Abstract
Aim
Interindividual epigenetic variation is likely to be an important mechanism contributing to the interindividual variability in the expression and function of ATP-binding cassette, sub-family B, member 1 (ABCB1). The aim of the present study was to explore the effect of interindividual epigenetic variability in the ABCB1 promoter on ABCB1 expression and function in healthy Chinese subjects.
Methods
Using bisulfite sequencing polymerase chain reaction (PCR) and chromatin immunoprecipitation assays, the DNA methylation and histone acetylation status of the ABCB1 promoter in stool DNA and exfoliated colonic epithelial cells of 157 healthy Chinese male volunteers was analysed. ABCB1 mRNA levels in colonic epithelial cells were detected by real-time PCR. The digoxin pharmacokinetics in subjects with different epigenetic profiles was investigated after a single oral administration of digoxin (0.5 mg).
Results
The methylation levels of ABCB1 promoter in stool DNA showed a significant interindividual variation, from 0.84% to 18.05%. A high methylation level of the ABCB1 promoter was closely related to the low levels of acetylated histone H3 and ABCB1 mRNA expression. In the high methylation group, the area under the concentration–time curves (AUC(0–4 h) and AUC(0–10 h)) of digoxin was increased by 19% [95% confidence interval (CI) 10%, 31%; P = 0.024] and 13% (95% CI 8%, 26%; P = 0.026), respectively, and the peak concentration (Cmax) of digoxin was increased by 30% (95% CI 12%, 41%; P = 0.021) compared with the low methylation group.
Conclusions
The epigenetic modifications of the ABCB1 promoter show high interindividual variability in healthy Chinese subjects, and are closely related to the interindividual variation in ABCB1 mRNA expression and digoxin 0–4 h plasma concentrations in vivo.
Keywords: ABCB1, digoxin, epigenetic, expression, interindividual variation
What is Already Known about this Subject
There is high interindividual variability in ABCB1 expression and function. The effect of genetic polymorphism of ABCB1 on this variation has been extensively studied, but the results to date are controversial. Besides genetic polymorphism, epigenetic modifications such as DNA methylation and histone acetylation can also influence the expression and function of ABCB1.
What this Study Adds
Epigenetic modifications of the ABCB1 promoter show high interindividual variability in healthy Chinese subjects.
The epigenetic modifications of the ABCB1 promoter are closely related to the interindividual variation in ABCB1 mRNA expression level and digoxin pharmacokinetics in vivo.
Introduction
ATP-binding cassette, sub-family B, member 1 (ABCB1), also named P-glycoprotein (P-gp), is a member of the ATP-binding cassette (ABC) transporter superfamily 1. ABCB1 is expressed in many normal tissues, such as the liver, kidney, colon, jejunum, blood–brain barrier, pancreas, adrenal cortex and placenta, where it plays a vital role in protecting organs from xenobiotics and toxins. Moreover, ABCB1 is also overexpressed in many solid tumours, and plays a major role in multidrug resistance (MDR) 2–5.
It is well recognized that there is high interindividual variability in ABCB1 expression and function 6–8. In the past few years, attention has been focused on ABCB1 genetic polymorphism – i.e. single nucleotide polymorphisms (SNPs), located in the ABCB1 coding region, and their association with ABCB1 expression and function. A total of 66 SNPs have been identified in the coding sequence of ABCB1 so far, but the most widely investigated for their clinical implications are C1236T, G2677A/T and C3435T. To date, however, there is no clear consensus on whether or not these SNPs, or combinations of these SNPs, can change the expression and/or function of ABCB1 9–12.
Besides genetic polymorphism, another potential source of phenotypic differences is epigenetic variation. Epigenetics refers to heritable changes in gene expression that occur without alteration in the primary DNA sequence. The most well-known epigenetic mechanisms in humans are DNA methylation, histone modification and modulation of gene expression by noncoding RNAs 13. The Human Epigenome Project (HEP) and many other investigations have provided strong evidence that there are considerable interindividual variations in DNA methylation and histone acetylation status in the human genome 14–17. Moreover, there exists an epigenetic cross-talk between DNA methylation and histone modification 18,19. Recently, a growing number of studies have indicated that epigenetic mechanisms are likely to play a significant role in the regulation of ABCB1 expression and function. These studies showed that ABCB1 promoter hypomethylation was associated with an increased ABCB1 level in cancer cells and a poor prognosis in patients with a malignant tumour 20–27. These studies also indicated that an elevated histone acetylation level at the ABCB1 promoter was associated with an increased ABCB1 level in cancer cells 28–30. However, the effects of epigenetic modifications of the ABCB1 promoter on ABCB1 expression and function in healthy subjects have not been investigated so far.
In the present study, the DNA methylation status of the ABCB1 promoter in the stool DNA of healthy male Chinese volunteers was examined, the histone acetylation status of the ABCB1 promoter in the colonic epithelial cells of subjects was assessed, and the influence of epigenetic variation on ABCB1 mRNA expression and the in vivo pharmacokinetics of a typical ABCB1 probe, digoxin, were also analysed. Our data suggested that the epigenetic profiles of the ABCB1 promoter display a high level of interindividual variability, and are closely related to ABCB1 expression and function in vivo.
Methods
Subjects
A total of 157 healthy male volunteers (aged 18–58 years; weight range 52–78 kg; body mass index range 20–25 kg m–2) with the ABCB1 C1236T, G2677T/A and C3435T wild-type allele were enrolled in the study after successful genotyping (Supplementary Table 1). All the participants were of Chinese Han origin, and proved to be in good health on the basis of medical history, physical examination and laboratory evaluation. After giving written informed consent, they were asked to refrain from taking any drugs, alcohol, nicotine or caffeine for at least 2 weeks before and throughout the study period. This study protocol was approved by the ethics committee board of Chongqing Medical University, Chongqing, P.R. China (No. 2013003), and was registered with the Chinese Clinical Trial Registry (No. ChiCTR-OO-14004892).
ABCB1 genotyping
Genomic DNA was isolated from the leukocytes of 513 healthy male volunteers using Wizard® Genomic DNA Purification Kit (Promega Corporation, Wisconsin, USA). Candidate SNPs (C1236T, G2677T/A and C3435T) in the ABCB1 gene were genotyped using PCR restriction fragment length polymorphism (PCR-RFLP) as described previously 31. All PCR reactions were run with a negative control (no genomic DNA) to ensure that there was no contamination of reagents. PCR products were digested using specific restriction enzymes: HaeIII for C1236T, BsrI/BanI for G2677T/A and Sau3AI for C3435T. All genotype analyses were performed three times. Confirmation of PCR-RFLP was performed using ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit and ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
Stool DNA purification and bisulfite sequencing PCR (BSP)
Stool samples were received within 4 h after defecation, and aliquots taken and stored at –80 °C until use. Genomic DNA was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). The quality of human DNA recovered from the stool samples was verified by PCR amplification of the human β-actin gene. BSP was conducted as described previously 32. Briefly, genomic DNA was bisulfite converted using the EZ DNA MethylationTM Kit (ZYMO Research, Orange, CA, USA). A 1.03 kilobase region of the ABCB1 promoter containing 59 CpG sites was amplified; the primer sequences used are shown in Table1. The PCR cycles were as follows: 94 °C for 2 min, 8 cycles of 94 °C for 10 s, 64 °C for 20 s, and 68 °C for 50 s, followed by 25 cycles of 94 °C for 15 s, 60 °C for 20 s and 68 °C for 60 s. The PCR products were ligated into the pTA2-T vector (Takara, Shiga, Japan) and transfected into competent Escherichia coli cells (strain DH5α). Ten bacterial colonies for each amplified fragment were picked and sequenced by the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit and ABI 3730 DNA analyzer (Applied Biosystems). The methylation level was calculated by the formula: number of C reads divided by the sum of C and T reads.
Table 1.
Primers used
| Primer set | Oligonucleotide sequence | Amplicon size(base pairs) |
|---|---|---|
| BSP assay | ||
| PCR1 sense | 5′- GACAGGTGTTAGGAAGTAGAAAGGTGAT-3′ | 406 (nt –251 to +155) |
| PCR1 antisense | 5′- TCTAAATCTAACCCCACTTAATCCCCAT-3′ | |
| PCR2 sense | 5′-GGGTTAGATTTAGATTTAGGAGTTT-3′ | 496 (nt +285 to +780) |
| PCR2 antisense | 5′-CAAACTAATTACCTTTTATTATTCAATTTA-3′ | |
| ChIP assay | ||
| CpG island 1 sense | 5′-GCAACGGAAGCCAGAACA-3′ | 117 (nt –188 to –72) |
| CpG island 1 antisense | 5′-CAGCCAATCAGCCTCACC-3′ | |
| CpG island 2 sense | 5′-CCGCTGTTCGTTTCCTTTA-3′ | 106 (nt +40 to +145) |
| CpG island 2 antisense | 5′-TTTGCGTGCCCCTACCTC-3′ | |
| CpG island 3 sense | 5′-GTGGGTGGGAGGAAGCAT-3′ | 120 (nt +399 to +518) |
| CpG island 3 antisense | 5′-TCCAGCATCTCCACGAAGG-3′ | |
| Reporter gene plasmid construction | ||
| Fragment 1 sense | 5′-TTTTCTTTCATTCCATTTATCATC-3′ | 234 (nt –519 to –286) |
| Fragment 1 antisense | 5′-TTCATATCCATATAACTACAGGAC-3′ | |
| Fragment 2 sense | 5′-GACTTATGTGAACTTTGAA-3′ | 149 (nt –285 to –137) |
| Fragment 2 antisense | 5′-GAAACTGCGAAACAGGTTGA-3′ | |
| Fragment 3 sense | 5′- TCGAGGAATCAGCATTCAG-3′ | 117 (nt –136 to +20) |
| Fragment 3 antisense | 5′- GGAAGAGCCGCTACTCGAATGAGCT-3′ | |
| Fragment 4 sense | 5′- AAGCTCAAAGAAGCAGAGGCCGC-3′ | 242 (nt +21 to +262) |
| Fragment 4 antisense | 5′- GGACTTGCCAGAGGACTTCACACTA-3′ | |
| Fragment 5 sense | 5′-ATGGGGACCAAGTGGGGTTAGAT-3′ | 260 (nt +263 to +522) |
| Fragment 5 antisense | 5′-GTCTCCAGCATCTCCACGAAGGCAG-3′ | |
| Fragment 6 sense | 5′-GAGACCCCGCGCACAGGAAAGCCCC-3′ | 224 (nt +517 to +740) |
| Fragment 6 antisense | 5′-TCTTCTTTGCTCCTCCATTGCGGTC-3′ | |
Abbreviations are as follows: BSP, bisulfite sequencing PCR; ChIP, chromatin immunoprecipitation; nt, nucleotide; PCR, polymerase chain reaction.
In vitro methylation of reporter gene plasmids and luciferase reporter assay
Fragments of the ABCB1 promoter, including –519 to –286, –285 to –137, –136 to +20, +21 to +262, +263 to +522 and +517 to +740, were amplified by PCR and fused upstream of the firefly luciferase gene in the pGL4.17 plasmid. The primer sequences used are shown in Table1. Reporter constructs were confirmed by DNA sequencing. Whole ABCB1 reporter plasmids were methylated using CpG methyltransferase (M.SssI) (New England Biolabs, Massachusetts, USA), and in a parallel control reaction the same plasmid was mock-methylated in the absence of S-adenosylmethionine. Methylated and mock-methylated plasmids were digested with restriction enzymes and relegated back to pGL4.17 (Promega). The extent of methylation was determined by digestion with a mixture of the methylation-sensitive restriction enzymes HpaII and HhaI. The unmethylated (mock-methylated) or methylated ABCB1 promoter/firefly luciferase fusion genes were transfected into COS-7 cells using LipofectamineTM 2000 (Invitrogen, New York, USA). In each experiment, the phRG-Basic plasmid, encoding Renilla luciferase, was cotransfected for the purpose of normalization. Luminescence was measured 48 h after transfection using the dual-luciferase reporter assay system (Promega). Reporter activity was normalized by calculating the ratio of firefly to Renilla luciferase values.
Separation of exfoliated colonic epithelial cells and chromatin immunoprecipitation (ChIP)
Exfoliated colonic epithelial cells were isolated using the SCSRTM Fecal Cell Isolation Kit (NonInvasive Technologies, Maryland, USA) as previously described 33–35. 1 × 106 cells were used for the ChIP assay, using the EZ-Magna ChIP™ HiSens Chromatin Immunoprecipitation Kit (Millipore, California, USA). Anti-acetyl histone H3 and anti-acetyl histone H4 antibody (Millipore) were used for detection of acetylated histones H3 and H4. Immunoprecipitated DNA was analysed using real-time PCR using the SYBR® Premix Ex Taq™ II Kit (Takara). The conditions for all reactions were as follows: 95 °C for 30 s, 95 °C for 5 s and 60 °C for 30 s. Primers were designed to evaluate the region spanning the three putative CpG islands of the ABCB1 promoter (Table1). Immunoprecipitation with nonspecific IgG was performed as a negative control. A sample representing linear amplification of the total input DNA was used as an input control.
Total RNA purification and real-time PCR of stool samples
Stool samples were received within 1 h after defecation. Total RNA was immediately extracted using the E.Z.N.A.® Stool RNA Kit (Omega, Georgia, USA), and was reverse transcripted into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, New York, USA). ABCB1 mRNA expression level was measured by real-time PCR using the SYBR® Premix Ex Taq™ II Kit, and the glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) was used as an endogenous reference. PCR amplification was carried out using forward primer 5′-GACAGGCATCTCCAAGCATT-3′ and reverse primer 5′-ATCTGGTTTGTGCCCACTCT-3′ for ABCB1, and forward primer 5′-ACCACAGTCCATGCCATCAC-3′ and reverse primer 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH. The reaction was performed over 40 cycles at 95 °C for 30 s, 95 °C for 5 s and 60 °C for 30 s. The relative mRNA expression was normalized to the GAPDH and the analysis was carried out using the 2-△△Ct method as previously described 36.
Digoxin pharmacokinetics assessment
All subjects received a 0.5 mg oral dose of digoxin (0.25 mg per tablet; Hangzhou Minsheng Pharmaceutical Group Co., Hangzhou, P.R. China) with 150 ml of water after an overnight fast. Serial blood samples were collected into ethylenediaminetetraacetic acid (EDTA) tubes immediately before and at 0.25, 0.5, 0.75, 1, 2, 4, 6, 8 and 10 h after digoxin administration. Serum was separated by centrifugation and stored at –80 °C until the quantitative drug analysis. Plasma digoxin concentrations were quantified by the liquid chromatography–tandem mass spectrometry (LC/MS/MS) method as previously described 37. D3-digoxin [purity ≥ 99%, high-performance liquid chromatography (HPLC), Toronto Research Chemicals, Ontario, Canada] was used as an internal standard. Standard analytical digoxin (purity ≥ 99%, HPLC) was purchased from the National Institutes for the Control of Pharmaceutical and Biological Products (Beijing, China). Briefly, samples were prepared by adding 20 μl of d3-digoxin solution (10 ng ml–1) to 200 μl of serum sample, and then extracted with methyl tert-butyl ether. LC/MS/MS analysis was performed using the Finnigan LCQ Deca XPplus (Finnigan, San Jose, CA, USA). A Capcell C18 MG III analytical column (100 mm × 2.0 mm I.D. 5 μm; Shiseido, Tokyo, Japan) and a mobile phase [acetonitrile:10 mmol l–1 ammonium acetate (including 0.1% formic acid) = 3:2] at a flow rate of 0.3 ml min–1 was applied. The lower limit of quantification was 0.1 ng ml–1. The ion transitions monitored were as follows: m/z 798.4 to 651.2 for digoxin and m/z 801.4 to 654.2 for d3-digoxin. The intra- and interday coefficients of variation were less than 10%. All samples from each subject were assayed in triplicate, together with calibration and quality controls. The peak concentration (Cmax) and time to Cmax (tmax) were obtained by inspection of the concentration–time data. The area under the concentration–time curves (AUCs) of digoxin were calculated by the linear trapezoidal rule.
Statistical analysis
All data were analysed using SPSS software for Windows (version 18.0; SPSS, Chicago, IL, USA). In vitro data were analysed using analysis of variance followed by a Dunnett's post hoc test or by Student’s t-test. The AUC values of digoxin were analysed using the paired Student’s t-test. The Wilcoxon signed-rank test was performed for the nonparametric paired two-group comparisons of Cmax and tmax. The 95% confidence interval (CI) of the difference between groups was calculated using a two-sample t-test. Results are plotted as mean ± standard error, unless otherwise stated. P < 0.05 indicated statistical significance.
Results
Interindividual variability in DNA methylation status of the ABCB1 promoter
We searched the ABCB1 promoter region for the location of CpG dinucleotides using MethPrimer software (http://www.urogene.org/methprimer/). CpG dinucleotides were found extensively located near the transcription start site (TSS). According to the location intensity of CpG dinucleotides, we observed three putative CpG islands (Figure1A). The average methylation level of the ABCB1 promoter region studied was about 7.41% in the stool DNA of all healthy volunteers. Moreover, the distribution of the individual methylation levels of the ABCB1 promoter showed a broad range of interindividual variation, from 0.84% to 18.05% (Figure1B). We also analysed the methylation level variation in each CpG island: CpG island 2 showed the greatest range of variation (0.71–33.86%), CpG island 3 the second highest range (0.67–13.33%) and CpG island 1 the smallest range of variation (0.69–10.23%). Although the natural log (LN)-transformed data fit a normal distribution, the median value was chosen to describe the middle of the CpG methylation status. The median methylation level of CpG island 1, 2 and 3 were 2.67%, 9.31% and 6.36%, respectively (Figure1C–E).
Figure 1.
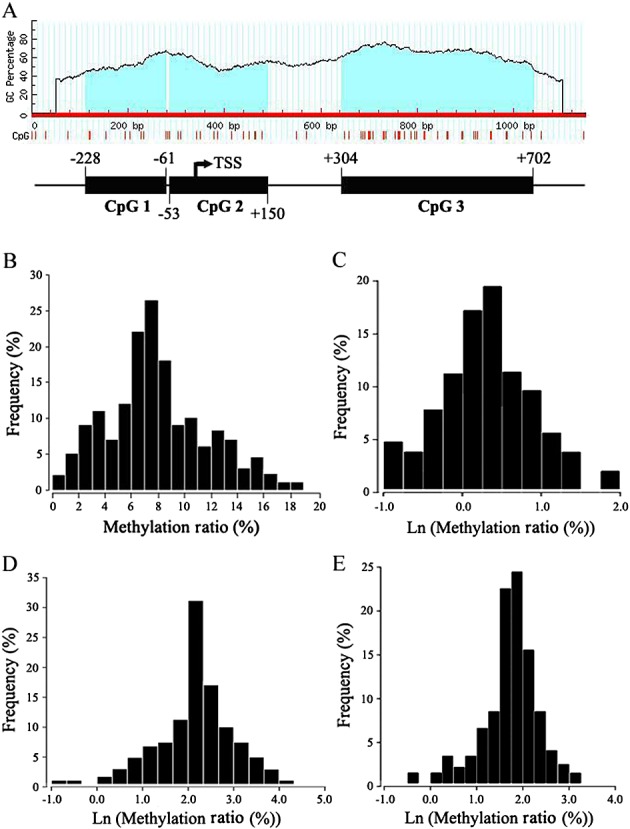
DNA methylation status of three CpG islands in the ABCB1 promoter region in the stool DNA of healthy male Chinese volunteers. (A) Computational analysis of the CpG island distribution in the ABCB1 promoter region; (B–E) Distribution of the methylation levels of the ABCB1 promoter in individuals in total studied region, CpG island 1, CpG island 2 and CpG island 3, respectively. TSS, transcriptional start site.
Effect of methylation status of different ABCB1 promoter regions on its transcriptional activity
Our results of the BSP assay indicated that the different ABCB1 promoter regions showed different degrees of interindividual variability. To study the influence of the methylation status of these promoter regions on ABCB1 transcription activity, we analysed the abilities of in vitro-methylated and mock-methylated ABCB1 promoter fragments to direct the reporter gene expression in transiently transfected COS-7 cells. Firstly, we assessed the effect of the –136 to +20 promoter region of ABCB1 on transcriptional activity, and the result showed that this region demonstrated good luciferase activity above the pGL4.17-Basic background. However, after in vitro methylation, the luciferase activity of this region was significantly decreased by 77.8% compared with mock-methylation treatment. We then inserted in vitro-methylated and mock-methylated fragments, including –519 to –286, –285 to –137, +21 to +262, +263 to +522, and +517 to +740, into the upstream or downstream of the –136 to +20 region, and observed the different effects of the methylation status of these regions on luciferase activity. Our results indicated that in vitro methylation reduced the transcriptional activities of all studied promoter regions. The most affected region was +21 to +262, where the activity was reduced by 40.4%, followed by +263 to +522 (36.21%), –285 to –137 (19.83%), –523 to –286 (7.80%), and +523 to +757 (5.91%). Thus, these findings demonstrate that the methylation status of different ABCB1 promoter regions can influence transcriptional activity to a varying degree (Figure2).
Figure 2.
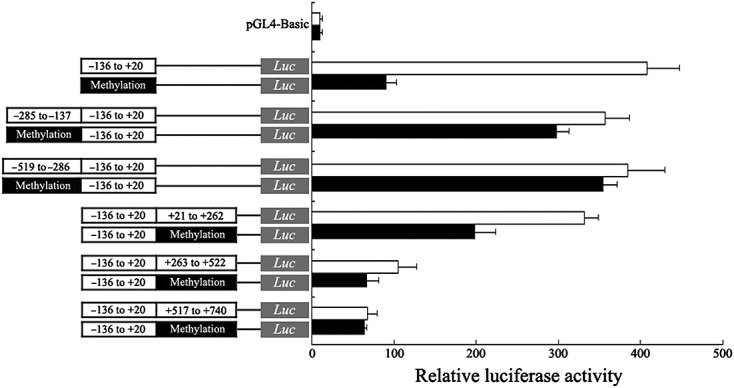
Methylation effects of different ABCB1 promoter regions on transcriptional activity in a luciferase reporter assay. The mean reporter activity ± standard error (Firefly/Renilla luciferase activity) from three independent experiments is presented.
Association between DNA methylation and histone acetylation status of the ABCB1 promoter
Different types of epigenetic modifications are often closely linked with each other 18,19. In order to study whether or not a relationship exists between ABCB1 promoter methylation and histone acetylation status in the colonic epithelial cells of healthy subjects, we divided the volunteers into three groups according to the methylation level of the ABCB1 promoter: the low methylation group (<10th percentile), middle methylation group and high methylation group (>80th percentile) according to a previous report 38. We then randomly selected 30 volunteers from the low and high methylation groups, each group with 15 volunteers, with a median age of 35 (22–49) years and a median weight of 65.9 (52–76.2) kg. Their methylation profiles of the ABCB1 promoter in stool DNA are shown in Figure3.
Figure 3.
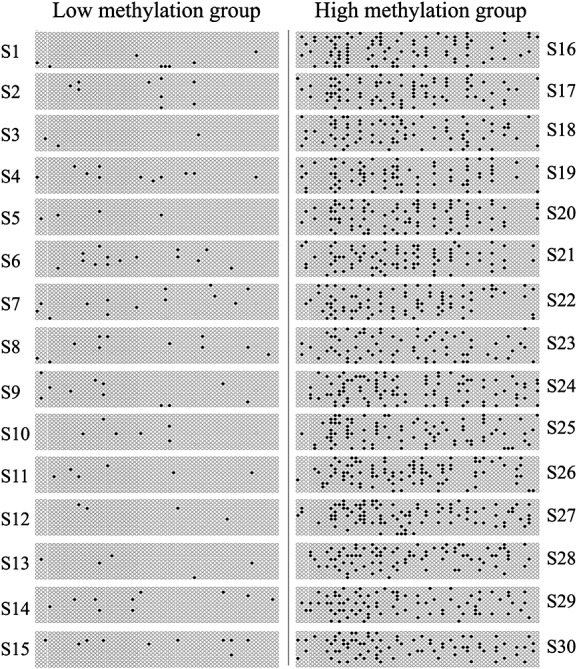
Bisulfite sequencing results of the ABCB1 promoter in stool DNA from healthy male Chinese subjects in the low and high methylation groups. Each row of circles represents a single cloned allele and each circle represents a single CpG dinucleotide. White circles represent unmethylated CpG dinucleotide, and black circles represent methylated CpG dinucleotide.
We carried out the ChIP assay using their separated colonic epithelial cells. Our results showed that the histone H3 acetylation level was significant increased at the CpG island 2 locus in the low methylation group (8.16% input; 95% CI 7.02%, 10.23%) compared with the high methylation group (6.92% input; 95% CI 5.03%, 7.21%) (P = 0.027). H3 acetylation levels at the other two CpG islands, and H4 acetylation levels at all three CpG islands did not show a significant difference between the two groups (Figure4). These data demonstrate that there is a significant association between the DNA methylation and histone acetylation status of the ABCB1 promoter.
Figure 4.
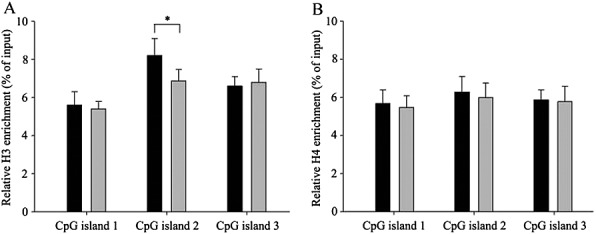
High ABCB1 promoter methylation is associated with H3 histone deacetylation in the colonic epithelial cells of healthy male Chinese subjects. (A) Relative acetylated H3 enrichment. (B) Relative acetylated H4 enrichment. The results are expressed as the percentage of immunoprecipitate over total input DNA utilized. Error bars show the standard error of three different experiments with independent chromatin preparations. N = 15 per group, *P < 0.05.
Effect of epigenetic status of the ABCB1 promoter on mRNA expression
Our results from the luciferase reporter assay indicated that the methylation status of the ABCB1 promoter region –136 to +262 had a significant influence on transcriptional activity. Moreover, this region completely covers the CpG island 2, showing the greatest interindividual variability in epigenetic patterns among three CpG islands. Therefore, we hypothesized that the interindividual epigenetic variation in the ABCB1 promoter might have an impact on the interindividual variability in ABCB1 mRNA expression. We therefore collected fresh stool samples from the above-mentioned 30 male volunteers in the low and high methylation groups, extracted the total RNA and analysed ABCB1 mRNA expression levels. ABCB1 mRNA was detected in all samples, and the expression levels showed considerable interindividual variability (Figure5A). Statistical analysis showed that an increased ABCB1 mRNA expression level was observed in the low methylation group (ABCB1/GAPDH mRNA ratio 0.81; 95% CI 0.66, 0.95) compared with the high methylation group (ABCB1/GAPDH mRNA ratio 0.39; 95% CI 0.24, 0.56) (P = 0.008) (Figure5B).
Figure 5.
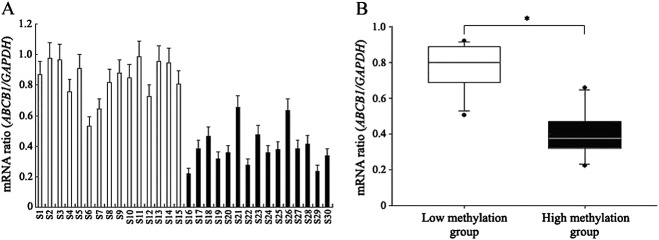
The relationship between epigenetic status of the ABCB1 promoter and ABCB1 mRNA expression in vivo. (A) Interindividual variability in ABCB1 mRNA expression in colonic epithelial cells. (B) The epigenetic status of the ABCB1 promoter is related to the ABCB1 mRNA expression level in colonic epithelial cells. Error bars show the standard error of three different experiments with independent chromatin preparations. N = 15 per group; *P < 0.05.
Effect of epigenetic status of the ABCB1 promoter on the pharmacokinetic profile of digoxin in vivo
The selected 30 healthy subjects in the low and high methylation groups were recruited for this study, with 15 volunteers in each group. The main pharmacokinetic parameters of digoxin in the two groups are summarized in Table2.
Table 2.
Pharmacokinetic parameters of digoxin in the low and high methylation groups after a single 0.5 mg oral dose
| Parameter | Low methylation group | High methylation group | Percentage change(95%CI) | P value |
|---|---|---|---|---|
| AUC(0–4 h) (ng·h ml–1) | 4.31 ± 1.03 | 5.12 ± 1.42 | 19% (10%, 31%) | 0.024 |
| AUC(0–10 h) (ng·h ml–1) | 6.83 ± 1.01 | 7.72 ± 1.24 | 13% (8%, 26%) | 0.026 |
| Cmax (ng ml–1) | 1.92 ± 0.26 | 2.49 ± 0.18 | 30% (12%, 41%) | 0. 021 |
| tmax (h) | 0.98 ± 0.22 | 0.93 ± 0.18 | NA | 0.236 |
Values show mean ± standard error. N = 15 per group. P < 0.05 indicated statistical significance. Abbreviations are as follows: AUC, area under the concentration–time curve; CI, confidence interval; Cmax, peak concentration; NA, not applicable; tmax, time to Cmax.
The mean digoxin plasma concentration profiles in the subjects in the two groups are shown in Figure6A. We observed a 19% (95% CI 10%, 31%; P = 0.024) and 13% (95% CI 8%, 26%; P = 0.026) increase in AUC(0–4 h) and AUC(0–10 h), and a 30% (95% CI 12%, 41%; P = 0.021) increase in the Cmax of digoxin in the high methylation group compared with the low methylation group. We did not find a statistically significant difference in tmax between the two groups. A large interindividual variation in the Cmax of digoxin among the subjects was also found (Figure6B). These findings indicate that there is a relationship between the epigenetic status of the ABCB1 promoter and the pharmacokinetic profiles of orally administered digoxin.
Figure 6.
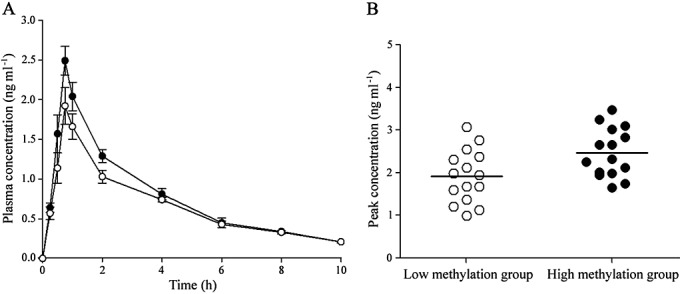
The relationship between epigenetic status of the ABCB1 promoter and digoxin pharmacokinetics. (A) The epigenetic status of the ABCB1 promoter is related to the concentration–time profile of digoxin in healthy Chinese subjects. (B) The epigenetic status of the ABCB1 promoter is related to peak concentration values for digoxin in healthy Chinese subjects. Data show mean ± standard error; N = 15 per group.
Discussion
To our knowledge, this is the first study to observe the interindividual variability in the epigenetic status of the ABCB1 promoter in healthy Chinese subjects, and the relationship between these variations and ABCB1 expression and function in vivo.
Recently, numerous studies have suggested that epigenetic factors play an important role in the regulation of ABCB1 expression and/or function 20–30. However, the epigenetic modification is tissue specific 39, and few studies have been carried out in healthy subjects. The intestinal epithelium is known to undergo constant and rapid renewal, resulting in millions of cells being shed into the fecal stream every day. It is estimated that one-sixth to one-third of these cells are shed every 24 h, giving rise to about 1010 exfoliated cells per day, most of which are colonic epithelial cells. Human stools are therefore an ideal source of viable colonic epithelial cells 33,40,41. Using appropriate methods, genomic DNA, total RNA and even viable colonic epithelial cells can be extracted or isolated from stool samples and used to carry out some relevant investigations. Nowadays, assay of methylated DNA markers in stools has been a promising noninvasive approach for colorectal cancer screening 42.
A CpG island is defined by a region of DNA greater than 200 base pairs, with a G/C content above 0.5 and an observed or expected presence of CpG dinucleotides above 0.6 43. According to these specifications, we find three CpG islands within the ABCB1 promoter region, including 59 CpG dinucleotides. Our study indicated that the DNA methylation status of the ABCB1 promoter shows considerable interindividual variability in the stool DNA of healthy Chinese subjects. Many previous studies have also demonstrated that there is significant interindividual variability in DNA methylation status either at the global level or at the level of some specific/selected genes 15–17,44–47. DNA methylation variation plays an important role in the development of many diseases, such as cancer, immune abnormalities, cardiovascular diseases, type-2 diabetes, obesity and mental disorders 48–51, although the causes of such variation have not been fully elucidated. Besides differences in genetic background, the variation in methylation can be attributed to many environmental factors, such as diet, smoking habits, physical activity, climate, pathogen exposure, etc. 52. Diet is thought to be one of the main causes of the alteration in the DNA methylation status occurring not only in the periconceptional period, but also in adulthood 44,53. Considering the physiological position, diet may have an important influence on epigenetic variation in genes expressed in intestinal epithelial cells, including ABCB1. Using exfoliated colonic epithelial cells, the effect of diet on ABCB1 epigenetic status needs to be studied intensively.
In the present study, we found that the lowest and highest methylation levels of the ABCB1 promoter in individual samples were 0.84% and 18.05%, respectively. The methylation profiles of some specific genes studied in healthy subjects also exhibit a similar degree of variation 17,47. The establishment of ABCB1 methylation status is a complex and dynamic process, strictly connected with various genetic and environmental factors in individuals. ABCB1 is abundantly expressed in the human intestine and plays an important role in the bioavailability and cell-toxicity limitation of drugs and xenobiotics 54,55. Abnormal methylation of the ABCB1 promoter may be closely related to the development, treatment and prognosis of many intestinal diseases. Therefore, it is crucial to assess the normal range of methylation variation of the ABCB1 promoter in order to associate unexpected methylation levels with related disease processes.
DNA methylation and histone acetylation are major epigenetic modifications that are most intensively studied in the context of gene transcription. DNA methylation is associated with the silencing of gene expression, while histone acetylation is associated with active transcription. Moreover, there is an epigenetic cross-talk between them during gene transcription 13,18,19. Consistent with this, our results indicate that the high acetylated level of H3 histone at the CpG island 2 locus of the ABCB1 promoter is closely related to the low methylation level in colonic epithelial cells.
We also found a high interindividual variability in ABCB1 mRNA expression in the colonic epithelial cells of healthy Chinese subjects, which is consistent with previous investigations demonstrating large interindividual variation in ABCB1 mRNA expression along the human intestinal tract 6,8,55. Variation in intestinal ABCB1 level has important clinical significance because it can influence the activity and bioavailability of some orally administered ABCB1 substrate drugs 12,56. To date, researchers have found many factors that may contribute to the regulation of the ABCB1 level in vivo, including transcription factors, reactive oxygen species, and some drugs such as rifampin and paclitaxel 10. However, in healthy people, the effect of ABCB1 genetic polymorphism on interindividual variation in ABCB1 level has attracted the most attention. Some studies have suggested that the ABCB1 C3435T SNP is correlated with the ABCB1 level; however, others have shown the exact opposite results. For example, the ABCB1 level appears to be unrelated to the C3435T genotype in the placenta, CD34+ haematopoietic stem cells and the liver. Besides C3435T, ABCB1 T-2410C, T-1910C and G2677A polymorphisms may also be associated with ABCB1 expression, but the results are still contradictory 9,10,12. In the present study, we suggest that the interindividual variability in epigenetic status, including DNA methylation and histone acetylation, may play an important role in interindividual variation in ABCB1 mRNA expression. Further investigation is needed to determine the impact of epigenetic variation on the level of the ABCB1 protein.
Digoxin is a cardiac glycoside used for the treatment of congestive heart failure and atrial fibrillation. Its unique mechanism of action, complicated pharmacokinetic properties, narrow therapeutic index and extensive toxicity profile make it a medication that requires individualized dosing based on multiple patient-specific factors 57. It is a substrate of intestinal and renal ABCB1, and does not undergo significant hepatic metabolism. As an in vivo ABCB1 probe, digoxin is more sensitive for intestinal ABCB1 than for renal ABCB1 58. Recently, evidence has shown that digoxin is probably also a substrate for other transporters, including organic anion transporting polypeptides (OATPs), sodium-dependent multivitamin transporter (SMVT) and apical sodium bile acid transporter (ASBT), but the results from these studies are still controversial 59. Despite the uncertainty regarding the involvement of other non-P-gp transporters in digoxin disposition, digoxin is still the best available and most practical in vivo ABCB1 probe at present 58. Since Hoffmeyer et al. 60 first reported that ABCB1 C3435T SNP was associated with increased plasma concentrations after oral administration of digoxin, numerous studies have been conducted concerning the effects of ABCB1 polymorphisms and haplotypes on digoxin pharmacokinetics. However, the results are still open to debate 9,10,12. To the best of our knowledge, all the available literature on the impact of ABCB1 polymorphisms on digoxin pharmacokinetics focus only on the functions of C1236T, G2677T/A and C3435T SNPs. Therefore, only the possible influences of these three SNPs on digoxin pharmacokinetics have been ruled out in this study. However, there is still a possibility that other ABCB1 SNPs can affect digoxin pharmacokinetics, and this issue requires further investigation.
In the present study, we demonstrated that the epigenetic status of ABCB1 expressed in the intestinal tract may be closely related to the pharmacokinetic profiles of orally administered digoxin. Previous investigations showed that the functional effect of ABCB1 on the pharmacokinetics of digoxin primarily occurs during the initial hours of absorption and is best reflected by AUCs covering the early hours after oral drug intake 61–63. Thus, we used AUC(0–4 h) and AUC(0–10 h) to measure the digoxin absorption in the present study. Our results showed that the AUC(0–4 h), AUC(0–10 h) and Cmax for digoxin were increased by 19%, 13% and 30%, respectively, in the high methylation group compared with the low methylation group. As there are many factors influencing the pharmacokinetics of digoxin, of which ABCB1 activity is only one, the differences in digoxin pharmacokinetics between the two groups observed in the present study are obvious 57. Moreover, digoxin has a narrow therapeutic index, and small variations in blood concentration may easily result in toxic or subtherapeutic concentrations 64. In the present study, although the values of Cmax and tmax might be inaccurate because they are dependent on the sampling times, the data suggest a relationship between the epigenetic status of ABCB1 and digoxin pharmacokinetics. Therefore, the epigenetic profile of ABCB1 is worth considering during the course of digoxin therapy.
As it is almost impossible to obtain epithelial cells from the intestinal tract, other than the colon, in healthy volunteers, we cannot analyse the epigenetic profiles of ABCB1 in other sites. However, for a given individual, the epigenetic profiles of ABCB1 in colonic epithelial cells at least partially reflect the epigenetic status of ABCB1 in the entire intestinal tract. To the best of our knowledge, this is the first report demonstrating the relationship between the epigenetic profiles of ABCB1 and the pharmacokinetics of digoxin in healthy subjects.
In conclusion, the present study indicates that the epigenetic modifications of the ABCB1 promoter show high interindividual variability in healthy Chinese subjects, and are closely relate to the interindividual variation in ABCB1 mRNA expression and digoxin pharmacokinetics in vivo. It also provides a new explanation for the mechanism behind the interindividual variability in ABCB1 expression and function in healthy people. Further investigations are needed to explore the combined influence of genetic and epigenetic factors on the interindividual variability in ABCB1 expression and function, as well as its molecular mechanism and clinical significance.
Acknowledgments
This work was supported by the National Scientific Foundation of China (No. 81102511, 81473284), Natural Science Foundation of Chongqing (No. cstc2011jjA10004), Postdoctoral Science Foundation of China (No.2011 M500669), Special Postdoctoral Research Foundation of Chongqing (No.Yu xm201102007), Medical Scientific Research Foundation of Chongqing, China (No. 2013-2-150) and Funds for outstanding young scholars in Chongqing Medical University (No. CYYQ201301).
Competing Interests
All authors have completed the ICMJE uniform disclosure form at http://www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare no support from any organization for the submitted work, no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years and no other relationships or activities that could appear to have influenced the submitted work.
Contributors
LXW, ZY, HHZ were responsible for the conception and design of the work. LXW, CJW, YL and XZ acquired the data. LXW, YL and YYS analysed and interpreted the data. LXW and CJW drafted and revised the article critically for intellectual content. LXW, CJW, YL, XZ, YYS, ZY and HHZ gave final approval of the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Table 1 Subjects characteristics
References
- Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61:682–99. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin Pharmacokinet. 2003;42:59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- Goda K, Bacsó Z, Szabó G. Multidrug resistance through the spectacle of P-glycoprotein. Curr Cancer Drug Targets. 2009;9:281–97. doi: 10.2174/156800909788166493. [DOI] [PubMed] [Google Scholar]
- Hodges LM, Markova SM, Chinn LW, Gow JM, Kroetz DL, Klein TE, Altman RB. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein) Pharmacogenet Genomics. 2011;21:152–61. doi: 10.1097/FPC.0b013e3283385a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KG, Sikic BI. Molecular pathways: regulation and therapeutic implications of multidrug resistance. Clin Cancer Res. 2012;18:1863–9. doi: 10.1158/1078-0432.CCR-11-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodahl EL, Ho RJ. The role of MDR1 genetic polymorphisms in interindividual variability in P-glycoprotein expression and function. Curr Drug Metab. 2004;5:11–9. doi: 10.2174/1389200043489108. [DOI] [PubMed] [Google Scholar]
- Meier Y, Pauli-Magnus C, Zanger UM, Klein K, Schaeffeler E, Nussler AK, Nussler N, Eichelbaum M, Meier PJ, Stieger B. Interindividual variability of canalicular ATP-binding-cassette (ABC)-transporter expression in human liver. Hepatology. 2006;44:62–74. doi: 10.1002/hep.21214. [DOI] [PubMed] [Google Scholar]
- Thörn M, Finnström N, Lundgren S, Rane A, Lööf L. Cytochromes P450 and MDR1 mRNA expression along the human gastrointestinal tract. Br J Clin Pharmacol. 2005;60:54–60. doi: 10.1111/j.1365-2125.2005.02389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaeda T. MDR1 genotype-related pharmacokinetics: fact or fiction? Drug Metab Pharmacokinet. 2005;20:391–414. doi: 10.2133/dmpk.20.391. [DOI] [PubMed] [Google Scholar]
- Marchetti S, Mazzanti R, Beijnen JH, Schellens JH. Concise review: Clinical relevance of drug drug and herb drug interactions mediated by the ABC transporter ABCB1 (MDR1, P-glycoprotein) Oncologist. 2007;12:927–41. doi: 10.1634/theoncologist.12-8-927. [DOI] [PubMed] [Google Scholar]
- Wolf SJ, Bachtiar M, Wang J, Sim TS, Chong SS, Lee CG. An update on ABCB1 pharmacogenetics: insights from a 3D model into the location and evolutionary conservation of residues corresponding to SNPs associated with drug pharmacokinetics. Pharmacogenomics J. 2011;11:315–25. doi: 10.1038/tpj.2011.16. [DOI] [PubMed] [Google Scholar]
- Brambila-Tapia AJ. MDR1 (ABCB1) polymorphisms: functional effects and clinical implications. Rev Invest Clin. 2013;65:445–54. [PubMed] [Google Scholar]
- Ivanov M, Kacevska M, Ingelman-Sundberg M. Epigenomics and interindividual differences in drug response. Clin Pharmacol Ther. 2012;92:727–36. doi: 10.1038/clpt.2012.152. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Hildmann T, Novik KL, Lewin J, Tost J, Cox AV, Andrews TD, Howe KL, Otto T, Olek A, Fischer J, Gut IG, Berlin K, Beck S. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, Coutifaris C, Sapienza C. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet. 2010;6:e1001033. doi: 10.1371/journal.pgen.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito J, Hirota T, Kikunaga N, Otsubo K, Ieiri I. Interindividual differences in placental expression of the SLC22A2 (OCT2) gene: relationship to epigenetic variations in the 5′-upstream regulatory region. J Pharm Sci. 2011;100:3875–83. doi: 10.1002/jps.22595. [DOI] [PubMed] [Google Scholar]
- Pirazzini C, Giuliani C, Bacalini MG, Boattini A, Capri M, Fontanesi E, Marasco E, Mantovani V, Pierini M, Pini E, Luiselli D, Franceschi C, Garagnani P. Space/population and time/age in DNA methylation variability in humans: a study on IGF2/H19 locus in different Italian populations and in mono- and di-zygotic twins of different age. Aging (Albany NY) 2012;4:509–20. doi: 10.18632/aging.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–8. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Baker EK, Johnstone RW, Zalcberg JR, El-Osta A. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene. 2005;24:8061–75. doi: 10.1038/sj.onc.1208955. [DOI] [PubMed] [Google Scholar]
- El-Khoury V, Breuzard G, Fourré N, Dufer J. The histone deacetylase inhibitor trichostatin A downregulates human MDR1 (ABCB1) gene expression by a transcription-dependent mechanism in a drug-resistant small cell lung carcinoma cell line model. Br J Cancer. 2007;97:562–73. doi: 10.1038/sj.bjc.6603914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K, Hembruff SL, Sprowl JA, Parissenti AM. The temporal relationship between ABCB1 promoter hypomethylation, ABCB1 expression and acquisition of drug resistance. Pharmacogenomics J. 2010;10:489–504. doi: 10.1038/tpj.2010.1. [DOI] [PubMed] [Google Scholar]
- Mishra DK, Chen Z, Wu Y, Sarkissyan M, Koeffler HP, Vadgama JV. Global methylation pattern of genes in androgen-sensitive and androgen-independent prostate cancer cells. Mol Cancer Ther. 2010;9:33–45. doi: 10.1158/1535-7163.MCT-09-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa T, Nutahara K, Higashihara E. Association of circulating tumor cells with tumor-related methylated DNA in patients with hormone-refractory prostate cancer. Int J Urol. 2010;17:466–75. doi: 10.1111/j.1442-2042.2010.02502.x. [DOI] [PubMed] [Google Scholar]
- Onda K, Suzuki R, Tanaka S, Oga H, Oka K, Hirano T. Decitabine, a DNA methyltransferase inhibitor, reduces P-glycoprotein mRNA and protein expressions and increases drug sensitivity in drug-resistant MOLT4 and Jurkat cell lines. Anticancer Res. 2012;32:4439–44. [PubMed] [Google Scholar]
- Dejeux E, Rønneberg JA, Solvang H, Bukholm I, Geisler S, Aas T, Gut IG, Børresen-Dale AL, Lønning PE, Kristensen VN, Tost J. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Mol Cancer. 2010;9:68. doi: 10.1186/1476-4598-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CJ, Wang F, Ren MF, Mi YJ, Yan YY, To KK, Dai CL, Wang YS, Chen LM, Tong XZ, Liang YJ, Fu LW. Up-regulation of ABCB1/P-glycoprotein by escaping promoter hypermethylation indicates poor prognosis in hematologic malignancy patients with and without bone marrow transplantation. Leuk Res. 2011;35:73–9. doi: 10.1016/j.leukres.2010.04.019. [DOI] [PubMed] [Google Scholar]
- Toth M, Boros IM, Balint E. Elevated level of lysine 9-acetylated histone H3 at the MDR1 promoter in multidrug-resistant cells. Cancer Sci. 2012;103:659–69. doi: 10.1111/j.1349-7006.2012.02215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique R, Oliveira AI, Costa VL, Baptista T, Martins AT, Morais A, Oliveira J, Jerónimo C. Epigenetic regulation of MDR1 gene through post-translational histone modifications in prostate cancer. BMC Genomics. 2013;17:898. doi: 10.1186/1471-2164-14-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyasu H, Goto-Koshino Y, Fujino Y, Ohno K, Tsujimoto H. Epigenetic regulation of the ABCB1 gene in drug-sensitive and drug-resistant lymphoid tumour cell lines obtained from canine patients. Vet J. 2014;199:103–9. doi: 10.1016/j.tvjl.2013.10.022. [DOI] [PubMed] [Google Scholar]
- Cascorbi I, Gerloff T, Johne A, Meisel C, Hoffmeyer S, Schwab M, Schaeffeler E, Eichelbaum M, Brinkmann U, Roots I. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169–74. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- Baker EK, El-Osta A. Epigenetic regulation of multidrug resistance 1 gene expression: profiling CpG methylation status using bisulphite sequencing. Methods Mol Biol. 2010;596:183–98. doi: 10.1007/978-1-60761-416-6_9. [DOI] [PubMed] [Google Scholar]
- Gireesh T, Nair PP, Sudhakaran PR. Studies on the bioavailability of the provitamin A carotenoid, beta-carotene, using human exfoliated colonic epithelial cells. Br J Nutr. 2004;92:241–5. doi: 10.1079/BJN20041175. [DOI] [PubMed] [Google Scholar]
- Kamra A, Kessie G, Chen JH, Kalavapudi S, Shores R, McElroy I, Gireesh T, Sudhakaran PR, Dutta SK, Nair PP. Exfoliated colonic epithelial cells: surrogate targets for evaluation of bioactive food components in cancer prevention. J Nutr. 2005;135:2719–22. doi: 10.1093/jn/135.11.2719. [DOI] [PubMed] [Google Scholar]
- Gireesh T, Sudhakaran PR. In vitro uptake of β-carotene by human exfoliated colonic epithelial cells. Int J Food Sci Nutr. 2009;60:109–18. doi: 10.1080/09637480701610525. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Li SJ, Liu G, Jia J, Miao Y, Gu S, Miao P, Shi X, Wang Y, Yu C. Therapeutic monitoring of serum digoxin for patients with heart failure using a rapid LC-MS/MS method. Clin Biochem. 2010;43:307–13. doi: 10.1016/j.clinbiochem.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Hu CY, Mohtat D, Yu Y, Ko YA, Shenoy N, Bhattacharya S, Izquierdo MC, Park AS, Giricz O, Vallumsetla N, Gundabolu K, Ware K, Bhagat TD, Suzuki M, Pullman J, Liu XS, Greally JM, Susztak K, Verma A. Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival. Clin Cancer Res. 2014;20:4349–60. doi: 10.1158/1078-0432.CCR-14-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Ghosh S. Epigenetics: differential DNA methylation in mammalian somatic tissues. FEBS J. 2008;275:1617–23. doi: 10.1111/j.1742-4658.2008.06330.x. [DOI] [PubMed] [Google Scholar]
- Iyengar V, Albaugh GP, Lohani A, Nair PP. Human stools as a source of viable colonic epithelial cells. FASEB J. 1991;5:2856–9. doi: 10.1096/fasebj.5.13.1655550. [DOI] [PubMed] [Google Scholar]
- Albaugh GP, Iyengar V, Lohani A, Malayeri M, Bala S, Nair PP. Isolation of exfoliated colonic epithelial cells, a novel, non-invasive approach to the study of cellular markers. Int J Cancer. 1992;52:347–50. doi: 10.1002/ijc.2910520303. [DOI] [PubMed] [Google Scholar]
- Labianca R, Merelli B. Screening and diagnosis for colorectal cancer: present and future. Tumori. 2010;96:889–901. [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Scoccianti C, Ricceri F, Ferrari P, Cuenin C, Sacerdote C, Polidoro S, Jenab M, Hainaut P, Vineis P, Herceg Z. Methylation patterns in sentinel genes in peripheral blood cells of heavy smokers: influence of cruciferous vegetables in an intervention study. Epigenetics. 2011;6:1114–9. doi: 10.4161/epi.6.9.16515. [DOI] [PubMed] [Google Scholar]
- Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, Putter H, Slagboom PE, Heijmans BT. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J. 2010;24:3135–44. doi: 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16:547–54. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- Schneider E, Pliushch G, El Hajj N, Galetzka D, Puhl A, Schorsch M, Frauenknecht K, Riepert T, Tresch A, Müller AM, Coerdt W, Zechner U, Haaf T. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38:3880–90. doi: 10.1093/nar/gkq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- Ballestar E. Epigenetics lessons from twins: prospects for autoimmune disease. Clin Rev Allergy Immunol. 2010;39:30–41. doi: 10.1007/s12016-009-8168-4. [DOI] [PubMed] [Google Scholar]
- Portha B, Fournier A, Kioon MD, Mezger V, Movassat J. Early environmental factors, alteration of epigenetic marks and metabolic disease susceptibility. Biochimie. 2014;97:1–15. doi: 10.1016/j.biochi.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of disease susceptibility. Transl Res. 2015;165:12–7. doi: 10.1016/j.trsl.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani C, Bacalini MG, Sazzini M, Pirazzini C, Franceschi C, Garagnani P, Luiselli D. The epigenetic side of human adaptation: hypotheses, evidences and theories. Ann Hum Biol. 2015;42:1–9. doi: 10.3109/03014460.2014.961960. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly S, Paine MF. P-glycoprotein increases from proximal to distal regions of human small intestine. Pharm Res. 2003;20:1595–9. doi: 10.1023/a:1026183200740. [DOI] [PubMed] [Google Scholar]
- Zimmermann C, Gutmann H, Hruz P, Gutzwiller JP, Beglinger C, Drewe J. Mapping of multidrug resistance gene 1 and multidrug resistance-associated protein isoform 1 to 5 mRNA expression along the human intestinal tract. Drug Metab Dispos. 2005;33:219–24. doi: 10.1124/dmd.104.001354. [DOI] [PubMed] [Google Scholar]
- Ieiri I. Functional significance of genetic polymorphisms in P-glycoprotein (MDR1, ABCB1) and breast cancer resistance protein (BCRP, ABCG2) Drug Metab Pharmacokinet. 2012;27:85–105. doi: 10.2133/dmpk.dmpk-11-rv-098. [DOI] [PubMed] [Google Scholar]
- Ehle M, Patel C, Giugliano RP. Digoxin: clinical highlights: a review of digoxin and its use in contemporary medicine. Crit Pathw Cardiol. 2011;10:93–8. doi: 10.1097/HPC.0b013e318221e7dd. [DOI] [PubMed] [Google Scholar]
- Nader AM, Foster DR. Suitability of digoxin as a P-glycoprotein probe: implications of other transporters on sensitivity and specificity. J Clin Pharmacol. 2014;54:3–13. doi: 10.1002/jcph.200. [DOI] [PubMed] [Google Scholar]
- Ma JD, Tsunoda SM, Bertino JS, jr, Trivedi M, Beale KK, Nafziger AN. Evaluation of in vivo P-glycoprotein phenotyping probes: a need for validation. Clin Pharmacokinet. 2010;49:223–37. doi: 10.2165/11318000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, Brinkmann U. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RB, Leake BF, Choo EF, Dresser GK, Kubba SV, Schwarz UI, Taylor A, Xie HG, McKinsey J, Zhou S, Lan LB, Schuetz JD, Schuetz EG, Wilkinson GR. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70:189–99. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- Gerloff T, Schaefer M, Johne A, Oselin K, Meisel C, Cascorbi I, Roots I. Br J Clin Pharmacol. 2002;54:610–6. doi: 10.1046/j.1365-2125.2002.01691.x. . MDR1 genotypes do not influence the absorption of a single oral dose of 1 mg digoxin in healthy white males. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal K, Weinbrenner A, Giessmann T, Stuhr M, Franke G, Zschiesche M, Oertel R, Terhaag B, Kroemer HK, Siegmund W. Oral bioavailability of digoxin is enhanced by talinolol: evidence for involvement of intestinal P-glycoprotein. Clin Pharmacol Ther. 2000;68:6–12. doi: 10.1067/mcp.2000.107579. [DOI] [PubMed] [Google Scholar]
- Currie GM, Wheat JM, Kiat H. Pharmacokinetic considerations for digoxin in older people. Open Cardiovasc Med J. 2011;5:130–5. doi: 10.2174/1874192401105010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1 Subjects characteristics


