Abstract
To understand whether the distinct VHDJH gene utilization by natural polyreactive Abs reflects the developmentally restricted Ig VHDJH rearrangements putatively expressed by B-1 cells, we generated 11 (8 IgM, 1 IgG3, 2 IgA1), 7 (6 IgM, 1 IgG1), and 7 (2 IgM, 3 IgG1, 2 IgG3) mAb-producing lines using B-1a (surface CD5+, CD45RAlow), B-1b (surface CD5−, CD45RAlow, CD5 mRNA+), and B-2 (surface CD5−, CD45RAhigh, CD5 mRNA−) cells, respectively, sorted from adult human peripheral blood. Most B-1a and B-1b, but no B-2, cell-derived mAbs were polyreactive; i.e., they bound different self and foreign Ags with different affinities. B-1a and B-2 mAbs preferentially utilized VH4 (p = 0.003) and VH3 (p = 0.010) genes, respectively. All three mAb populations utilized DXP, DLR, DN DH genes, and JH6, but no mAb utilized DHQ52. There were fewer unencoded nucleotide (N) additions in the VHDJH junctions of B-1b (3.00 ± 2.52, mean ± SD) than of B-1a (12.45 ± 3.93, p = 1.23 × 10−5) or B-2 (8.29 ± 4.75, p = 0.020) mAbs. Partly due to the fewer N additions and a paucity of D-D fusions, the B-1b mAb CDR3s were significantly shorter than the B-1a mAb CDR3s (p = 0.013), which contained a nonrandom Tyr distribution (p = 0.003). Finally, all but two B-1 cell-derived mAbs were mutated, in a fashion similar to that of the Ag-selected B-2 mAbs. Thus, in the human adult, B-1 cells that make natural polyreactive Abs may not be representative of the predominantly B-1 developmental waves of colonization of the fetal and neonatal B cell repertoires, and are somatically selected.
Numerous studies have suggested that in the mouse B-1a, B-1b, and B-2 cells originate from distinct developmental waves of B cell precursors (1–3). Murine B-1a cells (formerly known as Ly-1+ B cells), and, perhaps, the related B-1b cells (formerly known as Ly-1 sister B cells), arise early in ontogeny. They would persist in the adult as self-replenishing cells that derive from the replication of other surface (s)Ig+ B cells rather than from bone marrow sIg− progenitors. Along with their B-1b counterparts, they are highly represented in the peritoneal cavity. B-2 cells or “conventional” B cells develop late during ontogeny, emerge from adult bone marrow sIg− cells, and are the predominant clonotypes in the adult spleen and lymph nodes. Perhaps reflecting their distinct origins and/or clonal amplification due to somatic selection, B-1 (B-1a and B-1b) cells may express a selected repertoire of Ig H chain V, D, and/or J genes that differs from that expressed by B-2 cells (1–3).
The early murine B cell Ig H chain gene repertoire is the product of preferential rearrangements of members of JH-proximal VH-families in the absence of terminal deoxynucleotidyl transferase. The resulting absence of unencoded nucleotide (N) additions in the Ig VHDJH junctions creates a repertoire of limited diversity and restricted length (4, 5). The primarily germline restricted repertoire of expressed Ig VHDJH genes encode for Ag-binding sites with distinctive functional features. Most fetal and neonatal Abs appear capable of binding two or more Ags, including self and exogenous Ags, even highly different in nature (polyreactive Abs) (6). A similar pattern of polyreactivity has also been noted in Abs derived from human fetal B lymphocytes (7). Polyreactive Abs account for the vast majority of the Ig that have been traditionally referred to as natural autoantibodies or Abs. Because of their reactivity with self Ags, natural Abs would provide the core of the interclonal network of “connectivity” crucial in shaping the early B cell repertoire development, and may also provide the templates for autoantibodies arising later in life (8, 9). Because of their reactivity with multiple exogenous, particularly bacterial and viral, Ags, natural Abs may play a major role in the initial response to infection (8, 9). In both early and adult life, natural Abs are mainly the product of B-1 cells (3, 10).
In the human, first and early second trimester fetal B cells are mainly surface CD5− (11). They express IgM genes that frequently use VH3 family genes and contain H chain complementarity-determining region (CDR)38 sequences similar in length to those found in the mouse in spite of the presence of some N additions (12). Generation of short H chain CDR3s is abetted by frequent use of the short DHQ52 gene segment and the absence of the long JH6 gene segment (12). Neonatal B cells are mostly surface CD5+ (3, 10, 13). They express mainly IgM that frequently use VH3 genes with longer CDR3 sequences, which result from increased frequency of D-D joins, increased N addition, use of JH6, and use of DH gene segments that are longer than DHQ52 (14, 15). These longer VHDJH junctional sequences encode H chain CDR3 structures not seen in the mouse. Human adult B cells are mainly CD5− but also include a large CD5+ subpopulation. Unselected human adult peripheral blood B cells express mainly VH3 family genes with an IgM H chain CDR3 composition and length similar to those of the Ig described in the neonate (16).
The human adult B cell repertoire comprises three cell subsets similar to those seen in the mouse: B-1a, B-1b, and B-2 cells (17–20). B-1a cells, which are surface CD5+CD45RAlow, account for 15 to 30% of the circulating, splenic, and tonsillar B lymphocytes in the adult, and constitute the major B lymphocyte population in the fetus and neonate. B-1b cells are surface CD5− and CD45RAlow, but express CD5 mRNA at levels comparable with those of B-1a cells, and account for 2 to 6% of adult peripheral B lymphocytes. B-2 cells are surface CD5− and CD45RAhigh, include memory B cells, and constitute the major population of the adult B cell repertoire. Both B-1a and B-1b cells produce mainly IgM naturally occurring Abs, the majority of which are polyreactive, although polyreactive IgG and IgA have also been isolated (18–20). In contrast, B-2 cells produce mainly Ag-induced monoreactive IgG (18, 20).
We addressed the issue as to whether the utilization of distinct VHDJH gene rearrangements by polyreactive Abs reflects the developmentally restricted Ig VHDJH rearrangements putatively expressed by circulating human B-1 cells. We generated polyreactive IgM, IgG, and IgA mAbs from sorted human adult peripheral blood B-1a and B-1b cells, and monoreactive IgG mAbs from sorted B-2 cells. By cloning and sequencing the VHDJH gene segments, we found that the VH and JH gene utilization, and/or the patterns of junctional VHDJH gene sequences of the mAbs derived from B-1a and, to a lesser extent, B-1b cells differ from those of the mAbs derived from “conventional” B-2 cells, and therefore those expressed by the human adult B cell repertoire as a whole. We also found that the gene selection and junctional pattern of the VHDJH sequences of the natural polyreactive mAbs analyzed do not reflect those thought to be characteristic of the B-1 cell repertoire at large, as previously defined by the analysis of neonatally expressed Ig genes. These differences suggest that the polyreactive B-1 cell-derived Ab repertoire in the human adult is generated through somatic expansion, possibly dependent on Ag, of a minor clonotypic B cell population, rather than resulting from the mere germline encoded natural reactivity. Our conclusion is further supported by the high load of putative replacement (R) mutations in the B-1a and B-1b polyreactive Ig-binding sites with preferential segregation of the R mutations within the VH segment CDRs. These results should prompt a further re-evaluation of the forces that shape the composition of the adult human B cell repertoire in both normal and disease states.
Materials and Methods
Generation of mAb-producing cell lines from sorted human B-1a, B-1b, and B-2 cells
The generation and features of 18 of the 25 B-1a, B-1b, or B-2 cell-derived mAb-producing cell lines have been reported (20–23). The remaining seven cell lines (cell lines 417.F1.1.35, 417.7.3.1, 417.32.F4.8.5, 417.32.F24.7, 417.F33.3, 417.F32.36, and 417.F32.26) were generated for the purpose of these studies. B lymphocytes were enriched by T cell and monocyte depletion from the PBMCs of healthy donors A (cell line 416.F2.2.34) (20), B (cell lines 417.F25, 417.F9.5, 417.F21.5, 417.F14, 417.F22, 417.F23, 417.F29, 417.F28, 417.F1.1.35, 417.7.3.1, 417.32F4.8.5, 417.32.F24.7, 417.F33.3, 417.F32.36, 417.F32.26) (20), C (cell line 418.F63.35) (20), D (cell line 18) (22), E (cell line 21) (22), F (cell line 17) (22), G (cell line 44) (22), H (cell line 26) (22), and the rheumatoid arthritis patient I (cell lines 63, 65, and 67) (23). Enriched B cells were reacted with specific mouse mAbs to CD20, CD5, and CD45RA (Coulter Immunology, Hialeah, FL) and applied to an EPICS Elite FACS (Coulter Immunology) to yield purified B-1a, B-1b, and B-2 cells (20, 24). In some experiments, only CD5+ (B-1a) cells were sorted using mAbs to CD20 and CD5 (19, 22). Sorted B-1a, B-1b, and B-2 cells were infected with EBV for 1 h and then immediately distributed into 96-well plates (18–24). The Igs produced by these microcultures were analyzed using appropriate ELISA (18–24). Microcultures originally seeded with B-1a or B-1b cells were selected for production of natural IgM Abs binding to self and/or foreign Ags, including IgG Fc fragment, ssDNA, insulin, thyroglobulin, actin, phosphorylcholine, tetanus toxoid, Escherichia coli β-galactosidase, and/or E. coli LPS (18–24). Microcultures originally seeded with B-2 cells were selected for the production of mainly IgG, irrespective of their Ag-binding activity. The B-1b cell-derived IgG mAb 417.F1.1.35, and all B-2 cell-derived IgM and IgG mAbs did not bind any of the above nine Ags tested, and were operationally defined as monoreactive Abs, specific for Ags yet to be identified. EBV-transformed B cells were cloned by sequential subculturing (18–24), and then, with the exception of the cell line 417.7.3.1, fused with human-mouse heterohybridoma F3B6 cells (25). The Ag-binding activities of the mAbs were analyzed in specific dose-saturable binding and competitive inhibition ELISAs using the nine Ags listed above, and the dissociation constant (Kd) for Ag was calculated as reported (18–27). The Kd for the different Ags were different in each mAb; the Kd for the same Ag were different in the different mAbs, and ranged from 10−5 to 10−8 M. EBV-transformed cells and/or EBV-transformed B cell hybrids were used for mAb VHDJH gene cloning and CD5 mRNA analysis.
Analysis of CD5-specific mRNA expression
The levels of cellular CD5 mRNA were analyzed using the PCR method we devised ad hoc (20, 24, 26). Briefly, poly(A)+ RNA from each mAb-producing cell line (0.3 μg), freshly isolated T cells (0.3–0.009 μg), and F3B6 cells (0.3 μg) were purified by affinity absorption to oligo(dT)-cellulose using the Mini RiboSep ULTRA mRNA Isolation Kit (Becton Dickinson Labware, Bedford, MA), and then reverse transcribed using M-MLV reverse transcriptase (SuperScript Preamplification System for First Strand cDNA Synthesis, Life Technologies, Gaithersburg, MD) in conjunction with a poly(dT)12–18 primer. The cDNA were used as templates in PCR amplifications utilizing the β-actin-specific sense (5′-GTACCACTGGCATCGTGATGGACT-3′) and antisense (5′-ATCCACACGGAGTACTTGCGCTCA-3′) oligonucleotide primers, or the CD5-specific sense (5′-AGACGGATGGCACATGGTTT-3′) and antisense (5′-TTGTCCTGGGCCTCATAGCT-3′) oligonucleotide primers. When amplifying cDNA, the β-actin-specific and CD5-specific sense and antisense primers yielded ~0.6 kb and ~0.45 kb products, respectively. Each PCR was performed in a 50-μl vol and consisted of 25 cycles of: denaturation, 94°C for 1 min; annealing, 60°C for 1 min; and extension, 72°C for 2 min. The amplified cDNA was applied to a 1.2% agarose gel containing 1 μg/ml of ethidium bromide, fractionated, and transferred onto a nylon membrane (GeneScreen Plus, Du Pont, NEN, Boston, MA). Hybridization with the internal β-actin specific oligonucleotide, (5′-GACTGACTACCTCATGAAGATCCT-3′) or the internal CD5-specific oligonucleotide (5′-CCAGAAGACAACACCTCCAA-3′) probe, labeled with [γ-32P]ATP (NEN), was performed overnight at 43°C. The filters were washed twice with 2× SSC/0.1% SDS at room temperature for 30 min, and twice with 1× SSC/0.1% SDS at 52°C for 30 min. Autoradiography was performed overnight using Kodak XAR-5 film (Eastman Kodak Co., Rochester, NY).
Cloning and sequencing of the mAb VHDJH gene segments
mRNA was isolated from the mAb-producing cell lines and reverse transcribed as detailed above. Second-strand cDNA synthesis and amplification were performed by PCR using 100 ng of first-strand cDNA as template in a 50-μl reaction volume. The degenerate sense oligonucleotide primers consisted of sequences encompassing an area of the leader region of the different VH gene families plus an EcoRI site, as follows: VH1 (5′-GGGAATTCATGGACTGGACCTGGAGG(AG)TC(CT)TCT(GT)C-3′); VH2 (5′-GGGAATTCATGGACATACT(GT)TG(GT)T(CT)CACGCT(CT)CT(GC)C-3′); VH3 (5′-GGGAATTCATGGAG(CT)TTGGGCTGA(CG)CTGG(CG)TTT(CT)T-3′); VH4 (5′-GGGAATTCATGAA(AG)CA(TC)CGTGGTTCTT(CT)(AC)T(CT)CT(CG)C-3′); VH5 (5′-ATGGGGTCAACCGCCATCCTCGCCCT-3′); and VH6 (5′-GGGAATTCATGTCTGTCTCCTTCCTCATCTTCC-3′). The antisense Cμ (5′-CCGAATTCAACGAGGGGGAAAAGGGTT-3′) and Cγ (5′-CCGAATTCTAGTCCTTGACCAGGCAGCC-3′) oligonucleotide primers (27) consisted of the reverse complements of the 5′ portion of the Cμ and Cγ sequences (20 nucleotides), respectively, plus an EcoRI site. VHDJH cDNA was amplified by 30 cycles of PCR in a 50-μl vol. Each cycle consisted of: denaturation, 94°C for 1 min; annealing, 52°C for 1 min; and extension, 72°C for 2 min. Amplified cDNA products were purified by fractionation in 1.2% Ultra Pure Low Melting Point Agarose (Life Technologies), and then ligated into the pCR II plasmid vector (Invitrogen Corp., San Diego, CA). Sequencing was performed by the dideoxynucleotide chain termination method using [α-35S]dATP (NEN) and the TaqTrack Sequencing System (Promega Corp., Madison, WI). Each VHDJH gene sequence was derived from the analysis of at least four independent recombinant clones. Differences in nucleotide sequences among different recombinant clones were rarely observed (<0.0005/base, which is consistent with the error rate of the Taq polymerase). The BLAST algorithm, as found in the NCBI World Wide Web home page accessed through the Netscape Navigator, was used to analyze the GenBank DNA sequence database. The MacVector version 5.0 sequence analysis software (International Biotechnologies, New Haven, CT) was used to analyze the current human Ig gene V-BASE database (MRC Centre for Protein Engineering, Cambridge, U.K.).
Statistical methods
The observed frequencies of VH or JH gene utilization by the mAbs produced by B-1a, B-1b, and B-2 cells were compared with the expected frequencies using the χ2 goodness-of-fit test (28). When the overall differences were significant at p < 0.050, the frequencies of VH or JH gene utilization were compared with those expected using the exact binomial distribution (28). The expected frequencies were based on the genomic complexity and distribution of the 7 different VH gene families or six JH genes. The adopted Ig gene terms of comparison were based on the family genomic complexities reported by Walter et al. (29), Matsuda et al. (30), Cook et al. (31), and Cook and Tomlinson (32), who place the numbers of the functional genes for the seven different VH families as follows: VH1, 11 members; VH2, 3 members; VH3, 22 members; VH4, 11 members; VH5, 2 members; VH6, 1 member; and VH7, 1 member. The JH gene terms of comparison were based on an equal representation of the genomic JH genes, as reported by Ravetch et al. (33), and/or on reported frequencies of use of individual gene segments at different stages of ontogeny. The reported p values for the significant comparisons were based on the two-tailed t test.
To test whether the VH and JH genes utilized by the mAbs from the different B cell subsets were different, the χ2 test for homogeneity was performed (28). When the overall differences were significant at p < 0.050, the frequencies of VH and JH gene utilization by the mAbs produced by B-1a, B-1b, and B-2 cells were compared between populations using the χ2 test for independent proportions (28). Exact methods were applied when sample sizes were small. The reported p values for the significant comparisons were based on the two-tailed t test. The χ2 test for homogeneity and the χ2 test for independent proportions were applied to test whether the total number of putative R + silent (S) mutations and the total number of putative R mutations in the mAb VH genes of the three B cell subsets were significantly different. The comparison of the total number of R + S mutations in the CDRs and in the FRs of the mAb VH genes of the three B cell subsets was also assessed by the same statistical analysis method. The average number of unencoded N nucleotides found at the VHDJH junctions, and the average number of amino acids comprising the CDR3 of the mAbs produced by B-1a, B-1b, and B-2 cells were compared between the three populations by analysis of variance (28). Pairwise comparisons were evaluated using the Tukey method for multiple comparisons at an α = 0.05 level (28). Nonparametric analyses, based on rank-transformed data, were also performed; but no significant differences were observed. The reported p values for the significant comparisons were based on the two-tailed t test.
Results
Generation and characterization of human mAbs from B-1a, B-1b, and B-2 cells
We generated 11, 7, and 7 mAbs from B-1a, B-1b, and B-2 cells, respectively, sorted from the peripheral blood of nine adult subjects. The cell origin, the H chain isotype, the L chain type, and the Ag-binding activity of these mAbs are listed in Table I. Eight of the B-1a cell-derived mAbs were IgM, one IgG3, and two IgA1. Six of the B-1b cell-derived mAbs were IgM, and one IgG1. Two of the B-2 cell-derived mAbs were IgM, and five IgG (three IgG1, and two IgG3). All B-1a and B-1b cell-derived mAbs—with the exception of mAb 417.F1.1.35, for which a specific Ag was not determined, and mAb 417.F9.5, which bound to ssDNA only—were polyreactive. They bound with different affinities to nine different self- and foreign Ags, as detailed in Materials and Methods. None of the B-2 cell-derived mAbs bound to any of the nine Ags tested. These mAbs were therefore operationally defined as monoreactive to an unidentified Ag (20). Thus, the classes and the Ag reactivity of the mAbs generated were representative of the predominant classes and functional features of the Abs produced by each B cell subset.
Table I.
Features of the mAbs derived from sorted human B-1a, B-1b, and B-2 cells
| Clone | Donor | Chain
|
Ag Reactivity | VH Genea
|
Identityb | Nucleotide Differences
|
D Genec | JH Gened | GenBank Accession No. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDR
|
FR
|
|||||||||||
| H | L | Family | Member | R:S | R:S | |||||||
| B-1a cell-derived mAbs | ||||||||||||
| mAb 18 | D | μ | κ | Polyreactive | 3 | V3-23 | 98 | 2:0 | 0:1 | DLR4, DMX | JH2 | L23931 |
| mAb 21 | E | μ | κ | Polyreactive | 3 | V3-23 | 91 | 7:0 | 6:7 | DM1, DMX | JH6b | L23932 |
| mAb 417.F25 | B | μ | λ | Polyreactive | 3 | V3-33 | 97 | 3:3 | 1:2 | DLR3 | JH4b | L23557 |
| mAb 17 | F | γ3 | κ | Polyreactive | 4 | V4-34 | 88 | 7:3 | 6:6 | D6, DLR1 | JH4b | L23930 |
| mAb 44 | G | α1 | λ | Polyreactive | 4 | V4-34 | 96 | 2:1 | 2:5 | DXP′1, DLR1 | JH6b | L23934 |
| mAb 63 | I | μ | λ | Polyreactive | 4 | V4-34 | 100 | 0:0 | 0:0 | DXP4 | JH4b | X54441 |
| mAb 416.F2.2.34 | A | μ | λ | Polyreactive | 4 | V4-39 | 96 | 4:0 | 3:6 | DK1, DN2 | JH3b | L23556 |
| mAb 417.F9.5 | B | μ | λ | Monoreactivee | 4 | VHIV-4 | 96 | 3:1 | 4:3 | DN1 | JH4b | L23570 |
| mAb 65 | I | α1 | λ | Polyreactive | 4 | V4-31 | 91 | 6:1 | 7:6 | DXP4, D21-9 | JH6b | X54443 |
| mAb 67 | I | μ | γ | Polyreactive | 4 | V4-4 | 99 | 0:0 | 1:0 | DN4, DXP4, DK4 | JH4b | X54445 |
| mAb 26 | H | μ | λ | Polyreactive | 4 | V4-28 | 87 | 4:4 | 11:12 | D21-9 | JH2 | L23933 |
| B-1b cell-derived mAbs | ||||||||||||
| mAb 417.F21.5 | B | μ | κ | Polyreactive | 1 | V1-2 | 97 | 3:0 | 4:3 | DXP′1 | JH4b | L23558 |
| mAb 417.F14 | B | μ | κ | Monoreactivef | 1 | V1-2 | 93 | 4:2 | 10:3 | DXP4 | JH4b | L23559 |
| mAb 417.F22 | B | μ | λ | Polyreactive | 1 | V1-46 | 96 | 2:3 | 2:3 | D21-9 | JH6b | L23560 |
| mAb 417.F1.1.35 | B | γ1 | λ | Monoreactiveg | 1 | V1-69 | 97 | 1:1 | 4:3 | DXP′1 | JH6b | L23561 |
| mAb 418.F63.35 | C | μ | λ | Polyreactive | 3 | V3-33 | 100 | 0:0 | 0:0 | DXP′1 | JH5b | L23571 |
| mAb 417.F23 | B | μ | λ | Polyreactive | 3 | H11 | 95 | 5:0 | 5:2 | D4, DN2 | JH3b | L23562 |
| mAb 417.7.3.1h | B | μ | λ | Polyreactive | 4 | V4-59 | 96 | 2:0 | 7:2 | DN1 | JH5b | L23563 |
| B-2 cell-derived mAbs | ||||||||||||
| mAb 417.32F4.8.5 | B | γ1 | κ | Monoreactiveg | 1 | DP-3 | 94 | 2:2 | 7:4 | DXP1 | JH6b | L23555 |
| mAb 417.F29 | B | μ | λ | Monoreactiveg | 3 | V3-33 | 99 | 2:1 | 1:0 | DM1 | JH6b | L23564 |
| mAb 417.32.F24.7 | B | γ3 | κ | Monoreactiveg | 3 | V3-9 | 96 | 3:1 | 5:1 | DLR4 | JH6b | L23565 |
| mAb 417.F33.3 | B | γ3 | κ | Monoreactiveg | 3 | V3-30 | 97 | 2:2 | 4:1 | DXP4 | JH5b | L23566 |
| mAb 417.F32.36 | B | γ1 | κ | Monoreactiveg | 3 | V3-33 | 98 | 1:0 | 2:1 | DN2 | JH1 | L23567 |
| mAb 417.F32.26 | B | γ1 | λ | Monoreactiveg | 3 | V3-30.3 | 93 | 4:1 | 8:3 | DXP4, DXP1 | JH3b | L23568 |
| mAb 417.F28 | B | μ | κ | Monoreactiveg | 3 | V3-23 | 79 | 13:2 | 20:8 | DMX | JH6b | L23569 |
Compared with the genomic germline sequences.
The complete sequences of the germ-line and expressed JH genes are reported in References 16 and 33.
Monoreactive to SSDNA.
Monoreactive to β-galactosidase.
Ag not identified.
mAb 417.7.3.1 was a cloned mAb-producing EBV-transformed B cell line. All other mAb-producing cell lines were B cell hybrids.
CD5 mRNA expression by the mAb-producing B-1a, B-1b, and B-2 cell-derived lines
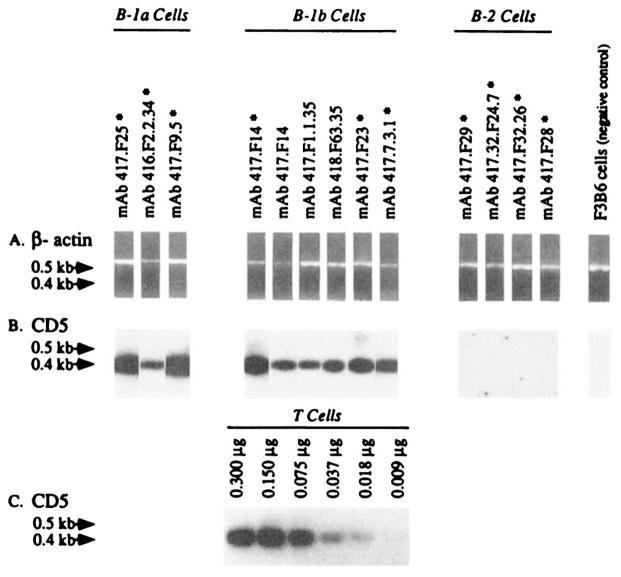
We have shown that B-1a and B-1b cells, but not their B-2 counterparts, express high levels of CD5 mRNA (20, 24, 26), and that the density of CD5 on the surface of B-1a cells is directly proportional to the level of cellular CD5 mRNA expression (20). To further verify the origin of the mAb-producing cell lines generated using sorted B cell subsets, we analyzed the level of CD5 mRNA expression in three B-1a, three B-1b, and four B-2 cell-derived mAb-producing EBV-transformed lines progenitors of the respective somatic cell hybrids, as well as three B-1b cell-derived somatic hybrids. Poly(A)+ RNA were individually extracted from each of the 12 mAb-producing cell lines, from the negative control F3B6 cells, and from freshly purified T cells, and independently reverse transcribed. cDNA was utilized as a template for CD5 or β-actin specific DNA amplification. The reverse transcription of the different poly(A)+ samples yielded similar amounts of cDNA, as indicated by the PCR-amplification of comparable amounts of β-actin cDNA (Fig. 1A). The PCR amplification of CD5 mRNA-derived cDNA was dose dependent as shown by the amplification of increasing amounts of cDNA from T cells (Fig. 1C). The validity of analyzing a hybridoma rather than an EBV transformant in the cases of the cell lines 417.F14, 417.F1.1.35, and 418.F63.35 was indicated by 1) the consistent failure to amplify CD5 mRNA from the negative control F3B6 cells (Fig. 1B), and 2) the detection of comparable amounts of CD5 mRNA in the mAb 417.F14-producing EBV-transformed B cell hybrid and its EBV-transformed clonal progenitor (Fig. 1B). All of the B-1a and B-1b tested, but none of the B-2 cell-derived mAb-producing cell lines, expressed significant amounts of CD5 mRNA (Fig. 1B), thus confirming the respective B cell origin of the different cell lines.
FIGURE 1.
Expression of CD5 mRNA by the B-1a, B-1b, and B-2 cell-derived mAb-producing lines. Poly(A)+ RNA (0.300 μg) extracted from mAb-producing EBV-transformed cell lines (asterisks), mAb-producing EBV-transformed somatic cell hybrid lines, freshly isolated T cells, or from the human-mouse heterohybridoma F3B6 cells, were individually reverse transcribed. The cDNA was individually amplified by PCR using β-actin-specific or CD5-specific oligonucleotide primers and fractionated on a 1.2% agarose gel (see Materials and Methods). A, Ethidium bromide-stained gel containing amplified β-actin DNA (~0.6 kb); B, Hybridization of the 32P-labeled “internal” CD5-specific oligonucleotide probe with fractionated CD5 DNA amplified from cDNA reverse transcribed from the mAb-producing cell lines or the fusion partner F3B6 cells; C, hybridization of the 32P-labeled “internal” CD5-specific oligonucleotide probe with fractionated CD5 DNA amplified from cDNA individually reverse transcribed from different amounts (0.009 to 0.300 μg) of purified T cell mRNA.
VH genes of the mAbs produced by B-1a, B-1b, and B-2 cells
The VH gene sequences of mAb 18, mAb 21, mAb 17, mAb 44, mAb 63, mAb 65, mAb 67, and mAb 26 have been previously reported (22, 23). Their respective GenBank accession numbers are listed in Table I. The VH genes of the mAbs produced by the remaining three B-1a, seven B-1b, and seven B-2 cell-derived lines were cloned and sequenced for the purpose of these studies and are available in the GenBank data base under accession numbers L23555 through L23571 (Table I). The deduced amino acid sequences of all 25 mAb VH genes are depicted in Figure 2. The degree of identity of the mAb VH segment nucleotide and deduced amino acid sequences to those of the closest reported germline VH genes ranged from 79% (mAb 417.F28 VH gene sequence compared with that of the germline V3-23 gene) to 100.0% (mAb 63 and mAb 418.F63.35 VH gene sequences compared with those of the germline V4-34 and V3-33 genes, respectively) and are summarized in Table I. The mAbs produced by B-1a and B-2 cells utilized genes of the VH4 and VH3 families, respectively, at significantly higher frequency than that theoretically expected (p = 0.003 and p = 0.010, χ2 goodness-of-fit and exact binomial distribution), while the mAbs produced by B-1b cells did not show any significant bias in VH family utilization (p = 0.240). The mAbs produced by B-1a and B-1b cells differed significantly in utilization of VH1 (p = 0.010, χ2 test for homogeneity and the χ2 test for independent proportions) and VH4 genes (p = 0.050); those produced by the B-1a and B-2 cell subsets differed significantly in utilization of VH4 genes (p = 0.009); and those produced by the B-1b and B-2 cell subsets did not differ significantly in utilization of the VH genes (p = 0.135).
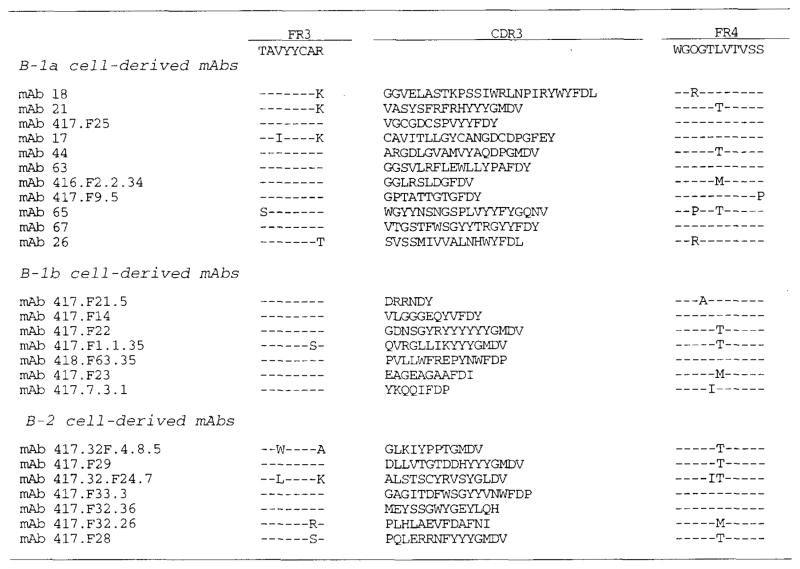
FIGURE 2.
Deduced amino acid sequences of the VH genes utilized by the mAbs produced by B-1a, B-1b, and B-2 cells. In each cluster, the top sequence is given for comparison and represents the reported germ-line VH gene displaying the highest degree of identity to the expressed VH genes. The V3-23, V3-30, V3-33, H11, and V3-9 genes are members of the VH3 gene family. The V4-34, V4-39, VIV-4, V4-31, V4-4, V4-28, and V4-59 genes are members of the VH4 gene family. The V1-2, V1-46, V1-69, and DP-3 genes are members of the VH1 gene family. Dashes indicate identities. Solid lines on the top of each sequence depict CDR. These sequence data are available from EMBL/GenBank/DDBJ under the accession numbers listed in Table I.
D and JH genes and junctional VHDJH sequences of the mAbs produced by B-1a, B-1b, and B-2 cells
The nucleotide sequences of the D genes of the mAbs produced by the B-1a, B-1b, and B-2 cells were compared with those of the reported germline D gene segments (34–38) (Fig. 3). The best fit D gene segments were identified based on the following criteria: 1) priority was given to identity with a VH or JH gene sequence when VH and D, or D and JH region sequences overlapped (16); 2) if more than one candidate D gene was identified, the germline D gene sequence displaying the longest stretch of identity (with a minimum match of six nucleotides in a stretch of 7 bp or with a minimum match of five nucleotides in a row) (16) to that of the mAb D gene was assigned (16). In those gene segments with more than one D gene segment, the germline position of each gene segment relative to its partner was taken into account. D gene segments were identified in all 25 mAb sequences.
FIGURE 3.
Nucleotide sequences of the VHDJH junctions (CDR3-FR4 areas) of the mAbs produced by B-1a, B-1b, and B-2 cells including unencoded nucleotides. The nucleotide sequences of the relevant portions of the germ-line D genes are given for comparison, and appear above the expressed mAb D segments. Dashes indicate identity with germline D segment sequences. Asterisks indicate the reverse complement of the known germline D segment. Unencoded nucleotide additions are between VH and D, D and D, and D and JH gene segments. The TGG triplet encoding the invariant Trp that marks the beginning of the 3′ conserved portion of the JH gene is listed under FR4. These sequences are available from EMBL/GenBank/DDBJ under the accession numbers of the respective VH genes as listed in Table I.
* Denotes a D gene that is utilized in an inverted position.
The mAb D gene utilization is listed in Table I. The degree of identity of the mAb gene sequences to those of the putative D genes ranged from 75 (mAb 21) to 100% in seven mAbs (mAb 417.7.3.1, mAb 417.F23, mAb 417.F28, mAb 417.F29, mAb 417.F32.36, mAb 417.F33.3, and mAb 26). Fifteen, seven, six, five, and two mAb D gene sequences could be assigned to portions of those of members of the germ-line DXP, DLR, DN, DM, and DK families, respectively. The mAbs produced by the B-1a, B-1b and B-2 cells did not differ significantly in the utilization of the DXP family (p = 0.53, χ2 test for homogeneity and the χ2 test for independent proportions). Within the DXP family, six, four, three, and two mAb D segment sequences could be assigned to portions of those of the germ-line DXP4, DXP1′, DXP D21/9, or DXP1 genes, respectively. Within the DLR family, two, two, one, one, and one mAb D gene segment sequences could be assigned to portions of those of germline DLR4, DLR1, DLR3, D4, and D6 genes, respectively. Within the DN family, three, two, and one mAb D gene segment sequences could be assigned to portions of germline DN2, DN1, and DN4 genes, respectively. Within the DM family, three and two sequences could be assigned to portions of those of the germline DMX and DM1 genes, respectively. Finally, within the DK family, one mAb D gene sequence could be assigned to portions of the DK1 gene segment, and a second mAb D sequence gene segment sequence could be assigned to portions of the DK4 gene segment. No mAb D segment bore resemblance to sequences of the germline DA or DHQ52 genes.
Sixteen mAbs utilized a single germline D gene; eight utilized two different germline D genes, likely as D-D gene fusions (e.g., mAb 65); and one mAb sequence (mAb 67) utilized three different D genes (Table I and Fig. 3). In five mAbs, the D segments could be accounted for by the utilization of portions of the reverse complements of germline D genes (e.g., mAb 18). All five of the mAbs with inverted D segments utilized two or more D genes, including two sequences with two inverted D gene segments. The mAbs produced by B-1a cells differed from the mAbs produced by B-1b and B2 cells in the use of multiple D gene segments (7 of 11 vs 1 of 7 and 1 of 7, respectively, p = 0.04, χ2 test for independent proportions). Thus, the B-1a, B-1b, and B-2 cell-derived mAbs utilized a heterogeneous selection of germline D genes in conventional, fused, or inverted configurations, but the B-1a-derived Abs differed from the B-1b and B-2 Abs in the frequent use of more than one D gene segment.
The germline JH repertoire consists of six functional gene segments and three pseudogenes (33). The sequences of the mAb JH genes were compared with those of the germline JH genes and their-allelic forms (16). The region of the mAb JH gene sequences depicted in Figure 3 spans from the 5′ end of the open reading frame of the JH gene to the first TGG triplet, coding for the invariant Trp. Nine mAbs (three from B-1a, three from B-1b, and three from B-2) utilized JH genes in full-length form, each with zero to six mutations. Seven, four, and four mAbs from B-1a, B-1b, and B-2 cells, respectively, utilized JH genes in a truncated form, each with zero to six mutations. Consistent with the findings of Yamada et al. (16) and Schroeder et al. (39), the sites of truncation for the 15 mAb JH genes were at or 5′ of the first TG doublet (Fig. 3). In the germline JH3b gene sequence, the first TG doublet appears at position 2, and correspondingly, the JH genes utilized by mAb 416.F2.2.34 (B-1a), mAb 417.F23 (B-1b), and mAb 417.F32.26 (B-2) were truncated by zero to three nucleotides. In the JH1, JH2, JH5b, and JH4b germline genes, the first TG doublet appears at nucleotide positions 3, 4, 6, and 8, respectively, and thus the JH genes of mAbs utilizing the JH1, JH2, JH5b, and JH4b gene segments were truncated by a maximum of zero, one, eight, and eight nucleotides, respectively (e.g., mAb 417.F32.36 from B-2, mAb 26 from B-1a, mAb 417.7.3.1 from B-1b, and mAb 417.F21.5 from B-1b cells, respectively). The first TG doublet in the JH6b germline gene appears at position 22, and is consistent with the wide heterogeneity of truncation sites in the expressed JH6b segments. For example, the JH gene of mAb 417.F22 (B-1b) was untruncated, whereas that of mAb 44 (B-1a) was truncated by 16 nucleotides. The structure of the 3′ sequences of the mAb JH genes (beginning with the invariant TGG codon) was highly conserved. The JH genes of 18 mAbs were unmutated in this region, while those of mAb 416.F2.2.34 (B-1a), mAb 417.F9.5 (B-1a), mAb 65 (B-1a), mAb 417.F21.5, (B-1b), mAb 418.F63.35 (B-1b), mAb 417.7.3.1 (B-1b), and mAb 417.32.F24.7 (B-2) displayed one or two somatic point mutations. The mAbs from B-1a cells utilized the JH6b gene three times, the JH4b gene five times, the JH3b gene once, and the JH2 gene twice; those from B-1b cells utilized the JH6b, JH5b, and JH4b genes each twice, and the JH3 gene once; and those from B-2 cells utilized the JH6b gene four times, and the JH5b, JH3b, and the JH1 genes each once (Table I). The mAbs produced by B-1a and B-2 cells utilized at significantly higher frequency than that theoretically expected JH4b (p = 0.020, χ2 goodness-of-fit test and exact binomial distribution analyses) and JH6b (p = 0.009) genes, respectively, while the mAbs produced by B-1b cells did not show any apparent bias in JH gene utilization (p = 0.550). In addition, JH gene utilization by the mAbs produced by the B-1a and B-1b (p = 0.731, χ2 test for homogeneity), the B-1b and B-2 (p = 0.170), and the B-1a and B-2 (p = 0.120) subsets was not significantly different.
The junctional VHDJH nucleotide sequences of the mAbs produced by B-1a, B-1b, and B-2 cells are depicted in Figure 3. All the residues that could not be attributed to the sequence of D genes according to the aforementioned criteria were considered unencoded nucleotide (N) additions. No N additions were found in the VHD junctions of B-1b cell-derived mAb 417.F21.5 and mAb 417.F32, and the DJH junctions of mAb 417.F2.2.34 and mAb 417.F14, but a minimum of two N nucleotides were found in all of the sequences. In the B-1a cell-derived mAbs, the total number of unencoded VHDJH junction nucleotides ranged from 7 (mAb 21) to 45 (mAb 18) (15.5 ± 10.5, mean ± SD); in the B-2 cell-derived mAbs, it ranged from 9 (mAb 417.F32.36) to 19 (mAb 417.32.F26) nucleotides (13.4 ± 3.3); and in the B-1b cell-derived mAbs it ranged from 2 (mAb 417.F21.5) to 20 (mAb 417.F14) (7.7 ± 5.9). Although there was a trend for fewer N additions in the VHDJH junctions of the mAbs produced by B-1b cells, such a trend failed to achieve statistical significance (B-1b vs B-1a, p = 0.142; B-1b vs B-2, p = 0.059; and B-1a vs B-2, p = 0.886, one way analysis of variance).
The difference in the mean number of N nucleotide additions between the mAbs generated from the different B cell subsets was in part due to the N nucleotides added during D-D gene fusion, but differences were also detected in the VHD and the DJH gene junctions. In the VHD junctions, the average number of N nucleotide additions was: B-1a, 5.9 ± 1.3 (mean ± SD); B-1b, 4.6 ± 1.7; and B-2, 6.3 ± 1.7. In the DJH junctions the average number of N nucleotide additions was: B-1a, 8.1 ± 1.7; B-1b, 2.9 ± 2.2; and B-2, 5.9 ± 2.2. However, these differences did not reach statistical significance.
H chain CDR3 structure of the mAbs from B-1a, B-1b, and B-2 cells
The deduced amino acid sequences of the DJH genes of the mAbs produced by B-1a, B-1b, and B-2 cells were segregated into CDR3 and FR4 (Fig. 4), as outlined in the compilation by Kabat et al. (40). The FR4 sequences displayed minimal codon diversity. In contrast, the CDR3 sequences were highly divergent in composition and length, ranging from 11 (mAb 416.F2.2.34) to 26 (mAb 18) (17.64 ± 4.25, mean value ± SD) codons for B-1a, 6 (mAb 417.F21.4) to 17 (mAb 417.F22) (12.00 ± 4.00) codons for B-1b, and 12 (mAb 417.32F.4.8.5) to 18 (mAb 417.F33.3) (14.86 ± 2.12) codons for B-2 cell-derived mAbs. Partially due to the paucity of D-D fusions, and to fewer N segment additions, the H chain CDR3s of the mAbs produced by B-1b cells were significantly shorter than those of the mAbs produced by B-1a (p = 0.013, analysis of variance and Tukey method for multiple comparisons), although they were not shorter than those of the mAbs from B-2 cells (p = 0.120). The H chain CDR3s of the mAbs produced by B-1a and B-2 cells did not significantly differ in length (p = 0.130).
FIGURE 4.
FR3-CDR3-FR4 (deduced) amino acid sequences of the mAbs derived from B-1a, B-1b, B-2 cells. The FR3 and FR4 are depicted as consensus sequences. Dashes indicate identity.
JH6 differs from the other JH elements in the presence of a series of Tyr residues in the CDR3 portion of the gene product, enriching the carboxyl terminal portion of the CDR3 for an amino acid whose combined properties of hydrophilicity, hydrophobicity, space-filling shape, and flexibility allow it to play a key role in a number of Ag-Ab interactions (41, 42). We compared the distribution of Tyr to Gly in the amino and carboxyl termini of the H chain CDR3s of the B-1a-, B-1b-, and B-2-derived mAbs. Each H chain CDR3 was divided in half (when present, the odd residue was attributed to the amino terminus) and the number of Gly and Tyr residues in each portion of the CDR3 determined. Among the B-2 cell-derived mAbs, there were 4 Gly and 3 Tyr in the amino terminus, and 3 Gly and 11 Tyr in the carboxyl terminus (7 of which derived from the JH6 gene segment). These differences in amino acid distribution were not statistically significant (p = 0.6, χ2 test for homogeneity). Among the B-1b cell-derived mAbs, there were 6 Gly and 4 Tyr in the amino terminus, and 2 Gly and 11 Tyr in the carboxyl terminus (seven of which were derived from JH6 gene segments). These differences in amino acid distribution just failed to reach statistical significance (p = 0.074, χ2 test for homogeneity). However, among the B-1a cell-derived mAbs, there were 16 Gly and 9 Tyr in the amino terminus, and only 5 Gly with 21 Tyr (7 derived from the JH6 gene segment) in the carboxyl terminus. The probability that this biased amino acid distribution was not due to chance alone was highly significant (p = 0.003, χ2 test for homogeneity), consistent with the positive selection in the H chain CDR3 structure of an amino acid (Tyr) that is known for the promiscuity of its molecular interactions.
Somatic point mutations in the mAb VH gene segments
The analysis of the B-1 cell-derived murine CH lymphoma has shown that these cells express Ig genes in unmutated configuration and has led to the view that some if not all B-1 cells may be equipped with a primordial, possibly ineffective mutational machinery (43). As a result of the affinity maturation process which entails enhancement of the Ag-binding potential and preservation of the overall structure of the combining site, nucleotide differences yielding amino acid replacements, R mutations, accumulate in Ab V segment CDRs rather than FRs, resulting in high and low CDR R:S mutation ratios, respectively (44). To verify whether human B-1 cells mutate less effectively their expressed Ig genes and, therefore, differ in their ability to sustain a process of Ag-driven selection as compared to their “conventional” B-2 counterparts, we analyzed the number and distribution of the mAb VH gene R and S mutations using the respective putative germline VH gene sequences as references. All but one (mAb 63) of the B-1a, all but one (mAb 418.F63.35) of the B-1b, and all B-2 cell-derived mAbs were mutated (Table I). Furthermore, 1) the mAbs produced by B-1a and B-1b cells displayed a load of total (R + S) mutations (4.3 × 10−2 and 3.5 × 10−2 change/base, respectively) comparable with that of the mAbs produced by B-2 cells (4.9 × 10−2 change/base) (B-1a vs B-2, p = 0.779; B-1b vs B-2, p = 0.460; B-1a vs B-1b, p = 0.485, χ2 test for homogeneity); 2) the mAbs produced by B-1a and B-1b cells displayed a load of R mutations (2.5 × 10−2 and 2.4 × 10−2 R change/base, respectively) comparable with that of the mAbs produced by B-2 cells (3.6 × 10−2 R change/base) (B-1a vs B-2, p = 0.450; B-1b vs B-2, p = 0.431; B-1a vs B-1b, p 0.941, χ2 test for homogeneity); and 3) the mAbs produced by B-1a and B-1b cells displayed a preferential distribution of R mutations within the CDRs as compared with the FRs (B-1a, 5.3 × 10−2 vs 1.6 × 10−2 R change/base, p = 1.26 × 10−7; B-1b, 3.7 × 10−2 vs 2.0 × 10−2 R change/base, p = 0.001, χ2 test for homogeneity) and high CDR R:S mutation ratios (Table I), in a fashion similar to that of the mAbs derived from B-2 cells (5.8 × 10−2 vs 2.9 × 10−2 R change/base, p = 2.73 × 10−7). Thus, the B-1a and B-1b cells mutated thoroughly their expressed Ig V genes, and likely sustained a process of Ag-driven selection similar to that of their “conventional” B-2 counterparts.
Discussion
To gain insight into the mechanisms that underlie the generation of the human B-1 cell-derived polyreactive Ab repertoire, we generated 11 (8 IgM, 1 IgG3, 2 IgA1), 7 (6 IgM, 1 IgG1), and 7 (2 IgM, 3 IgG1, 2 IgG3) mAb-producing EBV-transformed cell lines using B-1a, B-1b, and B-2 cells, respectively, sorted from adult peripheral blood. We have previously shown that B-1 cells are transformed by EBV as effectively as their B-2 counterparts (18–21). Consistent with the discrete functional features of the Ig produced by the different B cell subsets, most B-1a and B-1b cell-derived mAbs were polyreactive, whereas all B-2 cell-derived mAbs were monoreactive. The origin of the mAb-producing cells was confirmed by the expression of CD5 mRNA in three B-1a and five B-1b cell-derived lines, including a B-1b cell line expressing IgG1; and the lack thereof in four B-2 cell-derived lines, including two cell lines that expressed IgM. Thus, the fidelity of CD5 mRNA expression continued after transformation. The VH, D, and JH gene sequence analysis suggested that natural polyreactive Abs derived from adult human B-1a and B-1b cells are produced by a selected assortment of clonotypes that not only differs from that of their monoreactive Ab-producing “conventional” B-2 counterparts, but that also bears little resemblance to the clonotypic composition characteristic of the fetal or neonatal B repertoire, where B-1 cells predominate. It also showed that in addition to their distinctive clonotypic makeup, the natural polyreactive Ab-producing B-1a and B-1b cells analyzed here expressed Ig VHDJH genes that contain a load of R mutations that is comparable in magnitude and distribution with that of the VHDJH genes expressed by the monoreactive Ab-producing “conventional” B-2 cells, and that is consistent with positive clonal selection by Ag.
Preferential use of specific VH gene families by fetal and neonatal B lymphocytes in the mouse would yield an Ab repertoire containing Ag-binding sites supported by similar FR structures (12, 42). Due to the lack of N additions, fetal Ig H chains that contain the same VH gene segment often share H chain CDR3s of similar germline-encoded sequence and structure, thereby restricting the diversity of the repertoire (4–6, 45, 46). The absence of N additions limits not only the diversity, but also the range of CDR3 lengths that can be created. Thus, Ag-binding structures that are common in the adult may be rare or may not even exist in the fetus (12). It has been suggested that the structural limitations imposed on fetal H chain CDR3s may play a critical role in establishing the polyreactivity that is characteristic of the fetal Ab repertoire (42). Molecular modeling indicates that limitations in the size of H chain CDR3 results in a relatively “concave” or “flat” Ab-binding site (12, 41). Such a “flat” site could maximize the number of different interactions possible between the residues of the Ag-binding site and potential Ags, allowing for the binding of different Ags, as characteristic of the natural (polyreactive) Ig population expressed in the fetal and neonatal B cell repertoires. It might also prevent a tight conformational fit between Ag and Ab, consistent with the observed low to moderate affinity also characteristic of the fetal and neonatal Ig repertoires. The preferential use of JH-proximal VH family genes, limited or absent N additions in the VHDJH gene junctions, and an absence or relative paucity of somatic mutations similar to those seen in fetal and neonatal Ig have been documented in the natural autoantibodies or Abs that occur in adult murine life and that, like those in early life, are mainly polyreactive and represent the products of B-1 cells (6). Thus, the identity and/or strong similarities of VHDJH gene sequences expressed by B-1 cells in the adult to those expressed early in murine life to encode for natural Ab-binding sites support the results of cell transfer experiments suggesting that adult B-1 cells are self-replenishing from sIg+ lymphocytes that first emerged during fetal life (1, 3, 5). Indeed, should the mature B lymphocyte population be ablated by experimental means, only under exceptional circumstances can B-1 cells regenerate in the adult mouse (3).
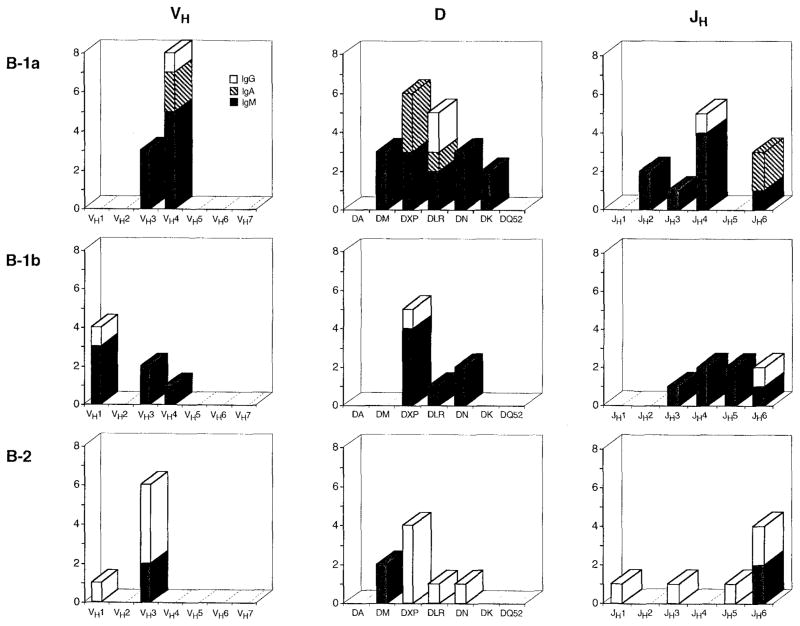
The results of the present VH, D, and JH gene analyses, as summarized in Figure 5, suggest that like their murine equivalents, human B-1a and, perhaps, B-1b cells that are committed to the production of natural polyreactive Abs consist of a clonotypic population distinct from that of their monoreactive Ab-producing “conventional” B-2 counterparts. The natural mAb-producing B-1a cells studied here preferentially utilized VH4 and JH4 genes, while the monoreactive mAb-producing B-2 cells preferentially utilized VH3 and JH6 genes. The fact that the biased Ig VH and JH gene expression by the CD5−, CD45RAhigh cells analyzed here closely reflects the biased Ig VH and JH gene expression that is characteristic of the adult human B cell repertoire at large is consistent with notion that B-2 cells, as defined by surface CD5−, CD45RAhigh lymphocytes, are representative of the predominant population in the human adult B cell repertoire (10, 20), and further emphasizes the distinctiveness of the B-1 cell population. The natural polyreactive B-1b cell-derived mAbs may also utilize selected VH and/or JH genes, but not to an extent that is significantly different from B-1a or B-2 cell-derived mAbs.
FIGURE 5.
Graphic representation of the selection of VH, D, and JH genes utilized by the B-1a, B-1b, and B-2 cell-derived mAbs. IgM Abs are depicted by solid bars, IgA Abs are depicted by cross-hatched bars, and IgG Abs are depicted by white bars.
The present findings also support the contention that adult B-1 cells that are committed to the production of natural polyreactive Abs are not representative of the predominantly B-1 developmental waves of colonization of the fetal and/or neonatal B cell repertoire. Major defining characteristics of Abs generated during the fetal life of the human are a preferential use of members of the VH3 family, biased usage of DH and JH gene segments and limits in the extent of N additions, especially between DH and JH (12). DHQ52, which is both the most JH-proximal and the shortest DH gene segment, is used in up to 40% of first and second trimester fetal Ig transcripts, but in less than 1% of adult Ig rearrangements (12, 47). Members of the DXP, DLR and DN families, which are the longest conventional DH gene segments, predominate in the adult repertoire. Fetal Ig transcripts rarely contain more than one DH gene segment, whereas approximately 10 to 15% of cord blood and the adult peripheral blood transcripts contain multiple Ds. JH4 is the most commonly used JH gene segment at all stages of ontogeny. The short JH3 is the second most commonly used JH gene segment in the fetus; JH6, the longest human JH gene segment, is rarely utilized in fetal Ig transcripts but is the second most commonly used JH gene in the adult B cell repertoire. Finally, although DHQ52 rearrangements from 8 weeks of gestation on display extensive N additions between D and JH, VHDJH rearrangements that use other DH gene segments, such as DXP, DN, and DLR, often lack N additions between D and JH. Similar gene rearrangements, however, display extensive N additions between D and JH in the expressed adult B cell repertoire.
In the fetus, differences in D and JH gene utilization, and N addition lead to a repertoire of Ig H chain CDR3 intervals that is distinctly shorter than that found in the adult. Few H chain CDR3 intervals of greater than 16 amino acids have been described and the average CDR3 interval in the fetus measures 10.5 ± 2.5 residues (mean ± SD). In cord blood, the average is 13.4 ± 4.5 residues; and in the adult, approximately 50% of CDR3 intervals are 16 amino acids or longer. The distribution of CDR3 lengths and the lesser number of N additions in the DJH junctions of the mAbs of the B-1b repertoire is similar to that seen in fetal life, but neither the B-1b nor B-1a natural Ig repertoires match the pattern of DH or JH utilization seen in second trimester fetal liver. Thus, although some of the natural mAbs analyzed here displayed features reminiscent of those of the early B cell repertoire, e.g., short CDR3 intervals with minimal N additions in the DJH junctions of the Abs expressed by B-1b cells, neither the B-1a nor the B-1b cell-derived Abs were encoded by selections of Ig VHDJH gene sequences overlapping with those of the fetal or neonatal human B cell repertoire.
The feature of the B-1a and B-1b cell-derived mAbs most reminiscent of the fetal repertoire is polyreactivity. Analysis of the mAbs reported here suggests that germline-encoded limitations in the sequence and in the length of H chain CDR3, as seen in B-1b cell-derived Ig, cannot per se account for polyreactivity, as polyreactivity is also a feature of the B-1a cell-derived mAbs. The B-1a mAbs frequently used multiple D segments, and contained, on average, more unencoded nucleotide additions in their VHDJH junctions (17.0 ± 10.5, mean ± SD) than the B-1b cell-derived mAbs did (7.7 ± 5.9). The preferential use of multiple D elements by the B-1a cell-derived mAbs and the relative lack of N additions in the B-1b cell-derived mAbs resulted in significant differences in CDR3 length, with the H chain CDR3s of B-1a cell-derived mAbs, on average, five to six amino acids larger than those found in B-1b cell mAbs. The use of multiple D-D joins leaves open the possibility that the polyreactivity of B-1a-derived Abs reflects fine differences in the mechanisms of Ig gene rearrangement inherent to different human B cell lineages or populations. Accordingly, preferential usage of selected VH and and/or JH genes could merely reflect preferential rearrangement of given family members or genes by precursors of different populations. Alternatively, restricted expression of given VH family genes may be brought about or magnified by selection by a “superantigen” for unconventional Ag-binding sites encoded partially or entirely by the FR structures that define the family (48), as seen in the binding of Staphylococcus protein A by most human VH3 family gene products (49), and in the binding of the I/i Ag by the VH4 family V4-34 gene product in human cold agglutinins (50). Such broad selection processes are likely to have a major impact on the assortment of the B cell repertoire at different stages of ontogeny. However, amplification of a highly restricted population of B cell clonotypes is a typical feature of the mechanism of Ag selection and depends on driving forces that impact primarily on somatically generated structures, and mainly the H chain CDR3, as suggested by the crystallographic analysis of different Ag-Ab complexes (51). A primary role of the H chain CDR3 in providing the main structural correlates for polyreactivity, and therefore Ag selection in B-1 cell expansion is suggested by in vitro human Ig V(D)J gene shuffling and site mutagenesis experiments by us and others (52–56). Expansion of clonotypes bearing polyreactive but functionally discrete surface F(ab′)2 receptors for Ag could be driven by a heterogeneous assortment of naturally occurring Ags, as suggested by the structural heterogeneity of the H chain CDR3s of the present B-1 cell-derived natural mAbs, and that of other human polyreactive IgM, IgA, and IgG mAbs (8, 19–27, 57–59) with multiple but yet discrete Ag-binding activities. This contention is also consistent with the asymmetrical enrichment for Tyr residues in the present mAb H chain CDR3s. Preference for a promiscuous Ag-binding residue in the Ag-binding site could likely occur as a result of selection by Ag.
The hypothesis that, as suggested for murine B-1 cells (60–61),9 Ag receptor-based selection plays an important role in shaping the B-1 repertoire and may be at the basis of the emergence and/or expansion of most natural polyreactive Ab-producing cell clonotypes in the human adult is further supported by the high degree of somatic diversification of the B-1 cell-derived polyreactive mAb VHDJH segments analyzed here. Somatic diversification is underpinned by cellular events that are triggered by Ag selection and that lead to Ab affinity maturation. All but 2 (1 from B-1a and 1 from B-1b cells) of the 18 B-1 cell-derived mAbs sequenced displayed a somatic R mutation load comparable with that found in the putatively monoreactive B-2 cell-derived mAbs, with preferential distribution of R mutations within the VH segment CDRs as compared with the FRs (4.97 × 10−2 vs 2.10 × 10−2 R change/base, respectively, p = 0.004), and high CDR R:S mutation ratios (Table I). In selection by Ag of “conventional” B-2 cells, Ig somatic mutation and class-switching are in general concurrent although mechanistically independent processes. The presence of somatic mutations in the majority of B-1 cell-derived IgM (present data), IgG and IgA (58, 59) strongly suggests that these B cells can undergo a process of clonal expansion and selection mediated by occupancy of the surface F(ab′)2 receptor for Ag. Such an Ag-dependent clonal selection would be consistent with the oligoclonality detected in human adult B-1a cells by other investigators (62).
Although limited in scope, the present findings suggest that in the adult human, natural Ab-producing B-1a cell precursors are distinct, and possibly derive through Ag-driven selection from a restricted set of clonotypes that accounts for a negligible proportion of the early or adult B cell repertoire. The latter possibility is at variance with the early ontogenic origin of B-1a cells, as defined by many experiments in the mouse (3, 5), but would be supported by the recent suggestion—also in the mouse—that adult bone marrow B cells can progress along two discrete differentiation pathways, B-1 or B-2, in response to different stimuli (2, 61): 1) B cells induced to proliferate by cross-linking of surface IgM by soluble anti-μ F(ab′)2 fragments express surface CD5; 2) whereas B cells induced to proliferate in response to thymus-dependent inductive signals, e.g., CD40L, display high density of surface CD23 and no CD5. Cross-linking of surface IgM in absence of engagement of CD40 by CD40L on Th cells is characteristic of thymus-independent type 2 (TI-2) Ags, including ubiquitous self Ags or commonly occurring bacterial Ags, and would be highly effective in B cells expressing Ig genes encoding for natural polyreactive autoantibodies. A TI Ab response, defined as a response that can be induced in vivo in absence of T cells, should arise in the absence of Ig somatic diversification. However, like the polyreactive B-1a and B-1b cell-derived mAbs analyzed here, Abs to prototypic TI-2 Ags, as those on certain human pathogenic bacteria, e.g., Haemophilus influenzae type B polysaccharide, have been shown to undergo extensive somatic mutation and diversification (63, 64). Accordingly, murine Ab responses to similar formally TI-2 Ags, e.g., Pneumococcus polysaccharides, have been shown to be under the influence of regulatory T cells (65). Thus, most of the B-1 cells found in human adults may derive from the expansion of limited clonotypes selected through TI-2 mechanisms.
Taken collectively, the present findings suggest that in the adult human, B-1 cells are distinct from their monoreactive “conventional” B-2 counterparts, and likely emerge through Ag-dependent expansion of a minor clonotypic B cell population, possibly through a somatic diversification mechanism similar to the one that drives the affinity maturation of a “conventional” B-2 Ab response. They, however, cannot provide any indication as to whether the restricted B cell clonotypes to which antigenic pressure is applied are part of the predominantly B-1 developmental waves of colonization of the fetal and neonatal B cell repertoire or emerge through the B-1 differentiation pathway that is putatively available to a mature virgin B cell in the adult bone marrow, as discussed above. These two hypothetically distinct patterns of B-1 cell developmental selection are not mutually exclusive, and would rely on a common stimulus for their initiation, i.e., the engagement of the polyreactive surface F(ab′)2 receptor for Ag by structures borne on self-components or common microbial agents. The nature of the selection factors impinging on B lymphocytes and eventually leading to the expansion of natural Ab-producing B-1 cell precursors remains to be elucidated.
Acknowledgments
We are grateful to Dr. Mimi Kim (Laboratory of Epidemiology and Biostatistics, Institute of Environmental Medicine, New York University School of Medicine) for her help with the statistical analysis. We thank Dr. Hideyuki Ikematsu (Division of Molecular Immunology) for his collaboration in some of these experiments.
Footnotes
This work was supported by United States Public Health Service Grants AR 40908 (P.C.) and AI 33621 (H.W.S.). This is publication 15 from the Division of Molecular Immunology.
The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL Data Bank with accession numbers L23555 to L23571.
Abbreviations used in this paper: CDR, complementarity-determining region; FR, framework region; Kd, Ab dissociation constant for Ag; N additions, unencoded nucleotide additions; R, replacement (mutation); S, silent (mutation); H chain, heavy chain; L chain, light chain; TI, thymus independent.
M. Nouri-Shirazi and A. M. Stall. CD40 signaling induces lineage-specific B cell differentiation in neonatal mice. Submitted for publication.
References
- 1.Herzenberg LA, Kantor AB. B-cell lineages exist in the mouse. Immunol Today. 1993;14:79. doi: 10.1016/0167-5699(93)90063-Q. [DOI] [PubMed] [Google Scholar]
- 2.Haughton G, Arnold LW, Whitmore AC, Clarke SH. B-1 cells are made, not born. Immunol Today. 1993;14:84. doi: 10.1016/0167-5699(93)90064-R. [DOI] [PubMed] [Google Scholar]
- 3.Hardy RR, Hayakawa K. CD5 B cells, a fetal B cell lineage. Adv Immunol. 1994;55:297. doi: 10.1016/s0065-2776(08)60512-x. [DOI] [PubMed] [Google Scholar]
- 4.Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med. 1990;172:1377. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H, Forster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VH-D-JH joining: implications for the joining mechanism and the ontogenetic timing of LY1 B cell and B-CLL progenitor generation. EMBO J. 1990;9:2133. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin F, Chen X, Shu F, Kearney JF. Development of the mouse B cell repertoire. Ann NY Acad Sci. 1995;764:207. doi: 10.1111/j.1749-6632.1995.tb55829.x. [DOI] [PubMed] [Google Scholar]
- 7.Guigou V, Builbert B, Moinier D, Tonnelle C, Boubli L, Avrameas S, Fougereau M, Fumoux F. Immunoglobulin repertoire of human polyspecific antibodies and B cell ontogeney. J Immunol. 1991;146:1368. [PubMed] [Google Scholar]
- 8.Avrameas S. Natural autoantibodies: from horror autotoxicus to gnothi seauton. Immunol Today. 1991;12:154. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 9.Schettino EW, Ichiyoshi Y, Casali P. Structure-function relation in natural and disease-associated human autoantibodies. In: Zanetti M, Capra JD, editors. The Antibodies. Vol. 2. Harwood Academic Publishers; Amsterdam: 1996. pp. 155–203. [Google Scholar]
- 10.Casali P, Kasaian MT, Haughton G. B-1 (CD5) cells. In: Coutinho A, Kazatchkine MD, editors. Autoimmunity: Physiology and Disease. John Wiley and Sons; New York: 1994. pp. 57–88. [Google Scholar]
- 11.Solvason N, Chen X, Shu F, Kearney JF. The human fetal omentum: a site of B cell generation. J Exp Med. 1992;175:397. doi: 10.1084/jem.175.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder HW, Jr, Mortari F, Shiokawa S, Kirkham PM, Elgavish RA, Bertrand FE., III Developmental regulation of the human antibody repertoire. Ann NY Acad Sci. 1995;764:242. doi: 10.1111/j.1749-6632.1995.tb55834.x. [DOI] [PubMed] [Google Scholar]
- 13.Kipps TJ. The CD5 B cell. Adv Immunol. 1989;47:117. doi: 10.1016/s0065-2776(08)60663-x. [DOI] [PubMed] [Google Scholar]
- 14.Sanz I. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J Immunol. 1991;147:1720. [PubMed] [Google Scholar]
- 15.Mortari F, Newton JA, Wang JY, Schroeder HW., Jr The human cord blood antibody repertoire: frequent use of the VH7 gene family. Eur J Immunol. 1992;22:241. doi: 10.1002/eji.1830220135. [DOI] [PubMed] [Google Scholar]
- 16.Yamada M, Wasserman R, Reichard BA, Shane S, Caton AH, Rovera G. Preferential utilization of specific immunoglobulin heavy chain diversity and joining segments in adult human peripheral blood B lymphocytes. J Exp Med. 1991;173:395. doi: 10.1084/jem.173.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy RR, Hayakawa K, Shimizu M, Yamasaki K, Kishimoto T. Rheumatoid factor secretion from human Leu-1+ B cells. Science. 1987;236:81. doi: 10.1126/science.3105057. [DOI] [PubMed] [Google Scholar]
- 18.Casali P, Burastero SE, Nakamura M, Inghirami G, Notkins AL. Human lymphocytes making rheumatoid factor and antibody to ssDNA belong to the Leu-1+ B-cell subset. Science. 1987;236:77. doi: 10.1126/science.3105056. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura M, Burastero SE, Notkins AL, Casali P. Human monoclonal rheumatoid factor-like antibodies from CD5 (Leu-1+) B cells are polyreactive. J Immunol. 1988;140:4180. [PubMed] [Google Scholar]
- 20.Kasaian MT, Ikematsu H, Casali P. Identification and analysis of a novel human surface CD5− B lymphocyte subset producing natural antibodies. J Immunol. 1992;148:2690. [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura M, Burastero SE, Ueki Y, Larrick JW, Notkins AL, Casali P. Probing the normal and autoimmune B cell repertoire with Epstein-Barr virus: frequency of B cells producing monoreactive high affinity autoantibodies in patients with Hashimoto’s disease and systemic lupus erythematosus. J Immunol. 1988;141:4165. [PubMed] [Google Scholar]
- 22.Sanz I, Casali P, Thomas JW, Notkins AL, Capra JD. Nucleotide sequences of eight human natural antibody VH regions reveals apparent restricted use of VH families. J Immunol. 1989;142:4054. [PubMed] [Google Scholar]
- 23.Harindranath N, I, Goldfarb S, Ikematsu H, Burastero SE, Wilder RL, Notkins AL, Casali P. Complete sequence of the genes encoding the VH and VL regions of low and high affinity monoclonal IgM and IgA1 rheumatoid factors produced by CD5+ B cells from a rheumatoid arthritis patient. Int Immunol. 1991;3:865. doi: 10.1093/intimm/3.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasaian MT, Casali P. B-1 cellular origin and VH segment structure of IgG, IgA, and IgM anti-DNA autoantibodies in patients with systemic lupus erythematosus. Ann NY Acad Sci. 1995;764:410. doi: 10.1111/j.1749-6632.1995.tb55856.x. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura M, Casali P. Generation of human monoclonal antibody-producing cell lines by Epstein-Barr virus (EBV)-transformation of autoreactive B lymphocytes and by somatic cell hybridization techniques: application to the analysis of the autoimmune B cell repertoire. Immunomethods. 1992;1:159. doi: 10.1016/S1058-6687(05)80012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantovani L, Wilder RL, Casali P. Human rheumatoid B-1a (CD5+ B) cells make somatically hypermutated high affinity IgM rheumatoid factors. J Immunol. 1993;151:473. [PMC free article] [PubMed] [Google Scholar]
- 27.Kasaian MT, Ikematsu H, Balow JE, Casali P. Structure of the VH and VL segments of monoreactive and polyreactive IgA autoantibodies to DNA in patients with systemic lupus erythematosus. J Immunol. 1994;152:3137. [PMC free article] [PubMed] [Google Scholar]
- 28.Zar JH. Biostatistical Analysis. Prentice-Hall; Englewood Cliffs, NJ: 1984. [Google Scholar]
- 29.Walter MA, Surti U, Hofker MH, Cox DW. The physical organization of the human immunoglobulin heavy chain gene complex. EMBO J. 1990;9:3303. doi: 10.1002/j.1460-2075.1990.tb07530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda F, Shin EK, Nagaoka H, Matsumura R, Haino M, Fukita Y, Takaishi S, Imai T, Riley JH, Anand R, Soeda E, Honjo T. Structure and physical map of 64 variable segments in the 3′ 0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet. 1993;3:88. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- 31.Cook GP, I, Tomlinson M, Walter G, Riethman H, Carter NP, Buluwela L, Winter G, Rabbitts TH. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat Genet. 1994;7:162. doi: 10.1038/ng0694-162. [DOI] [PubMed] [Google Scholar]
- 32.Cook GP, I, Tomlinson M. The human immunoglobulin VH repertoire. Immunol Today. 1995;16:237. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]
- 33.Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Structure of the human immunoglobulin μ locus: characterization of embryonic and rearranged J and D genes. Cell. 1981;27:583. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- 34.Siebenlist U, Ravetch JV, Korsmeyer S, Waldmann T, Leder P. Human immunoglobulin D segments encoded in tandem mutigenic families. Nature. 1981;294:631. doi: 10.1038/294631a0. [DOI] [PubMed] [Google Scholar]
- 35.Ichihara Y, Matsuoka H, Kurosawa Y. Organization of human immunoglobulin heavy chain diversity gene loci. EMBO J. 1988;7:4141. doi: 10.1002/j.1460-2075.1988.tb03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buluwela L, Albertson DG, Sherrington P, Rabbitts PH, Spurr N, Rabbitts TH. The use of chromosomal translocations to study human immunoglobulin gene organization: mapping DH segments with 35 kb of the Cμ gene and identification of a new DH locus. EMBO J. 1988;7:2003. doi: 10.1002/j.1460-2075.1988.tb03039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichihara Y, Abe M, Yasui H, Matsuoka H, Kuroswawa Y. At least five DH genes of human immunoglobulin heavy chains are encoded in 9-kilobase DNA fragments. Eur J Immunol. 1988;18:649. doi: 10.1002/eji.1830180426. [DOI] [PubMed] [Google Scholar]
- 38.Sanz I, Wang SS, Meneses G, Fischbach M. Molecular characterization of human Ig heavy chain DIR genes. J Immunol. 1994;152:3958. [PubMed] [Google Scholar]
- 39.Schroeder HW, Jr, Hillson JL, Perlmutter RM. Early restriction of the human antibody repertoire. Science. 1987;238:791. doi: 10.1126/science.3118465. [DOI] [PubMed] [Google Scholar]
- 40.Kabat EA, Wu TT, Perry HM, Bottesman KS, Foeller C. Sequences of proteins of immunological interest. United States Department of Health and Human Services; Bethesda, MD: 1991. [Google Scholar]
- 41.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 42.Kirkham PM, Schroeder HW., Jr Antibody structure and the evolution of immunoglobulin V gene segments. Semin Immunol. 1994;6:1. doi: 10.1006/smim.1994.1045. [DOI] [PubMed] [Google Scholar]
- 43.Pennel CA, Arnold LW, Haughton G, Clarke SH. Restricted Ig variable region gene expression among Ly-1+ lymphomas. J Immunol. 1988;141:2788. [PubMed] [Google Scholar]
- 44.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15:367. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perlmutter RM, Kearney JF, Chang SP, Hood LE. Developmentally controlled expression of immunoglobulin VH genes. Science. 1985;227:1597. doi: 10.1126/science.3975629. [DOI] [PubMed] [Google Scholar]
- 46.Alt FW, Blackwell TK, Yancopoulos GD. Development of the primary Ab repertoire. Science. 1987;238:1079. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 47.Pascual V, Verkruyse L, Casey ML, Capra JD. Analysis of Ig H chain gene segment utilization in human fetal liver: revisiting the “proximal utilization hypothesis”. J Immunol. 1993;151:4164. [PubMed] [Google Scholar]
- 48.Silverman GJ. Superantigens and spectrum of unconventional B-cell antigens. Immunologist. 1994;2:51. [Google Scholar]
- 49.Potter KN, Li Y, Capra JD. Staphylococcal protein A simultaneously intereacts with framework region 1, complementary-determining region 2, and framework region 3 on human VH3-encoded Igs. J Immunol. 1996;157:2982. [PubMed] [Google Scholar]
- 50.Li Y, Spellerberg MB, Stevenson FK, Capra JD, Potter KN. The binding specificity of human V4-34 (VH 4-21) encoded antibodies is determined by both VH framework 1 and complementarity determining region 3. J Immunol. 1996;256:577. doi: 10.1006/jmbi.1996.0110. [DOI] [PubMed] [Google Scholar]
- 51.Wilson IA, Stanfield RL. Antibody-antigen interactions. Curr Opin Struct Biol. 1993;3:113. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 52.Martin T, Duffy SF, Carson DA, Kipps TJ. Evidence for somatic selection of natural autoantibodies. J Exp Med. 1992;175:983. doi: 10.1084/jem.175.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin T, Crouzier R, Weber JC, Kipps TJ, Pasquali JL. Structure-function studies on a polyreactive (natural) autoantibody. J Immunol. 1994;152:5988. [PubMed] [Google Scholar]
- 54.Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin H and L chain V segments. J Exp Med. 1994;180:885. doi: 10.1084/jem.180.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ichiyoshi Y, Zhou M, Casali P. A human anti-insulin IgG autoantibody apparently arises through clonal selection from an insulin-specific “germ-line” natural Ab template. J Immunol. 1995;154:226. [PMC free article] [PubMed] [Google Scholar]
- 56.Crouzier R, Martin T, Pasquali JL. H chain variable region, L chain variable region, and H chain CDR3 influences on the mono- and polyreactivity and on the affinity of human monoclonal rheumatoid factors. J Immunol. 1995;154:4526. [PubMed] [Google Scholar]
- 57.Harindranath N, Ikematsu H, Notkins AL, Casali P. Structure of the VH and VL regions of polyreactive and monoreactive human natural antibodies to HIV-1 and E. coli β-galactosidase. Int Immunol. 1993;5:1523. doi: 10.1093/intimm/5.12.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikematsu H, Harindranath N, Ueki Y, Notkins AL, Casali P. Clonal analysis of a human antibody response. II. Sequences of the VH genes of human IgM, IgG, and IgA to rabies virus reveal preferential utilization of VH3 segments and somatic hypermutation. J Immunol. 1993;150:1325. [PMC free article] [PubMed] [Google Scholar]
- 59.Ikematsu H, Schettino EW, Casali P. Structure of the VHDJH segments of human natural polyreactive IgM and IgG antibodies. Ann NY Acad Sci. 1995;764:362. [PubMed] [Google Scholar]
- 60.Arnold LW, Pennell CA, McCray SK, Clarke SH. Development of B-1 cells: segregation of phosphatidyl choline-specific B cells to the B-1 population occurs after immunoglobulin gene expression. J Exp Med. 1994;179:1585. doi: 10.1084/jem.179.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wortis HH, Teutsch M, Higer M, Zheng J, Parker DC. B-cell activation by crosslinking of surface IgM or ligation of CD40 involves alternative signal pathways and results in different B-cell phenotypes. Proc Natl Acad Sci USA. 1995;92:3348. doi: 10.1073/pnas.92.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber JC, Blaison G, Martin T, Knapp AM, Pasquali JL. Evidence that the VκIII gene usage is nonstochastic in both adult and newborn peripheral B cells and that peripheral CD5+ adult B cells are oligoclonal. J Clin Invest. 1994;93:2093. doi: 10.1172/JCI117204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adderson EE, Shackelford PG, Insel RA, Quinn A, Wilson PM, Carroll WL. Immunoglobulin light chain variable region gene sequences for human antibodies to Haemophilus influenzae type b capsular polysaccharide are dominated by a limited number of V kappa and V lambda segments and VJ combinations. J Clin Invest. 1992;89:729. doi: 10.1172/JCI115649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adderson EE, Shackelford PG, Quinn A, Wilson PM, Cunningham MW, Insel RA, Carroll WL. Restricted immunoglobulin VH usage and VDJ combinations in the human response to Haemophilus influenzae type b capsular polysaccharide: nucleotide sequences of monospecific anti-Haemophilus antibodies and polyspecific antibodies cross-reacting with self antigens. J Clin Invest. 1993;91:2734. doi: 10.1172/JCI116514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker PJ. Regulation of magnitude of antibody-response to bacterial polysaccharide antigens by thymus-derived lymphocytes. Infect Immun. 1990;58:3465. doi: 10.1128/iai.58.11.3465-3468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]