Abstract
Studying changes in nuclear architecture is a unique approach toward the understanding of nuclear remodeling during tumor development. One aspect of nuclear architecture is the orientation of chromosomes in the three-dimensional nuclear space. We studied mouse chromosome 11 in lymphocytes of [T38HxBALB/c]N mice with a reciprocal translocation between chromosome X and 11 (T38HT(X;11)) exhibiting a long chromosome T(11;X) and a short chromosome T(X;11) and in fast-onset plasmacytomas (PCTs) induced in the same strain. We determined the three-dimensional orientation of chromosome 11 using a mouse chromosome 11 specific multicolor banding probe. We also examined the nuclear position of the small translocation chromosome T(X;11) which contains cytoband 11E2 and parts of E1. Chromosomes can point either with their centromeric or with their telomeric end toward the nuclear center or periphery, or their position is found in parallel to the nuclear border. In T38HT(X;11) nuclei, the most frequently observed orientation pattern was with both chromosomes 11 in parallel to the nuclear border (“PP”). PCT cells showed nuclei with two or more copies of chromosome 11. In PCTs, the most frequent orientation pattern was with one chromosome in parallel and the other pointing with its centromeric end toward the nuclear periphery (“CP”). There is a significant difference between the orientation patterns observed in T38HT(X;11) and in PCT nuclei (P < .0001).
Introduction
Chromosomes are organized in evolutionary conserved chromosome territories [1]. Their nonrandom three-dimensional (3D) positions were previously described [2], e.g., the localization of the active and inactive chromosome X and their respective genes [3], [4]. Euchromatin of rod photoreceptor cells in nocturnal mammals is found in the periphery, whereas it is found in the center in diurnal mammals [5]. Not only chromosome territories are in the focus of research but also the localization of telomeric regions [6], [7].
Tumor development is greatly influenced by genomic instability [8], and telomere dysfunction plays an important role in genomic instability [9]. Therefore, it is essential to study nuclear architecture in normal and tumor cells. Movement of telomeric regions during the cell cycle was observed in living ECV-TRF1 and -TRF2 cells [10] and in human osteosarcoma U2OS cells [11]. Chromosomes of primary human fibroblasts alter their positions within 15 minutes after they are made quiescent due to a removal of serum from the culture medium. This repositioning is probably dependent on nuclear myosin 1β [12]. Further changes of chromosome positions can be found during adipocyte differentiation [13] or T-cell differentiation [14].
Telomere lengthening is a method to prevent genomic instability of rapidly dividing cells [15]. This can occur due to telomerase [16] or due to cycles of homologous recombination during the process of alternative telomere lengthening [17]. In fast-onset plasmacytomas (PCTs), the telomere length is significantly increased for the translocation chromosome T(X;11) carrying 11E2 [18].
In the current study, we used a [T38HxBALB/c]N congenic mouse model with a reciprocal translocation between chromosomes X and 11 (rcpT(X;11). This unique mouse model exhibits a long chromosome T(11;X) and a short chromosome T(X;11). The short chromosome T(X;11) contains cytoband 11E2 and parts of cytoband E1 (Figure 1) [19]. To determine the chromosome orientation in cancer cells and in the same cell lineage, we studied mouse PCT induced in this unique mouse model. There are slow- and fast-developing PCTs. Slow-onset PCTs are induced only by pristane (2,6,10,14-tetramethylpentadecane) [20]; fast-onset PCTs are induced by pristane and v-abl/myc [19], [21]. In the current study, we focused on the fast-onset PCTs. These exhibit a nonrandom duplication of chromosome 11, cytoband 11E2, associated with the overexpression of genes within 11E2 [19]. Cytoband 11E2 is syntenic to human chromosome 17q25 and rat 10q32 [22]. It is frequently altered in tumors of lymphoid and nonlymphoid origin [23], [24], [25]. The mean latency of fast-onset PCTs is only 45 days [19], [21]. We compared these fast-onset PCT cells with control B lymphocytes of [T38HxBALB/c]N mice with the rcpT(X;11) translocation (T38HT(X;11)).
Figure 1.
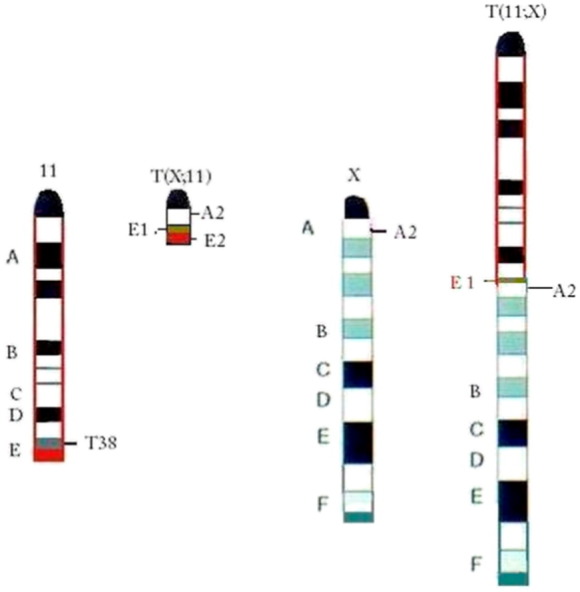
Graphical illustration of the chromosomal constitution of chromosomes 11 in the (BALB/c x T38H) F1 N backcross generation mouse. Chromosome (Chr) 11 with the breakpoint T38H in the telomeric cytoband 11E1 (brown), while the breakpoint in Chr X is located in the centromeric A2 band. The cytoband 11 E2 is colored in red. The reciprocally translocted T(11;X) chromosome resulted from the fusion of the ABCD bands of Chr 11 proximal to the T38H breakpoint with the centromeric A2 band of Chr X. The T(X;11) chromosome was generated by the translocation of the X-derived A2 sub-band onto the 11E1 cytoband of Chr 11. This figure has been published in Genes Cancer. 2010;1(8):847–858 [19] and is reprinted here with permission.
Our aim was to determine the orientation of chromosome 11 in PCTs and lymphocytes of [T38HxBALB/c]N rcpT(X;11) mice. In a previous study, we determined the orientation of chromosome 11 in 3D nuclei of PreB lymphocytes of BALB/c origin and of [T38HxBALB/c]N wild-type mice without the rcpT(X;11) translocation [26] and found a distinct difference between the frequency of the observed orientation patterns in both cell types. Both normal lymphocyte types studied showed a preference in chromosome 11 orientation, where both chromosomes 11 were observed in parallel to the nuclear border [26]. In the current study, we investigated potential changes in the orientation that occur during the process of PCT development.
Originally, multicolor banding (mBANDing) was developed to detect intrachromosomal changes in metaphases [27]. In our previous study, we used mBANDing for the first time in 3D interphase nuclei to determine the orientation of chromosome 11. Chromosome 11 is labeled by four overlapping fluorochromes (Texas Red, GOLD, DEAC, and FITC). This enabled us to analyze whether the centromeric or the telomeric end was orientated toward the nuclear center or periphery. Only one other group used mBANDing on interphase nuclei before. They studied the grade of condensation of human chromosome 5 [28].
We analyzed the orientation patterns of chromosome 11 in PCT cells and [T38HxBALB/c]N rcpT(X;11) lymphocytes. There was a significant difference noted with respect to their chromosome 11 orientation (P < .0001). The nuclear position of the small translocation T(X;11) was also studied visually. It was most frequently found in the intermediate region of the nucleus. There was no significant change in position of T(X;11) detected between the two cell types (P = .06).
Material and Methods
Cell Harvest
Primary lymphocytes were harvested from spleens of 6- to 8-week-old congenic [T38HxBALB/c]N rcpT(X;11) mice [19]. PCT cells were harvested from the ascites of fast-onset PCT mice. The [T38HxBALB/c]N rcpT(X;11) mice were pretreated with pristane intraperitoneally and after 5 days infected with a v-abl/myc virus also administered intraperitoneally. The mean latency of fast-onset PCTs is 45 days [19], [21]. Procedures were performed in accordance to Animal Protocol 11-019 approved by Central Animal Care Services, University of Manitoba (Winnipeg, MB, Canada).
3D Nuclear Hybridizations
For 3D nuclei fixation, lymphocytes were centrifuged at 1000 rpm for 10 minutes. After resuspension of the pellet, cells were carefully placed onto slides and fixed with 3.7% formaldehyde/1 × PBS for 20 minutes at room temperature. Next, the slides underwent washing steps in 1 × PBS shaking. Subsequently, the slides were washed in 0.5% Triton-X-100 for 10 minutes. The slides were incubated for 1 to 2 hours in 20% glycerol and were then subjected to four freeze-thaw cycles in liquid nitrogen afterward. Next, the slides were washed 3 × in 1 × PBS and then incubated in fresh 0.1 M HCl for 5 minutes. After washing the slides in 1 × PBS, they were placed for at least 1 hour in 70% formamide/2 × SSC.
Multicolor Banding
The mBANDing probe for mouse chromosome 11 (Metasystems, Altussheim, Germany) was developed by Benedek et al. (2004) [29]. The slides were equilibrated in 2 × SSC, treated with RNAase A (100 μg/ml) in 2 × SSC at 37°C for 1 hour, and then incubated in freshly prepared 0.01 M HCl with 100 μg/ml pepsin for 2 minutes. After washing the slides in 1 × PBS, they were pretreated in 1% formaldehyde in 1 × PBS/50 mM MgCl2, followed by washing in 1 × PBS. Next, the slides were incubated in 0.1 × SSC and then transferred into 2 × SSC at 70°C for 30 minutes for denaturation. After cooling the solution to 37°C, the slides were transferred to 0.1 × SSC and then subjected to 0.07 M NaOH at room temperature for 1 minute. Afterward, the slides were placed in 0.1 × SSC and then 2 × SSC at 4°C followed by dehydration in ethanol (30%, 50%, 70%, and 90%). Next, the mBANDing probe was applied. The slides were sealed with rubber cement and incubated for 2 days at 37°C. After hybridization, the slides were washed in 1 × SSC at 75°C and in 4 × SSC/0.05% Tween20. The cells were counterstained with 4’6’-diamidino-2-phenylindole and mounted with ProLong Gold antifade (Invitrogen/Gibco, Burlington, ON, Canada).
Image Acquisition
For the two-dimensional image acquisition, an Axioplan 2 microscope (Carl Zeiss Ltd., Toronto, ON, Canada) with a 63 ×/1.4 oil objective lens (Carl Zeiss Ltd., Toronto, ON, Canada) and the ISIS-FISH imaging system 5.0 SR 3 (Metasystems Group Inc., Boston, MA) were used. The chromosomal counterstain was visualized with the help of a 4’6’-diamidino-2-phenylindole filter. To detect the four regions of chromosome 11 that were labeled with different fluorochromes (DEAC, FITC, Gold, and Texas Red), narrow band-pass filters were used (Chroma Technologies) as described by our group previously.
3D image acquisition was conducted using an AxioImager Z2 microscope (Carl Zeiss Inc. Canada) equipped with the same filters and an AxioCam MRm (Carl Zeiss Inc. Canada), combined with the Axiovision Release 4.8 software (Carl Zeiss Inc. Canada). Z-stacks of 80 slices, with 200-nm axial distance and 102-nm lateral pixel size, were acquired to reconstruct a 3D image. Using Axiovision Release 4.8 software (Carl Zeiss Inc. Canada), deconvolution was conducted with the constrained iterative algorithm (Schaefer et al., 2001).
Image Analysis
The results presented in this paper were analyzed by visual inspection. The chromosome 11 mBAND probe is composed of four different fluorochromes labeling four different overlapping regions of the whole chromosome 11. The telomeric end is labeled with FITC (green), the centromeric end with Texas Red (magenta), and the intermediate bands with DEAC (cyan blue) and Gold (red) (Figure 2). DEAC was not always detectable. The mBAND paint made it possible for the visual observer to determine the orientation of chromosome 11. To analyze the position of the small translocation chromosome T(X;11) labeled only with FITC (green), we divided the nucleus visually into three regions: periphery, intermediate, and center.
Figure 2.
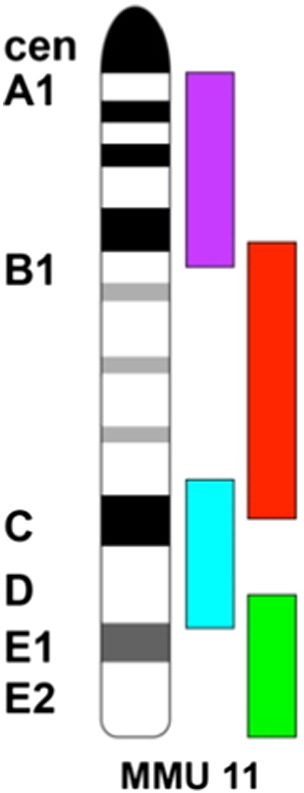
mBAND labeling scheme of mouse chromosome 11. Mouse chromosome 11 is divided into four overlapping segments. The telomeric end is labeled with FITC (green), the centromeric end with Texas Red (magenta), and the intermediate bands with DEAC (cyan blue) and Gold (red). Note cytoband 11E2 at the telomeric end. This figure has been published in BMC Cell Biology 2014, 15:22 http://dx.doi.org/10.1186/1471-2121-15-22[26] and is reprinted here with permission.
In addition, we used novel automated software to confirm our visual results [30]. In short, the nucleus was segmented first with an isodata threshold after some smoothing and out-of-focus blur subtraction. The chromosome bands were then segmented after the recorded images were blurred based on the band sizes. These bands were then linked together to chromosome territories based on a utility function determined by overlap and distance between the segmented bands. The orientation of each CT was then determined by calculating the eigenvectors of the inertia tensor; the orientation is indicated by the more outlying band. Consequently, the chromosome is pointing either with its telomeric end or with its centromeric end toward the nuclear periphery. Because an exact measurement is performed, no parallel orientation category is needed anymore.
The automatic analysis was only performed for the diploid cells. Automation of chromosome orientation by this program was not implemented for tri- and tetraploid cells. Therefore, we present the results assessed by visual inspection. Over 300 nuclei per cell type were acquired, and we were able to determine the orientation pattern in 224 PCT and 257 T38HT(X;11) nuclei.
Statistical Analysis
The visually assessed orientation patterns were analyzed by chi-square, likelihood ratio chi-square, and Mantel-Haenszel chi-square tests. The nuclear positions of T(X;11) were compared by the same tests. They all led to the same result; only chi-square is shown in the paper.
The automatically measured orientation distributions were compared to each other with two-sample, two-sided Kolmogorov-Smirnov test. The two cell types that display each of the observed orientation patterns were compared using chi-square, likelihood ratio chi-square and Mantel-Haenszel chi-square tests. They yielded the same results.
Results
In this study, we analyzed chromosome 11 orientation patterns in lymphocytes of PCTs and of [T38HxBALB/c]N rcpT(X;11) mice. We performed mBANDing and analysis on more than 300 nuclei of each cell type. The mouse chromosome 11 mBAND probe labels four overlapping segments with four fluorescing colors. The telomeric end is labeled with FITC Green, the centromeric end with Texas Red, and the regions in between with Gold and DEAC (Figure 2). The segment labeled with DEAC was not always detectable. The mBANDed nuclei were imaged using Axiovision 4.8 Software (Carl Zeiss Inc. Canada) and deconvolved with a constrained iterative algorithm [31]. By visual inspection, we analyzed the orientation of chromosome 11 of the respective cell types and subsequently determined orientation patterns. Nuclei of T38HT(X;11) lymphocytes consistently showed a diploid chromosome constitution, whereas nuclei of PCTs either were diploid or showed an increase in chromosome 11 copy numbers. Moreover, we identified the position of the small translocation chromosome T(X;11) in all nuclei of T38HT(X;11) and PCT.
By visual inspection, we observed three different orientations in nuclei of T38HT(X;11) and PCTs: 1) chromosome 11 points with its telomeric end to the nuclear periphery and with its centromeric end to the nuclear center (“T”); 2) it points with its centromeric end to the nuclear periphery and with its telomeric end to the center (“C”); and 3) chromosome 11 is in parallel to the nuclear border (“P”). Combining the observed orientations of all chromosomes in one nucleus, we determined an orientation pattern. All observed orientation patterns are shown in Table 1. The most frequently observed orientation pattern in T38HT(X;11) was with both chromosomes located in parallel to the nuclear border (“PP”) (35.0%). In all PCTs, “PP” was only observed in 10.7% (P < .01). The orientation pattern “CP” with one homolog pointing with its centromeric end toward the nuclear periphery and the other homolog being in parallel was found most frequently in PCTs (13.4% of all PCTs) and in 23.7% of T38HT(X;11) (P = .83). Both chromosomes pointing with their centromeric ends toward the periphery (“CC”) was observed in 18.7% of T38HT(X;11) and in 8.5% of all PCTs (P = .0001). The third most common orientation pattern in PCTs is “PT,” with one homolog in parallel and the other pointing with its telomeric end to the periphery (8.5%). This orientation pattern was found in 13.6% of T38HT(X;11) (P < .01). Representative images of diploid PCT and T38HT(X;11) nuclei analyzed visually and automatically are illustrated in Figure 3.
Table 1.
Orientation Patterns of Chromosome 11 and Their Frequency in Diploid Cells of Congenic [T38HxBALB/c]N Mice Showing T(X;11) and in PCTs
| T38H T[X;11] |
Diploid PCTs |
Triploid PCTs |
Tetraploid PCTs |
All PCTs |
|
|---|---|---|---|---|---|
| Both homologs in parallel to the nuclear border (PP) | 90 | 24 | 24 | ||
| One copy points with telomeric end to the nuclear center; the other copy is in parallel (PT) | 35 | 19 | 19 | ||
| Both homologs point with their telomeric end to the nuclear periphery (TT) | 8 | 7 | 7 | ||
| One copy points with its telomeric end and the other copy with its centromeric end to the nuclear periphery (CT) | 15 | 7 | 7 | ||
| Both copies point with their centromeric ends to the nuclear periphery (CC) | 48 | 13 | 13 | ||
| One homolog points with its centromere to the nuclear periphery; the other is parallel to the nuclear border (CP) | 61 | 30 | 30 | ||
| CCC | 1 | 1 | |||
| CCP | 8 | 8 | |||
| CPP | 12 | 12 | |||
| CPT | 19 | 19 | |||
| CCT | 9 | 9 | |||
| CTT | 2 | 2 | |||
| TTT | 2 | 2 | |||
| PTT | 7 | 7 | |||
| PPT | 15 | 15 | |||
| PPP | 17 | 17 | |||
| CCCP | 1 | 1 | |||
| CCPP | 4 | 4 | |||
| CPPP | 4 | 4 | |||
| PPPP | 5 | 5 | |||
| CPPT | 3 | 3 | |||
| CPTT | 3 | 3 | |||
| CCPT | 3 | 3 | |||
| PPTT | 3 | 3 | |||
| PPPT | 5 | 5 | |||
| PTTT | 1 | 1 |
The table shows the orientation patterns observed in the diploid T38HT(X;11) cells and in PCTs with two, three, or four copies of chromosome 11. The last column lists the orientation patterns in all PCT cells together (C = centromere points to periphery, T = telomere points to periphery, P = chromosome is parallel to nuclear periphery).
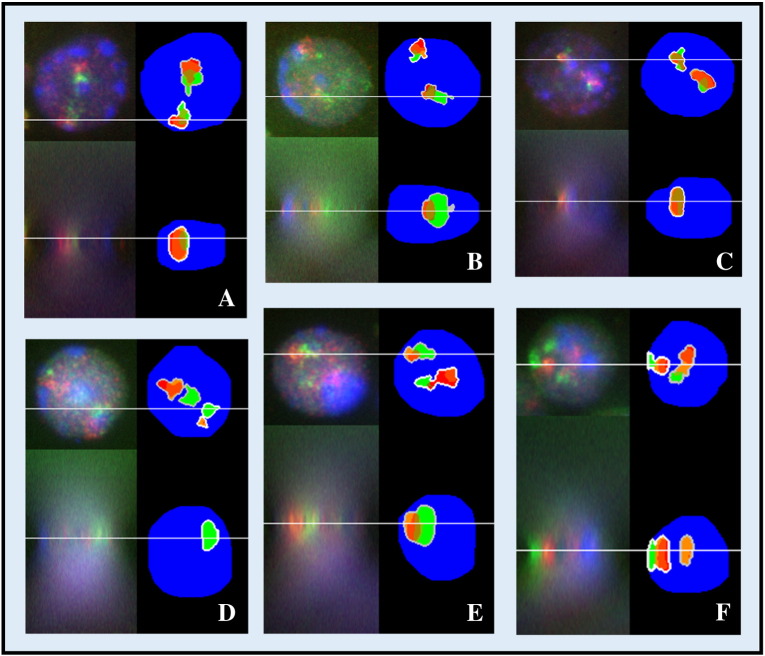
Figure 3.
3D images of diploid PCT and T38HT(X;11) nuclei analyzed by the automatic program (C = centromere (magenta) points to periphery, T = telomere (green) points to periphery). The acquired nuclei, respectively, the same nuclei demonstrated with false colors after segmentation, are displayed in the xy-axis (at the top) and in the xz-axis (at the bottom). (A) PCT nucleus showing the orientation “CC.” (B) PCT nucleus with the orientation pattern “CT.” (C) PCT nucleus showing the orientation pattern “TT.” (D) T38HT(X;11) nucleus showing the orientation “CC.” (E) T38HT(X;11) nucleus with the orientation pattern “CT.” (F) T38HT(X;11) nucleus showing the orientation pattern “TT.”
Ninety-two of 224 (41.1%) PCT nuclei showed three copies of chromosome 11 (Table 1). The orientation pattern “CPT,” with one copy pointing with its telomeric end and another copy with its centromeric end toward the nuclear periphery and one copy located in parallel, was also observed in 8.5% of all PCTs. The second most frequently orientation pattern in PCTs with three copies of chromosome 11 is “PPP,” with all homologs located in parallel (7.6%).
Twenty-three of 224 (10.3%) PCT nuclei showed four copies of chromosome 11 (Table 1). Images of PCT nuclei with more than two copies of chromosome 11 can be seen in Figure 4.
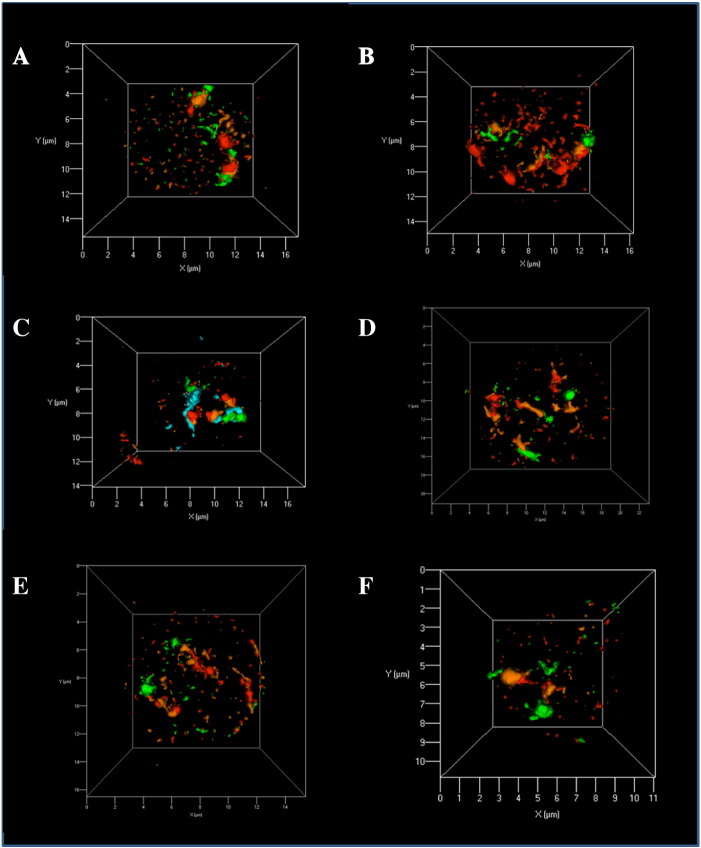
Figure 4.
Representative 3D images of PCT and T38HT(X;11) nuclei with more than two copies of chromosome 11 and one image with T(X;11) present. The scale of the x- and y-axis is in [μ]. The telomeric end is labeled with FITC Green, the centromeric end with Texas Red, and the regions in between with Gold (orange) and DEAC (aqua blue). DEAC was not always detectable (C = centromere (magenta) points to periphery, T = telomere (green) points to periphery, P = chromosome is parallel to nuclear periphery).
(A) PCT nucleus showing the orientation pattern “CPT.” (B) PCT nucleus with the orientation pattern “CCT.” (C) PCT nucleus with orientation pattern “CTT.” (D) PCT nucleus with the orientation pattern “CCPP.” (E) PCT nucleus showing the orientation pattern “TTT.” (F) T38HT(X;11) nucleus with the orientation pattern “TT” and the small translocation chromosome T(X;11) in the center.
When comparing T38HT(X;11) nuclei to all PCT nuclei with respect to their orientation patterns, a significant difference was noted (P < .0001). Regarding only diploid cells of PCTs and T38HT(X;11), there was no significant difference (P = .10). However, comparing diploid PCTs to PCTs with three or four copies of chromosome 11, a significant difference was noted (P < .0001).
For unknown reasons, the small translocation chromosome T(X;11) carrying cytoband 11E2 was only detected in 31.3% of all 224 mBANDed PCT nuclei and in 34.6% of the 257 mBANDed T38HT(X;11) nuclei. The most frequently observed position was in the intermediate region of the nucleus (61.8% of T38HT(X;11) and 74.3% of PCTs, respectively; P = .10) (Table 2). There is no significant difference between the two cell types regarding the position of T(X;11) (P = .0612). An image showing T(X;11) is demonstrated in Figure 4.
Table 2.
Nuclear Position of the Small Translocation Chromosome T(X;11) in Cells of Congenic [T38HxBALB/c]N Mice Showing T(X;11) and in Diploid, Triploid, and Tetraploid PCTs
| Nuclear Position | T38H T[X;11] | All PCTs | Chi-Square |
|---|---|---|---|
| Periphery | 14 (15.7%) | 12 (17.1%) | .81 |
| Intermediate | 55 (61.8%) | 52 (74.3%) | .10 |
| Central | 20 (22.5%) | 6 (8.6%) | .02 |
The nuclear positions of T(X;11) were compared by chi-square analysis. There is no significant difference between the two cell types regarding the nuclear position of T(X;11) (P = .06). A chi-square value of P > .05 indicates that the frequency of the T(X;11) position is similar between the two cell types.
Discussion
We used mBANDing to determine the orientation of chromosome 11 in PCTs and lymphocytes of [T38HxBALB/c]N rcpT(X;11) mice, and we defined orientation patterns. In diploid nuclei of PCTs and T38HT(X;11), we found six distinct orientation patterns (Table 1). Most frequently observed was the orientation pattern “PP” (both chromosomes 11 in parallel to the nuclear border) in 35.0% of T38HT(X;11) and “CP” (one chromosome 11 is pointing with its centromeric end toward the nuclear periphery, whereas the other homolog is in parallel) in 13.4% of all PCTs. With respect to diploid nuclei of PCTs and diploid nuclei of T38H T[X;11] studied in this paper, we did not find a significant difference in the frequency of orientation patterns (P = .10). Analyzing PCT nuclei with three or four copies of chromosome 11, we found various orientation patterns (Table 1), e.g., the orientation pattern “CPT” (one homolog is pointing with its centromeric end and another with its telomeric end toward the nuclear center, and a third is in parallel) in 8.5% of all PCTs. When comparing PCT nuclei with three or four chromosome 11 copies to diploid PCT nuclei, a significant difference was noted (P < .0001). Furthermore, when comparing T38HT(X;11) to all PCT nuclei, we identified different orientation patterns (P < .0001).
The small translocation chromosome T(X;11) was analyzed visually and found in the intermediate region of the nucleus in 74.3% of the PCTs and in 61.8% of T38HT[X;11] (P = .10) (Table 2).
In our previous paper, we presented nonrandom orientation patterns for chromosome 11 in 3D nuclei of PreB lymphocytes of BALB/c origin and of [T38HxBALB/c]N wild-type mice [26]. There was a distinct difference between the frequency of the observed orientation patterns, and this was found in both cell types. The orientation pattern most frequently observed was with both chromosomes 11 in parallel to the nuclear periphery (“PP”). The second most common pattern was with one homolog in parallel and the other homolog pointed with its centromeric end toward the nuclear periphery (“CP”).
The focus of the current study is the 3D nuclear orientation of chromosome 11 in mouse PCTs. We investigated changes in the nuclear orientation during the process of PCT development.
Nuclear architecture is important for nuclear function [5], [9]. It is known that telomere dysfunction leads to genomic instability and therefore to tumorigenesis. Key factors of telomere dysfunction are the shortening of telomeres, breakage-bridge-fusion cycles, and the formation of telomeric aggregates (TAs) [32], [33], [34]. A trigger for TA formation is c-Myc deregulation [35]. Louis et al. (2005) described not only that c-Myc deregulation leads to TA formation resulting in breakage-bridge-fusion cycles but also that changes of nuclear positions lead to closer proximity of telomeres, resulting in chromosomal rearrangements [35]. Changes in chromosome orientation may also lead to closer proximity of telomeres and could therefore be linked to telomere aggregation.
Rotation is a way of movement and a possible way to change nuclear positions. The mechanisms of a possible rotation are currently unknown. One may hypothesize that chromosomes rotate to access transcription factories. The transcription of genes within the telomeric end 11E2 might be enhanced due to telomeric orientation toward the nuclear center. Future studies will elucidate these questions.
In conclusion, we found distinct 3D orientation patterns of mouse chromosome 11 in diploid lymphocytes of [T38HxBALB/c]N rcpT(X;11) mice and of PCTs. How and whether the changes of the orientation patterns in PCT nuclei with three or four chromosomes 11 impact on tumor progression will be the focus of future studies.
Acknowledgements
This study was supported by the Canadian Institutes of Health Research. A. K. S. received a travel award from Bayer. We thank Mary Cheang for statistical analysis. The authors declare that they have no competing interests.
Footnotes
This study was supported by the Canadian Institutes of Health Research.
References
- 1.Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat Res. 2002;504:37–45. doi: 10.1016/s0027-5107(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyer KA, Canfield TK, Gartler SM. Molecular cytological differentiation of active from inactive X domains in interphase: implications for X chromosome inactivation. Cytogenet Cell Genet. 1989;50:116–120. doi: 10.1159/000132736. [DOI] [PubMed] [Google Scholar]
- 4.Dietzel S, Schiebel K, Little G, Edelmann P, Rappold GA, Eils R, Cremer C, Cremer T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- 5.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 6.Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quina AS, Parreira L. Telomere-surrounding regions are transcription-permissive 3D nuclear compartments in human cells. Exp Cell Res. 2005;307:52–64. doi: 10.1016/j.yexcr.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Mai S. Initiation of telomere-mediated chromosomal rearrangements in cancer. J Cell Biochem. 2010;109:1095–1102. doi: 10.1002/jcb.22501. [DOI] [PubMed] [Google Scholar]
- 10.De Vos WH, Hoebe RA, Joss GH, Haffmans W, Baatout S, Van Oostveldt P, Manders EM. Controlled light exposure microscopy reveals dynamic telomere microterritories throughout the cell cycle. Cytometry A. 2009;75:428–439. doi: 10.1002/cyto.a.20699. [DOI] [PubMed] [Google Scholar]
- 11.Molenaar C, Wiesmeijer K, Verwoerd NP, Khazen S, Eils R, Tanke HJ, Dirks RW. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 2003;22:6631–6641. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda M, Tanabe H, Yoshida K, Oikawa K, Saito A, Kiyuna T, Mizusawa H, Mukai K. Alteration of chromosome positioning during adipocyte differentiation. J Cell Sci. 2004;117:5897–5903. doi: 10.1242/jcs.01508. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, McQueen PG, Lichtman MK, Shevach EM, Parada LA, Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet Genome Res. 2004;105:292–301. doi: 10.1159/000078201. [DOI] [PubMed] [Google Scholar]
- 15.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 16.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 17.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 18.Kuzyk A, Mai S. Selected telomere length changes and aberrant three-dimensional nuclear telomere organization during fast-onset mouse plasmacytomas. Neoplasia. 2012;14:344–351. doi: 10.1593/neo.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiener F, Schmälter AK, Mowat MR, Mai S. Duplication of subcytoband 11E2 of chromosome 11 is regularly associated with accelerated tumor development in v-abl/myc–induced mouse plasmacytomas. Genes Cancer. 2010;1:847–858. doi: 10.1177/1947601910382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter M, Wiener F. Plasmacytomagenesis in mice: model of neoplastic development dependent upon chromosomal translocations. Carcinogenesis. 1992;13:1681–1697. doi: 10.1093/carcin/13.10.1681. [DOI] [PubMed] [Google Scholar]
- 21.Wiener F, Coleman A, Mock BA, Potter M. Nonrandom chromosomal change (trisomy 11) in murine plasmacytomas induced by an ABL-MYC retrovirus. Cancer Res. 1995;55:1181–1188. [PubMed] [Google Scholar]
- 22.Koelsch BU, Rajewsky MF, Kindler-Rohrborn A. A 6-Mb contig-based comparative gene and linkage map of the rat schwannoma tumor suppressor region at 10q32.3. Genomics. 2005;85:322–329. doi: 10.1016/j.ygeno.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107:76–94. doi: 10.1159/000046636. [DOI] [PubMed] [Google Scholar]
- 24.Lastowska M, Chung YJ, Cheng Ching N, Haber M, Norris MD, Kees UR, Pearson AD, Jackson MS. Regions syntenic to human 17q are gained in mouse and rat neuroblastoma. Genes Chromosomes Cancer. 2004;40:158–163. doi: 10.1002/gcc.20031. [DOI] [PubMed] [Google Scholar]
- 25.Turhan N, Yurur-Kutlay N, Topcuoglu P, Sayki M, Yuksel M, Gurman G, Tukun A. Translocation (13;17)(q14;q25) as a novel chromosomal abnormality in acute myeloid leukemia-M4. Leuk Res. 2006;30:903–905. doi: 10.1016/j.leukres.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Schmälter AK, Kuzyk A, Righolt CH, Neusser M, Steinlein OK, Muller S, Mai S. Distinct nuclear orientation patterns for mouse chromosome 11 in normal B lymphocytes. BMC Cell Biol. 2014;15:22. doi: 10.1186/1471-2121-15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chudoba I, Plesch A, Lorch T, Lemke J, Claussen U, Senger G. High resolution multicolor-banding: a new technique for refined FISH analysis of human chromosomes. Cytogenet Cell Genet. 1999;84:156–160. doi: 10.1159/000015245. [DOI] [PubMed] [Google Scholar]
- 28.Lemke J, Claussen J, Michel S, Chudoba I, Muhlig P, Westermann M, Sperling K, Rubtsov N, Grummt UW, Ullmann P. The DNA-based structure of human chromosome 5 in interphase. Am J Hum Genet. 2002;71:1051–1059. doi: 10.1086/344286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benedek K, Chudoba I, Klein G, Wiener F, Mai S. Rearrangements of the telomeric region of mouse chromosome 11 in Pre-B ABL/MYC cells revealed by mBANDing, spectral karyotyping, and fluorescence in-situ hybridization with a subtelomeric probe. Chromosome Res. 2004;12:777–785. doi: 10.1007/s10577-005-5264-z. [DOI] [PubMed] [Google Scholar]
- 30.Righolt CH, Schmälter AK, Kuzyk A, Young I, van Vliet L, Mai S. Measuring murine chromosome orientation in interphase nuclei. Cytometry A. 2015;87(8):733–740. doi: 10.1002/cyto.a.22674. [Epub 2015 Apr 17] [DOI] [PubMed] [Google Scholar]
- 31.Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J Microsc. 2001;204:99–107. doi: 10.1046/j.1365-2818.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 32.Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mai S, Garini Y. Oncogenic remodeling of the three-dimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell Cycle. 2005;4:1327–1331. doi: 10.4161/cc.4.10.2082. [DOI] [PubMed] [Google Scholar]
- 34.Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. [DOI] [PubMed] [Google Scholar]
- 35.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z, Kuttler F, Chuang TC, Moshir S, Mougey V. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci U S A. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]