Abstract
Aim of the study
Matrix metalloproteinases (MMPs) are a zinc-dependant endopeptidase family that can degrade extracellular matrix components. Their dysregulation has been proven in several diseases, including cancer. Genetic variations in MMP promoter regions can alter their expression. The aim of the present study is to investigate the correlation of MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G) single nucleotide polymorphisms (SNPs) with oesophageal squamous cell carcinoma (ESCC) initiation and progression susceptibility in Iranian patients.
Material and methods
MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G) SNPs were detected using polymerase chain reaction-restriction fragment-length polymorphism (PCR-RFLP) technique in 70 patients and 60 healthy controls. The genotypes and allele distributions were statistically compared in patients and controls. The correlation of MMP-2 (-1306C/T) and MMP-9 (-1562C/T) polymorphisms with clinicopathological features were investigated in 53 patients.
Results
No statistically significant differences were observed in genotype and allele frequencies of MMP-2 (-1306C/T) and MMP-9 (-1562C/T) between patients and controls (p > 0.05). In addition, no relevance was observed in MMP-2 (-1306C/T) and MMP-9 (-1562C/T) SNPs and clinicopathological features. There was no nucleotide variation in MMP-12 (-82) in the case and control groups.
Conclusions
This study indicates that these three SNPs may have no significant association in ESCC risk in Iranian patients.
Keywords: matrix metalloproteinase, single nucleotide polymorphism, esophagus squamous cell carcinoma
Introduction
Oesophageal cancer is the world's eighth most prevalent malignancy and the sixth most common cause of cancer death [1]. It is characterised by poor prognosis, rapid progression, and low survival – 5-year survival is less than 5% [2]. Histologically, oesophageal carcinoma occurs in two pathological forms: oesophagus squamous cell carcinoma (ESCC) in the middle or upper part of the oesophagus, and gastric cardia adenocarcinoma (GCA) in the lower part or junction of the oesophagus and the stomach. The incidence of oesophageal neoplasia varies considerably from high-risk areas like Africa and some parts of Asia to low-risk areas in western and central Africa and Central America [3]. North and northeast of Iran are regions with high occurrence of oesophageal cancer, and ESCC encompasses > 90% of all oesophageal malignancies in these areas [4]. Its rapid progression and invasive trait give the fatal property to ESCC. Tumour invasion and metastasis are multistep processes that are greatly facilitated by protease activities for destruction and remodelling of extracellular matrix (ECM) and basement membrane barriers. Studies in the past 50 years have considered Matrix metalloproteinases (MMPs) as primary molecules aiding tumour cells in metastasis processes [5]. These degradative enzymes are a family of Zn2+-dependent endopeptidases totally capable of decomposing all extracellular components [6]. Matrix metalloproteinases can implicate in tumour development by degradation of ECM and also by release of several ECM-bound biomolecules such as growth factors and agents involved in angiogenesis [7]. Among 24 members of the MMP family, MMP-2 and MMP-9, known as Gelatinases, by degradation of type IV collagen, the major component of basement membrane, have crucial roles in the early stages of tumour invasion [8]. Gelatinases also participate in tumour angiogenesis via enhancement of the bioavailability of pro-angiogenic factors such as VEGF [9, 10]. MMP-12, human macrophage metalloelastase, degrades elastin, type IV collagen, opening the way for migration of tumour cells [11].
Matrix metalloproteinase expression, initially regulated at the transcriptional level, and alteration in cis element binding sites in their promoter can convert their expression. Multiple SNPs have been identified on MMP-2, MMP-9, and MMP-12 promoters [6]. Several studies performed in different populations of the world have demonstrated the influence of MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G) polymorphisms on different malignant cancers, such as digestive cancers [12, 13]. In addition, some studies have shown overexpression of MMP-2 and MMP-9 in ESCC tumour cells and their association with tumour invasion [14, 15]. Consequently, the present study was planned to evaluate whether these three polymorphisms are good prognostic markers for recognition and screening of susceptible individuals in high-risk areas of Iran.
Material and methods
Study participants
In this study, polymorphisms were analysed in two groups: case and control. The case group comprised 70 patients with ESCC, 37 (52.9%) men and 33 (47.1%) women, aged 34-81 years with a mean age of 59.5 years. Pathological data of 55 patients were available and are described in Table 1. Control participants included 60 healthy individuals having no history or diagnosis of cancer or any serious disease, 30 (50%) men and 30 (50%) women, aged 28–75 with a mean age of 46.53 years.
Table 1.
Pathological data of oesophageal squamous cell carcinoma patients
| Case number | Differentiation | LMN |
|---|---|---|
| 1 | * | + |
| 2 | Moderately differentiated | + |
| 3 | Well differentiated | – |
| 4 | Moderately differentiated | – |
| 5 | Well differentiated | – |
| 6 | Poorly differentiated | + |
| 7 | Moderately differentiated | + |
| 8 | Poorly differentiated | – |
| 9 | Moderately differentiated | – |
| 10 | Poorly differentiated | + |
| 11 | Poorly differentiated | + |
| 12 | Moderately to poorly differentiated | – |
| 13 | Poorly differentiated | + |
| 14 | Moderately differentiated | + |
| 15 | Well differentiated | – |
| 16 | Moderately differentiated | – |
| 17 | Well differentiated | + |
| 18 | Moderately differentiated | + |
| 19 | Moderately differentiated | + |
| 20 | Moderately differentiated | – |
| 21 | Poorly differentiated | * |
| 22 | Moderately differentiated | – |
| 23 | Well differentiated | – |
| 24 | Poorly differentiated | – |
| 25 | Moderately differentiated | + |
| 26 | Moderately differentiated | + |
| 27 | Well differentiated | + |
| 28 | Well differentiated | + |
| 29 | Moderately to poorly differentiated | – |
| 30 | Well differentiated | + |
| 31 | Poorly differentiated | + |
| 32 | Moderately differentiated | + |
| 33 | Well differentiated | + |
| 34 | Moderately differentiated | + |
| 35 | Well differentiated | + |
| 36 | Well differentiated | + |
| 37 | Moderately differentiated | + |
| 38 | Moderately differentiated | – |
| 39 | Well differentiated | + |
| 40 | Moderately differentiated | + |
| 41 | Poorly differentiated | + |
| 42 | Moderately differentiated | + |
| 43 | Moderately differentiated | + |
| 44 | Well differentiated | + |
| 45 | Well differentiated | – |
| 46 | Well differentiated | – |
| 47 | Well differentiated | + |
| 48 | Moderately differentiated | + |
| 49 | Moderately differentiated | – |
| 50 | Well differentiated | + |
| 51 | Well differentiated | + |
| 52 | Moderately differentiated | + |
| 53 | Moderately differentiated | + |
| 54 | Well differentiated | * |
| 55 | Well differentiated | + |
data not available
LMN – lymph nod metastasis
The study was approved by the Ethics Committee of Tehran University of Medical Sciences.
DNA extraction
Tumour tissues were collected immediately after surgery in liquid nitrogen and stored at –70°C until the time of use. In order to extract DNA, the tissue specimens were dissected and adipose and connective tissues were removed. Then the specimens were fragmented and suspended in buffer containing 0.15 mM NaCl, 10 mM EDTA, and 10 mM Tris-HCl, pH 7.5, and the lysis step was followed by the addition of 50–100 µg/ml proteinase K and SDS with a final concentration of 0.5%. The lysate was incubated over night at 37°C. It was then mixed with an equal volume of phenol and after gentle mixing, centrifuged at 14 000 rpm for five minutes. The supernatant was mixed with equal volumes of chloroform and isoamyl alcohol (24: 1) and centrifuged at 14 000 rpm for five minutes. Subsequently, the supernatant was diluted with 2.2 volume of absolute ethanol. The precipitated DNA was washed with 70% ethanol. It was dried at room temperature. Finally it was dissolved in 100–200 µl TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5) and stored at –20°C [16]. In order to collect saliva from healthy volunteers, they were given screw-top containers and asked about an hour after brushing their teeth to spit vigorously about 30 ml into the containers. Then samples were kept at –20°C until the time of use. DNA was extracted from saliva by using the method of Lum and Marchand (1998) [17]. The collected DNA concentrations were evaluated by UV spectroscopy method, and then for evaluation of their molecular weight status they were run on 0.8% agarose gel stained with ethidium bromide.
MMP-2 (-1306C/T), MMP-9 (-1562C/T) and MMP-12 (-82A/G) genotyping
The MMP-2, MMP-9, and MMP-12 SNPs were determined using PCR-Restriction fragment length polymorphism (PCR-RFLP) assay. The amplification primers, their annealing temperatures, and the size of each fragment is shown in Table 2. Each PCR was performed in a total volume of 50 µl, containing 12.5 ng DNA, 5 µl 10X PCR buffer, 3.75 mM MgCl2, 0.31 mM dNTPs, 2.5 U Taq DNA polymerase, and 0.62 µM forward and reveres primers. The PCR thermo cycling conditions for MMP-2 and MMP-12 were as follows: 5 minutes at 94°C at initial step followed by 35 cycles of 45 seconds at 94°C, 45 seconds at 58°C for MMP-2 and 53°C for MMP-12, 45 seconds at 72°C, with the final step at 72°C for 10 minutes. The cycling conditions for MMP-9 were started at 95°C for 5 minutes, followed by 35 cycles of 1 minute at 93°C, 30 seconds at 61.8°C, 50 seconds at 72°C, and completed extension at 72°C for 5 minutes. 10 µl of each PCR product was digested at 37°C overnight by 10 U of appropriate restriction enzymes. FspBl, Sphl, and Pvull were used for MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G), respectively. Products were separated on 3% agarose gel staining with ethidium bromide. The fragment lengths of digested products are summarised in Table 3.
Table 2.
Polymorphisms, primers, annealing temperatures, and size of fragments
| Polymorphism | Primer | Annealing temperature (°C) | Size (bp) |
|---|---|---|---|
| MMP-2 (-1306C/T) | 5′-CTTCCTAGGCTGGTCCTTACTGA-3′ Forward 5′-CTGAGACCTGAAGAGCTAAAGAGCT-3′ Reveres |
66 | 193 bp |
| MMP-9 (-1562C/T) | 5′-GCCTGGCACATAGTAGGCCC-3′ Forward 5′-TCTCTCAGCCGGCATC-3′ Reveres |
65 | 435 bp |
| MMP-12 (-82A/G) | 5′-GAGATAGTCAAGGGATGATATCA-3′ Forward 5′-AAGAGCTCCAGAAGCAGTGG-3′ Reveres |
60 | 199 bp |
Table 3.
Restriction enzymes and fragment lengths
| Polymorphism | Restriction enzyme | Fragment length |
|---|---|---|
| MMP-2 (-1306C/T) | FspBl | 188+5 bp C 162+26+5 bp T |
| MMP-9 (-1562C/T) | Sphl | 247+188 bp T 435 bp C |
| MMP-12 (-82A/G) | Pvull | 199 bp A 175+24 bp G |
Statistical analysis
Statistical analysis was carried out using the SPSS 18 software package. The difference in distribution of alleles and genotypes between patients and controls was compared by two-sided chi-square test. The association of genotype and allele distributions with ESCC risk was evaluated by OR (odds ratio) with 95% confidence interval (CI). The correlation between genotypes and clinicopathological characteristics, the tumour differentiation, and lymph node metastasis, was analysed by Fisher exact test. A 5% probability value was considered significant.
Results
The detection of MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G) polymorphisms was performed well. The results in Table 4 show that this study did not demonstrate any statistically significant differences in allele and genotype distributions in MMP-2 (-1306C/T) or MMP-9 (-1562C/T) between ESCC patients and controls (p > 0.05). The statistical analysis of these two polymorphisms did not show any correlation between genotype frequencies and ESCC risk (OR = 0.862, 95% CI: 0.385–1.927 for MMP-2 and OR = 1.092, 95% CI: 0.541–2.206 for MMP-9). Pathological information, tumour differentiation, and lymph node metastasis of 55 patients was available in the current study; Fisher exact test was performed to determine the association of genotype frequency with tumour differentiation and lymph node metastasis (Table 5). There was no association between MMP-2 (-1306C/T) and MMP-9 (-1562C/T) genotype frequencies and tumour development (p > 0.05).
Table 4.
Genotype and allele distributions of MMP-2 and MMP-9 single nucleotide polymorphisms.
| Controls n (%) | Patients n (%) | OR (95% CI) | P value (2-tail) | |
|---|---|---|---|---|
| MMP-2 (-1306 C/T)† | ||||
| CC | 46 (76.7) | 51 (73.9) | ||
| CT | 10 (16.7) | 17 (24.6) | ||
| TT | 4 (6.7) | 1 (1.4) | ||
| (CC, CT+TT) | 0.862 (0.385–1.927) | 0.718 χ2 = 0.1305 | ||
| C | 102 (85) | 119 (86.2) | ||
| T | 18 (15) | 19 (13.8) | ||
| C, T | 1.105 (0.55–2.219) | 0.778 χ2 = 0.0792 | ||
| MMP-9(-1562C/T)‡ | ||||
| CC | 34 (56.7) | 40 (58.8) | ||
| CT | 23 (38.3) | 25 (36.8) | ||
| TT | 3 (5) | 3 (4.4) | ||
| (CC, CT+TT) | 1.092 (0.541–2.206) | 0.805 χ2 = 0.0608 | ||
| C | 91 (75.8) | 105 (77.2) | ||
| T | 29 (24.2) | 31 (22.8) | ||
| C, T | 1.079 (0.605–1.926) | 0.796 χ2 = 0.0669 | ||
One tumour sample was missed
Two tumour samples were missed
Table 5.
Association of genotype distributions with clinicopathological characteristics
| MMP-2 (-1306C/T) | MMP-9 (-1562C/T) | |||
|---|---|---|---|---|
| Differentiation | CC | CT, TT | CC | CT, TT |
| Well | 15 | 4 | 13 | 7 |
| Moderately | 17 | 6 | 11 | 10 |
| Poorly | 7 | 4 | 8 | 3 |
| p value (2-tail)* | 0.664 | 0.575 | ||
| LMN | ||||
| Positive | 26 | 10 | 22 | 14 |
| Negative | 13 | 4 | 10 | 6 |
| p value (2-tail)* | 1.00 | 1.00 | ||
LMN – lymph node metastasis
p value for Fisher's exact test
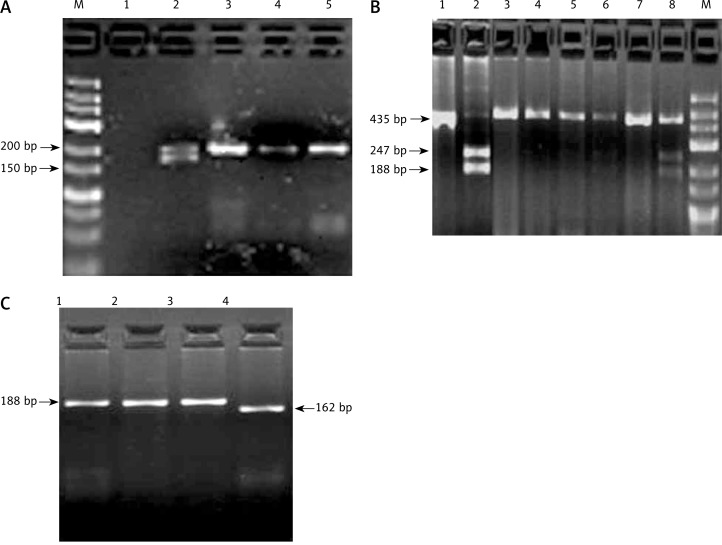
No single nucleotide polymorphisms were detected in MMP-12 (-82), and all the samples showed A allele in this site. The separated bands of each polymorphism on agarose gel are shown in Fig. 1.
Fig. 1.
Separated bands of MMP-2 (-1306C/T), MMP-9 (-1562C/T) polymorphisms on agarose gel. A) MMP-2 (-1306C/T) genotypes. Lane M DNA marker, lane 2 CT genotype, lanes 3, 4, 5 CC genotype. B) MMP-9 (-1562C/T) genotypes. Lane M DNA marker, lanes 1, 3, 4, 5, 7CC genotype, lane 2 TT genotype, lanes 6, 8 CT genotype. C) MMP-2 (-1306C/T) genotypes, lane 1, 2, 3 CC genotype, lane 4 TT genotype
Discussion
Single nucleotide polymorphisms (SNPs) are the most frequently inherited sequence variations in a particular gene, and they occur in every 100–200 base pairs [18]. Some SNPs on MMP-2 promoter have been identified; among these, MMP-2 (-1306C/T) has been reported by several studies to be more involved in many cancer types [19]. In MMP-2 at -1306, substitution of T allele instead of C disrupts the Sp1-binding site and leads to lower promoter activity, and investigations have shown that the presence of C allele in this region enhances the promoter activity by as much as two times [19, 20]. In our study the genotype distribution of MMP-2 (-1306C/T) was CC 73%, CT 24%, and TT 1.4% in patients and CC 76%, CT 16.7%, and TT 6.7% in the control group, i.e. there was no statistically significant difference between patients and controls (p > 0.05). Various studies on MMP-2 (-1306C/T) SNP and risk of different types of cancer did not report homogenous results in different populations of the world. Studies in China reported that, compared with CT and TT genotypes, CC genotype significantly increases susceptibility to oesophageal, lung, and breast cancers [12, 21, 22]. Additionally, a study in China on gastric cardia adenocarcinoma found that patients with CC genotype are at three-fold higher risk than those with CT and TT genotypes [13]. However, our investigation is in agreement with some studies from Japan, Turkey, and Sweden, which did not find any correlation between MMP-2 (-1306C/T) polymorphism and cancer [23–25]. Two Chinese meta-analysis studies demonstrated that T allele frequency is significantly lower in the Asian population (13.6%) than in Europeans (23.3%), which is in agreement with the results obtained from our study (13–15%) [26, 27].
MMP-9 C to T transition at -1562 has been proven to decrease the affinity of the transcriptional repressive protein to bind to its site at MMP-9 promoter, which subsequently causes higher promoter function. The current study did not find any significant differences in distribution of genotypes and alleles in MMP-9 (-1562C/T) between patients and healthy individuals (CC 58.8%, CT 36.8%, and TT 4.4% and CC 56.7%, CT 38.3%, and TT 5% in patients and controls, respectively). Unfortunately a few investigations were carried out analysing the association of MMP-9 (-1562C/T) polymorphism with oesophageal cancer risk, but the outcome obtained in the present study was consistent with other studies on different cancer types in Sweden, China, Tunisia, and France [25, 28–30].
We also analysed the association of these two polymorphisms with pathological characteristics, tumour differentiation, and lymph node metastasis; our findings did not show any association between genotype distribution of MMP-2 and MMP-9 SNPs and these pathological features.
The MMP-12 is located at chromosome 11q22, and the A to G transition at the -82 region, the recognition site of AP-1 transcriptional factor, decreases the binding potential of AP-1 protein and modifies the MMP-12 expression in vitro [31]. All the samples in this study showed A allele in MMP-12 (-82) and no nucleotide variation was identified in MMP-12 (-82). Some previous studies have shown the influence of GG genotype in MMP-12 (-82) site on the risk of some cancers; one study in the US indicated that GG genotype in MMP-12 (-82) increased the risk of invasive bladder cancer about 4.5 fold [32]. The high frequency of A allele at position -82 has been reported in some studies. In the previous research in northern China the frequency of A allele has been reported to be more than 90% [12]. This study suggested that the occurrence of MMP-12 (-82) A to G transition may differ between variant populations.
Both environmental and genetic factors play roles in oesophageal squamous cell carcinoma outbreak [33]. Since individuals exposed to environmental risk factors show different susceptibility to ESCC, it seems that some genetic variations like genetic polymorphisms have been associated with oesophageal cancer development in Iran, and the discovery of influential polymorphisms on ESCC can be valuable markers for the identification of predisposed individuals in high-risk areas. Consequently, for the first time in this manuscript we analysed the role of MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G) polymorphisms in the development of ESCC in Iran.
We concluded that MMP-2 (-1306C/T), MMP-9 (-1562C/T), and MMP-12 (-82A/G) single nucleotide polymorphisms are not useful prognostic markers in the identification and screening of susceptible individuals in areas with high occurrence of oesophageal squamous cell carcinoma. Even if a larger sample size results in significance, it may still not be useful enough for the identification and screening of ESCC.
Acknowledgments
We would like to thank Dr Nasrin Shojaee for her worthwhile advice.
The authors declare no conflict of interest.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Kollarova H, Machova L, Horakova D, Janoutova G, Janout V. Epidemiology of esophageal cancer – an overview article. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2007;151:17–28. doi: 10.5507/bp.2007.003. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Kamangar F, Malekzadeh R, Dawsey SM, Saidi F. Esophageal Cancer in Northeastern Iran: A Review. Arch Iran Med. 2007;10:70–82. [PubMed] [Google Scholar]
- 5.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 6.Fanjul-Fernandez M, Folgueras AR, Cabrera S, Lopez-Otin C. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. BBA. 2010;1803:3–19. doi: 10.1016/j.bbamcr.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and theirpharmacological targeting. FEBS J. 2010;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 8.Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rundhaug Matrix metalloproteinases, angiogenesis, and cancer. Clin Cancer Res. 2003;9:551–4. [PubMed] [Google Scholar]
- 10.Klein T, Bischoff R. Physiology and pathology of matrix metalloproteases. Amino Acids. 2010;10:689–726. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amălinei C, Caruntu Id, Giuşcă Se, Anca Bălan R. Matrix metalloproteinases involvement in pathologic conditions. Rom J Morphol Embryol. 2010;51:215–28. [PubMed] [Google Scholar]
- 12.Li Y, Sun D, Duan Y, Zhang X, Wang N. Association of functional polymorphisms in MMPs genes with gastric cardia adenocarcinoma and esophageal squamous cell carcinoma in high incidence region of North China. Mol Biol Rep. 2010;37:197–205. doi: 10.1007/s11033-009-9593-4. [DOI] [PubMed] [Google Scholar]
- 13.Miao X, Yu C, Tan W, Xiong P, Liang G, Lu W, Lin D. A Functional Polymorphism in the matrix metalloproteinase-2 gene promoter (-1306C/T) is associated with risk of development but not metastasis of gastric cardia adenocarcinoma. Cancer Res. 2003;63:3987–90. [PubMed] [Google Scholar]
- 14.Li Y, Ma J, Guo Q, Duan F, Tang F, Zheng P, Zhao Z, Lu G. Overexpression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:664–7. doi: 10.1111/j.1442-2050.2008.00928.x. [DOI] [PubMed] [Google Scholar]
- 15.Samantaray S, Sharma R, Chattopadhyaya TK, Gupta SD, Ralhan R. Increased expression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2004;130:37–44. doi: 10.1007/s00432-003-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane DP, Midgley C, Hupp TR, Lu X, et al. On the regulation of the p53 tumor suppressor, and its role in the cellular response to DNA damage. Philos Trans R Soc Lond B Biol Sci. 1995;347:83–7. doi: 10.1098/rstb.1995.0013. [DOI] [PubMed] [Google Scholar]
- 17.Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–24. [PubMed] [Google Scholar]
- 18.Zhai X, Wang H, Zhu X, Miao H, et al. Gene polymorphisms of ABC transporters are associated with clinical outcomes in children with acute lymphoblastic leukemia. Arch Med Sci. 2012;4:659–71. doi: 10.5114/aoms.2012.30290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhary AK, Singh M, Bharti AC, Asotra K, Sundaram S, Mehrotra R. Genetic polymorphisms of matrix metalloproteinases and their inhibitors in potentially malignant and malignant lesions of the head and neck. J Biomed Sci. 2010;17:1–13. doi: 10.1186/1423-0127-17-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price SJ, Greaves DR, Watkins H. Identification of Novel, Functional Genetic Variants in the Human Matrix Metalloproteinase-2 Gene. J Biol Chem. 2001;276:7549–58. doi: 10.1074/jbc.M010242200. [DOI] [PubMed] [Google Scholar]
- 21.Yu C, Pan K, Xing D, Liang G, Tan W, Zhang L, Lin D. Correlation between a single nucleotide polymorphism in the matrix metalloproteinase-2 promoter and risk of lung cancer. Cancer Res. 2002;62:6430–3. [PubMed] [Google Scholar]
- 22.Zhou Y, Yu C, Miao X, Tan W, Liang G, Xiong P, Sun T, Lin D. Substantial reduction in risk of breast cancer associated with genetic polymorphisms in the promoters of the matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 genes. Carcinogenesis. 2004;25:399–404. doi: 10.1093/carcin/bgh020. [DOI] [PubMed] [Google Scholar]
- 23.Ohtani H MNaMY. Functional polymorphisms in the promoter regions of matrix metalloproteinase-2, -3, -7, -9 and TNF-alpha genes, and the risk of colorectal neoplasm in Japanese. Yonago Acta Medica. 2009;52:47–56. [Google Scholar]
- 24.Ayşegül B, Veysi GH, Muzaffer M, Irfan D, Azra A, Hulyam K. Is a single nucleotide polymorphism a risk factor for lung cancer in the matrix metalloproteinase-2 promoter? Mol Biol Rep. 2011;38:1469–74. doi: 10.1007/s11033-010-0253-5. [DOI] [PubMed] [Google Scholar]
- 25.Elander N, Söderkvist P, Fransén K. Matrix metalloproteinase (MMP) -1, -2, -3 and -9 promoter polymorphisms in colorectal cancer. Anticancer Res. 2006;26(1B):791. [PubMed] [Google Scholar]
- 26.Langers AM, Verspaget HW, Hommes DW, Sier CF. Single-nucleotide polymorphisms of matrix metalloproteinases and their inhibitors in gastrointestinal cancre. World J Gastrointest Oncol. 2011;3:79–98. doi: 10.4251/wjgo.v3.i6.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng B, Cao L, Ma X, Wang W, Wang D, Yu L. Meta-analysis of association between matrix metalloproteinases 2, 7 and 9 promoter polymorphisms and cancer risk. Mutagenesis. 2010;25:371–9. doi: 10.1093/mutage/geq015. [DOI] [PubMed] [Google Scholar]
- 28.Xu E, Xia X, Lu B, Huang Q. Association of matrix metalloproteinase-2 and -9 promoter polymorphisms with colorectal cancer in Chinese. Mol Carcinog. 2007;46:924–9. doi: 10.1002/mc.20323. [DOI] [PubMed] [Google Scholar]
- 29.Nasr HB, Mestiri S, Chahed K, Bouaouina N, et al. Matrix metalloproteinase-1 (-1607) 1G/2G and -9 (–1562) C/T promoter polymorphisms: Susceptibility and prognostic implications in nasopharyngeal carcinomas. Clin Chim Acta. 2007;384:57–63. doi: 10.1016/j.cca.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Rollin J, Regina S, Vour'h P, Iochmann S. Influence of MMP-2 and MMP-9 promoter polymorphisms on gene expression and clinical outcome of non-small cell lung cancer. Lung Can. 2006;56:273–80. doi: 10.1016/j.lungcan.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Jormsjö S, Ye S, Moritz J, et al. Allele-specific regulation of matrix metalloproteinase-12 gene activity is associated with coronary artery luminaldimensions in diabetic patients with manifest coronary artery disease. Circ Res. 2000;86:998–1003. doi: 10.1161/01.res.86.9.998. [DOI] [PubMed] [Google Scholar]
- 32.Kader AK, Shao L, Dinney CP, et al. Matrix metalloproteinase polymorphisms and bladder cancer risk. Cancer Res. 2006;66:11644–8. doi: 10.1158/0008-5472.CAN-06-1212. [DOI] [PubMed] [Google Scholar]
- 33.Sadjadi A, Marjani H, Semnani S, Nasseri-Moghaddam S. Esophageal cancer in Iran: a review. MEJC. 2010;1:5–14. [Google Scholar]