98.9 % K. pneumoniae, 100 % P.mirabilis, one E.aerogenes isolate, and one E.cloacae isolate. Molecular-genetic mechanism of MDR in K.pneumoniae and P.mirabilis isolates were based on carrying of epidemic extended-spectrum beta-lactamase blaCTX-M-15 gene (87.2 and 90.6 % accordingly), carbapenemase blaOXA-48-like gene (55.3 and 23.3 % accordingly), and class 1 (54.8 and 31.3 % accordingly) and class 2 (90.6 % P. mirabilis) integrons. The blaOXA-48-like-positive K. pneumoniae were collected during whole two-year surveillance period, while P. mirabilis and Enterobacter spp. carrying blaOXA-48-like genes were detected only after four and 18 months after the research start, respectively. The blaOXA-48-like gene acquisition was shown for P. mirabilis isolates collected from five patients and for E. cloacae isolate collected from one patient during their stay in the ICU, presumably from blaOXA-48-like-positive K. pneumoniae. The source of the blaOXA-244 gene acquired by E. aerogenes isolates and the time of this event were not recognized.
Conclusions
The expanding of CPE in the surveyed ICU was associated with the spread of blaOXA-48 and blaOXA-244 carbapenemase genes documented not only among K.pneumoniae, well-known bacterial host for such genes, but among P.mirabilis, E.aerogenes, and E. cloacae.
Keywords: Enterobacteriaceae, Klebsiella pneumoniae, Proteus mirabilis, Enterobacter aerogenes, Enterobacter cloacae, OXA-48-like carbapenemase, Hospital pathogens, Antibacterial resistance, Horizontal gene transfer
Background
One of the main problems of healthcare worldwide is hospital-acquired infections (HAI) caused by multi drug resistant (MDR) pathogens [1, 2] including carbapenemase-producing Enterobacteriaceae (CPE) [3]. The spread of CPE over the last decades is a great danger because carbapenems are the last treatment options for infections caused by MDR bacteria. Recently carbapenemases belonged to three Ambler-classes of beta-lactamases [4] were reported from CPE isolated in Russia: the KPC-type (class A), the NDM-type (class B), and the OXA-48-type (class D) [5]. The OXA-48 carbapenamase is enzyme that was first identified in Klebsiella pneumoniae clinical strains isolated in Turkey, 2003 [6]. Currently OXA-48-producers are described among many species of enterobacteria: K. pneumoniae, Escherichia coli, Enterobacter cloacae, Citrobacter freundii, Serratia marcescens, Morganella morganii and Proteus mirabilis [7–9]. The endemic areas of OXA-48-producers are Turkey, the North Africa and India. Nosocomial outbreaks caused by OXA-48-producers are registered in many European countries including France, Germany, Switzerland, Spain, Holland, and Great Britain. Sporadic cases of such infections are observed in North and South America, China, Australia, and in the Middle East countries [10]. Wide spread of blaOXA-48-like genes among enterobacteria is explained by their localization on the 62 kb conjugative plasmid belonged to IncL/M incompatibility group. Such plasmids are characterized by a high rate of transfer and a wide range of hosts. Their high conjugative potential is due to inactivation of tir gene coding the repressor of conjugative plasmid transfer, by the inserting of the composite transposon Tn1991/Tn1991.2 carrying the blaOXA-48-like gene [11]. In several studies is shown that blaOXA-48-carrying plasmids provide the ability of both clonal and horizontal transfer [7]. There are several variants of blaOXA-48-like genes coding enzymes differ from each other on some amino acid substitutions [12]. For instance, the blaOXA-244 gene discovered in Spain in 2013 differs from the blaOXA-48 gene in a single nucleotide substitution A640G that result in a single amino acid substitution Arg214Gly [13]. It should be noted that carbapenemase activity of all OXA-48-like enzymes are low, so they cannot provide the high levels of carbapenem resistance without additional contribution of other factors such as porin mutations resulting in decrease of cell wall permeability [14].
In this paper we present phenotypic and genotypic characterization of antibacterial resistant K. pneumoniae, Proteus mirabilis, Enterobacter aerogenes, and E. cloacae isolates carrying blaOXA-48 and blaOXA-244 carbapenemase genes, and discuss K.pneumoniae as possibly source for blaOXA-48-like genes spreading in the ICU.
Methods
Bioethical requirements
The materials used in the work do not contain personal data of patients, because of clinical isolates were marked without name, date of birth, address, number of the disease history, personal documents and other personal materials. At the same time, in accordance with the Requirements of the Russian Federation Bioethical Committee, each patient signed a contract with hospital at admission to the clinic. The contract contained consent to treatment and laboratory examination, including a detailed investigation using instrumental methods.
Bacterial isolates
Antibacterial resistant hospital isolates of Klebsiella pneumoniae (n = 95) from 54 patients, Proteus mirabilis (n = 32) from 20 patients, Enterobacter aerogenes (n = 6) from 4 patients, and Enterobacter cloacae (n = 4) from 4 patients with mechanical ventilation in neurosurgical intensive care unit (ICU) of the Burdenko Neurosurgery Institute, Moscow, were collected from January, 2013 to October, 2014. Bacterial cultures were grown on the Nutrient Medium No. 1 (SRCAMB, Obolensk, Russia), Luria–Bertani (LB) broth (Difco, USA) and Muller-Hinton broth (Himedia, India) at 37 °C. Bacterial isolates were stored in 10 % glycerol at minus 70 °C.
Bacterial identification
Bacterial identification was done by Vitek-2 Compact (BioMerieux, France) and MALDI-TOF Biotyper (Bruker Daltonik, Germany).
Susceptibility to antibacterial agents
Minimal inhibitory concentrations (MICs) of antibacterials were determined by Vitek-2 device (BioMerieux, France) using VITEK-2 AST N-101 and AST N-102 cards: amoxicillin/clavulanic acid (AMC), ampicillin/sulbactam (SAM), cefuroxime (CXM), cefoxitin (FOX), cefotaxime (CTX), ceftriaxone (CRO), ceftazidime (CAZ), cefoperazone/sulbactam (CFS), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), tetracycline (TET), tigecycline (TGC), ciprofloxacin (CIP), chloramphenicol (CHL), gentamicin (GEN), tobramycin (TOB), amikacin (AMK), trimethoprim (TMP), trimethoprim/sulfamethoxazole (SXT), nitrofurantoin (NIT), and colistin (CST). Results were interpreted according to the 2014 European Committee on Antimicrobial Susceptibility Testing Recommendations (http://www.eucast.org/clinical_breakpoints/). E.coli strains ATCC 25922 and ATCC 35218 were used for quality control.
Conjugation experiment
Conjugation was performed using previously described method [15]. Donor and recipient overnight cultures were grown under aeration conditions at 120 rpm/min during 18 h at 37 °C on LB broth containing 100 mg/L cefotaxime (Sigma, USA) for donor strain, and 200 mg/L rifampicin (Sigma, USA) for recipient strain. Five milliliters of fresh LB broth without antibiotic were inoculated by 0.05 ml overnight cultures and incubated with aeration to OD600 = 0.6. Then 1 ml each donor and recipient cultures were combined into one tube and incubated at 37 °C for 3 h without shaking. After that the conjugating mixture was diluted with tenfold steps and 0.1 ml was plated in triplicate on LB agar containing 100 mg/L cefotaxime and 200 mg/L rifampicin. Bacteria were incubated at 37 °C for 48 h. To determine the efficiency of conjugation the conjugation mixture was inoculated in parallel on cefotaxime + rifampicin and rifampicin. Frequency of plasmid transfer was calculated as the number of transconjugant cells per number of recipient cells.
PCR detection of the resistance genes
PCR was performed using previously described oligonucleotide primers to detect blaCTX-M, blaTEM, blaSHV, blaOXA-48-like, blaNDM, blaKPC beta-lactamase genes and class 1 and 2 integrons [16–21], repA and traU genes which are the genetic markers of the IncL/M plasmid [22], and the ompK36 gene [23]. The PCR was carried using GradientPalmCycler (Corbert Research, Australia) and Thercyc cycler (DNA-Technology, Russia). PCR products were analysed by electrophoresis in 1.5 % agarose gel in a Sub-Cell GT apparatus (BioRad, USA).
Strain genotyping
Intra-species genotyping of K. pneumoniae, P. mirabilis, and Enterobacter spp. strains was done by Random Amplified Polymorphic DNA (RAPD-PCR) using «random» primers OPA11 and Wil 2 accordingly previously described method [24].
DNA sequencing
Cycle sequencing reactions were performed using the ABI PRISM BigDye Terminator v.3.1 kit. Purified products were analysed on an ABI PRISM 3100-Avant automated DNA Sequencer in the SINTOL Center for collective use (Moscow, Russia).
Bioinformatic analysis
A computer analysis of DNA sequences was performed using Vector NTI9 software (Invitrogen, USA) and BLAST web resource (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Class 1 and 2 integrons were analysed using INTEGRAL database (http://integrall.bio.ua.pt/?).
GenBank accession numbers
The accession numbers of K.pneumoniae DNA sequences are follows: 27 blaCTX-M-15 genes [GenBank: KJ187476, KJ187477, KM058748, KM058751, KM058752, KJ469366, KC817480, KJ363319, KJ363321, KJ481796, KM085432, KM871847, KP205559, KP205560, KP205561, KP205562, KP205563, KP214528, KP214529, KP214530, KP214531, KP214532, KP214533, KP214534, KP214535, KP214536, KP214537]; one blaCTX-M-3 gene [GenBank: KP214538]; 16 blaOXA-48 genes [GenBank: KJ481797, KJ481798, KM085437, KP100448, KP100449, KP198287, KP198288, KP198289, KP198290, KP198291, KP198292, KP198293, KP198294, KP205554, KP205555, KP205556]; four blaOXA-244 genes [GenBank: KJ187475, KJ481795, KJ481799, KM058746]; six class 1 integron gene cassettes arrays [GenBank: KJ363320, KM009101, KF952266, KM009102, KC862254, KF971879]; three ompC genes [GenBank: KJ469369,KJ579290, KJ579291]; one ompK36 gene [GenBank: KJ579289].
The accession numbers of P. mirabilis nucleotide sequences are follows: 10 blaCTX-M-15 genes [GenBank: KM009108, KM058749, KM058750, KC822920, KJ633803, KM085431, KM085433, KM085435, KM085436, KP271997]; one blaCTX-M-3 gene [GenBank: KM871848]; six blaOXA-48 genes [GenBank: KJ579286, KJ696733, KM058747, KP205551, KP205552, KP205553]; one blaOXA-244 gene [GenBank: KJ579285]; one class 1 integron gene cassettes array [GenBank: KM085438]; four class 2 integron gene cassettes arrays [GenBank: KJ579284, KM085439, KM085440, KP271998].
The accession numbers of E. aerogenes nucleotide sequences are follows: three blaOXA-244 genes [GenBank: KM357271, KP056309, KP205557]; the accession number of E. cloacae nucleotide sequence is one blaOXA-48 gene [GenBank: KP056311].
Results and discussion
Isolate sources and resistance phenotypes
Antibacterial resistant enterobacterial hospital isolates collected on the period of January, 2013 to October, 2014 from the patients with mechanical ventilation in neurosurgical intensive care unit (ICU) of the Burdenko Neurosurgery Institute, Moscow, belonged to Klebsiella pneumoniae (n = 95), Proteus mirabilis (n = 32), Enterobacter aerogenes (n = 6), Enterobacter cloacae (n = 4), Escherichia coli (n = 4), Serratia marcescens (n = 3), and Morganella morganii (n = 2). Carriers of OXA-48-like genes have been identified among them: 52 K.pneumoniae, 7 P.mirabilis, 3 E.aerogenes, and 1 E. cloacae isolates.
K. pneumoniae isolates were predominantly collected from respiratory system—endotracheal aspirate, bronchial lavage, and maxillary sinus (46.3 %), fewer isolates were obtained from the urine (40.0 %), wounds (6.3 %), lumbar and ventricular liquor (4.2 %), blood (2.1 %) and rectal swab (1.1 %). The major P. mirabilis isolates were isolated from urine (65.6 %), fewer isolates were obtained from the respiratory system (25.0 %), wounds (6.3 %) and blood (3.1 %). E. aerogenes isolates were collected from liquor (n = 5) and endotracheal aspirate (n = 1). E. cloacae isolates were collected from endotracheal aspirate (n = 3) and urine (n = 1). Major antibacterial resistant bacteria were collected from the respiratory system and from the urine and were associated with invasive equipment and instruments, namely mechanical ventilation and urinary catheters.
The most K.pneumoniae and P.mirabilis isolates in this study were resistant to the majority of used antibacterial agents: AMC, SAM, CXM, CTX, CRO, CIP, CHL, GEN, TOB, TMP, SXT and NIT. Additionally, the K. pneumoniae isolates were resistant to FOX, CAZ, CFS, FEP, ETP, MEM and TET, and the P. mirabilis isolates were resistant to IPM, TET, TGC and CST (Fig. 1). It is noteworthy the high proportion of the pathogens were resistant to TGC (32.4 % K.pneumoniae and 50.0 % P. mirabilis isolates) and to CST (87.5 % P. mirabilis isolates), that are the drugs using for the treatment of severe hospital infections. So, only one antibiotic, CST remains good activity against K.pneumoniae (89.5 % isolates), and only three drugs, CFS (100 % isolates), ETP (100 % isolates) and CAZ (87.1 % isolates) were effective against P. mirabilis (Fig. 1). The antibacterials used in this study belong to the eight functional classes: beta-lactams (penicillins, cephalosporins, and carbapenems), tetracyclines, fluoroquinolones, phenicoles, aminoglycosides, polymyxins, sulfonamides, and nitrofurans. According to Magiorakos et al. [25] many strains under study were categorized as multi drug resistant (MDR) pathogens because of 98.9 % K. pneumoniae and 100 % P. mirabilis isolates were resistant to three or more functional classes of drugs simultaneously (Fig. 2). It should be noted that a large part of both K. pneumoniae (35.9 %) and P. mirabilis (54.8 %) strains were resistant to seven functional antibacterial classes. One E. aerogenes isolate was estimated as an extremely drug resistant because of resistance to all used antibiotics except TGC; three isolates were resistant to cefalosporins, carbapenems and NIT; two isolates were resistant to NIT only. Among E. cloacae isolates one isolate was MDR with sensitivity to IPM and AMK only; other isolates were resistant only to penicillins and cephalosporins.
Fig. 1.
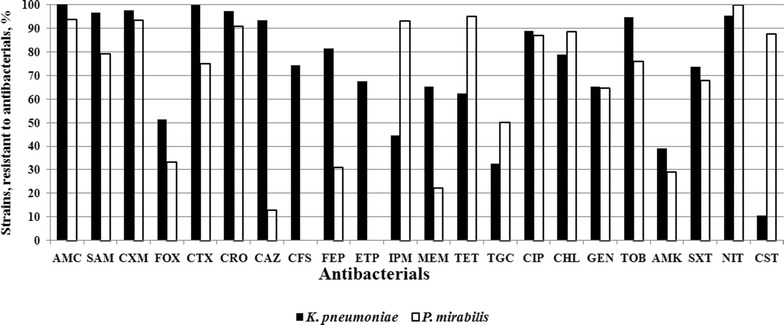
The proportion of isolates those are resistant to antibacterials: AMC amoxicillin/clavulanic acid, SAM amoxicillin/sulbactam, CXM cefuroxime, FOX cefoxitin, CTX cefotaxime, CRO ceftriaxone, CAZ ceftazidime, CFS cefoperazone/sulbactam, FEP cefepime, ETP ertapenem, IPM imipenem, MEM meropenem, TET tetracycline, TGC tigecycline, CIP ciprofloxacin, CHL chloramphenicol, GEN gentamicin, TOB tobramycin, AMK amikacin, TMP trimethoprim, SXT trimethoprim/sulfamethoxazole, NIT nitrofurantoin, CST colistin
Fig. 2.
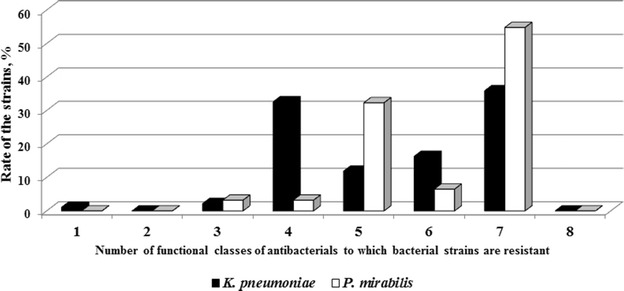
The proportion of bacterial isolates those are resistant to 1–8 functional classes of antibacterials simultaneously: beta-lactams (penicillins, cephalosporins, and carbapenems) tetracyclines, quinolones, fenicoles, aminoglycosides, polymyxins, sulfonamides, nitrofuranam
Resistance genotypes
Detection of the blaCTX-M, blaTEM, blaSHV, blaOXA-48-like, blaNDM, and blaKPC beta-lactamase genes as well as class 1 and class 2 integrons in the genomes of the strains has been done for study of the resistance molecular genetic mechanisms (Fig. 3). The blaCTX-M genes were detected in 87.2 % K. pneumoniae and 90.6 % P. mirabilis isolates. Among 38 blaCTX-M genes major (n = 36) were identified by sequencing as pandemic cephalosporinase blaCTX-M-15 gene and two blaCTX-M-3 genes. The blaSHV-type genes were detected only in the K. pneumoniae strains. No blaNDM and blaKPC genes have been obtained in our study. The blaOXA-48-like carbapenemase genes were detected in 55.3 % of K. pneumoniae, in 23.3 % of P. mirabilis, and in 20.0 % of Enterobacter spp. isolates. DNA sequencing of 28 blaOXA-48-like genes revealed two alleles: the blaOXA-48 gene was detected in K. pneumoniae (n = 17), P. mirabilis (n = 6) and E. cloacae (n = 1); the blaOXA-244 gene was detected in K. pneumoniae (n = 4), P. mirabilis (n = 1) and E. aerogenes (n = 1).
Fig. 3.
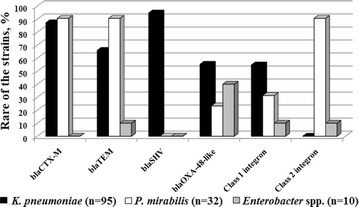
The proportion of bacterial isolates those are carrying the antibacterial resistance genes: bla CTX-M, bla TEM, bla SHV, bla OXA-48-like, class 1 and class 2 integrons
Mobile genetic elements, class 1 integrons were detected in 54.8 % K. pneumoniae and in 31.3 % P. mirabilis isolates (Fig. 3). Variable regions of class 1 integrons identified in K. pneumoniae carried three types of the gene cassette array: (dfrA1-orfC), (dfrA17-aadA5), and (dfrA12-orfF-aadA2). The class 1 integrons detected in P. mirabilis carried only one type of the gene cassette array (dfrA17-aadA5). Class 2 integrons obtained in P. mirabilis (90.6 %) isolates carried a gene cassette array (dfrA12-sat2-aadA1). However, one class 1 integron and one class 2 integron revealed in E.cloacae and E.cloacae isolates respectively did not have any gene cassette arrays, that might be considered as a reserve for future accumulating of the resistance genes. The integron identification numbers have been obtained in accordance with the nomenclature described by Moura et al. [26] using a specialized database INTEGRAL. The (dfrA1-orfC) gene cassette array was attributed to the In263 integron, the (dfrA17-aadA5) array to the In392–395 integron, the (dfrA12-orfF-aadA2) array to the In27 integron, and the (dfrA12-sat2-aadA1) array to the In2–4 integron. According to the report of Partridge et al. from the University of Sydney, Australia, the vast majority of gene cassettes submitted into the GenBank database to date are extremely rare (presented less than 10 times), while a small group of gene cassettes are extremely frequent, they were named “successful” cassettes [27]. All gene cassettes identified in clinical isolates collected from the ICU in our study belong to this group of “successful” cassettes. Thus, on the date Feb 6, 2015, the aadA1 cassette has been presented in the GenBank database 910 times, the aadA2 cassette 601 times, the aadA5 cassette 248 times, the dfrA1 cassette 348 times, the dfrA12 cassette 245 times, the orfF cassette 218 times, the dfrA17 cassette 209 times, the orfC cassette 135 times, and the sat2 cassette 79 times. These data suggest the high prevalence of the gene cassettes detected in our study among bacterial pathogens.
Since the permeability of bacterial cell wall is important for multi drug resistance [23], we carried out the detection of porin gene ompK36 and obtained this gene in 92.4 % K. pneumoniae isolates.
Diversity of RAPD-genotypes
Great genetic heterogeneity of the pathogen populations was shown by RAPD-PCR genotyping for K. pneumoniae (n = 72) and P. mirabilis (n = 32). It was revealed 36 K. pneumoniae genotypes and 8 P. mirabilis genotypes. Four K. pneumoniae genotypes were prevalent in different periods of the study. The K23 genotype (n = 14) was major in 2013, while the K26 genotype (n = 5) was major in the first half of 2014, and the K30 (n = 6) and the K31 (n = 5) genotypes were prevalent in the second half of 2014. Interestingly, K. pneumoniae isolates belonged to K23, K26 and K31 genotypes carried the blaOXA-48-like genes, whereas isolates belonged to K30 genotype did not have these genes. Among all P. mirabilis isolates collected in our study, the P1 RAPD-PCR-genotype (n = 23) was prevalent. It is noteworthy that all P. mirabilis isolates carried blaOXA-48-like genes belonged to this prevalent genotype (Table 1). So, great variety of K. pneumoniae and P. mirabilis genotypes indicates no single source for dissemination of these pathogens. Perhaps presence of blaOXA-48-like genes in bacterial genome provides some populational advantage for certain genotypes. Two RAPD-genotypes (E1 and E2) were identified for E. aerogenes isolates, but all three blaOXA-244-positive isolates were attributed to one genotype E1. Only one RAPD-genotype (E3) was determined for all E. cloacae isolates, including blaOXA-48-positive one.
Table 1.
The features of K. pneumoniae, P. mirabilis and Enterobacter spp. isolates that were collected from the patients of the ICU in 2013–2014
| Microbe | Strain | Isolation datea | Isolation source | Patient | bla TEM | bla SHV | bla CTX-M | bla OXA-48-like | int1 | ins1 | int2 | ins2 | Antibacterial resistance | RAPD type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae | B-38 | 15.01.13 | U | A | + | + | + | bla OXA-48 | + | + | – | – | Pn Cf Cp Tc Qn Cm Su Nf | K7 |
| K. pneumoniae | B-500 | 24.04.13 | EA | B | + | + | + | bla OXA-244 | + | – | – | – | Pn Cf Tc Qn Cm Su Nf | K23 |
| K. pneumoniae | B-567 | 21.05.13 | U | C | + | + | + | bla OXA-244 | + | – | – | – | Pn Cf Cp Qn Su Nf | K23a |
| K. pneumoniae | B-600 | 21.05.13 | EA | K | + | + | + | − | − | − | − | − | Pn Cf Qn Ag Su Nf | K29 |
| K. pneumoniae | B-757 K | 13.06.13 | U | C | + | + | + | bla OXA-244 | + | − | − | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | K23 |
| K. pneumoniae | B-811 K | 16.07.13 | IS | D | + | + | + | bla OXA-244 | + | + | − | − | Pn Cf Cp Qn Su Nf | K1 |
| K. pneumoniae | B-1667 | 31.10.13 | U | K | + | + | + | bla OXA-48 | − | − | − | − | Pn Cf Qn Cm Ag Su Nf | K29 |
| K. pneumoniae | B-1896 | 12.12.13 | U | K | + | + | + | bla OXA-48 | − | − | − | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | K29 |
| K. pneumoniae | B-481/14 | 25.03.14 | U | E | − | + | + | bla OXA-48 | + | − | − | − | Pn Cf Cp Qn Cm Ag Nf | K23 |
| K. pneumoniae | B-617/14 | 25.04.14 | U | E | − | + | + | bla OXA-48 | + | − | − | − | Pn Cf Cp Qn Cm Ag Nf | K23 |
| K. pneumoniae | B-690/14 K | 06.05.14 | U | E | − | + | + | bla OXA-48 | + | − | − | − | Pn Cf Cp Qn Cm Ag Nf | K23 |
| K. pneumoniae | B-767/14 | 27.05.14 | EA | N | − | + | + | bla OXA-48 | + | + | − | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | K28 |
| K. pneumoniae | B-941/14 | 24.06.14 | EA | F | + | + | − | bla OXA-48 | + | − | − | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | K36 |
| K. pneumoniae | B-R1a | 10.07.14 | RS | F | − | + | + | bla OXA-48 | + | + | − | − | Pn Cf Cp Tc Cm Ag Su | K31 |
| K. pneumoniae | B-1699/14 K | 23.10.14 | EA | L | − | + | + | bla OXA-48 | − | − | − | − | Pn Cf Cp Qn Cm Ag Nf | K31 |
| P. mirabilis | B-123 | 07.02.13 | U | G | + | − | + | − | + | + | + | + | Pn Cf Cp Qn Cm Ag Su Nf Co | P3 |
| P. mirabilis | B-239 | 27.02.13 | EA | H | + | − | + | − | + | + | + | + | Pn Cf Cp Qn Cm Ag Su Nf Co | P11 |
| P. mirabilis | B-371 | 28.03.13 | U | E | + | − | + | − | − | − | + | + | Pn Cf Cp Qn Cm Nf Co | P11 |
| P. mirabilis | B-431 M | 08.04.13 | EA | H | + | − | + | bla OXA-48 | + | + | + | + | Pn Cf Cp Qn Cm Ag Su Nf Co | P11 |
| P. mirabilis | B-757 M | 13.06.13 | U | C | + | − | + | bla OXA-244 | − | − | + | + | Pn Cf Cp Tc Qn Cm Ag Nf | P11 |
| P. mirabilis | B-1939 | 17.12.13 | W | M | + | − | + | − | − | − | + | + | Pn Cf Cp Qn Cm Ag Su Nf | P6 |
| P. mirabilis | B-416/14 | 13.02.14 | U | E | + | − | + | bla OXA-48 | − | − | + | − | Pn Cf Cp Tc Qn Cm Nf | P11 |
| P. mirabilis | B-690/14 M | 06.05.14 | U | E | + | − | + | bla OXA-48 | − | − | + | + | Pn Cf Cp Tc Qn Cm Nf | P11 |
| P. mirabilis | B-712/14 M | 12.05.14 | U | E | + | − | + | − | − | − | + | + | Pn Cf Tc Qn Cm Nf | P12 |
| P. mirabilis | B-682/14 | 06.05.14 | EA | I | + | − | + | − | + | + | + | + | Pn Cf Cp Tc Qn Cm Ag Su Nf | P12 |
| P. mirabilis | B-799/14 | 26.05.14 | EA | I | + | − | + | bla OXA-48 | + | + | + | + | Pn Cf Cp Tc Qn Cm Ag Su Nf | P12 |
| P. mirabilis | B-828/14 | 02.06.14 | EA | I | + | − | + | bla OXA-48 | + | + | + | + | Pn Cf Cp Tc Qn Cm Ag Su Nf | P12 |
| P. mirabilis | B-1578/14 | 01.09.14 | U | J | + | − | + | − | − | − | + | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | P15 |
| P. mirabilis | B-1672/14 M | 20.10.14 | U | J | + | − | + | bla OXA-48 | − | − | + | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | P15 |
| E. aerogenes | B-2012/14 | 17.06.14 | L | F | + | − | − | bla OXA-244 | − | − | − | − | Pn Cf Cp Nf | E1 |
| E. aerogenes | B-2137/14E | 25.06.14 | L | O | − | − | − | bla OXA-244 | − | − | − | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | E1 |
| E. aerogenes | B-2212/14 | 30.06.14 | L | F | − | − | − | bla OXA-244 | − | − | − | − | Pn Cf Cp Nf | E1 |
| E. cloacae | B-1530/14 | 30.08.14 | U | N | − | − | − | bla OXA-48 | + | − | − | − | Pn Cf Cp Tc Qn Cm Ag Su Nf | E3 |
EA endotracheal aspirate, IS intracranial sinus, L liquor, RS rectal swab, U urine, W wound, A, B, C, D, E, F, G, H, I, J, K, L, M, N and O patients, bla OXA-244, bla CTX-M, bla TEM, bla SHV beta-lactamase genes, int1 class 1 integrase gene, ins1 genetic cassette array of class 1 integron, int2 class 2 integrase, ins2 genetic cassette array of class 2 integron, Pn penicillins, Cf cephalosporins, Cp carbapenems, Tc tetracyclines, Qn fluoroquinolones, Cm phenicols, Ag aminoglycosides, Su sulfonamides, Nf nitrofurans, «+» positive, «−» negative
aDay, month, year
Spread of blaOXA-48-like genes in ICU
The case of the blaOXA-48 gene acquisition by K. pneumoniae within ICU was demonstrated for the Patient K. The first isolate belonged to K29 genotype (May 21, 2013) had no the blaOXA-48 gene; the next two isolates of K29 genotype (Oct 31, 2013 and Dec 12, 2013) have already acquired the blaOXA-48 gene (Table 1; Fig. 4). The first case of the blaOXA-48 gene emergence in a new bacterial host, P. mirabilis, was registered in the Patient H (Apr 08, 2013). It was happened four months after the start of the study, while such genes in K. pneumoniae isolates were detected during the entire two year period. Two months later, Jun 13, 2013, the blaOXA-244 gene was identified in P. mirabilis isolate collected from the Patient C. Unfortunately, a retrospective epidemiological analysis has not allowed concluding relatedness of occurrence the blaOXA-244 gene in Russia and in Spain where this gene was first described in 2013 [13]. The first case of the blaOXA-244 gene emergence in a bacterial host E. aerogenes was registered in the Patient F (Jun 17, 2014), i.e. 18 months after the start of the study. The first case of the blaOXA-48 gene identification in E. cloacae was obtained in the Patient O (Aug 30, 2014), i.e. 20 months after the start of the study. So, two blaOXA-48-like alleles have disseminated in the ICU simultaneously.
Fig. 4.
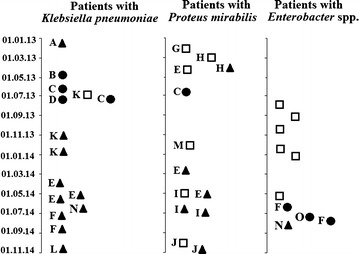
Emergence of K. pneumoniae, P. mirabilis and Enterobacter spp. clinical isolates carrying bla OXA-48-like genes on the period from 01.01.2013 to 01.11.2014: square no gene, filled triangle bla OXA-48 gene, filled circle bla OXA-244 gene; A, B, C, D, E, F, G, H, I, J, K, L, M, N and O patients
Comparative analysis of two or more P. mirabilis isolates collected from the same patient has been done for four patients: E, H, I, and J. All of these patients had no the blaOXA-48-like gene in their first isolates. The second isolates (Patients H and J) and the second and the third isolates (Patients E and I) carried the blaOXA-48-like genes. So, the blaOXA-48-like genes acquisition by P. mirabilis occurred in the ICU, presumably from blaOXA-48-like-positive K. pneumoniae which is half of all K. pneumoniae isolates under study. Additional proof of this hypothesis is the concurrent isolation of blaOXA-244-positive K. pneumoniae and P. mirabilis from urine of the Patient C (Fig. 4), as well as isolation of blaOXA-48-positive K. pneumoniae and E. cloacae from endotracheal aspirate and urine correspondently of the Patient N. Unfortunately, such clear hypothesis cannot to explain the fact of the blaOXA-48-positive K. pneumoniae isolation from endotracheal aspirate and feces of the Patient F and—on the same period—the blaOXA-244-positive E. aerogenes isolates from liquor of this patient (Table 1).
Plasmid localization of blaOXA-48-like genes
It is known that the blaOXA-48-like genes localized on the IncL/M conjugative plasmids [11, 22]. PCR-detection of the specific IncL/M plasmid markers—repA and traU—showed the presence of the plasmid in all blaOXA-48-like-positive P. mirabilis (n = 7) and Enterobacter spp. (n = 4) isolates, and the absence of such plasmid in the blaOXA-48-like-negative isolates. The same correlation between blaOXA-48-like gene and the markers of the IncL/M plasmid was revealed for K. pneumoniae strains. So the blaOXA-48-like genes in bacterial isolates under study were located on IncL/M conjugative plasmids.
The acquisition of the blaOXA-48-like gene consisting plasmid IncL/M has been demonstrated by the experimental conjugation at intra- and inter-species transfer. Two clinical K. pneumoniae strains, B-500 and B-757 K, resistant to cefotaxime were used as donors. The K. pneumoniae M-9 Rif strain was used as a recipient at intraspecies conjugation, and the E. coli HB101 Rif was used as a recipient at interspecies conjugation. Selection of transconjugants was done by selective markers cefotaxime and rifampicin. The efficiency of conjugation was 2.0–8.5 × 10−4 at intra- and 2.0–3.0 × 10−3—at inter-species conjugation. The IncL/M transfer from the donor to the recipient was detected by the plasmid markers repA and traU appearance in the transconjugants. The blaCTX-M, blaTEM, blaSHV, and class 1 integrase genes were detected in the transconjugant cells (Table 2). The distribution of genetic markers in the transconjugants shows that both donor K. pneumoniae strains carried two conjugative plasmids: the IncL/M plasmid (bla+OXA-244repA+traU+) and the unidentified plasmid (bla+CTX-Mbla+TEMbla+SHV) (Table 2).
Table 2.
Donor, recipient and transconjugant K. pneumoniae and E. coli strains features at intra- and inter-species conjugation
| Role at conjugation | Strain | Conjugation effectiveness | Genetic markers | Plasmids | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| repA | traU | bla OXA-244 | bla CTX-M | bla TEM | bla SHV | int1 | ompK36 | ||||
| Donor | K. pneumoniae B-500 | + | + | + | + | + | + | + | + | L/M, UP | |
| Donor | K. pneumoniae B-757 K | + | + | + | + | + | + | + | + | L/M, UP | |
| Recipient | K. pneumoniae 9 Rif | − | − | − | − | − | + | − | + | − | |
| Recipient | E. coli HB-101 Rif | − | − | − | − | − | − | − | − | − | |
| Transconjugant | K. pneumoniae 9/500-1 | 2.0 × 10−4 | + | + | + | + | + | + | + | + | L/M, UP |
| Transconjugant | K. pneumoniae 9/500-2 | 2.0 × 10−4 | − | − | − | + | + | + | + | + | UP |
| Transconjugant | E. coli HB/500-1 | 2.0 × 10−3 | + | + | + | − | − | − | − | − | L/M |
| Transconjugant | K. pneumoniae 9/757-1 | 8.5 × 10−4 | + | + | + | + | + | + | + | + | L/M, UP |
| Transconjugant | K. pneumoniae 9/757-2 | 8.5 × 10−4 | − | − | − | + | + | + | + | + | UP |
| Transconjugant | E. coli HB/757-1 | 3.0 × 10−3 | + | + | + | − | − | − | − | − | L/M |
repA, traU genetic markers of the IncL/M plasmid, bla OXA-244, bla CTX-M, bla TEM, bla SHV beta-lactamase genes, int1 class 1 integrase gene, ompK36 K. pneumoniae porin protein gene, «+» positive, «−» negative, L/M conjugative plasmid IncL/M, UP unidentified conjugative plasmid
Conclusions
The molecular genetic mechanisms for multi-drug resistance of K.pneumoniae, P.mirabilis, E.aerogenes and E. cloacae isolates collected from a Moscow neurosurgical ICU in 2013–2014 were shown associated with blaCTX-M, blaTEM, blaSHV beta-lactamase genes, class 1 and class 2 integrons, as well as with blaOXA-48-like carbapenemase genes. Two alleles genes, the blaOXA-48 and the blaOXA-244, have been identified in the isolates under study. Moreover, P. mirabilis, E. aerogenes and E. cloacae are first determined as bacterial hosts for the of blaOXA-48-like genes in Russia. The blaOXA-48-like genes were found to be integrated into the well-known conjugative plasmids of IncL/M incompatibility group that probably were transferred from K. pneumoniae to P. mirabilis and to E. cloacae during the period of the patient stay in the ICU. The source of the blaOXA-244 gene acquired by E. aerogenes isolates and the time of this event were not recognized.
So, this study highlights ongoing expanding of CPE pathogens in Russia. Obtained knowledge is important for an adequate evaluation of the epidemiological situation, for predicting the development of the situation in the future, and for the correct choice of the optimal strategy for the antibacterial therapy.
Authors’ contributions
NKF, EIA and ONE planned this study. ONE and NVK analyzed information about the patients and obtained the clinical samples. IAA, SYS, AIK and NNK collected bacterial isolates, performed bacterial identification and susceptibility to antibacterial agents. EIA, AIK, NNK and ESL performed the conjugation experiment, PCR detection of the resistance genes, strain genotyping and DNA sequencing—under the supervision of NVV. NKF carried bioinformatic analysis. NKF, EIA and ONE wrote the manuscript while EAS and IAD made significant contributions to the manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
The study was financially supported by the State Research Center for Applied Microbiology and Biotechnology, Obolensk, and The Burdenko Neurosurgery Institute, Moscow. The work of NKF, AIK, EAS, and NVV consisting in genotyping Klebsiella pneumoniae was also supported by the Russian Science Foundation (Grant No. 15-15-00058).
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Nadezhda K. Fursova, Email: n-fursova@yandex.ru
Eugeny I. Astashkin, Email: ast.ev@mail.ru
Anastasia I. Knyazeva, Email: meta-homo@mail.ru
Nikolay N. Kartsev, Email: karcev.nikolay@mail.ru
Ekaterina S. Leonova, Email: e.leonova@mail.ru
Olga N. Ershova, Email: oershova@nsi.ru
Irina A. Alexandrova, Email: ialexandrova@nsi.ru
Natalia V. Kurdyumova, Email: nkurdyumova@nsi.ru
Svetlana Yu. Sazikina, Email: sazikina@nsi.ru
Nikolay V. Volozhantsev, Email: nikvol@obolensk.org
Edward A. Svetoch, Email: svetoch@obolensk.org
Ivan A. Dyatlov, Email: dyatlov@obolensk.org
References
- 1.Rosenthal VD, Bijie H, Maki DG, et al. International nosocomial infection control consortium (INICC) report, data summary of 36 countries, for 2004–2009. Am J Infect Control. 2012;40(5):396–407. doi: 10.1016/j.ajic.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Glasner C, Albiger B, Buist G, et al. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill. 2013;18(28):20525. doi: 10.2807/1560-7917.es2013.18.28.20525. [DOI] [PubMed] [Google Scholar]
- 3.Birgand G, Armand-Lefevre L, Lepainteur M, et al. Introduction of highly resistant bacteria into a hospital via patients repatriated or recently hospitalized in a foreign country. Clin Microbiol Infect. 2014;20(11):O887–O890. doi: 10.1111/1469-0691.12604. [DOI] [PubMed] [Google Scholar]
- 4.Ambler RP. The structure of-lactamases. Philos Trans R Soc Lond B Biol Sci. 1036;1980(289):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 5.Ageevets VA, Partina IV, Lisitsyna ES, et al. Emergence of carbapenemase-producing Gram-negative bacteria in Saint Petersburg, Russia. Int J Antimicrob Agents. 2014;44(2):152–155. doi: 10.1016/j.ijantimicag.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Heritier C, Tolun V, et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arana DM, Saez D, García-Hierro P, et al. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin Microbiol Infect. 2015;21(2):148.e1–148.e4. doi: 10.1016/j.cmi.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Hammoudi D, Moubareck CA, Aires J, et al. Countrywide spread of OXA-48 carbapenemase in Lebanon: surveillance and genetic characterization of carbapenem-non-susceptible Enterobacteriaceae in 10 hospitals over a one-year period. Int J Infect Dis. 2014;29:139–144. doi: 10.1016/j.ijid.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Al Laham N, Chavda KD, et al. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob Agents Chemother. 2015;59(7):4305–4307. doi: 10.1128/AAC.00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20(9):821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 11.Potron A, Poirel L, Nordmann P. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother. 2014;58(1):467–471. doi: 10.1128/AAC.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Martínez L, González-López JJ. Carbapenemases in Enterobacteriaceae: types and molecular epidemiology. Enferm Infecc Microbiol Clin. 2014;32(Suppl 4):4–9. doi: 10.1016/S0213-005X(14)70168-5. [DOI] [PubMed] [Google Scholar]
- 13.Oteo J, Hernández JM, Espasa M, et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother. 2013;68(2):317–321. doi: 10.1093/jac/dks383. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54(1):24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fursova NK, Pryamchuk SD, Abaev, et al. Genetic environments of bla(CTX-M) genes located on conjugative plasmids of Enterobacteriaceae nosocomial isolates collected in Russia within 2003–2007. Antibiot Khimioter. 2010;55(11-12):3–10. [PubMed] [Google Scholar]
- 16.Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006;57(1):14–23. doi: 10.1093/jac/dki398. [DOI] [PubMed] [Google Scholar]
- 17.Edelstein M, Pimkin M, Palagin I, et al. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47(12):3724–3732. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priamchuk SD, Fursova NK, Abaev, et al. Genetic determinants of antibacterial resistance among nosocomial Escherichia coli, Klebsiella spp., and Enterobacter spp. isolates collected in Russia within 2003–2007. Antibiot Khimioter. 2010;55(9-10):3–10. [PubMed] [Google Scholar]
- 19.Hujer KM, Hujer AM, Hulten EA, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006;50(12):4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Chen Y, Jia X, et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect. 2012;18(12):E506–E513. doi: 10.1111/1469-0691.12035. [DOI] [PubMed] [Google Scholar]
- 21.Rasheed JK, Biddle JW, Anderson KF, et al. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J Clin Microbiol. 2008;46(6):2066–2069. doi: 10.1128/JCM.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56(1):559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J-H, Siu LK, Fung C-P, et al. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother. 2010;65:986–990. doi: 10.1093/jac/dkq056. [DOI] [PubMed] [Google Scholar]
- 24.Zimmer M, Barnhart H, Idris U, et al. Detection of Campylobacter jejuni strains in the water lines of a commercial broiler house and their relationship to the strains that colonized the chickens. Avian Dis. 2003;47(1):101–107. doi: 10.1637/0005-2086(2003)047[0101:DOCJSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 26.Moura A, Soares M, Pereira C, et al. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25(8):1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 27.Partridge SR, Tsafnat G, Coiera E, et al. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33(4):757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]


