Summary
Objective
Neisseria meningitidis serogroup W135 has been associated with global outbreaks since the 2000 Hajj. Considering that N. meningitidis serogroup W135 is the third most prevalent serogroup isolated in Brazil in the last 10 years, and the possibility that the Hajj-related N. meningitidis serogroup W135 clone has been causing disease in Brazil, the present study characterized invasive N. meningitidis serogroup W135 isolates recovered in Brazil from 1990 to 2005.
Methods
The isolates were characterized by serotyping, PorA and PorB VR typing, FetA and 16S rRNA typing, multilocus sequence typing (MLST) and pulsed field gel electrophoresis (PFGE).
Results
Based on MLST, 73% of the isolates were clustered in one major clone of ST-11 complex/ET37 complex. Strains of this clone had the same STs, serotypes and PorA VR types as found in Hajj-related N. meningitidis serogroup W135 clone. One of these strains had the Hajj-2000 outbreak strain genotype, including 16S rRNA gene sequence 31 and 84% relatedness by PFGE.
Conclusion
Taken together, these data suggest that the Hajj-related N. meningitidis serogroup W135 clone is present in Brazil but has not yet caused a substantial number of infections. Given the emergence of N. meningitidis serogroup W135 globally and the unpredictability of meningococcal disease epidemiology, continued surveillance for this invasive N. meningitidis serogroup W135 clone is needed for control and prevention strategies.
Keywords: Neisseria meningitidis, Serogroup W135, Epidemiology
Introduction
Meningococcal disease (MD) is an important cause of morbidity and mortality and a leading cause of bacterial meningitis and septicemia in children and young adults worldwide.1 In Brazil, around 3200 new cases of sporadic and outbreak-associated MD are reported each year, making this disease an important concern for Brazilian public health authorities (http://www.saude.gov.br/sinanweb).
The distribution of meningococcal serogroups in Brazil has changed with time Brazil but Neisseria meningitidis serogroups A, B, and C have been the most prevalent since the 1920s.2–5 We have approximately 17,000 N. meningitidis strains in our collection isolated from 1970 to 2005. N. meningitidis serogroups B (n = 11,515) and C (n = 4915) represent 68% and 29% of the collection, respectively. Other serogroups, namely W135 (n = 396), Y (n = 84), A (n = 81), 29E (n = 4), Z (n = 2), and X (n = 3) represent 3.0% of isolates.
Although these less frequently isolated serogroups are generally considered to have low potential to cause invasive disease or outbreaks, in 2000, a N. meningitidis serogroup W135 outbreak was reported in Saudi Arabia during the Hajj pilgrimage.6 Since then, the Hajj-related N. meningitidis serogroup W135 clone has spread outside Saudi Arabia to other countries,7,8 including to Burkina Faso where it caused a large meningococcal disease epidemic in 2002.9 The Hajj-related N. meningitidis serogroup W135 clone is characterized by serotype 2a, PorA variable region (VR) VR1-5, VR2-2, sequence type (ST)-11 and 16S rRNA types 13, 14 or 31.10
The first reported N. meningitidis serogroup W135 disease in Brazil occurred in 1983 in Sao Paulo state from a fatal case in a 10 year old child, but the number of MD cases caused by this serogroup has been as low as 40 cases per year, with an incidence of approximately 0.1 per 100,000 inhabitants (Epidemiological Surveillance Office, personal communication). However, since 2000 there has been a significant trend (p ≤ 0.0001) in the percentage of meningococcal disease caused by serogroup W135 in Brazil (http://www.saude.cve.sp.gov.br).11,12 N. meningitidis serogroup W135 disease has also recently increased in Argentina, accounting for 28% of cases during the first 5 months of 2008.13
The molecular characteristics and strain relatedness of the N. meningitidis serogroup A, N. meningitidis serogroup B, and N. meningitidis serogroup C isolated during the last three epidemic periods in Greater Sao Paulo, have been established,3–5 but the molecular characteristics of Brazilian N. meningitidis-serogroup-W135 are unknown. Considering that N. meningitidis serogroup W135 is the third most prevalent serogroup isolated in Brazil in the last 10 years, and the possibility that the Hajj-related N. meningitidis serogroup W135 clone has been causing disease in Brazil, it is important to understand the clonal characteristics and relatedness of N. meningitidis serogroup W135 strains isolated in Brazil. Using a series of phenotypic and genotypic approaches, we characterized all available Brazilian N. meningitidis serogroup W135 strains isolated in Brazil from 1990 to 2005.
Material and methods
Bacterial strains
A total of 216 N. meningitidis serogroup W135 invasive strains from 15 Brazilian States were characterized in this study (Table 1).
Table 1.
Origin of 216 Neisseira meningitidis serogroup W135 strains isolated in Brazil by year.
| Brazilian states | Year | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||||
| 1990 | 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | |
| Alagoas | 1 | |||||||||||||||
| Bahia | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| Ceara | 1 | |||||||||||||||
| Distrito Federal | 3 | 2 | 1 | |||||||||||||
| Espirito Santo | 1 | 1 | ||||||||||||||
| Goais | 1 | |||||||||||||||
| Minas Gerais | 1 | 3 | 4 | |||||||||||||
| Mato Grosso do Sul | 1 | |||||||||||||||
| Pernambuco | 1 | 2 | ||||||||||||||
| Parana | 2 | 1 | 1 | 1 | 6 | 2 | 4 | 2 | 2 | 2 | 1 | |||||
| Rio de Janeiro | 2 | 1 | 1 | 1 | 2 | 2 | 6 | 1 | ||||||||
| Rio Grande do Sul | 2 | 1 | 1 | 3 | 2 | 3 | 1 | 3 | 1 | 4 | 1 | |||||
| Santa Catarina | 1 | 1 | 1 | 2 | 1 | 1 | ||||||||||
| Sergipe | 1 | |||||||||||||||
| São Paulo | 4 | 12 | 6 | 4 | 7 | 1 | 6 | 6 | 3 | 9 | 11 | 11 | 8 | 6 | 8 | 14 |
| Total | 14 | 16 | 9 | 7 | 12 | 5 | 8 | 16 | 13 | 19 | 18 | 15 | 11 | 11 | 24 | 18 |
Serological typing
Serotyping for all 216 isolates was performed by dot-blotting using whole cell suspensions as previously described.14 The serotyping was performed with a set of 18 PorB and 15 PorA murine MAbs specific for the variable regions. MAbs for serotypes 2a (F12-7B7/1E10), 2b (F1-9H10/1B3), 4 (F10-2H7/1F7), 7 (F22-8B5/1D10), 9 (F24-11F5/3B4), 17 (F4-3C1/1A6), 10 (F11-6D12/1C5) and 23 (F29-1G1/B4); and for serosubtypes P1.1 (F10-5G6/1B11), P1.4 (F11-2A9/1A4), P1.9 (F24-5E11/2H9), P1.15 (F8-8F12/1D6), P1.14-6 (F29-8H1/1E11), and P1.22-1 (F4-1F1/1F3), were produced at IAL by the authors. MAbs for serotypes 8 (2725H6) and 15 (1951C8), and for serosubtypes P1.2 (1649C7) were provided by C. E. Frasch, FDA, Bethesda, MD. MAbs for serotypes 2c (5-1-P2c), 5 (7BG5-H2), 11 (9-1-P11), and 19 (17-1-P19), and for serosubtypes P1.3 (12-1), P1.16 (3-1-P1.16), and P1.19 (7A2-11) were provided by W. D. Zollinger, WRAIR, Washington, D.C. MAb for serotype 22 (ATIA5A7/5) was provided by P. Kriz, NIPH, Prague, Czech Republic. MAbs for serotypes 1 (MN3C6B-95/680), 14 (MN5C8C-95/688), and 21 (6B11-F2-B5-95/692); and serosubtypes P1.5 (MN22A9.19-95/702), P1.7 (MN14C11.6-95/706), P1.10 (MN20F4.17-95/710), P1.12 (MN20A7.10-95/712), and P1.14 (MN21G3.17-95/716) were provided by the NIBSC, Potters Barr, England.
OMP gene sequencing
DNA sequences encoding PorA, PorB and FetA were performed as described elsewhere.15–19 The assignment of porA, porB2 and porB3, and fetA alleles was performed by querying the website (http://neisseria.org/nm/typing/mlst).
MLST
MLST was performed according to the methods of Maiden et al.20 Primers, determination of sequence alleles, and designation of sequence types are described on the MLST website (http://neisseria.org/nm/typing/mlst). Minimum spanning tree was constructed to infer evolutionary models, using Bionumerics software (version 5.10), which incorporates the BURST algorithm (Applied Maths).21
16S rRNA gene sequencing
A total of 142 N. meningitidis serogroup W135 isolates were selected for 16S rRNA gene typing based on serotyping, PorA VR typing and MLST results. We also included strain LNP17592, a N. meningitidis serogroup W135 isolated during the Hajj-2000 outbreak in France and strain LNP18359 a N. meningitidis serogroup W135 isolated in France in 2001 as Hajj-2000 outbreak negative control. 16S rRNA gene typing was carried out as previously described,22 with some minor modifications. The 16S rRNA genes from N. meningitidis isolates were amplified with primers 9R (5′TTCGGTTCTTCGCTGCT3′) and 2174F (5′ATGCGTTCGATA TTGCTATCT3′). Reaction mixtures were first incubated for 5 min at 94 °C. The mixture then underwent 30 cycles of 15 s at 94 °C, 15 s at the annealing temperature of 56 °C, and then 90 s at 72 °C. Finally, the reaction mixtures were incubated at 72 °C for 5 min. The amplified product of approximately 1480 bp was sequenced by using the previously described primers 357, 530, 790, 981 in the forward and reverse orientations, as well as primers 8F and 1492R described before.11 Six additional sequencing primers were also used, primers 1968F (5′GTCGTCAGCTCGTGTCGT GAG3′), 1083F (5′CGTGACATGTTGGGTTAAGTC3′), 1127F (5′ATTAGTTGCCATCATTCAGTT3′), 1333R-(5′CTAGCGATTCC GACTTCATGC3′), 180R (5′TCTCTCAAGACGTATGCGGTA3′), 591R (5′CATCCTGCTTAAGTAACCGTC3′).
PFGE
We further analyzed a total of fifteen isolates representative for each year, from 1990 to 2005, of serotype 2a, PorA VR1-5 and VR2-2 types, ST-11 and 16S type 13; and one strain (N.976/02) presenting 16S type 31, were chosen to be additionally analyzed by PFGE, as previously described, using the restriction enzyme SpeI.23,24 The strains LNP17592 and LNP18359 were also included. Low Range PFG Marker was used for intragel normalization. The restriction profiles were analyzed by visual comparison and by computer-assisted using the Bionumerics software (Applied Maths, St-Martens-Latem, Belgium). The Dice band-based similarity coefficient (SD) was used with a band position tolerance of 1.5% and an optimization of 0.5%.
For this study, we defined the Hajj-related N. meningitidis serogroup W135 clone as a group of isolates presenting serotype 2a, PorA VR-5,2, ST-11, and 16S types 13, 14, or 31 that is very closely (at least >80% similarity) related by PFGE with a Hajj-2000 outbreak strain.
Results
Serotyping
The distribution of serotype and serosubtype of the 216 isolates is shown in Table 2. Serotype 2a was the most frequent (149/216, 69%), followed by serotype 19,10 (41/216, 19%); all other serotypes occurred in fewer than 4.0% of isolates. Serosubtype P1.2 was the most frequent (113/216, 52%), followed by serosubtypes P1.16 (39/216, 18%) and P1.5,2 (33/216, 15%); all other serosubtypes occurred in fewer than 1.8% of the isolates. The 216 N. meningitidis serogroup W135 isolates displayed 28 different serotype-serosubtype antigen combinations. The three most common antigenic combinations were W135:2a:P1.2 (106/216, 49%), W135:19,10:P1.16 (36/216, 17%) and W135:2a:P.5,2 (30/216, 14%).
Table 2.
Results of serological and molecular typing of 216 Neisseria meningitidis serogroup W135 strains isolated in Brazil from 1990 to 2005.
| Groups | ST complex | ST | Serotype | Serosubtype | PorA VR types | 16S types | PorB VR types | FetA VR types | Total number of strains | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| VR1 | VR2 | |||||||||
| ST-11 n = 157 | ST-11 complex/ET37 complex | 11 | 2a | NST | 5 | 2 | 13 | 2-144 | F1-1 | 1 |
| ST-11 complex/ET37 complex | 11 | 2a | NST | 5 | 2 | 13 | 2-145 | F1-1 | 3 | |
| ST-11 complex/ET37 complex | 11 | 2a | NST | 5 | 2 | 13 | 2-69 | F1-1 | 6 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.19,15 | 19 | 15 | ND | 2-147 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-144 | F1-1 | 6 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2145 | F1-1 | 34 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 254 (GQ294475)a | 2145 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 255 (GQ294476) | 2145 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 256 (GQ294477) | 2145 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-146 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-150 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-2 | F1-1 | 11 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 257 (GQ294478) | 2-48 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-69 | F1-1 | 39 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 258 (GQ294479) | 2-69 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 259 (GQ294480) | 2-69 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-69 | F1-5 | 2 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5 | 2 | 13 | 2-69 | F4-12 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.2 | 5-1 | 2-2 | ND | 2-69 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.5 | 5-1 | 10-13 | ND | 2-145 | F1-1 | 1 | |
| ST-11 complex/ET37 complexb | 11b | 2ab | P1.5,2b | 5b | 2b | 31b | 2-145 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.5,2 | 5 | 2 | 13 | 2-2 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.5,2 | 5 | 2 | 13 | 2-69 | F1-1 | 11 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.5,2 | 5 | 2 | 13 | 2-145 | F1-1 | 15 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.5,2 | 5 | 2 | 13 | 2-148 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2a | P1.5,2 | 5 | 2 | 13 | 3-36 | F1-1 | 2 | |
| ST-11 complex/ET37 complex | 11 | 2b | P1.2 | 5 | 2 | ND | 2-3 | F1-1 | 3 | |
| ST-11 complex/ET37 complex | 11 | 2b | P1.2 | 5 | 2 | ND | 3-3 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2b | P1.5,2 | 5 | 2 | ND | 2-3 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | 2b | NST | 5 | 2 | ND | 2-3 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | NT | P1.2 | 5 | 2 | ND | 2-147 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 11 | NT | P1.2 | 5 | 2 | ND | 2-149 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 4344 | 2a | P1.2 | 5 | 2 | ND | 2-146 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 6308 | 2a | P1.2 | 5 | 2 | ND | 2-146 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 2956 | 2a | P1.2 | 5 | 2 | ND | 2-2 | F1-1 | 1 | |
| ST-11 complex/ET37 complex | 3419 | 2b | P1.2 | 5 | 2 | ND | 2-3 | F1-1 | 1 | |
| Non-ST-11 n = 59 | ST-22 complex | 6342 | 23 | P1.16 | 21 | 16 | ND | 2-23 | F4-1 | 1 |
| ST-22 complex | 3664 | 23 | P1.3 | 18-1 | 3 | ND | 2-23 | F4-1 | 1 | |
| ST-22 complex | 22 | 23 | P1.3 | 18-1 | 3 | ND | 2-23 | F4-1 | 1 | |
| ST-23 complex/Cluster A3 | 23 | 19,14 | NST | 18-1 | 30-3 | ND | 336 | F5-13 | 1 | |
| ST-23 complex/Cluster A3 | 6309 | 19,14 | P1.14 | 7-2 | 14 | ND | 3-215 | F1-18 | 1 | |
| ST-32 Complex/ET5 Complex | 639 | 4 | NST | 7-1 | 1 | ND | 3-79 | F5-13 | 1 | |
| ST-32 Complex/ET5 Complex | 3774 | 4,7 | 19.15 | 19 | 15 | ND | 3-1 | F5-1 | 1 | |
| ST-32 Complex/ET5 Complex | 3654 | 4,7 | 19.15 | 19 | 15 | ND | 3-1 | F5-1 | 1 | |
| ST-32 Complex/ET5 Complex | 6303 | 2a | NST | 7 | 16 | ND | 3-24 | F3-3 | 1 | |
| ST-41/44 complex/Lineage 3 | 6310 | 19.10 | NST | 12-1 | 16-8 | ND | 3-122 | F1-34 | 1 | |
| ST-41/44 complex/Lineage 3 | 2288 | 4,21 | NST | 7-4 | 1 | ND | 3-239 | F1-8 | 1 | |
| ST-167 complex | 3039 | 19,1 | P1.5 | 5-1 | 10-4 | ND | 3-348 | F3-9 | 2 | |
| ST-174 complex | 844 | 19,1 | P1.12 | 12-1 | 16-8 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 844 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F1-18 | 2 | |
| ST-174 complex | 844 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F1-19 | 1 | |
| ST-174 complex | 844 | 19,1 | P1.16 | 21 | 16 | ND | 3-236 | F1-29 | 1 | |
| ST-174 complex | 844 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F2-7 | 1 | |
| ST-174 complex | 844 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 25 | |
| ST-174 complex | 844 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-15 | 1 | |
| ST-174 complex | 844 | 19,1 | P1.5,2 | 5-20 | 2-2 | ND | 3-35 | F1-1 | 1 | |
| ST-174 complex | 844 | NT | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 844 | NT | NST | 7-2 | 13-2 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6353 | 7 | NST | 7-2 | 13 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6327 | 17,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6325 | 23 | P1.12 | 12-1 | 13 | ND | 2-9 | F6-2 | 1 | |
| ST-174 complex | 6302 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6307 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6311 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6328 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6329 | 19,1 | P1.16 | 21 | 16 | ND | 3-35 | F5-13 | 1 | |
| ST-174 complex | 6304 | 2b | P1.5,2 | 5-1 | 2-1 | ND | 3-3 | F1-5 | 1 | |
| ST-175 complex | 5770 | 17,7 | P1.5 | 5-1 | 10-22 | ND | 3-100 | F1-7 | 1 | |
| – | 6306 | 23 | P1.7 | 7-1 | 1 | ND | 3-9 | F3-9 | 1 | |
VR, Variable region; ST, Sequence type; NT, non-serotypeable; NST, non-serosubtypeable; STs, PorA and PorB written in bold were first described in this study.
Genbank accession number for 16S rRNA gene sequence.
This isolate strain (N.976-02) has all of the Hajj-2000 outbreak strain characteristics.
MLST, PorA, PorB2 and PorB3, FetA and 16S rRNA typing
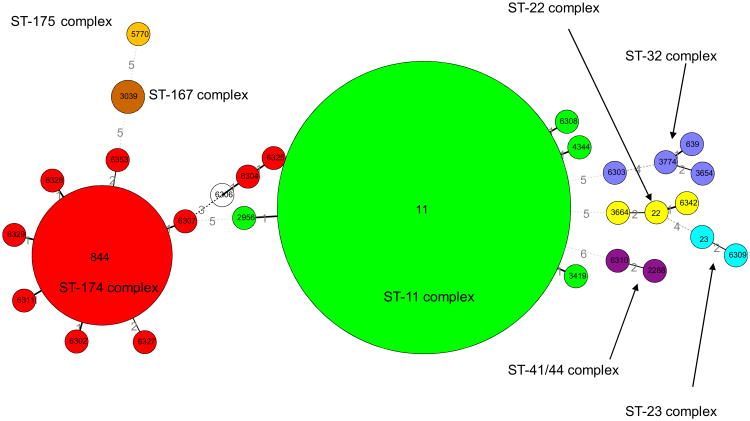
Twenty-nine different STs were found among the 216 N. meningitidis serogroup W135 isolates (Table 2) and the relationships between closely related isolates are displayed (Fig. 1). The 29 STs were grouped into eight different clonal complexes: ST-11 complex (n = 157/216, 73%), ST-174 complex (n = 44/216, 20%), ST-32 complex (n = 4/216, 2.0%), ST-22 complex (n = 3/216, 1.0%), ST-167 complex (n = 2/216, 1.0%), ST-23 complex (n = 2/216, 1.0%), ST-41/44 Lineage 3 (n = 2/216, 1.0%) and ST-175 complex (n = 1/216, 1.0%). The most prevalent clonal complex, ST-11 complex, was represented by the sequence type ST-11 (n = 153/157, 97%). The second most prevalence clonal complex, ST-174 complex was represented by sequence types ST-844 (n = 35/43, 81.0%) and nine STs represented by only one isolate. From the total of 29 STs found, 14 (48%) were novel STs, and 14 alleles were first described in this study. The novel STs were distributed among six different clonal complexes, and eight (57%) of them belonged to ST-174 complex. Four novel STs (ST-6302, ST-6307, ST-6308, ST-6342) were found being single locus variant (SLV) from ST-11, ST-174 or ST-22. The ST-6303, ST-6304, ST-6309, ST-6310, ST-6311, ST-6325, ST-6328, ST-6329 and ST-6353 and were found being double locus variant (DLV) from ST-32, ST-41-44, ST-174 or ST-23. The remaining ST-6306 was assigned without clonal complex.
Figure 1.
Minimum spanning tree analysis of N. meningitidis W135 strains. The large number within each oval represents the ST and the clonal complex are indicated.
Based on MLST data the studied collection was clustered into two major groups (ST-11 and Non-ST-11). ST-11 group is composed by 157 (73%) N. meningitidis serogroup W135 isolates belonging to ST-11 complex/ET37 complex. All strains in this group express the same STs, serotypes and PorAVR types as found on Hajj-related N. meningitidis serogroup W135 strains. Non-ST-11 group is composed by 59 (27%) N. meningitidis serogroup W135 isolates of different ST complexes with a large diversity of STs, serotypes and PorA VR types.
Among 216 isolates, we found 11 types of PorA VR1 and 15 types on PorA VR2. The most common PorA VR1 and VR2 combination was VR1-5 and VR2-2 (154/216, 71.3%). Thirteen alleles on PorB2 and PorB3 variants were found. The most prevalent alleles of PorB2 were 2-145 (57/216, 26.4%) and 2-69 (62/216, 28.7%), and the allele 3-35 (41/216, 19%) of PorB3. Seventeen alleles on FetA variants were grouped in six families of FetA. The most prevalent variant was F1-1 (155/216, 72.0%). The 16S rRNA gene typing revealed eight different types: 13 (135/142, 95.0%), type 31 (1/142, 0.7%) and six new types described in this study, (Table 2). Among all the isolates, only one strain (N.976/02) isolated in Bahia State in 2001 was found to have all the phenotypic and genotypic characteristics of the Hajj-2000 outbreak strain: W135:2a:P1.5,2, PorA VR type 5,2, ST-11, and 16S type 31.
PFGE
Among the 16 isolates selected for PFGE, three distinct PFGE groups composed by seven PFGE profiles were identified, with 79% overall relatedness (Fig. 2). Group 1 was composed by only two PFGE profiles, sharing 84% similarity, the LNP17592 (Hajj-2000 outbreak strain) and the Brazilian strain N.976/02. The larger group (group 2) contained 11 isolates and that included the LNP18359 (Hajj-2000 outbreak negative control) showed 93% similarity. Among the group 2, the PFGE patterns were seen that had <80% similarity to the group containing the Hajj-2000 outbreak strain. The remaining group 3 presented three PFGE profiles and included the oldest isolates, from 1990 to 1993, clustered with 94% similarity (Fig. 2).
Figure 2.
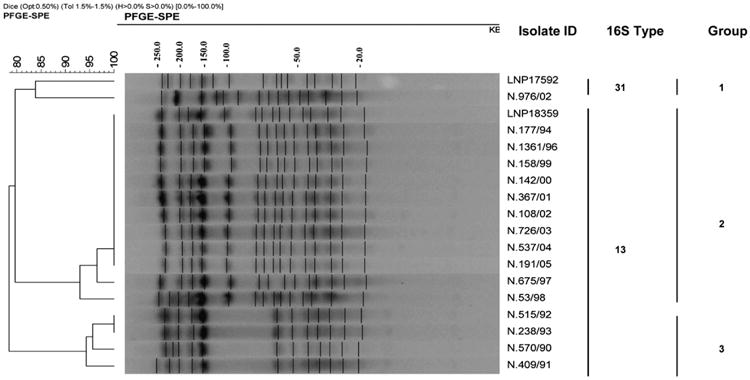
Dendrogram showing genetic relatedness of PFGE profiles among N. meningitidis serogroup W135 strains of serotype 2a, PorA VR type 5,2 and ST-11. The LNP17592 corresponds the Hajj-2000 outbreak strain. The strain identification number, 16S types and group are indicated. Size markers in kb are shown above.
Discussion
Over the past several decades, there have been several Hajj-related meningococcal outbreaks. Moore et al.25 showed that N. meningitidis serogroup A belonging to the III-1 clonal group was introduced into Mecca by South Asian pilgrims attending the 1987 Hajj. Hajjis further disseminated this strain globally to both developed and developing countries upon returning home.26 Since this outbreak, the Saudi government instituted a policy of requiring that all foreign pilgrims provide evidence of vaccination against MD and many pilgrims received a bivalent meningococcal A+C polysaccharide vaccine.27
N. meningitidis serogroup W135 has generally been an uncommon cause of invasive MD and had never been known to cause a major outbreak.28 An international outbreak of N. meningitidis serogroup W135 disease among pilgrims returning from Saudi Arabia and their contacts was reported in 2000 and 2001, demonstrating the epidemic potential of this serogroup.8,29 Subsequently, cases were reported in 16 countries.7,8 Therefore, the current recommendation is for quadrivalent polysaccharide vaccine against N. meningitidis serogroups A, C, W135, and Y for Hajj pilgrims in place of the commonly-used bivalent N. meningitidis A+C polysaccharide vaccine.8,29
From 1990 to 2005, 81,850 cases of MD were reported in Brazil, but only 33% (n = 27,010 cases) were laboratory confirmed (Epidemiological Surveillance Office, personal communication). Of those, N. meningitidis serogroups B, C and W135 isolates were responsible for 63.0% (17,016 cases), 34% (9183 cases) and 1.5% (396 cases), respectively. The remaining 1.5% (415 cases) was due to N. meningitidis Y, 29E and Z together (Instituto Adolfo Lutz, National Reference Laboratory, Brazil).
We previously documented four epidemic lineages circulating in Brazil in a period over three decades.3–5,30,31 We have characterized the hyperinvasive lineages ST-32 complex/ET-5 complex, ST-11 complex/ET-37 complex, ST-8 complex/Cluster A4, and ST-103 complex in both N. meningitidis serogroup B and N. meningitidis serogroup C isolates. Our rates of N. meningitidis serogroup W135 MD cases remain low, but we have observed a slight increase of this serogroup over 25 years of laboratory-based surveil-lance. Whether this is an artifact of surveillance or not is unclear but this serogroup is now the third most common in Brazil, after serogroups B and C (National Reference Laboratory, Brazil). To better understand the characteristics of N. meningitidis serogroup W135 isolated in Brazil, we used genotypic approaches to characterize our collection of isolates recovered during the last 16 years.
Previous studies demonstrated that a limited repertoire of antigen variants persists over time and these tend to be associated with particular invasive clones.32,33 Our findings also suggest this kind of association, 98% of FetA type F1-1 and 100% of PorB2-69:P1.5,2 and PorB2-145:P1.5,2 combinations are associated with ST-11 complex/ET37 complex isolates. We also found association between PorB3-35:P1.21,16 and ST-174 complex isolates.
Sacchi et al.22 identified 16S rRNA types that are exclusively associated with strains of certain hypervirulent clones: 16S type 5 with Subgroup III, 16S type 4 with ST-32 complex/ET5 complex and 16S types 12 and 13 with ST-11 complex/ET37 complex. A single 16S type (type 31) was identified among the 26 Hajj-2000 outbreak strains and one sporadic isolate.10 Our results showed that 136 (86.6%) of the 157 Brazilian N. meningitidis serogroup W135 ST-11 complex/ET37 complex strains are similar to the Hajj-related N. meningitidis serogroup W135 clone (serotype 2a, PorA VR type 5,2, ST-11 and type 13), and only one strain (N.976/02) presented 16S type 31 as the Hajj-2000 outbreak strain.
PFGE has previously been shown to discriminate between different members of the ST-11 complex/ET37 complex.10,24 The present study demonstrated seven different profiles among the 16 isolates of N. meningitidis serogroup W135 ST-11 complex/ET37 complex isolated in Brazil from 1990 to 2005. The genetic diversity found among these isolates can be related to the natural horizontal genetic exchange presented by N. meningitidis strains. Our PFGE data also shows that Brazilian N. meningitidis serogroup W135 16S type 13 strains were not related with the Hajj-2000 outbreak strain. However, with distinguishable PFGE profiles, one Brazilian isolate of 16S type 31 was clustered with the Hajj-2000 outbreak strain. But as previously shown, indistinguishable PFGE profile is not a condition for a particular strain to be related to the Hajj-2000 outbreak strain.34 We hypothesize that the different clones of N. meningitidis serogroup W-135 described in this study were introduced into Brazil over the years, possibly from other countries.
In this study, we show that, during the last 16 years in Brazil, MD caused by N. meningitidis serogroup W135 is mainly due to local isolates of the ST-11 complex/ET37 complex, similar to what has been reported in others countries.24,34,35 Although we have found an isolate closely related with the Hajj-2000 outbreak strain in our collection, we were not able to establish any epidemiologic link between this related Hajj-2000 outbreak strain and the Hajj pilgrimage. Unlike in other countries, this was an isolated case and the reasons why this strain, which has been a major cause of outbreaks elsewhere, did not cause similar problems in Brazil are unclear. Changes in virulence over time and immunity in the population may be some of the reasons. A continued laboratory-based surveillance is recommended to evaluate the dissemination of this clone in Brazil.
Supplementary Material
Acknowledgments
We thank Dr Mohamed Taha and Dr J. A Vazquez for providing the controls strains (LNP17592 and LNP18359); Dr Leonard W. Mayer for providing the controls strains (M7085 and M9261); the central laboratory of each Brazilian State for providing the meningococcal isolates; Maria Cecília O. Gorla and Maria Vaneide de Paiva for identification and sorogrouping the isolates; Orgali Marques for providing the epidemiological data; Vivian Salgueiro for excellent technical assistance on PFGE; Lucilaine Ferrazoli for assistance with Bionumerics software and Kathleen Shutt for construction of the minimum spanning tree. This study was supported in part by a Fogarty International Center Global Infectious Diseases Research Training Program grant, National Institutes of Health to the University of Pittsburgh (5D43TW006592).
Footnotes
Conflict of interest: Dr. Harrison receives funding from the Centers for Disease Control and Prevention and the National Institute of Allergy and Infectious Diseases. He receives research support and lecture fees from Sanofi Pasteur; lecture fees from Novartis Vaccines; and has served as a consultant to GlaxoSmithKline, Novartis Vaccines, Sanofi Pasteur, and Wyeth. Other authors: no disclosures.
Appendix.Supplementary data: Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.jinf.2009.11.014.
References
- 1.Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27S:B51–B63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 2.Moraes JC, Barata RB. Meningococcal disease in São Paulo, Brazil, in the 20th century: epidemiological characteristics. Cad Saúde Pública. 2005;21:1458–71. doi: 10.1590/S0102-311X2005000500019. [DOI] [PubMed] [Google Scholar]
- 3.De Lemos APS, Yara TI, Gorla MCO, Paiva MV, Souza AL, Gonçalves MIC, et al. Clonal distribution of invasive Neisseria meningitidis serogroup C strains circulating from 1976 to 2005 in Greater São Paulo, Brazil. J Clin Microbiol. 2007;45(4):1266–73. doi: 10.1128/JCM.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sacchi CT, Pessoa LL, Ramos SR, Milagres LG, Camargo MCC, Hidalgo NT, et al. Ongoing group B Neisseria meningitidis epidemic in São Paulo, Brazil, due to increased prevalence of a single clone of the ET- 5 complex. J Clin Microbiol. 1992;30(7):1734–8. doi: 10.1128/jcm.30.7.1734-1738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacchi CT, Zanella RC, Caugant DA, Frasch CE, Hidalgo NT, Milagres LG, et al. Emergence of a new clone of serogroup C Neisseria meningitidis in São Paulo, Brazil. J Clin Microbiol. 1992;30(5):1282–6. doi: 10.1128/jcm.30.5.1282-1286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Meningococcal disease, serogroup W135. Wkly Epidemiol Rec. 2001;19:141–2. [PubMed] [Google Scholar]
- 7.Popovic T, Sacchi CT, Reevers MW, Whitney AM, Mayer LM, Noble CA. Neisseria meningitidis serogroup W135 isolates associated with the ET-37 complex. Emerg Infect Dis. 2000;6(4):428–9. doi: 10.3201/eid0604.000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, et al. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet. 2000;356(9248):2159. doi: 10.1016/S0140-6736(00)03502-9. [DOI] [PubMed] [Google Scholar]
- 9.Bertherat E, Yada A, Djingarey MH, Koumare B. Premiere epidemie de grande ampleur provoquee par Neisseria meningitidis W135 en Afrique. Med Trop. 2002;62:301–4. [PubMed] [Google Scholar]
- 10.Mayer LW, Reeves MW, Al-Hamdan N, Sacchi CT, Taha MK, Ajello GW, et al. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but expansion within the electrophoretic type-37 complex. J Infect Dis. 2002;185(11):1596–605. doi: 10.1086/340414. [DOI] [PubMed] [Google Scholar]
- 11.Barroso DE, Rebelo MC. Recognition of the epidemiological significance of Neisseria meningitidis capsular serogroup W135 in the Rio de Janeiro region, Brazil. Mem Inst Oswaldo Cruz. 2007;102(6):773–5. doi: 10.1590/s0074-02762007005000104. [DOI] [PubMed] [Google Scholar]
- 12.Weidlich L, Baethgen LF, Mayer LW, Moraes C, Klein CC, Nunes LS, et al. High prevalence of Neisseria meningitidis hypervirulent lineages and emergence of W135:P1.5,2:ST-11 clone in Southern Brazil. J Infect. 2008;57:324–31. doi: 10.1016/j.infect.200807.014. [DOI] [PubMed] [Google Scholar]
- 13.Efron AM, Sorhouet C, Salcedo C, Vázquez JA, Regueira M. Significant increase of serogroup W135 invasive Neisseria meningitidis strains in Argentina: a new epidemiological feature of the region. Proceedings of the 16th International Pathogenic Neisseria Conference; 2008. [Google Scholar]
- 14.Wedege E, Hoiby EA, Rosenqvist E, Froholm LO. Serotyping and subtyping of Neisseria meningitidis isolates by co-agglutination, dot-blotting and ELISA. J Med Microbiol. 1990;31(3):195–201. doi: 10.1099/00222615-31-3-195. [DOI] [PubMed] [Google Scholar]
- 15.Feavers IM, Gray SJ, Urwin R, Russel JE, Bygraves JA, Kaczmarski EB, et al. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J Clin Microbiol. 1999;37(12):3883–7. doi: 10.1128/jcm.37.12.3883-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison LH, Jolley KA, Schutt KA, Marsh JW, O'Leary M, Sanza LT, et al. Antigenic shift and increased incidence of meningococcal disease. J Infect Dis. 2006;193(9):1266–74. doi: 10.1086/501371. [DOI] [PubMed] [Google Scholar]
- 17.Russel JE, Jolley KA, Feavers IM, Maiden MC, Suker J. PorA variable regions of Neisseria meningitidis. Emerg Infect Dis. 2004;10(4):674–8. doi: 10.3201/eid1004.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson EA, Feavers IM, Maiden MC. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology. 2003;149:1849–58. doi: 10.1099/mic.0.26131-0. [DOI] [PubMed] [Google Scholar]
- 19.Urwin R, Feavers IM, Jones DM, Maiden MC, Fox AJ. Molecular variation of meningococcal serotype 4 antigen genes. Epidemiol Infect. 1998;121(1):95–101. doi: 10.1017/s0950268898008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiden MC, Bygraves JA, Feil E, Morelli G, Russel JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95(6):3140–5. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBurst: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence data. J Bacteriol. 2004;186:1518–30. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacchi CT, Whitney AM, Reeves MW, Mayer LW, Popovic T. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J Clin Microbiol. 2002;40(12):4520–7. doi: 10.1128/JCM.40.12.4520-4527.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovic T, Schmink S, Rosenstein NA, Ajello GW, Reeves MW, Plikaytis B, et al. Evaluation of pulsed-field gel electrophoresis in epidemiological investigations of meningococcal disease outbreaks caused by Neisseria meningitidis serogroup C. J Clin Microbiol. 2001;39(1):75–85. doi: 10.1128/JCM.39.1.75-85.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taha MK, Giorgini D, Ducos-Galand M, Alonso JM. Continuing diversification of Neisseria meningitidis W135 as a primary cause of meningococcal disease after emergence of the serogroup in 2000. J Clin Microbiol. 2004;42(9):4158–63. doi: 10.1128/JCM.42.9.4158-4163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore PS, Reeves MW, Schhwartz B, Gelin B, Broome CV. Intercontinental spread of an epidemic group A Neisseria meningitidis strain. Lancet. 1989;2(8657):260–3. doi: 10.1016/s0140-6736(89)90439-x. [DOI] [PubMed] [Google Scholar]
- 26.Moore PS, Harrison LH, Telzak EE, Ajello GW, Broome CV. Group A meningococcal carriage in travelers returning from Saudi Arabia. J Am Med Assoc. 1998;260(11):2686–9. [PubMed] [Google Scholar]
- 27.Dull PM, Abdelwahab J, Sacchi CT, Becker M, Noble CA, Barnett GA, et al. Neisseria meningitidis serogroup W-135 carriage among US travelers to the 2001 Hajj. J Infect Dis. 2005;191:33–9. doi: 10.1086/425927. doi:1.1086/425927. [DOI] [PubMed] [Google Scholar]
- 28.Apicella MA. Extrameningeal complications of Neisseria meningitidis serogroup W135 infection. Clin Infect Dis. 2004;38:1638–9. doi: 10.1086/421030. [DOI] [PubMed] [Google Scholar]
- 29.Hahné SJ, Gray SJ, Aguilera JF, Crowcroft NS, Nichols T, Kaczmarski E, et al. W135 meningococcal disease in England and Wales associated with Hajj 2000 and 2001. Lancet. 2002;359(9306):582–3. doi: 10.1016/S0140-6736(02)07716-4. [DOI] [PubMed] [Google Scholar]
- 30.Sacchi CT, Lemos APS, Camargo MCC, Whitney AM, Melles CEA, Solari CA, et al. Meningococcal disease caused by Neisseria meningitidis serogroup B serotype 4 in São Paulo, Brazil, 1990 to 1996. Rev Inst Med Trop São Paulo. 1998;40(2):65–70. doi: 10.1590/s0036-46651998000200001. [DOI] [PubMed] [Google Scholar]
- 31.Sacchi CT, Tondella MLC, Lemos APS, Gorla MCO, Berto DB, Kumiochi NH, et al. Characterization of epidemic Neisseria meningitidis serogroup C strains in several Brazilian states. J Clin Microbiol. 1994;32(7):1783–7. doi: 10.1128/jcm.32.7.1783-1787.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russel JE, Urwin R, Gray SJ, Fox AJ, Feavers IM, Maiden MCJ. Molecular epidemiology of meningococcal disease in England and Wales 1975–1995, before the introduction of serogroup C conjugate vaccines. Microbiology. 2008;154:1170–7. doi: 10.1099/mic.0.2007/014761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urwin R, Russel JE, Thompson EAL, Holmes EC, Feavers IM, Maiden MCJ. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect Immun. 2004;72(10):5955–62. doi: 10.1128/IAI.72.10.5955-62.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira E, Dias R, Giorgini D, Caniça M, Taha MK. Neisseria meningitidis serogroup W135 in Portugal – presence of the ST-11/ET-37 clonal complex. Pathol Biol. 2007;56:94–6. doi: 10.1016/j.patbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Molling P, Backman A, Olcén P, Fredlund H. Comparison of serogroup W-135 meningococci isolated in Sweden during a 23-year period and those associated with a recent Hajj pilgrimage. J Clin Microbiol. 2001;39(7):2695–9. doi: 10.1128/JCM39.72695-2699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.