Abstract
Although recent progress in understanding the biology and optimizing the treatment of acute lymphoblastic leukemia (ALL) has improved cure rates of childhood ALL to nearly 90%, the cure rate in adult ALL remains less than 50%. The poor prognosis in adult ALL has in part been attributed to larger proportion of high-risk leukemia showing drug resistance. Thus, identifying novel therapeutic targets in ALL is needed for further improvements in treatment outcomes of adult ALL. Genetic aberration of chromatin-modifying molecules has been recently reported in subtypes of ALL, and targeting components of chromatin complexes has shown promising efficacy in preclinical studies. Suppressor of variegation 3-9 homologue 2 (SUV39H2), also known as KMT1B, is a SET-domain–containing histone methyltransferase that is upregulated in solid cancers, but its expression is hardly detectable in normal tissues. Here, we show that SUV39H2 is highly expressed in ALL cells but not in blood cells from healthy donors and also that SUV39H2 mRNA is expressed at significantly higher levels in bone marrow or blood cells from patients with ALL obtained at diagnosis compared with those obtained at remission (P = .007). In four ALL cell lines (Jurkat and CEM derived from T-ALL and RS4;11 and REH derived from B-ALL), SUV39H2 knockdown resulted in a significant decrease in cell viability (~ 77%, P < .001), likely through induction of apoptosis. On the other hand, SUV39H2 overexpression made cells more resistant to chemotherapy. We conclude that SUV39H2 is a promising therapeutic target and further investigation of this therapeutic approach in ALL is warranted.
Introduction
Acute lymphoblastic leukemia (ALL) is a hematological malignancy characterized by maturation arrest and overproduction of immature, aberrant lymphoid cells. Based on the immunophenotype of the leukemic cells, ALL is classified as either B-cell or T-cell lymphoblastic leukemia [1], [2]. The 5-year event-free survival rate is nearly 90% in childhood ALL but only approximately 40% in adult ALL [3], [4], [5], [6], [7]. Notably, the poor prognosis in adult ALL has, in part, been attributed to a higher proportion of high-risk leukemias that demonstrate eventual drug resistance and subsequent relapse. Furthermore, patients of all ages with primary resistant leukemia or whose leukemia relapses after a transient response have few effective therapeutic options and face a dismal prognosis [8], [9], [10]. Therefore, a better understanding of the pathogenesis and the basis of chemoresistance is critically important to further improve the prognosis of these high-risk leukemias [11], [12].
Several studies have shown that epigenetic dysregulation characterized by changes in DNA methylation and histone modification ultimately influences transcriptional regulation and oncogenic signaling pathways in ALL [13], [14]. Notably, whole-genome and whole-exome sequencing in addition to global chromatin profiling studies has identified recurrent somatic mutations in genes encoding epigenetic modifiers in ALL, including MLL-FP, CBP, and NSD2 [13], [14]. Furthermore, aberrant amplification of genes encoding chromatin modifiers such as EZH2, MLL5, SUV39H1, and BRD4 has also been attributed to resistance to targeted therapy in T-ALL [15]. Indeed, approaches focused on targeting components of chromatin complexes have emerged as promising therapeutic strategies particularly when combined with chemotherapy [14], [16]. Taken together, these emerging findings strongly suggest that chromatin modifiers may play an important role in the development and progression of ALL.
Suppressor of variegation 3-9 homologue 2 (SUV39H2), also known as KMT1B, is a SET-domain–containing histone methyltransferase that is expressed only in testis among normal tissues [17]. However, SUV39H2 has been found to be upregulated in lung, cervical, bladder, esophageal, and prostate cancers as well as osteosarcomas and soft tissue sarcomas [18]. SUV39H2, which has been found to be localized in the nucleus, is known to epigenetically regulate gene expression by selectively trimethylating histone H3 on lysine 9 (H3K9), a modification associated with heterochromatin formation and subsequent gene silencing [19], [20]. Our recent study has also shown that SUV39H2 methylates histone H2AX on lysine 134 and subsequently enhances the phosphorylation of H2AX on serine 139 [18]. Phosphorylated histone H2AX (γ-H2AX) is associated with activation of DNA-damage repair pathways and enhanced radio and chemoresistance of cancer cells [21], [22], [23], [24], [25].
Here, we hypothesized that an SUV39H2-dependent pathway might play a role in survival and proliferation of ALL cells and that targeting the gene would result in antileukemic activity. Our investigation aimed to characterize the expression of SUV39H2 in ALL and examine possible biological roles of this gene in the pathogenesis and progression of the disease.
Results
SUV39H2 Overexpression in ALL Primary Blasts and Cell Lines
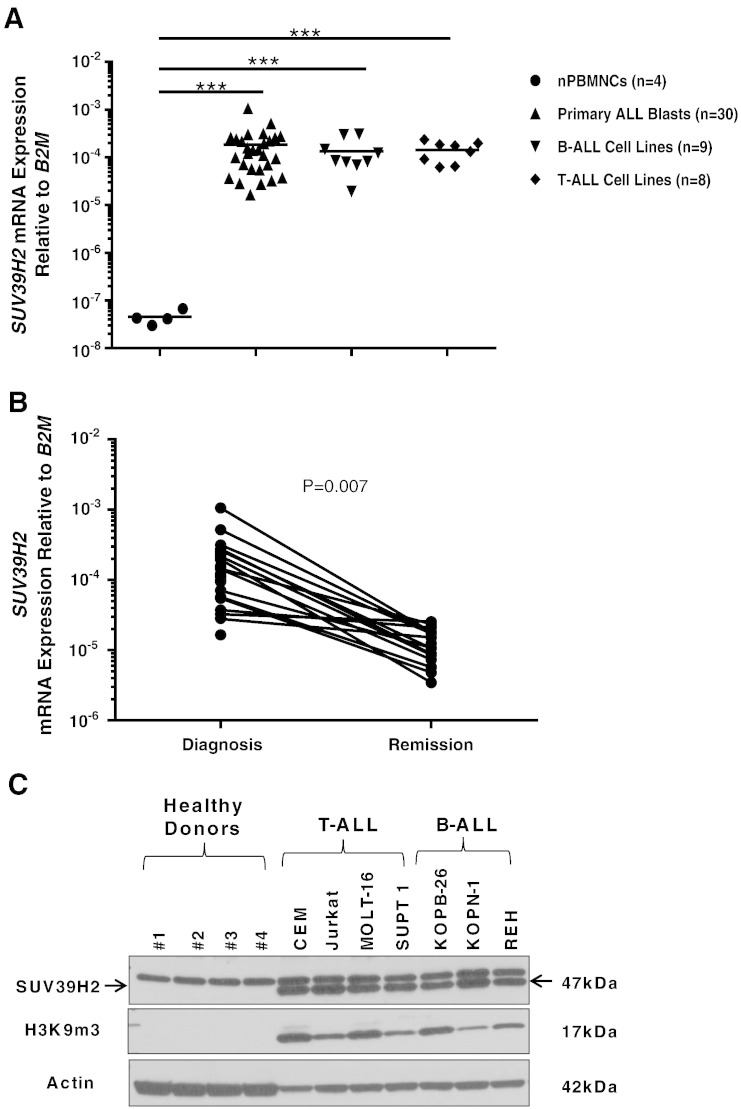
To examine the potential of SUV39H2 as a therapeutic target in ALL, we first compared SUV39H2 mRNA levels in primary cells obtained from 30 patients with ALL as well as 9 B-ALL and 8 T-ALL cell lines with those in normal peripheral blood mononuclear cells (nPBMNCs) obtained from 4 healthy donors. We found that SUV39H2 expression levels were significantly higher in ALL blasts and cell lines than in the nPBMNCs (P < .001; Figure 1A). No difference was observed in SUV39H2 mRNA levels between T-ALL and B-ALL cell lines. We also assessed SUV39H2 mRNA expression in paired bone marrow or peripheral blood samples obtained from 30 patients with ALL at diagnosis and remission. We found SUV39H2 expression to be significantly upregulated in cells obtained at the diagnosis compared with those obtained at the remission point (P = .007; Figure 1B). Furthermore, we analyzed SUV39H2 protein levels in nPBMNCs obtained from four healthy donors, and four T-ALL and three B-ALL cell lines, and found that SUV39H2 was only detectable in the ALL cell lines. Consistently, trimethylated H3K9 (H3K9me3) was detected in ALL cell lines but not in nPBMCs from healthy donors (Figure 1C).
Figure 1.
SUV39H2 mRNA expression is upregulated in ALL. (A) SUV39H2 mRNA levels in PBMNCs obtained from four healthy donors, primary blasts from 30 patients with ALL, as well as ALL cell lines (B-ALL, n = 9 and T-ALL, n = 8) as measured by RT-PCR. (B) SUV39H2 mRNA levels in paired samples obtained at diagnosis and at remission from patients with ALL (n = 30) as measured by RT-PCR. (C) Comparison of SUV39H2 and H3K9me3 protein expression in PBMNCs obtained from four healthy donors with that in ALL cell lines (B-ALL, n = 3 and T-ALL, n = 4) by Western blot analyses. mRNA levels were normalized to B2M expression levels. P values were calculated using Student’s t test (***P < .001).
The Effect of SUV39H2 Knockdown on Cell Viability in ALL Cell Lines
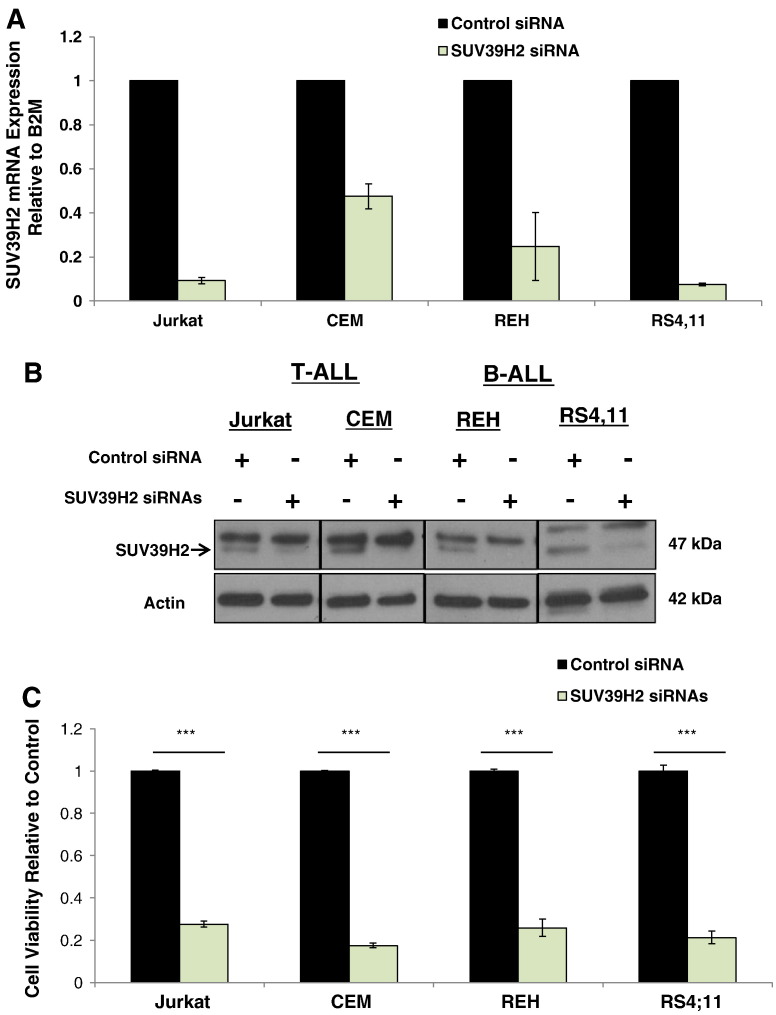
To assess the biological function of SUV39H2 in ALL cells, we first applied a loss-of-function approach using two T-ALL (Jurkat and CEM) and two B-ALL (REH and RS4;11) cell lines. Knockdown of SUV39H2 in these cells was confirmed by quantitative real-time polymerase chain reaction (qRT-PCR) (Figure 2A) and Western blot (Figure 2B) 20 and 30 hours after the transfection of siRNAs, respectively. Cells transfected with SUV39H2 siRNA showed a significant decrease in cell viability (~ 77%, P < .001) compared with those transfected with control siRNA as assessed by Cell Counting Kit–8 (CCK-8) assay 48 hours after the transfection (Figure 2C). These results were also validated in Jurkat cells using a second independent siRNA against SUV39H2 (Figure S1, A–C).
Figure 2.
SUV39H2 knockdown decreased cell viability in ALL cell lines. (A) SUV39H2 mRNA expression, (B) protein expression, and (C) cell viability in Jurkat, CEM, REH, and RS4;11 cells transfected with SUV39H2 siRNA(#2 and 4) or control siRNA. mRNA levels were normalized to B2M expression levels. Data are presented as mean ± SE; P values were calculated using Student’s t test (***P < .001).
The Effect of SUV39H2 Knockdown on Apoptosis in ALL Cell Lines
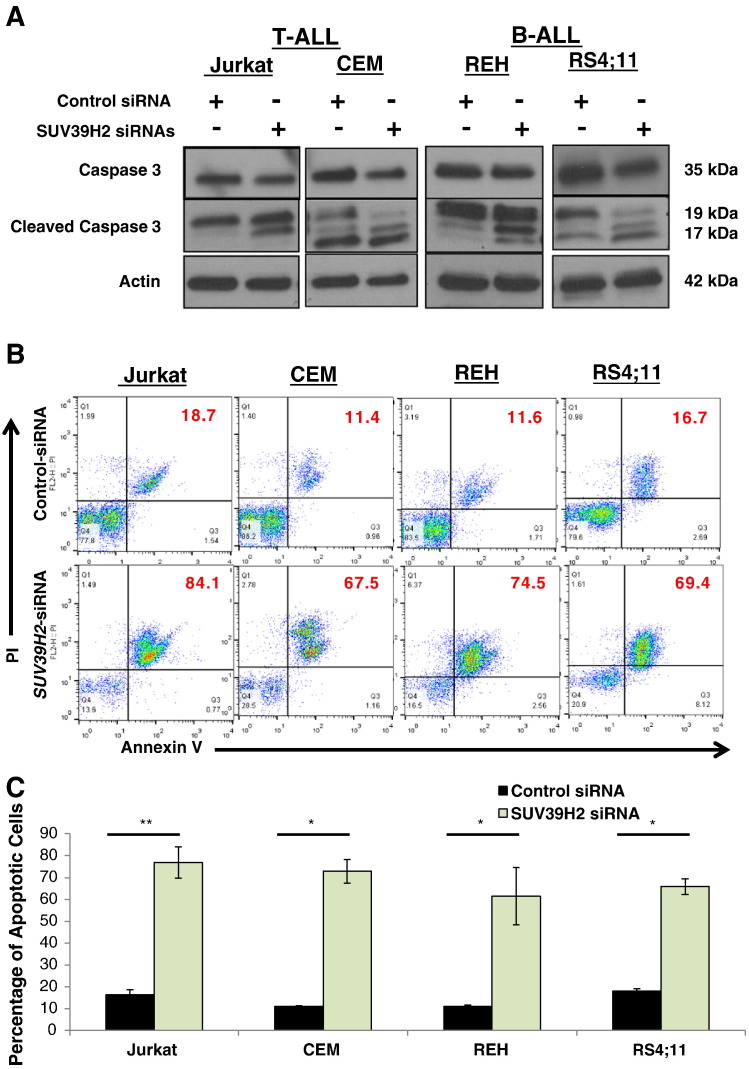
To further examine whether the decrease in cell viability with SUV39H2 siRNA in the four ALL cell lines (Jurkat, CEM, REH, and RS4;11) was caused by induction of apoptosis, we prepared protein lysates 30 hours after the transfection and examined the activation of caspase 3 by Western blot analysis. We observed an increase in cleaved caspase 3 in cells transfected with SUV39H2 siRNA compared with the cells transfected with control siRNA (Figure 3A). In addition, Annexin V and propidium iodide (PI) staining performed on the same set of cells at 48 hours after the transfection showed a significantly higher proportion of Annexin V/PI–positive cells in the cells transfected with SUV39H2 siRNA than cells transfected with control siRNA (69% vs 14%, P < .05; Figure 3, B and C).
Figure 3.
SUV39H2 knockdown induced apoptosis. (A) Assessment of caspase 3 activation by Western blot and (B) representative figure showing Annexin V and PI staining post SUV39H2 siRNA(#2 and 4) or control siRNA transfection in Jurkat, CEM, REH, and RS4;11 (the numbers represent the percentage of total cells). (C) Graphical quantification of average Annexin V and PI staining results in Jurkat, CEM, REH, and RS4;11 from two independent experiments. Data are presented as mean ± SE; P values were calculated using Student’s t test (*P < .05; ** P < .01; ***P < .001).
The Effect of SUV39H2 Overexpression on Cell Sensitivity to Chemotherapy
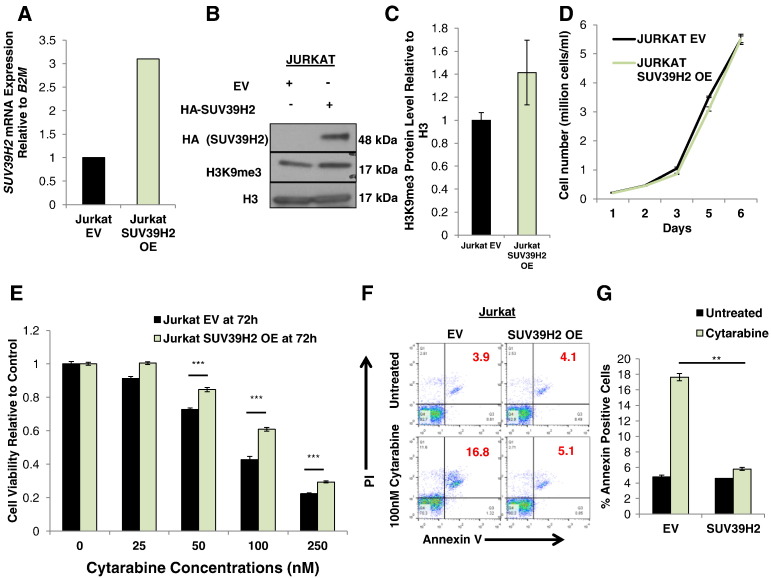
To further investigate the biological functions of SUV39H2 in ALL cells, we used 293T cells to generate lentiviral particles designed to express SUV39H2. We confirmed by Western blot analysis the ectopic expression of SUV39H2 and the increase of H3K9m3 in the 293T cells (Figure S2A). Jurkat cells were then transduced with the generated lentiviral particles, and SUV39H2 overexpression was confirmed by qRT-PCR (Figure 4A) and Western blot analyses (Figure 4B). We also confirmed the increase of H3K9m3 in SUV39H2-overexpressing (OE) Jurkat cells by Western blot analysis (Figure 4, B and C). In addition, we observed a reduction in the expression levels of downstream target genes (H19 and CD2) [26] in SUV39H2 OE Jurkat cells compared with Jurkat cells infected with empty vector (Jurkat EV) (Figure S3). Jurkat SUV39H2 OE and Jurkat EV cells showed no significant difference in their growth rate (Figure 4D). However, when cells were treated with cytarabine (Ara-C), Jurkat SUV39H2 OE cells were less sensitive to the treatment than Jurkat EV cells (Figure 4E). Notably, calculated IC50 values of cell viability after 72 hours of cytarabine treatment were 69 nM for Jurkat EV cells and 99 nM for Jurkat SUV39H2 OE cells (P < .001). A similar pattern of reduced sensitivity in Jurkat SUV39H2 OE cells was observed when cells were treated with either doxorubicin or dexamethasone (two other drugs used in the treatment of ALL) (Figure S4, B and C). In addition, Annexin V and PI staining showed significantly less proportion of dead cells in Jurkat SUV39H2 OE cells than in Jurkat EV cells following treatment with 100 nM cytarabine (P = .002; Figure 4, F and G).
Figure 4.
Effect of SUV39H2 overexpression on proliferation and chemosensitivity. (A) SUV39H2 mRNA expression and (B) protein expression of HA-SUV39H2 and H3K9m3 in Jurkat cells following lentiviral transduction. (C) Densitometry of H3K9me3 protein band obtained from triplicate experiments. (D) Comparison of proliferation between control Jurkat EV and Jurkat SUV39H2 OE. (E) Cell viability in Jurkat EV and Jurkat SUV39H2 OE cells following treatment with increasing concentrations of cytarabine. (F) Representative figure of Annexin V and PI staining and (G) graphical quantification of average Annexin V and PI staining results in Jurkat EV and Jurkat SUV39H2 OE cells after treatment with 100 nM cytarabine. Data are presented as mean ± SE; P values were calculated using Student’s t test (*P < .05; ** P < .01; ***P < .001).
Discussion
Emerging evidence has implicated aberrations in chromatin modifiers in the majority of cancer cells. In particular, dysregulation of histone H3 lysine methyltransferases and demethylases has been shown to play a role in leukemogenesis. Increase of H3K9 methylation as well as DNA methylation, partly attributed to inactivation of histone demethylases such as KDM4C, has been found to inhibit differentiation of hematopoietic cells [27]. Furthermore, mutations in histone methyltransferase enhancer of zeste homologue 2 (EZH2) has been documented in a wide variety of B- and T-cell lymphoproliferative disorders [28], [29]. Whole-genome and subsequent target sequencing studies of early T-cell precursor ALL have also identified high-frequency somatic alterations in histone-modyifying genes such as EZH2, EED, SUZ12, SETD2, and EP300 [30], [31], [32]. It is plausible that targeting components of chromatin repressor complexes that are upregulated in cancer cells may be potentially effective in treating lymphoid malignancies, including ALL. Indeed, components of chromatin complexes such as HDAC and EZH2 have been extensively investigated as drug targets in leukemia, and in vitro and in vivo results confirmed that they are promising targets for novel drug development [16], [29], [33], [34], [35], [36].
SUV39H2 governs methylation of H3K9 in heterochromatic regions as a part of repressor chromatin-remodeling complexes resulting in silencing of gene expression. SUV39H2 was originally identified as an embryonic- and testis-specific histone methyltransferase [19] but was found to be upregulated in lung cancer. It was also reported that SUV39H2 expression was hardly detectable in normal tissues except in the testis [18]. Here we have demonstrated that SUV39H2 was upregulated in the majority of ALL cell lines and in leukemia cells from adults with newly diagnosed ALL. We detected much higher expression levels of this gene in leukemia blasts obtained at diagnosis than in cells obtained from marrows or peripheral blood following induction of remission. ALL cells derived from both B-cell and T-cell lineages expressed significantly higher level of SUV39H2 than mononuclear cells obtained from healthy donors. These observations suggest that the upregulation of SUV39H2 in ALL cells is involved in leukemogenesis. Consistent with this hypothesis, we revealed that knockdown of SUV39H2 in ALL cell lines resulted in a significant decrease in cell viability associated with the enhancement of apoptosis. Although SUV39H2 overexpression in Jurkat cells did not result in increased cell proliferation, interestingly, we found that SUV39H2 overexpression resulted in cytarabine, doxorubicin, and dexamethasone chemoresistance in Jurkat cells. This finding is consistent with our recent report showing that knockdown of SUV39H2 increased the sensitivity of HeLa cells to doxorubicin by suppression of H2AX methylation and subsequent suppression of production of γ-H2AX [18], a DNA-damage marker that accumulates at sites of double strand breaks and triggers the DNA damage response signaling cascade [22], [37]. The potential for synergy using drugs targeting components of the repressor chromatin complexes, which are upregulated in cancer cells, with chemotherapy (for example, HDAC inhibitors in combination with cytarabine) has been demonstrated previously [16]. However, toxicity and selectivity hurdles facing clinical development of some of these combinational approaches suggest that identifying new targets may provide an alternative therapeutic strategy.
Given the upregulation of SUV39H2 in ALL and the dramatic increase of cell death upon its silencing, targeting SUV39H2 in ALL may be a promising therapeutic approach particularly in combination with DNA damage–inducing cytotoxic agents employed for treatment of this disease. Thus, clinical investigation of the potential of targeting SUV39H2 in ALL is warranted.
Materials and Methods
Cell lines for In Vitro Experiments
ALL cell lines were obtained from American Type Culture Collection. Cells are tested and authenticated by DNA profiling for polymorphic short tandem-repeat markers. Cells were passaged in our laboratory for fewer than 6 months after receipt. Cells were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% FBS (Life Technologies). List of cell lines used in this study is shown in the supplementary material (Table 1).
Patient Samples
RNA analysis was performed on paired pre-treatment and post-remission bone marrow/peripheral blood samples from 30 patients with ALL (age range, 17 to 73 years; median age, 41 years; 10 females and 20 males). Informed consent to use the tissue for investigational studies was obtained from each patient according to University of Chicago institutional guidelines and institutional review board approvals.
Transient Transfection, RNA Interference
siRNA oligonucleotide duplexes were purchased from Sigma-Aldrich for targeting the human SUV39H2 transcript, and sequences were as follows: SUV39H2 siRNA#1 (CACAGAUUGCUUCUUUCAA), siRNA#2 (CUUUGGUUGUUCAUGCACA), and siRNA#4 (CUGGAAUCAGCUUAGUCAA). For control, MISSION siRNA universal negative control 1 from Sigma-Aldrich was used.
Transient transfection of cells was performed using 1 nmol of siRNA and 100 μl of Gene Pulser buffer per reaction, and the cells were electroporated with a single electric pulse of 180 V for 12 milliseconds using the Bio-Rad Gene Pulse Xcell (Bio-Rad).
RNA Extraction, RNA Expression Quantification
Total RNA was extracted using Trizol reagent (Life Technologies). SUV39H2 mRNA expression in ALL cell lines and primary cells was measured by the ViiA 7 system according to the manufacturer's instructions. Each cDNA was synthesized using SuperScript III reagents (Life Technologies) according to the manufacturer's instructions. qRT-PCR was performed using commercially available TaqMan Gene Expression Assay probes or Sigma-Aldrich SYBR Green probes with the ViiA 7 system (Life Technologies). The expression levels were normalized to B2M gene.
Western Blot Analysis and Antibodies
Samples were prepared from the cells lysed with CelLytic M cell lysis reagent (Sigma-Aldrich) or NE-PER reagent (Thermo Scientific) containing a complete protease inhibitor cocktail (Roche Applied Science). Total proteins were separated by electrophoresis on 4% to 12% gradient PAGE gels (Bio-Rad) and transferred to polyvinylidine difluoride membrane (GE Healthcare). Proteins of interest were detected by incubating membrane with horseradish peroxidase–conjugated antibodies (Santa Cruz) and visualized with enhanced chemiluminescence (GE Healthcare) or SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific). The following antibodies were used: rabbit polyclonal SUV39H2 antibody (ab71683; Abcam), rabbit polyclonal H3 antibody (ab1791; Abcam), rabbit polyclonal H3K9me3 antibody (ab8898; Abcam), caspase-3 and cleaved caspase-3 (Cell Signaling), anti-hemagglutinin (HA) high affinity (clone 3F10; Roche Life Science), and mouse monoclonal β-actin (AC-15; Sigma-Aldrich).
Viability and Apoptosis Analyses
For viability analysis, CCK-8 assay (Dojindo Molecular Technologies, Inc.,) was performed in a 96-well plate. A total of 5 × 104 cells were plated per well.
For viability and apoptosis analyses, cells were collected, spun down then washed with PBS, and resuspended in 50 μl of binding buffer containing 2 μl of Annexin V-APC (eBioscience) and 5 μl of PI (eBioscience). After 20-minute incubation, fluorescence was quantified by flow cytometry on a FACSCalibur instrument (BD Biosciences).
Constructs and Viral Transduction
SUV39H2 was cloned in pLVX-Puro Vector (Clontech) containing a puromycin resistance gene. HA tag sequence was added downstream of the SUV39H2 gene. Sequence of the construct was checked by Sanger sequencing. For the virus production, 293T cells were transfected with a mix containing gag/pol vector (Addgene), pCMV-VSV-G vector (Addgene), and pLVX construct. Medium was changed after 12 hours, and infectious virions were harvested from the culture media at 48 and 72 hours posttransfection. Jurkat cells were then incubated with the harvested culture media for 72 hours before the selection of cells conferring resistance against puromycin (TOKU-E).
Proliferation Assay
Control Jurkat cells (containing an empty vector) and SUV39H2 OE Jurkat cells were suspended in 1 ml of RPMI medium supplemented with 10% FBS at a concentration of 1 × 105 cells/ml. Cells were then counted on days 1, 2, 3, 4, and 5 using Countess automated cell counter (Invitrogen).
Chemosensitivity Experiments
Jurkat cells containing an empty vector (Jurkat EV) or Jurkat cells stably overexpressing SUV39H2 (Jurkat SUV39H2 OE) were treated with increasing doses of cytarabine or doxorubicin or dexamethasone in a 96-well plate, each well containing 5 × 104 cells. At day 3 for cytarabine and doxorubicin, and day 7 for dexamethasone, CCK-8 reagent (Dojindo Molecular Technologies) was added to assess cell viability.
For viability and apoptosis analyses, Jurkat EV or Jurkat SUV39H2 OE cells were treated with 100 nM cytarabine. Cells were collected at 72 hours, and Annexin V and PI staining was performed as described in the viability and apoptosis subsection.
Statistical Analysis
Mechanistic and biological experiments were analyzed with paired and unpaired two-sided t tests. P values < .05 were considered statistically significant. Experiments were performed in duplicate or triplicate; results were presented as mean ± SE.
Acknowledgement
We thank the University of Chicago Cancer Research Foundation Women’s Board and Division of Biological Sciences and OncoTherapy Science for their support. This work was also supported by the following grants: National Institutes of Health grants T32GM007019, UM1 CA186705, and P30 CA14599-36.
Footnotes
This work was supported by the following grants: National Institutes of Health grants T32GM007019, UM1 CA186705, and P30 CA14599-36.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2015.07.003.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Pui C.H., Behm F.G., Crist W.M. Clinical and biologic relevance of immunologic marker studies in childhood acute lymphoblastic leukemia. Blood. 1993;82(2):343–362. [PubMed] [Google Scholar]
- 2.Pui C.H., Crist W.M. Treatment of childhood leukemias. Curr Opin Oncol. 1995;7(1):36–44. [PubMed] [Google Scholar]
- 3.Pui C.H., Relling M.V., Sandlund J.T., Downing J.R., Campana D., Evans W.E. Rationale and design of Total Therapy Study XV for newly diagnosed childhood acute lymphoblastic leukemia. Ann Hematol. 2004;83(Suppl. 1):S124–S126. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 4.Hoelzer D., Gokbuget N. New approaches to acute lymphoblastic leukemia in adults: where do we go? Semin Oncol. 2000;27(5):540–559. [PubMed] [Google Scholar]
- 5.Fielding A.K. The treatment of adults with acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2008;381–389 doi: 10.1182/asheducation-2008.1.381. [DOI] [PubMed] [Google Scholar]
- 6.Mullighan C.G., Willman C.L. Advances in the Biology of Acute Lymphoblastic Leukemia-From Genomics to the Clinic. J Adolesc Young Adult Oncol. 2011;1(2):77–86. doi: 10.1089/jayao.2011.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson S., Stock W. Progress in the treatment of adults with acute lymphoblastic leukemia. Curr Opin Hematol. 2008;15(4):400–407. doi: 10.1097/MOH.0b013e3283034697. [DOI] [PubMed] [Google Scholar]
- 8.Oudot C., Auclerc M.F., Levy V., Porcher R., Piguet C., Perel Y., Gandemer V., Debre M., Vermylen C., Pautard B. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: the FRALLE 93 study. J Clin Oncol. 2008;26(9):1496–1503. doi: 10.1200/JCO.2007.12.2820. [DOI] [PubMed] [Google Scholar]
- 9.Fielding A.K., Richards S.M., Chopra R., Lazarus H.M., Litzow M.R., Buck G., Durrant I.J., Luger S.M., Marks D.I., Franklin I.M. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood. 2007;109(3):944–950. doi: 10.1182/blood-2006-05-018192. [DOI] [PubMed] [Google Scholar]
- 10.Wetzler M., Stock W. Preface: acute lymphoblastic leukemia--quo vadis? Hematol Oncol Clin North Am. 2009;23(5):xi–xviii. doi: 10.1016/j.hoc.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Pui C.H., Evans W.E. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 12.Holleman A., Cheok M.H., den Boer M.L., Yang W., Veerman A.J., Kazemier K.M., Pei D., Cheng C., Pui C.H., Relling M.V. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 13.Greenblatt S.M., Nimer S.D. Chromatin modifiers and the promise of epigenetic therapy in acute leukemia. Leukemia. 2014;28(7):1396–1406. doi: 10.1038/leu.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaffe J.D., Wang Y., Chan H.M., Zhang J., Huether R., Kryukov G.V., Bhang H.E., Taylor J.E., Hu M., Englund N.P. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45(11):1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoechel B., Roderick J.E., Williamson K.E., Zhu J., Lohr J.G., Cotton M.J., Gillespie S.M., Fernandez D., Ku M., Wang H. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46(4):364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie C., Edwards H., Xu X., Zhou H., Buck S.A., Stout M.L., Yu Q., Rubnitz J.E., Matherly L.H., Taub J.W. Mechanisms of synergistic antileukemic interactions between valproic acid and cytarabine in pediatric acute myeloid leukemia. Clin Cancer Res. 2010;16(22):5499–5510. doi: 10.1158/1078-0432.CCR-10-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 18.Sone K., Piao L., Nakakido M., Ueda K., Jenuwein T., Nakamura Y., Hamamoto R. Critical role of lysine 134 methylation on histone H2AX for gamma-H2AX production and DNA repair. Nat Commun. 2014;5:5691. doi: 10.1038/ncomms6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Carroll D., Scherthan H., Peters A.H., Opravil S., Haynes A.R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20(24):9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Cao M., O'Sullivan R., Peters A.H., Jenuwein T., Blasco M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36(1):94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 21.Harper J.W., Elledge S.J. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5(7):675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 23.Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J. Genomic instability in mice lacking histone H2AX. Science. 2002;296(5569):922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassing C.H., Chua K.F., Sekiguchi J., Suh H., Whitlow S.R., Fleming J.C., Monroe B.C., Ciccone D.N., Yan C., Vlasakova K. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99(12):8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3(8-9):959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Mauger O., Klinck R., Chabot B., Muchardt C., Allemand E., Batsche E. Alternative splicing regulates the expression of G9A and SUV39H2 methyltransferases, and dramatically changes SUV39H2 functions. Nucleic Acids Res. 2015;43(3):1869–1882. doi: 10.1093/nar/gkv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C.R., Khanin R., Figueroa M.E., Melnick A. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lund K., Adams P.D., Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28(1):44–49. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 29.Hamamoto R., Saloura V., Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15(2):110–124. doi: 10.1038/nrc3884. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ntziachristos P., Tsirigos A., Van Vlierberghe P., Nedjic J., Trimarchi T., Flaherty M.S., Ferres-Marco D., da Ros V., Tang Z., Siegle J. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18(2):298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S.C., Phipson B., Hyland C.D., Leong H.S., Allan R.S., Lun A., Hilton D.J., Nutt S.L., Blewitt M.E., Smyth G.K. Polycomb repressive complex 2 (PRC2) suppresses Emu-myc lymphoma. Blood. 2013;122(15):2654–2663. doi: 10.1182/blood-2013-02-484055. [DOI] [PubMed] [Google Scholar]
- 33.Stubbs M.C., Kim W., Bariteau M., Davis T., Vempati S., Minehart J., Witkin M., Qi J., Krivtsov A.V., Bradner J.E. Selective Inhibition of HDAC1 and HDAC2 as a Potential Therapeutic Option for B-ALL. Clin Cancer Res. 2015;21(10):2348–2358. doi: 10.1158/1078-0432.CCR-14-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron E.E., Bachman K.E., Myohanen S., Herman J.G., Baylin S.B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 35.Klisovic M.I., Maghraby E.A., Parthun M.R., Guimond M., Sklenar A.R., Whitman S.P., Chan K.K., Murphy T., Anon J., Archer K.J. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia. 2003;17(2):350–358. doi: 10.1038/sj.leu.2402776. [DOI] [PubMed] [Google Scholar]
- 36.Odenike O., Halpern A., Godley L.A., Madzo J., Karrison T., Green M., Fulton N., Mattison R.J., Yee K.W., Bennett M. A phase I and pharmacodynamic study of the histone deacetylase inhibitor belinostat plus azacitidine in advanced myeloid neoplasia. Investig New Drugs. 2015;33(2):371–379. doi: 10.1007/s10637-014-0194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furuta T., Takemura H., Liao Z.Y., Aune G.J., Redon C., Sedelnikova O.A., Pilch D.R., Rogakou E.P., Celeste A., Chen H.T. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem. 2003;278(22):20303–20312. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.