Abstract
HuR, an RNA binding protein, binds to adenine- and uridine-rich elements (ARE) in the 3′-untranslated region (UTR) of target mRNAs, regulating their stability and translation. HuR is highly abundant in many types of cancer, and it promotes tumorigenesis by interacting with cancer-associated mRNAs, which encode proteins that are implicated in different tumor processes including cell proliferation, cell survival, angiogenesis, invasion, and metastasis. Drugs that disrupt the stabilizing effect of HuR upon mRNA targets could have dramatic effects on inhibiting cancer growth and persistence. In order to identify small molecules that directly disrupt the HuR–ARE interaction, we established a fluorescence polarization (FP) assay optimized for high throughput screening (HTS) using HuR protein and an ARE oligo from Musashi RNA-binding protein 1 (Msi1) mRNA, a HuR target. Following the performance of an HTS of ~6000 compounds, we discovered a cluster of potential disruptors, which were then validated by AlphaLISA (Amplified Luminescent Proximity Homogeneous Assay), surface plasmon resonance (SPR), ribonucleoprotein immunoprecipitation (RNP IP) assay, and luciferase reporter functional studies. These compounds disrupted HuR–ARE interactions at the nanomolar level and blocked HuR function by competitive binding to HuR. These results support future studies toward chemical probes for a HuR function study and possibly a novel therapy for HuR-overexpressing cancers.
NA-binding proteins (RBPs) are critical trans factors that associate with specific cis elements present in mRNAs, thereby regulating the fate of target mRNAs.1 The RBP Hu antigen R (HuR, also known as HuA; Hu references the patient's initials from whom an anti-HuR, autoinflammatory antibody was first isolated2) is a member of the embryonic lethal abnormal vision-like (ELAVL) protein family that binds to adenine- and uridine-rich elements (ARE) mainly located in the mRNA 3′-untranslated region (UTR).1,3,4 HuR is elevated in a broad range of cancer tissues compared with the corresponding normal tissues.5 In early reports, upregulated HuR in brain and colon cancers was linked to the enhanced expression of COX-2, VEGF, TGF-β, IL-8, and other cancer-associated proteins,6,7 Subsequent studies revealed that HuR was broadly overexpressed in virtually all malignancies tested, including cancers of the colon,5,8,9 prostate,10,11 breast,12 brain,6 ovaries,13 pancreas,14 and lung.15 Elevated cytoplasmic accumulation of HuR correlates with high-grade malignancy and serves as a prognostic factor of poor clinical outcome in those cancers.3,4,16 HuR is proposed to play a causal role in tumor development. Cultured carcinoma cells with elevated HuR produced significantly larger tumors than those arising from control populations in a mouse xenograft model,5 while reducing HuR by siRNA or microRNA led to decreased tumor size.5,17
HuR contains three RNA recognition motifs (RRM), of which RRM1 and RRM2 are involved in RNA binding, whereas RRM3 does not contribute to RNA binding but is needed for cooperative assembly of HuR oligomers on RNA.18 Many cytokine and proto-oncogene mRNAs have been identified as containing AREs within their 3′-UTRs, which confer a short mRNA half-life.19 Cytoplasmic binding of HuR to these ARE-containing mRNAs is generally accepted as leading to mRNA stabilization and increased translation.20,21 HuR promotes tumorigenesis by interacting with cancer-associated mRNAs which encode proteins implicated in different tumor processes including cell proliferation, cell survival, angiogenesis, invasion, and metastasis.3,4,16 HuR also promotes the translation of several target mRNAs encoding proteins that are involved in cancer treatment resistance.16,22–24 Taken together, these findings suggest that HuR is an attractive target for developing novel cancer therapies.
RBPs have been considered “undruggable targets” due to the lack of a well-defined binding pocket for target mRNA. Indeed, there has globally been limited success in finding small molecules that directly disrupt the HuR interaction with AREs of target mRNAs, with limited reports indicating several active hits arising from screening for HuR inhibitors.25–27 Those reported hits are structurally independent, so they cannot provide information for later structure–activity relationship (SAR) analysis to design more potent and specific HuR inhibitors. Currently, the most potent hit reported (MS-444) acts via inhibition of HuR homodimerization, leading to disruption of the HuR–ARE interaction.25 Here, we try to identify HuR inhibitors, which competitively bind to HuR and directly disrupt the HuR–ARE interaction.
In this study, we optimized a fluorescent polarization-based (FP-based) binding assay using human full-length HuR protein and an ARE region of Musashi1 (Msi1) 3′-UTR mRNA. HuR binds to and stabilizes the mRNA of Msi128 allowing for oncogenic overexpression of Msi1 and negative regulation of Numb and adenomatous polyposis coli (APC), which are involved in controlling Notch and Wnt signaling pathways.29 Using this FP-based HTS, we screened a library of ~6000 compounds and identified a set of HuR–ARE disruptors, which were validated by AlphaLISA assay, SPR, RNP IP, and luciferase reporter functional studies. The discovery of these inhibitors and related inactive compounds provides the impetus for rational design of more potent and specific HuR–ARE disruptors.
RESULTS AND DISCUSSION
FP Assay Setup and Optimization
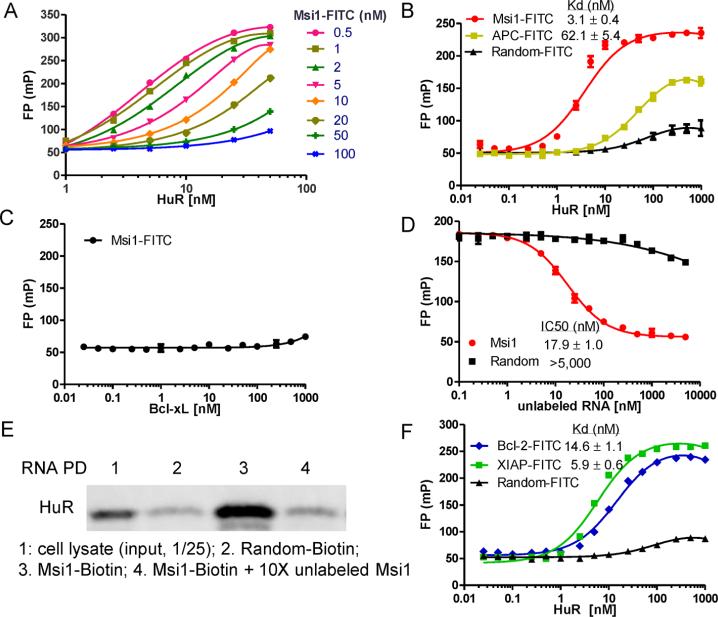
To identify small molecule disruptors of HuR–ARE interactions, we established a FP-based binding assay for HTS. We first optimized the assay by titrating both HuR protein (concentration range 1–50 nM) and a 16-nt ARE-containing fluorescein-labeled RNA oligo from Msi1 mRNA (AREMsi1, concentration range 0.5–100 nM). All test RNA concentrations showed dose–response curves with HuR protein (Figure 1A). Based on the fluorescence intensity, a final concentration of 2 nM AREMsi1 was used in optimization experiments and 1 nM was used for HTS. The equilibrium dissociation constant, Kd, of HuR binding to AREMsi1 was then determined in a saturation binding experiment using a 2 nM concentration of fluorescent ligand while varying concentrations of HuR protein. As shown in Figure 1B, HuR has high binding affnity to AREMsi1–FITC, with a Kd of about 3 nM and a dynamic range of 183 ± 5 mP (ΔmP = mP of bound RNA – mP of free RNA), but not to a 16-nt control RNA oligo with a random sequence. To further demonstrate specificity, HuR has a 20-fold lower binding affnity to a 16-nt oligo of the APC mRNA, which is not a predicted binding site for HuR (Figure 1B). A non-RNA binding protein Bcl-xL was used to determine the specificity of the AREMsi1 oligo binding to HuR. Bcl-xL has no binding to AREMsi1–FITC at the highest concentration tested (1000 nM, Figure 1C). The stability of the FP assay is critical for HTS and has been tested. By incubating the plate at RT for 24 h and reading the plate at set intervals, the stability of the FP assay was tested. Thus, the Kd value and the binding range were found to be reproducible throughout this period, which demonstrates the assay to be suitable for HTS (data not shown).
Figure 1.
Initial characterization of the FP assay. (A) The fluorescein-tagged ARE oligo derived from Msi1 mRNA (Msi1-FITC; 16 nt) and full length HuR protein were titered against each other to determine optimal assay concentrations. Based on the binding curve and fluorescence intensity, 2 nM Msi1-FITC RNA oligo was optimal. (B) HuR protein titration with 2 nM Msi1-FITC or control RNAs, a 16-nt fluorescein-labeled RNA oligo with random sequence (Random-FITC), and a 16-nt fluorescein-labeled RNA oligo of APC mRNA (APC-FITC). (C) Bcl-xL protein titration with 2 nM Msi1-FITC. (D) Competition experiment with unlabeled RNA oligos. Msi1-FITC RNA oligo and HuR protein were kept at 2 nM and 10 nM, respectively. (E) Pulldown analysis of AREMsi1 oligo binding to endogenous HuR. Cell lysate and random oligo were used as a positive control and negative control, respectively. (F) HuR protein titration with 2 nM 16-nt ARE oligo derived from Bcl-2 mRNA (Bcl-2-FITC), XIAP mRNA (XIAP-FITC), or Random-FITC. Mean and standard deviations are derived from three independent experiments.
In order to determine if this FP assay could be employed to evaluate competitive interactions, we performed a competitive binding experiment using the corresponding unlabeled RNA oligos. The signal/background window (the difference between the highest and lowest polarization values) and the sensitivity are two major factors that should be considered and could be affected by the concentration of labeled RNA and protein used in the experiment. Based on the saturation binding curves in Figure 1B, 10 nM HuR protein was used, which resulted in about 80% of maximal FP with AREMsi1 but no binding with random control RNA. Using the fixed concentration of labeled AREMsi1 and HuR protein and increasing doses of unlabeled RNA, the IC50 of unlabeled AREMsi1 and a random control RNA oligo was determined (Figure 1D). Unlabeled AREMsi1 competed for binding with an IC50 of 17.9 nM, while the random control RNA oligo did not compete with the binding at the highest concentration tested (5000 nM). These results indicated that this FP assay is suitable for detecting HuR–ARE disruptors.
After confirming the binding affnity and specificity of the 16-nt AREMsi1 to HuR purified from E. coli, we asked whether it binds to endogenous HuR by performing an RNA pulldown experiment. As shown in Figure 1E, biotinylated AREMsi1 effciently bound endogenous HuR in HCT-116 cells, which was attenuated by 10-fold unlabeled oligo. In addition, a biotinylated random control oligo was unable to pull down endogenous HuR, thereby demonstrating that the 16-nt AREMsi1 oligo specifically binds to endogenous HuR in HCT-116 cells.
We also tested the established FP assay using two other ARE oligos from the reported HuR target mRNAs, Bcl-230,31 and XIAP.23,32 As shown in Figure 1F, AREBcl-2 and AREXIAP exhibited similar binding curves with HuR protein and similar Kd values compared to AREMsi1. Based on this, the 16-nt AREMsi1 oligo was selected as a representative cognate ARE for development of the final HTS. We noted that in the natural context AREs are often longer and in proximal tandem repeats; therefore this 16-nt oligo can be regarded as an exemplar ARE element. Disruptors of the HuR–AREMsi1 interaction are also expected to inhibit binding of other ARE-containing RNAs (e.g., Bcl-2 and XIAP mRNAs).
FP-based High Throughput Screening
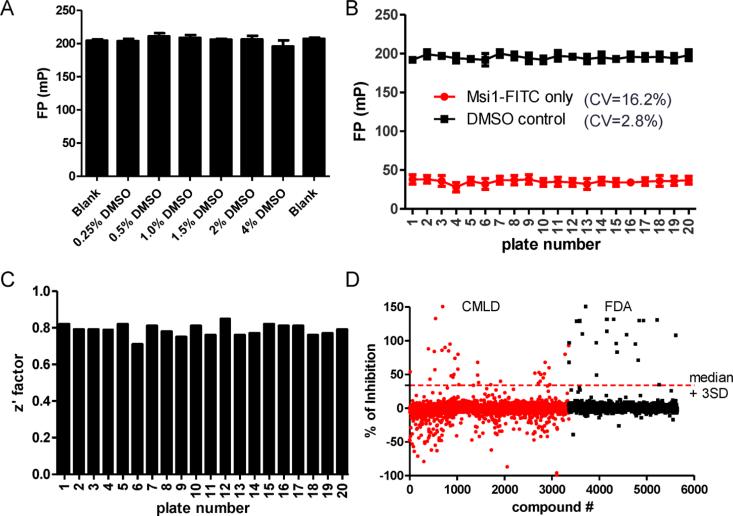
Prior to screening, we first verified that HuR–Msi1 binding is not affected by DMSO (Figure 2A). The HTS was performed in 384-well plate format, using DMSO only (no compound) for negative controls and FITC-labeled AREMsi1 only for positive controls. For the 20 plates screened, the coeffcient of variation (CV) of DMSO and AREMsi1 controls was 2.8% and 16.2%, respectively, with a signal to background window (S/B) of 5.6 (Figure 2B). The average Z′ factor was 0.79 ± 0.03 (Figure 2C). Together, these results indicated the assay was robust and reliable. We screened ~6000 compounds from two libraries: (1) an in-house library developed by the KU Chemical Methodologies and Library Development center (www.cmld.ku.edu) and (2) a library of FDA-approved drugs. The scattergram of the ~6000 compounds screened is shown in Figure 2D. Compounds were ranked according to their ability to disrupt the HuR–AREMsi1 interaction, and 58 compounds exhibiting significant inhibition (median +3 SD) were designated as the initial hits. After elimination of hits based on autofluorescent interference and suspected promiscuous behavior (as determined by PubChem searches), 38 candidate compounds were selected for further validation, resulting in a HTS hit rate of 0.7%.
Figure 2.
Assay optimization and screen execution. (A) The effect of DMSO on HuR–AREMsi1 binding was determined: DMSO (up to 4%, v/v) does not affect the binding in the FP assay. (B) FP values of positive controls (Msi1-FITC) and negative controls (DMSO) for the 20 screened plates and the corresponding CVs. Data points are mean ± SD. (C) The Z′ factors of the 20 plates were calculated using the following equation: 1 – [3(SDp + SDn)/|Mp – Mn|].39 The average Z′ factor was 0.79. (D) HTS was carried out with ~6000 compounds from the CMLD and FDA libraries. Shown here is the scattergram of compound activity expressed as % of inhibition. Median + 3SD was used as a threshold to pick initial hits.
Structure Clustering of Candidate Compounds
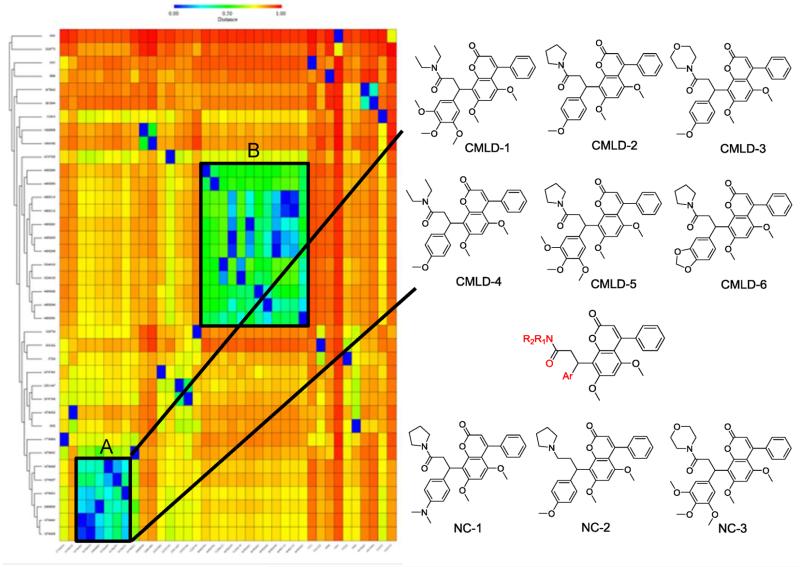
We applied the free ChemMine software (http://chemmine.ucr.edu/) to access the structure similarity of our hits. Application of hierarchical clustering based on pairwise compound similarities defined using atom pair descriptors and Tanimoto coeffcient comparisons resulted in a heat map of the distance matrix with the columns ranked by location in libraries and rows ranked by similarity. This heat map indicated two major clusters, A and B, containing 6 and 12 compounds, respectively, with the rest of the hits being singletons or occurring in pairs (Figure 3). Cluster A compounds exhibited greater potency in disrupting HuR–ARE interactions in the screening; therefore cluster A compounds were chosen for further validation.
Figure 3.
Structure clustering of candidate compounds. Left panel: initial 38 hits were clustered based on structure similarity using the ChemMine tool. The hits are represented by their CID in PubChem or ID. Two clusters (A and B) that contain related structures are highlighted. Right panel: chemical structures of compounds in the Cluster A, the putative pharmacophore, and three negative controls.
The structures of the six cluster A compounds (CMLD1–6) share a coumarin-derived core with differences at the aryl and amide substituents. Examination of the screened library showed that there were an additional 51 compounds having a similar scaffold, including numerous examples in which the amide group was replaced by an analogous amino substituent (Supporting Information Table 1). Since these compounds did not disrupt the HuR–AREMsi1 interaction in the initial screen, three representatives (NC1–3, Figure 3) were selected as negative controls for validation assays.
Validation of Cluster A Compounds by Biochemical Assays
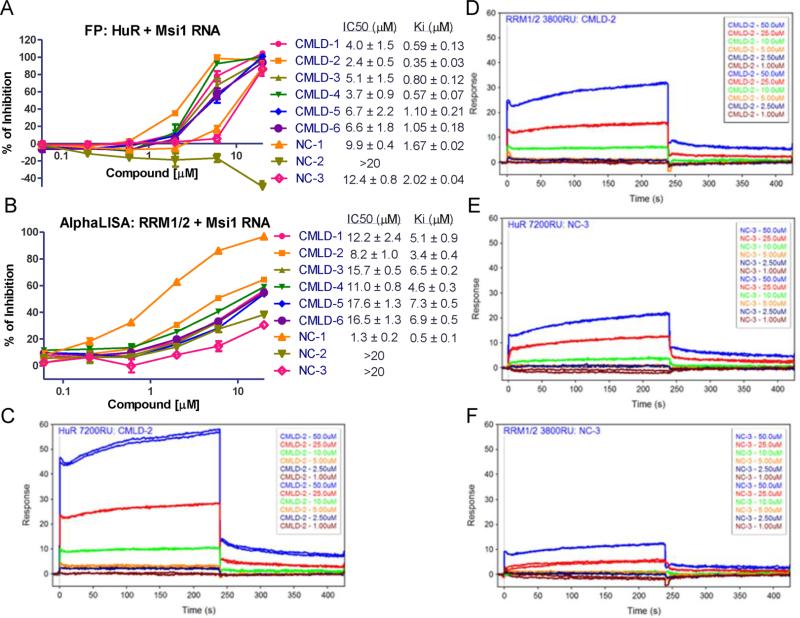
The FP assay was again used to validate the ability of cluster A compounds to disrupt the HuR–AREMsi1 interaction (Figure 4A). Compounds CMLD1–6 displayed a dose-dependent inhibitory effect with compound CMLD-2 being the most potent, with a Ki of 0.35 ± 0.3 μM (n = 3). NC-1 and NC-3 turned out to be weakly active while NC-2 did not disrupt the HuR–AREMsi1 interaction. Similar results were obtained using AREBcl-2 and AREXIAP (data not shown).
Figure 4.
Validation of cluster A compounds binding to HuR. (A). Dose–responses curve of Cluster A compounds and negative controls disrupting HuR–ARE Msi1 binding in FP assay using 10 nM HuR protein and 2 nM fluorescein-labeled Msi1 RNA. (B). Dose–response curve of Cluster A hits and NCs disrupting HuR–ARE Msi1 binding in ALPHA assay using 100 nM HuR RRM1/2 protein and 25 nM biotin-labeled Msi1 RNA. A and B are representative of three independent experiments; IC50 and Ki values are mean ± SD from three independent experiments. SPR analysis of CMLD-2 binding to immobilized HuR (C) and RRM1/2 (D) proteins. Six doses were used (1–50 μM) and duplicated. SPR analysis of NC-3 binding to immobilized HuR (E) and RRM1/2 (F) proteins. Six doses were used (1–50 μMM) and duplicated.
In order to determine that these inhibitory effects were not assay specific, another biochemical assay, the AlphaLISA, was employed to evaluate the HuR protein–RNA complex formation as performed previously.27 This assay includes four components: His6-tagged HuR RRM1/2 domain, biotinylated AREMsi1oligo, streptavidin coated donor beads, and nickel coated acceptor beads. The interaction of RRM1/2 and AREMsi1 brings the two beads close enough to generate a signal following excitation. The AlphaLISA assay was optimized by titration of RRM1/2 protein and AREMsi1, and a Kd value of 75 nM was calculated (Figure S1A,B). CMLD1–6 and NC1–3 compounds were tested using 25 nM RNA and 100 nM RRM1/2 protein (Figure 4B). Although the resulting Ki values are higher than corresponding values in FP assays, the rank order is the same with the exception of NC-1. The high affnity of NC-1 was due to a false positive result determined using the AlphaLISA TruHits kit (Figure S1C). The higher IC50 and Ki values in the AlphaLISA assay as compared to those in the FP assay was a result of different HuR fragments used in two assays. It was found that compounds also gave higher IC50 and Ki values in FP assay when RRM1/2 protein was used compared to full-length HuR (Figure S2).
Data from the FP and AlphaLISA assays do not indicate whether compounds are binding to protein or RNA. Therefore, SPR was used to verify the direct binding of compounds to HuR protein. As shown in Figure 4C–F, both CMLD-2 and NC-3 exhibit dose-dependent binding to full-length HuR protein and RRM1/2 fragment, but NC-3 had a relatively lower response to two proteins. Moreover, the same compound displayed a higher response to full-length HuR comparing to RRN1/2; this is consistent with the finding in the above two assays that compounds show less potency with RRM1/2 versus full-length HuR protein. Validation of cluster A hits by three biochemical assays supports our hypothesis that small molecule compounds disrupt the HuR–ARE interaction through directly binding to HuR protein.
HuR–ARE Disruptors Block HuR Function
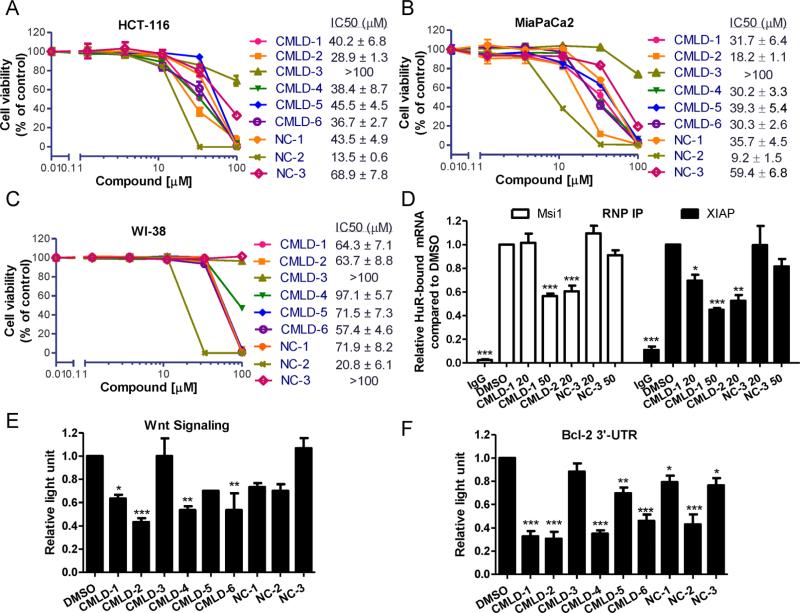
In order to determine the functional consequence HuR–ARE disruptors have on downstream targets and cancer cell growth, RNP IP, luciferase-based reporter assays, and cytotoxicity assays were performed. The cytotoxicity of CMLD1–6 and NC1–3 compounds on human cancer cells, normal fibroblast cell line WI-38, and normal human colon epithelial cell line CCD 841 CoN was examined first by a MTT-based cytotoxicity assay. Except for CMLD-3, the CMLD compounds 1–6 show moderate cytotoxicity against HCT-116 colon cancer cells and MiaPaCa2 pancreatic cancer cells. These same compounds show decreased cytotoxicity against normal cell WI-38 and CCD 841 CoN as demonstrated by ~2-fold greater IC50 values (Figure 5A–C and Figure S3). (Note that the accuracy of IC50 determination in WI-38 and CCD 841 CoN cells may be an underestimate, since with most compounds inhibitory activity was observed at the highest dose.) This suggests that these compounds are disrupting an oncogenic pathway preferentially active in cancer cells and resulting in decreased cell viability.
Figure 5.
HuR-mRNA disruptors block HuR function. The cytotoxicity of CMLD1–6 and NC1–3 against the human colon cancer cell line HCT-116 (A), pancreatic cell line MiaPaCa2 (B), and normal fibroblast cell line WI-38 (C). Figures are representative of three independent experiments, IC50 values are mean ± SD from three independent experiments. (D) RNP IP analysis of HuR bound mRNA affected by CMLD compounds. CMLD-1 and CMLD-2 at indicated doses, but not NC-3, potently inhibited HuR-bound Msi1 and XIAP mRNAs in HCT-116 cells compared to DMSO (set as 1). IgG was used as a negative control of the HuR antibody. Values are mean ± SD from two independent experiments. Luciferase reporter assays determining the effect the disruptors have on Wnt signaling pathway (E) and Bcl-2 3′-UTR (F). Values are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, ***P < 0.001, one-way ANOVA, comparing to DMSO control.
To address whether these CMLD compounds can disrupt endogenous HuR-mRNA interactions, we performed RNP IP assay to test the two most potent compounds CMLD-1 and CMLD-2, as well as negative control NC-3 (Figure 5D). The 50 μM CMLD-1 and 20 μM CMLD-2 significantly blocked HuR bound Msi1 mRNA in HCT-116 cells compared to DMSO control. Similar results were obtained with XIAP mRNA, while NC-3 did not significantly impact HuR-Msi1 or XIAP interactions at the two doses tested.
As mentioned above, Msi1 plays a major role in controlling the Wnt signaling pathway through negative regulation of APC. Moreover, another HuR target mRNA CTNNB15,33 which encodes β-catenin protein, is downstream of canonical Wnt signaling, where it serves as a transcriptional coactivator when translocated to the nucleus. To determine if CMLD compounds could impact oncogenic Wnt signaling through specific disruption of HuR–mRNA interactions, luciferase reporter assays were performed. HCT-116 cells stably expressing a Wnt responsive 7xTcf promoter/luciferase reporter were stimulated with 20 mM LiCl, an inhibitor of GSK3, which activates Wnt signaling, in combination with treatment of CMLD1–6 and NC1–3 compounds (20 μM) for 24 h. This concentration is below the IC50 values of these compounds in cytotoxicity assay in which cells are incubated with compounds for 4 days and was selected in order to inhibit the HuR effect on Wnt signaling without being cytotoxic. We found that CMLD-1, -2, -4, and -6 significantly impacted Wnt signaling but not NC compounds, as compared to DMSO control (Figure 5E). Interestingly, CMLD-5 and especially CMLD-3 showed less inhibition of Wnt signaling that reflected their effects on cell viability compared to other CMLDs.
HuR functions by binding to the 3′-UTR of target mRNAs, stabilizing the mRNAs of target genes and promoting translation. Another luciferase reporter assay was used to determine whether CMLDs affected HuR's ability to stabilize target mRNAs. HCT-116 cells were transiently transfected with a luciferase reporter construct bearing the ARE (NM_000633, 1274–1634) of Bcl-2 3′-UTR and treated with 20 μM compounds for 24 h. As shown in Figure 5F, all tested compounds except CMLD-3 reduced luciferase activity with differing effciency compared to the DMSO control. Further studies to determine the mechanism of low potency of CMLD-3 in cell-based assays need to be carried out. Interestingly, NC-2 also shows significant inhibition, which suggests that NC-2 acts upon the Bcl-2 3′-UTR through another mechanism since it has the strongest cytotoxicity among the nine tested compounds.
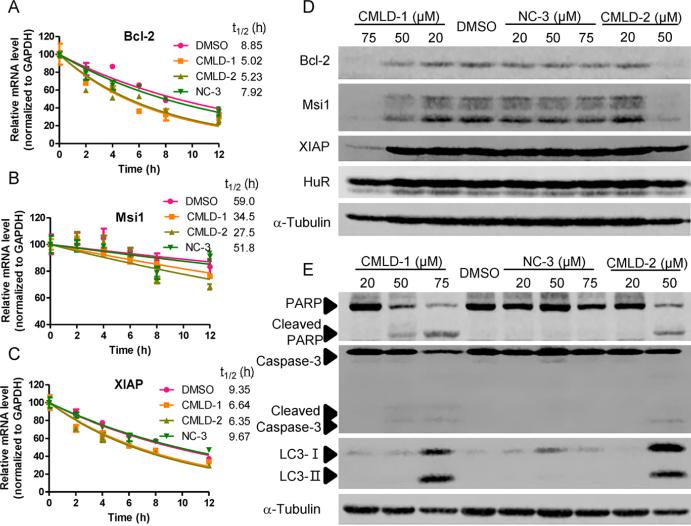
To further confirm that these disruptors can block the HuR function on stabilizing target mRNAs, the stability of Bcl-2, Msi1, and XIAP mRNAs was examined in HCT-116 cells after treatment with a transcription inhibitor actinomycin D. The 20 μM CMLD-1 and CMLD-2 shortened the half-lives of all three mRNAs as compared to a DMSO control, while 20 μM NC-3 treatment did not (Figure 6A–C). Consequently, the protein levels of Bcl-2, Msi1, and XIAP were decreased by CMLD-1 and CMLD-2 dose-dependently, but not by NC-3, indicating that CMLD-1 and CMLD-2 also block the HuR function on promoting translation (Figure 6D). Taken together, these studies demonstrate that many of the top compounds tested disrupt the HuR–ARE interaction, block HuR function, and reduce expression of HuR target mRNAs.
Figure 6.
CMLD-1 and CMLD-2 reduced HuR target mRNA stability and translation. Half-lives of Bcl-2 (A), Msi1 (B), and XIAP (C) mRNA in HCT-116 cells treated with 5 μg/mL actnomycin D together with 20 μM CMLD-1, CMLD-2, NC-3, or DMSO control were determined. Data are representative of two independent experiments. Protein levels of Bcl-2, Msi1, XIAP, HuR (D) and PARP, capsase-3, LC3 (E) in HCT-116 cells treated with CMLD-1, CMLD-2, NC-3, or DMSO at the indicated doses for 48 h were measured using Western Blot. α-Tubulin is a loading control.
To explore the cancer cell death mechanisms of CMLD compounds, we measured protein levels of cleaved PARP (poly ADP ribose polymerase) and cleaved caspase-3, two apoptosis markers, and LC3 conversion, a marker for induction of autophagy, in HCT-116 cells. As shown in Figure 6E, 75 μM CMLD-1 and 50 μM CMLD-2 induced obvious PARP cleavage and subtle caspase-3 cleavage. LC3-I conversion to LC3-II was detected when cells were treated with 75 μM CMLD-1 and 50 μM CMLD-2. These results suggest that CMLD-1 and CMLD-2 induce cancer cell death via apoptosis by inhibiting HuR stabilization of targets Bcl-2 and XIAP, two well-studied antiapoptotic proteins. In addition, these compounds also induce autophagy-associated cell death by inhibiting Bcl-2, as previously reported by other Bcl-2 inhibitors.34 Further studies to investigate whether these compounds kill cancer cells via other pathways need to be performed, and the HuR inhibitors may provide novel chemical probes for dissecting the molecular interplay of various cell death pathways.
CONCLUSION
A FP assay to identify small molecule disruptors of the HuR-mRNA interaction was developed. The ARE-containing RNA oligos were designed to probe with specificity and binding affnity. The optimized FP assay is sensitive (low Kd value) and stable and has a wide signal/background window suitable for HTS. Screening of ~6000 small molecule compounds resulted in an average plate Z′ factor of 0.79 and produced 38 initial hits. Structural clustering identified Cluster A, which contains six positive compounds and 51 negative compounds with a similar scaffold. These inhibitory compounds were validated as potent HuR–ARE disruptors by three biochemical assays and additional functional reporter assays. CMLD-2 was identified as a new HuR-mRNA disruptor, with a Ki of about 350 nM. In addition, this compound displayed cytotoxic selectivity on cancer cells and blocked HuR protein bound to Msi1 and XIAP mRNAs, thereby decreasing target mRNA stability and protein levels as well as inhibiting the Wnt signaling pathway. Although studies here on this cluster of compounds shared a coumarin-derived core are not suffcient to summary clear SAR, they do provide clues for future rational design and lead optimization efforts that could lead to the development of more potent and specific HuR-mRNA disruptors.
MATERIALS AND METHODS
Cell Culture and Reagents
Human colon cancer cell line HCT-116, pancreatic cancer cell line MiaPaCa2, normal lung fibroblast cell line WI-38, and normal human colon epithelial cell line CCD 841 CoN were purchased from American Type Culture Collection and cultured in DMEM (Mediatech) supplemented with 10% fetal bovine serum (v/v, FBS; Sigma-Aldrich) and 1% antibiotics (v/v, Mediatech) in a 5% CO2 humidified incubator at 37 °C. The MTT-based cytotoxicity assay and Western Blot were performed as described previously.35 The details of protein expression and purification, AlphaLISA assay, surface plasmon resonance, and mRNA stability assay were described in the Supporting Methods.
Fluorescence Polarization Assay
RNA oligos with the following sequences (with or without 3′ fluorescein) were purchased from Dharmacon: Msi1 RNA, 5′-GCUUUUAUUUAUUUUG-3′; Bcl-2 RNA, 5′-AAAAGAUUUAUUUAUU-3′; XIAP RNA, 5′-UAGUUAUUUUUAUGUC-3′; APC RNA, 5′-UAUUUGAUAGUACACU-3′. A 16-nt random sequenced RNA (a mixture of random sequences) was used as the negative control. Initial optimization experiments were performed in 96-well black plates (Corning) using the BioTek Synergy H4 plate reader. RNAs were pretreated by heating at 95 °C for 5 min and immediately cooling on ice for 5 min. For assay optimization and determination of the equilibrium dissociation constant (Kd), full-length HuR and 2 nM fluorescein labeled RNA were added to the assay buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 1 mM DTT and 0.05% (v/v) pluronic F-68) with a final volume of 100 μL and incubated at RT for 30 min. For assay stability testing, a plate was measured at different time points over a 24-h period. The Kd was determined from nonlinear regression fits of the data according to a one-site binding model in Prism 5.0 (GraphPad). In RNA competition assays, increasing concentrations of unlabeled RNAs were added to the preformed HuR–AREMsi1 (10 nM HuR and 2 nM Msi1 RNA) complex. For the compound competition assay, compounds with six doses (60 nM–20 μM) were added to the wells prior to the protein–RNA complex. Anisotropy measurements were taken after incubation at RT for 2 h. IC50, the concentration causing 50% inhibition, was calculated via sigmoid fitting of the dose response curve using Prism 5.0. Ki was calculated using free online software (http://sw16.im.med.umich.edu/software/calc_ki/). The percent of inhibition was calculated by comparing to a DMSO control. The FP value of the HuR–AREMsi1 complex with DMSO was defined as 0% inhibition; the FP value of labeled Msi1 RNA only was defined as 100% inhibition. The HTS was performed in 384-well black plates with a final volume of 50 μL, and the screened concentration of compounds was 20 μM (CMLD library) or 1 μM (FDA library).
RNA Pulldown and RNP IP
Two assays were carried out as reported previously with minor modifications.36 HCT-116 cell lysate was isolated using the Immunoprecipitation Kit (Protein G, Roche). For RNA pull down, cell lysate was incubated with 16-nt biotinylated Msi1 RNA oligo (1 μM) or random RNA oligo (1 μM) for 30 min with or without 10 μM unlabeled Msi1 oligo. Streptavidin beads (Roche) were then added to pull-down HuR protein bound to RNAs. Western blot analysis was performed to probe HuR protein using the HuR antibody according to our previous publication.35 For RNP IP, cell lysate was incubated with CMLD-1, CMLD-2, NC-3, or DMSO for 30 min. HuR antibody or mouse IgG (BD Biosciences) was added and incubated for another 60 min. Protein G agarose was used to pull-down bound HuR protein. After IP, RNA was isolated, and qRT-PCR was performed as we reported previously37 using the primers listed in Supporting Information Table 2.
Luciferase Reporter Assay
For Wnt signaling assays, HCT-116 cells were infected with 7TFP lentiviral vector38 (Addgene) to stably express the Wnt-responsive 7xTcf promoter/luciferase construct. HCT-116 stable cells were plated in a 96-well plate, and following attachment overnight, cells were stimulated with 20 mM LiCl and treated with 20 μM test compounds or DMSO for 24 h. Cells were lysed in reporter lysis buffer and assayed using a luciferase assay system (Promega). Reporter gene activities were normalized to total protein activity. For the Bcl-2 3′-UTR study, HCT-116 cells were seeded in a six-well plate and transfected with 1.6 μg of luciferase reporter construct bearing the Bcl-2 ARE (NM_000633, ntds 1274–1634) or luciferase reporter control; a renilla luciferase plasmid was cotransfected to control for transfection effciency. Sixteen hours after transfection, cells were trypsinized and plated in a 48-well dish. Following attachment, cells were treated with 20 μM test compounds or DMSO. Cells were harvested and assayed using the Dual-Glo Luciferase Assay (Promega) 24 h after treatment. All firefly luciferase values were normalized to the renilla control. The relative light unit was calculated for each treated sample by dividing normalized luciferase activity by that of the DMSO control, arbitrarily set as 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Porubsky in KU Specialized Chemistry Center for his help with providing Cluster A compounds and NCs for validation assays.
Funding
This study was supported in part by National Institutes of Health grant R01 CA178831 (to L.X., J.A.), R01 CA134609 (to D.A.D.), NIH COBRE-CCET P30 RR030926 and COBRE-PSF P30 GM110761 Pilot Projects (to L.X.), Kansas Bioscience Authority Rising Star Award, University of Kansas Cancer Center Pilot Grant, LESTR (Laboratory for Early Stage Translational Research) award (to L.X.), and K-INBRE (P20 GM103418) Postdoctoral Award (to X.W.)
Footnotes
Supporting Information
Supplementary figures, tables, and methods. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moll JW, Vecht CJ. Immune diagnosis of paraneoplastic neurological disease. Clin. Neurol. Neurosurg. 1995;97:71–81. doi: 10.1016/0303-8467(95)00007-7. [DOI] [PubMed] [Google Scholar]
- 3.Srikantan S, Gorospe M. HuR function in disease. Front. Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev.: RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- (6).Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 7.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo PS, Sullivan CA, Kiang S, Gao W, Uchio EM, Chung GG, Cha CH. Tissue microarray analysis of 560 patients with colorectal adenocarcinoma: high expression of HuR predicts poor survival. Ann. Surg. Oncol. 2009;16:200–207. doi: 10.1245/s10434-008-0209-3. [DOI] [PubMed] [Google Scholar]
- 10.Niesporek S, Kristiansen G, Thoma A, Weichert W, Noske A, Buckendahl AC, Jung K, Stephan C, Dietel M, Denkert C. Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression. Int. J. Oncol. 2008;32:341–347. [PubMed] [Google Scholar]
- 11.Barbisan F, Mazzucchelli R, Santinelli A, Lopez-Beltran A, Cheng L, Scarpelli M, Montorsi F, Montironi R. Overexpression of ELAV-like protein HuR is associated with increased COX-2 expression in atrophy, high-grade prostatic intraepithelial neoplasia, and incidental prostate cancer in cystoprostatectomies. Eur. Urol. 2009;56:105–112. doi: 10.1016/j.eururo.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 12.Heinonen M, Fagerholm R, Aaltonen K, Kilpivaara O, Aittomaki K, Blomqvist C, Heikkila P, Haglund C, Nevanlinna H, Ristimaki A. Prognostic role of HuR in hereditary breast cancer. Clin. Cancer Res. 2007;13:6959–6963. doi: 10.1158/1078-0432.CCR-07-1432. [DOI] [PubMed] [Google Scholar]
- 13.Denkert C, Weichert W, Pest S, Koch I, Licht D, Kobel M, Reles A, Sehouli J, Dietel M, Hauptmann S. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 2004;64:189–195. doi: 10.1158/0008-5472.can-03-1987. [DOI] [PubMed] [Google Scholar]
- 14.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, Keen JC, Yeo CJ, Gorospe M, Brody JR. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–4572. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Zhao W, Guo Y, Zhang B, Xie Q, Xiang D, Gao J, Wang B, Chen Z. The expression of RNA-binding protein HuR in non-small cell lung cancer correlates with vascular endothelial growth factor-C expression and lymph node metastasis. Oncology. 2009;76:420–429. doi: 10.1159/000216837. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple Functions of the RNA-Binding Protein HuR in Cancer Progression. Treatment Responses and Prognosis. Int. J. Mol. Sci. 2013;14:10015–10041. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, Cabo R, Gorospe M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle. 2010;9:1354–1359. doi: 10.4161/cc.9.7.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J. Biol. Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell. Signal. 2008;20:2165–2173. doi: 10.1016/j.cellsig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, Wang B, Bi J, Zhang C, Guo Y, Chu H, Liang X, Zhong C, Wang J. Cytoplasmic HuR expression correlates with P-gp, HER-2 positivity, and poor outcome in breast cancer. Tumour Biol. 2013;34:2299–2308. doi: 10.1007/s13277-013-0774-3. [DOI] [PubMed] [Google Scholar]
- 21.Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MC, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J. Biochem. 2012;151:423–437. doi: 10.1093/jb/mvs010. [DOI] [PubMed] [Google Scholar]
- 22.Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol. Cancer Res. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene. 2011;30:1460–1469. doi: 10.1038/onc.2010.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, Romeo C, Jimbo M, Norris ZA, Yeo CJ, Sawicki JA, Winter JM, Rigoutsos I, Yen TJ, Brody JR. HuR Posttranscriptionally Regulates WEE1: Implications for the DNA Damage Response in Pancreatic Cancer Cells. Cancer Res. 2014;74:1128–1140. doi: 10.1158/0008-5472.CAN-13-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S, Harrer N, Woisetschlaeger M, Buehler C, Uhl V, Auer M. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 26.Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH, Kim S, Park WY. Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA. Exp. Mol. Med. 2009;41:824–831. doi: 10.3858/emm.2009.41.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Agostino VG, Adami V, Provenzani A. A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation. PLoS One. 2013;8:e72426. doi: 10.1371/journal.pone.0072426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, Gelfond JA, Brenner AJ, Patel V, Trageser D, Scheffler B, Gorospe M, Penalva LO. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol. Cancer Res. 2012;10:143–155. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spears E, Neufeld KL. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J. Biol. Chem. 2011;286:4946–4950. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 31.Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ, Spicer EK. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol. Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Lian J, Wu X, He F, Karnak D, Tang W, Meng Y, Xiang D, Ji M, Lawrence TS, Xu L. A natural BH3 mimetic induces autophagy in apoptosis-resistant prostate cancer via modulating Bcl-2-Beclin1 interaction at endoplasmic reticulum. Cell Death Differ. 2011;18:60–71. doi: 10.1038/cdd.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Li M, Qu Y, Tang W, Zheng Y, Lian J, Ji M, Xu L. Design and synthesis of novel Gefitinib analogues with improved anti-tumor activity. Bioorg. Med. Chem. 2010;18:3812–3822. doi: 10.1016/j.bmc.2010.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr., Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.