Abstract
Background
IL-17A and IL-19 are highly expressed in chronic inflammatory diseases, such as psoriasis and asthma. IL-19 plays a significant role in the enhancement of TH2 cytokine secretion in allergic diseases, but its cellular source in asthmatic patients remains unknown.
Objective
Our aims were to determine whether the epithelium is a major source of airway mucosal IL-19 and to elucidate the mechanism of gene expression regulation.
Methods
Immunofluorescent staining was used to determine IL-19 protein expression in tracheal tissue sections of various airway diseases. Well-differentiated primary human bronchial epithelial cultures and a corresponding cell line were used as in vitro models to study gene regulation.
Results
We found significantly higher IL-19 expression in airway epithelia of asthmatic patients than in epithelia of patients with other diseases. Using a cytokine panel, we demonstrated the upregulation of IL-19 expression in cultures by two TH2 cytokines, IL-4 and IL-13, in addition to the previously found TH17 cytokine IL-17A. Moreover, cotreatment of IL-17A and IL-4/IL-13 synergistically upregulated IL-19 expression. Using siRNA and chemical inhibitor approaches, we demonstrated a transcriptional regulation of IL-19 by nuclear factor κB and signal transducer and activator of transcription (STAT) 6. The addition of IL-13 to IL-17A stimulation triggers a shift from nuclear factor κB–dependent transcriptional regulation to one that is STAT6 based. Using chromatin immunoprecipitation assays, we demonstrated the presence of STAT6-binding elements in the IL-19 promoter region.
Conclusion
We propose that an IL-17A– and IL-13–induced synergism in IL-19 stimulation in airway epithelia occurs through a STAT6-dependent pathway.
Keywords: IL-19, IL-17A, TH2 cytokine, IL-4, IL-13, airway epithelium, nuclear factor κB, signal transducer and activator of transcription 6, asthma
IL-19 is a member of the IL-10 cytokine family, which includes IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26.1–4 IL-19, IL-20, IL-22, IL-24, and IL-26 share structural characteristics and make up the IL-20 subfamily. Although IL-10 is immunosuppressive in the immune system, IL-20 subfamily cytokines are likely to be proinflammatory.2,5,6 In monocytes IL-19 secretion in response to LPS and GM-CSF has been shown to induce monocyte production of proinflammatory cytokines.7 IL-19 expression is also upregulated in psoriatic lesions,8–10 in sera from asthmatic patients, and in the urine of uremic patients, although the source of IL-19 production has not yet been identified.11,12
Very little is known about the roles and the cellular sources of IL-19 in the lung. Allergic asthma mouse models have suggested that IL-19 can induce the expression of TH2 cytokines from T cells, indicating that IL-19 might play an essential role in the dysregulation of mucosal immunity by shifting the TH1/TH2 balance.11,13
Although the primary source of IL-19 appears to be inflammatory cells, especially activated macrophages, other cell types, such as keratinocytes and bronchial epithelia, have also been shown to produce IL-19 in vitro under stimulatory conditions. The increased expression in keratinocytes might contribute to psoriasis.6,10,14–16 5′-(N-ethylcarboxamido)-adenosine has been reported to stimulate airway epithelial IL-19.17 Although IL-19 induction in asthmatic patients’ sera might be related to the induction of a TH2 response, its cellular source has not yet been identified.12,13
There is emerging evidence that points to an increase in both IL-17A and IL-19 levels in asthmatic patients.11,18 We recently demonstrated that IL-17A is a potent inducer of IL-19 expression in vitro by well-differentiated primary normal human bronchial epithelial (NHBE) cells.19 To investigate whether airway epithelial cells are the primary cellular sources of IL-19 in vivo, we examined IL-19 expression in human tracheal tissues of patients with various diseases by means of immunofluorescent staining and conducted in vitro studies to find potential IL-19 inducers and the mechanism of its expression. Through a panel of cytokines (IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12, IL-13, IL-15, IL-16, IL-17A, IFN-γ, TNF-α, and GM-CSF), we demonstrated the stimulatory effects of 2 of the TH2 cytokines, IL-4 and IL-13, but not IL-5 and IL-9, on IL-19 expression by NHBE cells. Moreover, IL-13 acts synergistically with IL-17A to further induce IL-19 expression through a signal transducer and activator of transcription (STAT) 6–dependent transcriptional pathway.
METHODS
Human airway tissue sources and culture conditions
All human bronchial tissues were obtained with informed consent from the University of California, Davis, Medical Center (Sacramento, Calif) and the National Disease Research Interchange (Philadelphia, Pa). Tissues from patients without diagnosed lung-related diseases were used for primary NHBE cultures. NHBE cells were maintained in air-liquid interface culture for full mucociliary differentiation.20,21 The immortalized NHBE cell line HBE1 was cultured under the same conditions as NHBE cells.19
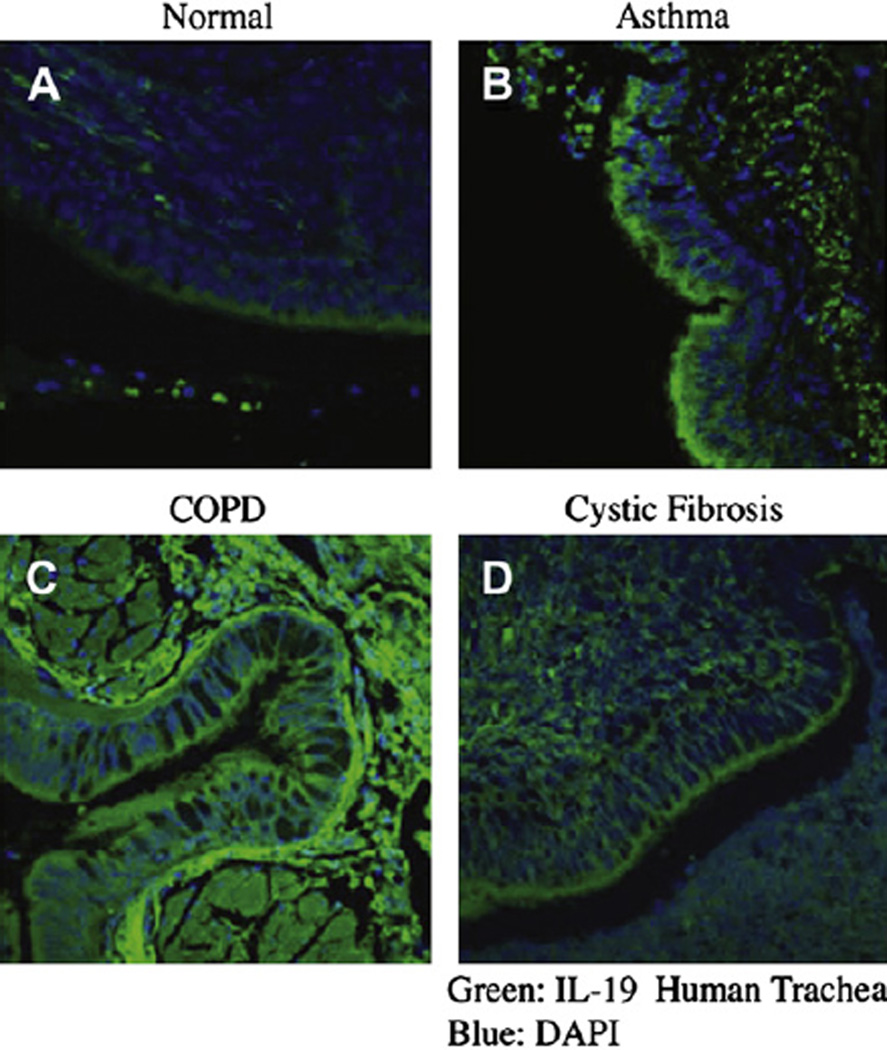
Tracheobronchial tissue sections from patients with asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis (CF) and from patients with no obvious airway diseases were obtained with consent from the University of Miami Medical Center. Paraffin tissue sections were prepared for immunohistochemistry. The ages (and sexes) of the patients with normal tracheal sections were as follows: 25 years (male), 50 years (male), and 71 years (male). These patients had no history of smoking or asthma. The ages (and sexes) of asthmatic patients were as follows: 15 years (female), 48 years (male), and 80 years (female). The ages (and sexes) of patients with CF (cystic fibrosis transmembrane conductance regulator: deltaF508) were as follows: 20 years (male), 30 years (male), and 18 years (male). The ages (and sexes) of patients with COPD were as follows: 63 years (male), 59 years (female), and 70 years (female). The selected representatives from each disease were as follows: control subject, 25 years (male); asthmatic patient, 15 years (female); patient with CF, 20 years (male); and patient with COPD, 63 years (male).
siRNA transfections
siRNAs for STAT6 and p65 from Ambion (Austin, Tex) were transiently transfected into NHBE and HBE1 cells with Oligofectamine (Invitrogen, Inc, Carlsbad, Calif), as described previously.19
Cytokine inhibitor treatment
All recombinant human cytokines were from R&D Systems, Inc (Minneapolis, Minn), and were used at 10 ng/mL, unless otherwise specified. The working concentrations of IL-13 and IL-17A were 25 ng/mL and 20 ng/mL, respectively.22 Actinomycin D (3 µg/mL), cycloheximide (5 µg/mL), and isohelenin (20 µmol/L) from Calbiochem, Inc (San Diego, Calif), were dissolved in dimethyl sulfoxide before use. Inhibitors and vehicle control were added to cells 3 hours before cytokine treatment, unless otherwise specified.
RNA isolation and real-time RT-PCR
Total RNA was extracted with RNA TRIzol reagent (Invitrogen, Inc) and was reverse transcribed with MoMLV-reverse transcriptase (Promega, Madison, Wis). SYBR Green–based real-time PCR was used to quantify the gene of interest, as described previously.19,23 Relative expressions of genes of interest were normalized to glyceraldehyde-3-phosphate dehydrogenase or β-actin transcripts.
Immunohistochemistry, immunoblotting, and ELISAs
Tracheal tissue sections underwent immunostaining with anti-IL-19 antibody or control IgG, according to our previous publications.24,25 Alexa Fluor 488-conjugated antibody (Invitrogen) and mounting solution with 4′-6-diamidino-2-phenylindole dihydrochloride were used for visualization by means of confocal microscopy (LSM 5 PASCAL; Carl Zeiss, Inc, Thornwood, NY).
The nuclear translocation of the transcription factors was analyzed by means of SDS-PAGE and Western blotting, according to our previous publication.22 Western blot antibodies were as follows: anti-p65 mAb and anti-p50 mAb (Biolegend, San Diego, Calif), anti-nucleolin mAb (Nventa, San Diego, Calif), and rabbit polyclonal anti-STAT6 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif).
A double-sandwich ELISA was developed for IL-19 secretion. Goat anti-IL-19 antibody (R&D Systems) coated onto a MaxiSorp plate (Nunc, Rochester, NY) was used as a capture antibody. The detection antibody was biotin-labeled goat anti-IL-19 (R&D Systems). Horseradish peroxidase–conjugated NeutrAvidin antibody (Invitrogen, Inc) and TMB substrates (Pierce, Inc, Rockford, Ill) were used for color development, with an absorbance of 650 nm. Recombinant IL-19 in a series of dilutions was used for absolute IL-19 quantification. The sensitivity of IL-19 detection was 1 ng per well.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were carried out according to the ChIP protocol from Millipore (Billerucam, Mass), with minor modifications. Cells were harvested and cross-linked with 1% formaldehyde at room temperature for 10 minutes. Nuclei were sonicated with a Bioruptor (Diagenode, Philadelphia, Pa) to fragment chromatin to approximately 500 bp. The sonicated nuclear fractions were divided for input control and for overnight incubation with either anti-STAT6 antibody or the negative control IgG. The antibody-protein-DNA complex was then pulled down with salmon sperm DNA–conjugated protein A agarose beads. Finally, genomic DNA recovered from the ChIP assay was PCR amplified with primers specific to the STAT6-binding elements of the IL-19 promoter region: IL-19–STAT6-I/II forward −3347, 5′-CAGTAGCCAGGATTGCCTGT-3′; IL-19–STAT6-I/II reverse –3183, 5′-GCCCCTGGATCTATGCTGTA-3′; IL-19–STAT6-III/IV forward −2299, 5′-TCCAGTCCACCCTCTCCTC–3′; and IL-19–STAT6-III/IV reverse −2127, 5′-AGGCGTGTCAGGAGGAACT-3′. The specificity of each primer set was verified by sequencing the amplicons and analyzing the dissociation curve of each gene-specific PCR product.
Statistical analyses
All NHBE cell experiments have been carried out separately from cells derived from at least 3 donors. Data shown in each figure were from experiments performed on 1 representative individual in triplicate, unless otherwise indicated. HBE1 cell experiments were done in 3 independent experiments. Values in each figure are presented as means ± SE. Group differences were calculated by using the Student t test, as described in the figures. P values of less than .05 were considered significant.
RESULTS
Is IL-19 expressed in asthmatic airway epithelia?
Three sets of paraffin tracheobronchial sections from individuals given diagnoses of COPD, CF, and asthma and with no detectable lung abnormalities were used for IL-19 immunofluorescence staining to assess the potential cellular sources for IL-19 in various airway diseases. A representative disease set is shown in Fig 1. Anti-IL-19 antibody staining (Alexa Flour 488, green) was prominently observed in the epithelial layer of tissue from asthmatic patients (Fig 1, B), and, to a lesser extent, in tissue sections from patients with CF (Fig 1, D) compared with that seen in normal control tissue (Fig 1, A). For sections from patients with COPD (Fig 1, C), IL-19 staining was primarily located underneath the epithelial layer, indicating a possible production of IL-19 by inflammatory cells rather than epithelial cells. The intensity of epithelial anti-IL-19 staining in sections from patients with CF was much less than that observed in airway epithelia from asthmatic patients. No staining was observed for sections treated with PBS or control IgG, secondary antibody, or both (data not shown). This result shows that airway epithelium is an important source of IL-19, which potentially plays a role in the mucosal immunity and might be upregulated in asthma.
FIG 1.
Immunofluorescent staining on tracheal tissue sections from patients with no identifiable lung disease (A; normal), asthma (B), COPD (C), and CF (D). Deparaffinated sections were stained with anti-IL-19 antibody followed with Alexa Fluor 488–conjugated secondary antibody (green) and 4′-6-diamidino-2-phenylindole dihydrochloride (DAPI; blue, nucleus). Original images were at ×100 magnification by means of confocal microscopy. The results shown here are representative of at least 3 patients in each disease category.
Is IL-19 expression regulated by cytokines?
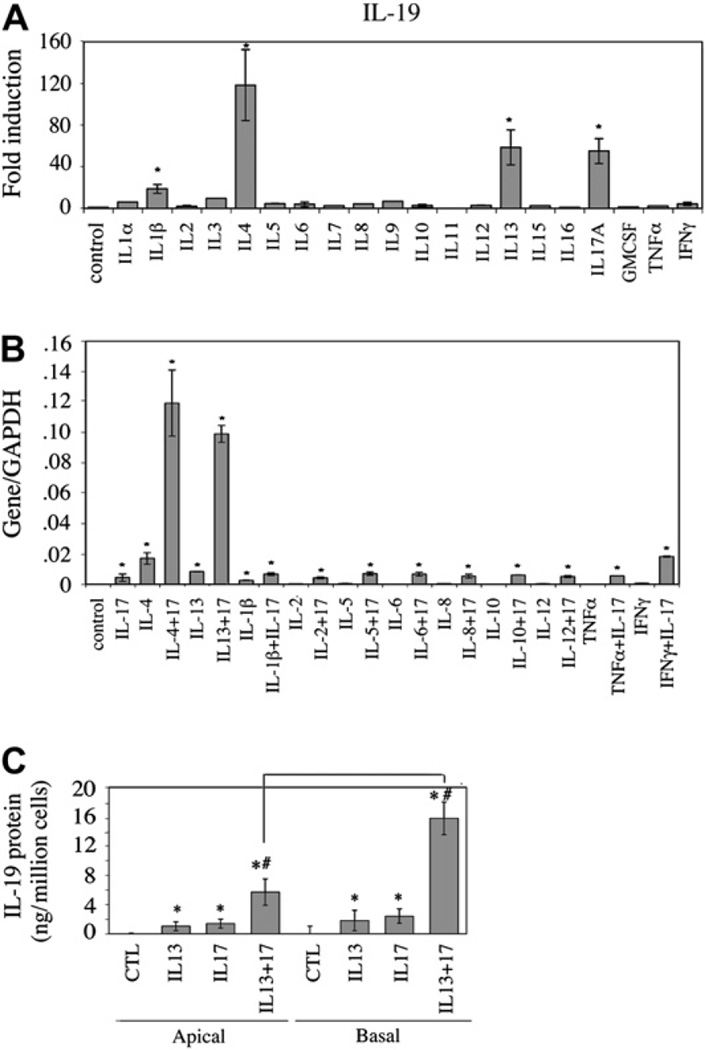
Well-differentiated NHBE cells from 3 individuals were treated with a panel of cytokines (IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12, IL-13, IL-15, IL-16, IL-17A, TNF-α, GM-CSF, and IFN-γ) to determine which cytokines stimulate IL-19 expression in airway epithelial cells. The fold induction of IL-19 mRNA by various cytokines was determined by using real-time PCR (Fig 2, A). In addition to the previously described IL-17A,19 the TH2 cytokines IL-4 and IL-13 were also able to significantly upregulate IL-19 gene expression. IL-1β was also able to stimulate IL-19 expression, albeit to a lesser extent. No IL-19 upregulation was observed for the TH2 cytokines IL-5 and IL-9 or for TH1 and other proinflammatory cytokines. From this cytokine panel, only IL-4 and IL-13 were capable of synergistically upregulating IL-17A–induced IL-19 expression (Fig 2, B). In this study IL-13/IL-17Acotreatment resulted in a 934-fold upregulation of IL-19 over that seen in untreated cells compared with that seen after IL-17A alone (42-fold) and IL-13 alone (79-fold).
FIG 2.
Induction of IL-19 in NHBE cells: A, enhanced IL-19 message by IL-1β, IL-4, IL-13, and IL-17A; B, synergistic stimulation of IL-19 message by IL-17A and IL-4 or IL-13; C, bidirectional secretion of IL-19 in cultures. Data were from 3 independent experiments. *P < .01 compared with the control. #P < .05 for apical and basal secretion comparison. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
This synergism could also be measured for IL-19 protein secretion. In Fig 2, C, all treatments of air-liquid interface cultures resulted in a polarized secretion of IL-19, which favored the basolateral chamber by more than 2-fold. IL-13 or IL-17A alone increased basolateral IL-19 secretion from undetectable levels to 3 to 4 ng, whereas IL-13/IL-17A cotreatment increased IL-19 secretion to 19 ng/106 NHBE cells in 24 hours.
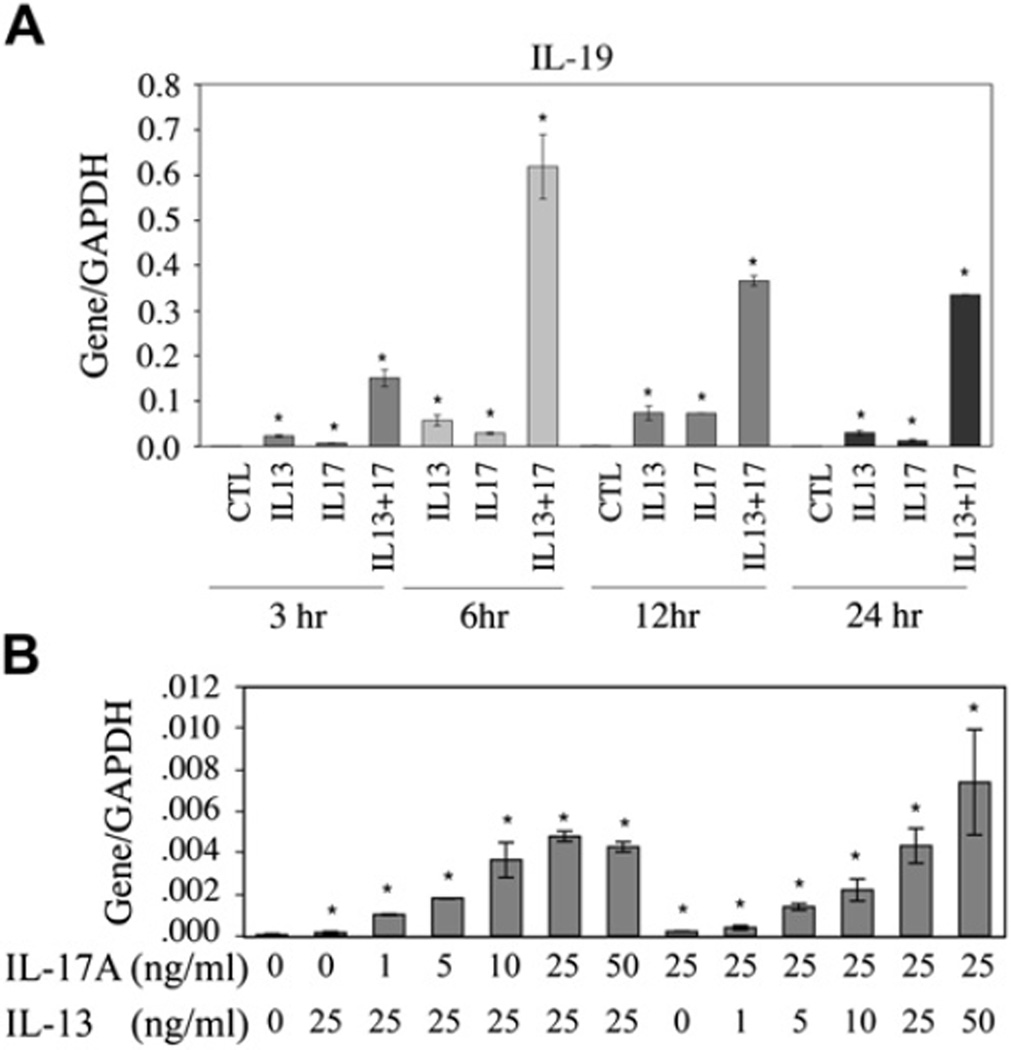
The synergistic effects of IL-13 and IL-17A on IL-19 expression were both time and concentration dependent. As shown in Fig 3, A, IL-19 mRNA levels increased above background values within 3 hours, reaching a plateau at between 6 and 12 hours. In addition to time dependence, we observed a dose-dependent synergism in IL-19 stimulation by IL-17A or IL-13 when the other cytokine was used at a constant concentration (Fig 3, B).
FIG 3.
Time- and concentration-dependent studies in NHBE cells: A, IL-19 induction by IL-13, IL-17A, or both in time course; B, various doses of IL-17A or IL-13 for IL-19 induction in the presence of a constant dose of IL-13 (25 ng/mL) or IL-17A (25 ng/mL), respectively. The experiments were repeated 3 times. *P < .05 compared with controls (no cytokine treatment). CTL, Control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
How is IL-19 expression regulated by IL-13 and IL-17A?
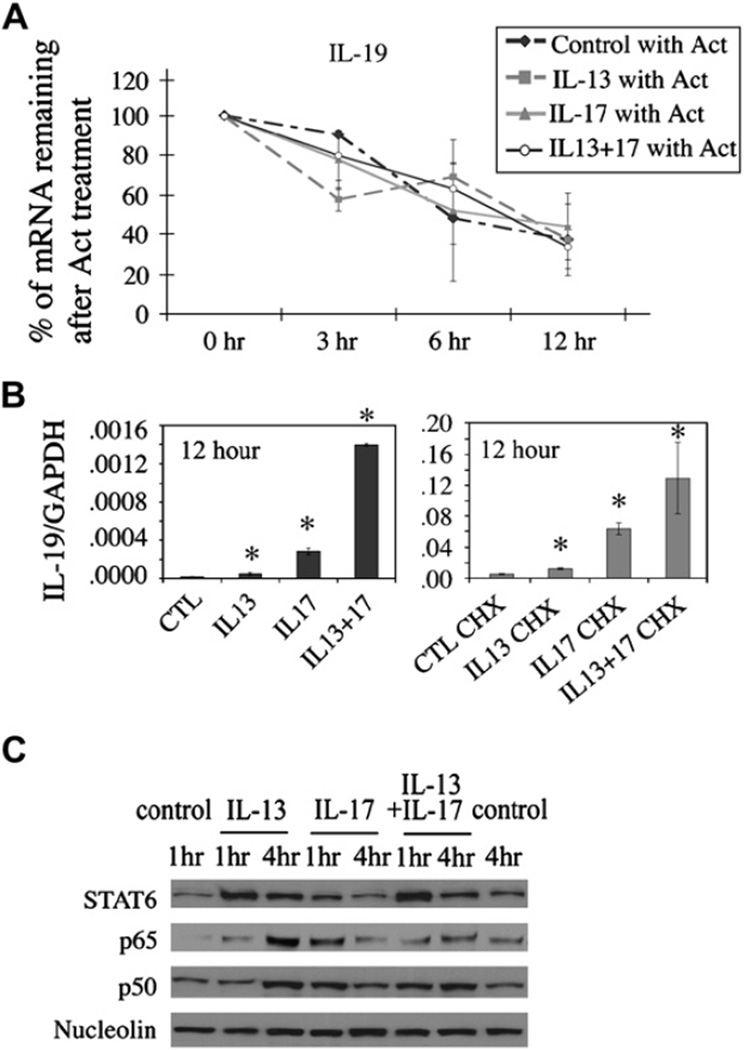
Cells were treated with actinomycin D after cytokine treatment or pretreated with cycloheximide followed by cytokine treatment to determine whether the synergistic induction of IL-19 in NHBE cells occurred at a transcriptional or posttranscriptional level. In primary NHBE cells treated with actinomycin D, IL-19 transcripts had a half-life of approximately 6 to 8 hours, which did not vary with vehicle, IL-13, and/or IL-17A treatment (Fig 4, A), indicating that these cytokines do not increase IL-19 expression through posttranscriptional mRNA stabilization.
FIG 4.
NHBE cells were treated with cytokines 24 hours before actinomycin D (Act; A) or cycloheximide (CHX; right) or vehicle (left; B) 30 minutes before cytokine treatments. RNA were harvested at indicated times. C, Nuclear translocation of STAT6, p65, and p50 in cytokine-treated HBE1 cells. The experiments were done 3 times. *P < .05 compared with controls. CTL, Control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine whether IL-19 gene induction is a direct response to IL-13 signaling, IL-17A signaling, or both, we used cycloheximide to block de novo protein synthesis. Pretreatment of cells with cycloheximide had no effect on IL-13 and IL-17 induction of IL-19 mRNA (Fig 4, B), suggesting that de novo protein synthesis is dispensable for IL-13–induced, IL-17A–induced, or both IL-19 expression and that IL-19 expression is a direct response to IL-13 stimulation, IL-17A stimulation, or both. Interestingly, cycloheximide treatment alone resulted in a 200-fold increase in IL-19 mRNA levels over that seen in untreated cells, regardless of cytokine treatment (compare left and right panels of Fig 4, B). This superinduction phenomenon by cycloheximide might suggest a potential posttranscriptional regulation of IL-19 mRNA expression independent of IL-13 or IL-17A signaling.
Based on these data, we hypothesize that the induction of IL-19 is through transcriptional activation, possibly through transcriptional factors, such as nuclear factor κB (NF-κB) and STAT6, which are known to be activated by IL-17A and IL-13, respectively.19,26 As shown in Fig 4, C, Western blot analysis of nuclear extracts showed increased nuclear translocation of p65/p50 after 4 hours of IL-13 treatment and 1 hour of IL-17Atreatment. IL-13/IL-17A cotreatment did not further enhance p65/p50 nuclear translocation. IL-13 was also able to induce STAT6 nuclear translocation at 1 and 4 hours, whereas IL-17A alone had no significant effect.
Is NF-κB involved in IL-19 induction by IL-13, IL-17A, or both?
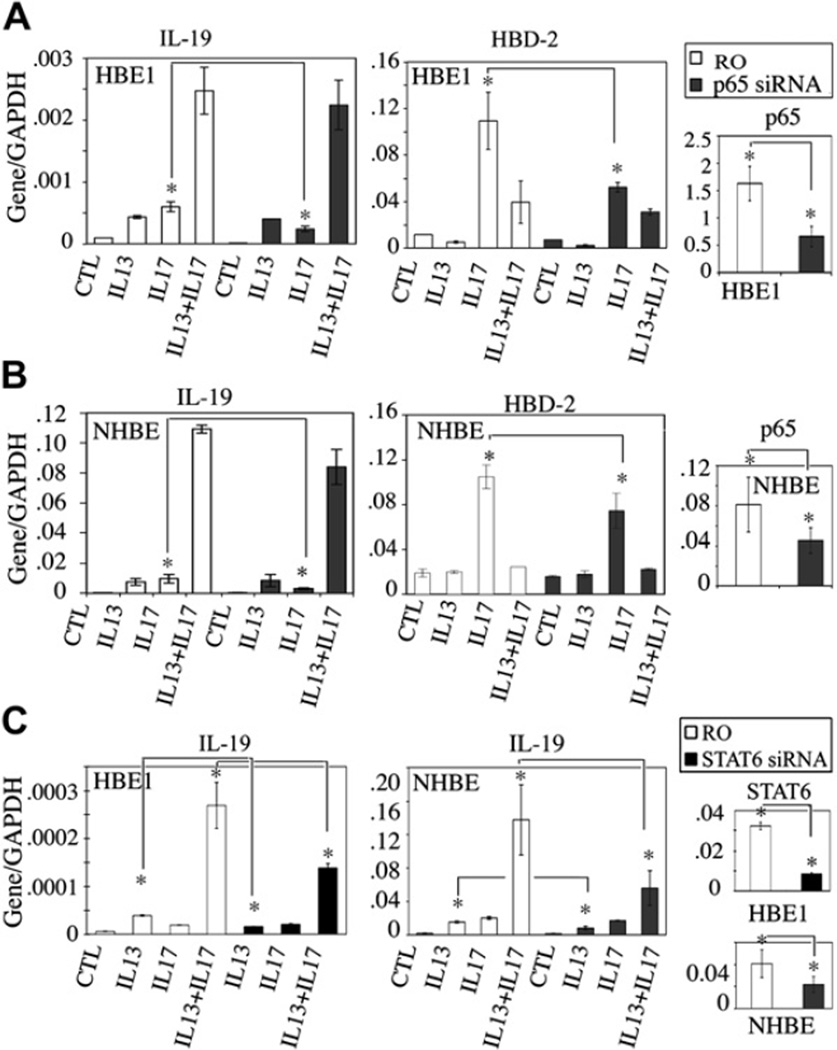
siRNA knockdown of the p65 subunit was performed to determine the role of IL-17A–induced p65 nuclear translocation in cytokine-induced IL-19 expression. As shown in Fig 5, A and B, respectively, for HBE1 and NHBE cells, siRNA knockdown resulted in 50% reduction of p65 mRNA. This lead to the suppression of IL-17A–induced IL-19 expression, but not of IL-13– or IL-13/IL-17A–induced IL-19 expression, compared with that seen with random oligomer negative controls. Human β-defensin 2 (HBD2) expression, which is inducible by IL-17A alone but not synergistically induced by IL-13/IL-17A cotreatment, could also be attenuated by p65 siRNA. This result emphasizes the uniqueness of the synergistic induction of IL-19 by IL-13 and IL-17A cotreatment. The random oligomer negative controls used in this study were also compared with non-siRNA–transfected cells and determined to have no effect on IL-19 induction (see Fig E1 in the Online Repository at www.jacionline.org). To support the p65 siRNA results, isohelenin, an IκB-α degradation inhibitor, can also suppress IL-17A–induced IL-19 and HBD-2 expression. It had no effect on IL-13– or IL-13/IL-17A cotreatment–induced IL-19 expression (see Fig E2 in the Online Repository at www.jacionline.org).
FIG 5.
A and B, Inhibition of IL-17A–induced, but not IL-13– or IL-13/IL-17A–induced, IL-19 expression by p65 siRNA. IL-13 did not induce or potentiate IL-17A–induced p65-dependent HBD2 expression. C, Inhibition of IL-13– and IL-13/IL-17A–induced, but not IL-17A–induced, IL-19 expression by STAT6 siRNA. Data were from 3 independent experiments. *P < .05. RO, Random oligomer; CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Is a STAT6-based transcriptional mechanism responsible for the synergism?
The same siRNA approach was also used to examine the role of STAT6 in IL-19 induction. As shown in Fig 5, C, STAT6 knockdown significantly attenuated both IL-13– and IL-13/IL-17A–induced IL-19 expression in both HBE1 and NHBE cells, as well as its own mRNA, compared with that seen with random oligomer negative controls (Fig 5, C). No significant inhibition was observed for IL-17A–induced IL-19. These results suggest a switch from an NF-κB to a STAT6-based mechanism in IL-17A–induced IL-19 expression when IL-13 is added.
Are there STAT6-binding cis elements in the promoter region of IL-19?
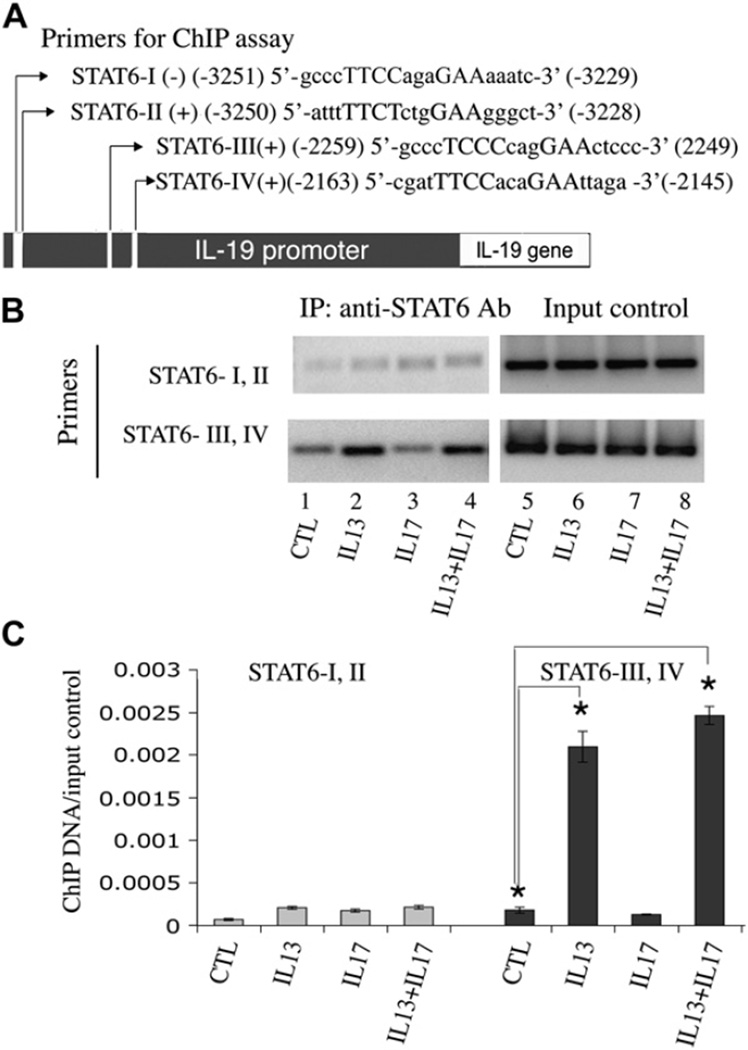
The putative promoter region of the IL-19 gene has been identified by others.7 By using Genomatix-MatInspector software (Genomatix Software GmbH, München, Germany), 4 putative STAT6 consensus-binding sites (TTC[N]2–4GAA) were identified within the 3800-bp region upstream of the IL-19 isoform 2 transcription start site (Fig 6, A). These are as follows: STAT6-I, antisense (−) 5′ gcccTTCCagaGAAaaatc 3′ (−3251 to −3229); STAT6-II, sense (+) 5′ atttTTCTctgGAAgggct 3′ (−3250 to −3228); STAT6-III, sense (+) 5′ gcccTCCCcagGAActccc 3′ (−2259 to −2249); and the most proximal STAT6-IV, sense (+) 5′ cgatTTCCacaGAAttaga 3′ (−2163 to −2145). PCR primers were designed specifically to amplify these sites for ChIP analysis. Because of the extensive overlap of the STAT6-I and STAT6-II sequences and the proximity of the STAT6-III and STAT–IV sequences, the PCR primers used could only differentiate STAT6-I/II from STAT6-III/IV. As shown in Fig 6, B, there was significant STAT6 binding to the STAT6-III/IV region after IL-13 and IL-13/IL-17A cotreatment in HBE1 cells, and the bindings were not significantly different in the 2 treatment groups. The binding of STAT6 to the STAT6-I/II region after treatment was much weaker in comparison. IL-17A treatment alone resulted in no or very low STAT6 and DNA binding. This result was further confirmed by means of real-time PCR (Fig 6, C). Data analysis and normalization are described in detail in Fig E3 (available in the Online Repository at www.jacionline.org). These results confirm the presence of STAT6-binding cis elements in the IL-19 promoter region and the induced binding of STAT6 to the STAT6-III/IV promoter region in cells after IL-13 and IL-13/IL-17A treatments.
FIG 6.
ChIP assays: A, identification of putative STAT6 sites in the 5′-flanking region of IL-19; B, PCR analysis of anti-STAT6 antibody–precipitated chromatins from HBE1 cells treated with cytokines; C, quantitative PCR analysis of anti-STAT6 ChIPs from preparations seen in Fig 6, B. Data were from 3 independent experiments. *P < .05. Ab, Antibody; IP, immunoprecipitation; CTL, control.
DISCUSSION
In this study we demonstrated that the airway epithelium is a major cellular source of IL-19, which has been shown to enhance TH2 cytokine expression.11 This conclusion is based on 2 main findings. First, IL-19 is highly expressed in the epithelia of asthmatic airways compared with expression in normal and other nonasthmatic diseased airways. CF airways express a lower level of IL-19 compared with asthmatic airways, which is possibly a result of increased IL-17A in airways of patients with CF.27 IL-19 staining in airways of patients with COPD is in the interstitial regions, likely because of production by inflammatory cells, such as macrophages.1
The second important finding is that IL-13, a TH2 cytokine, can strongly induce IL-19 mRNA and protein production in airway epithelial cells. Moreover, IL-19 is synergistically induced by cotreatment with IL-13 and IL-17A. This synergism appears to be specific to IL-19 because no synergistic effect was observed for another IL-17A−inducible gene, HBD-2 (Fig 5, A and B), or with any other IL-20 family members (unpublished data). This is the first demonstration that a TH2 cytokine can coordinate with a proinflammatory cytokine, such as IL-17A, to potentiate a TH2 response.11,18,28–31 This discovery emphasizes the uniqueness of epithelia-derived IL-19 in allergic diseases and suggests important roles for IL-13 and IL-17A in IL-19 regulation.32–34 We hypothesize that airway epithelial IL-19 is induced by IL-17A and TH2 cytokines, mainly IL-13, to further potentiate a TH2-dominant response in a positive feedback manner in the context of allergic airway diseases, such as asthma.
The roles of IL-4, IL-13, and IL-17A as cytokine mediators for inflammatory lung diseases are well established in numerous clinical and animal studies. In allergic asthma IL-4 and IL-13 are central mediators of many pathologic responses, such as airway hyperreactivity, mucous cell hyperplasia, and eosinophil recruitment.32–36 IL-17A also plays a role in human asthmatic patients and in airways of allergic animals.18,28,30,37–39 Studies have found that in patients with moderate-to-severe asthma, neutrophils are found to be more abundant than eosinophils in the bronchoalveolar lavage fluid.30,31,40 IL-17A can promote neutrophil recruitment, which might contribute to certain asthmatic phenotypes characterized by both high neutrophil counts and IL-17A expression. Other potential roles of IL-17A in asthmatic airways might include the stimulation of mucus cell differentiation25,41 and induction of chemokine and nitric oxide release.22,23,30,42
Our study suggests that coexistence of TH2 cytokines and IL-17A might have a significant effect on the pathogenesis of asthma by positively modulating the existing TH2-dominant response through airway epithelial IL-19 secretion. Previous studies have shown that IL-19 can promote the differentiation of TH cells toward a TH2 lineage.11 Thus IL-19 can act in a pathogenic manner to promote and amplify a TH2 inflammatory response in the lungs in response to IL-13 stimuli, IL-17A stimuli, or both.
Using actinomycin D and cycloheximide, we demonstrated that the upregulation of IL-19 by IL-13, IL-17A, or both works mainly through transcriptional activation and does not require cytokine-induced de novo protein synthesis. siRNA knockdown and ChIP assays demonstrated that NF-κB and STAT6 are important transcription factors for IL-17A– and IL-13–stimulated IL-19 gene expression, respectively. Although NF-κB–mediated transcription dominates IL-17A activation of IL-19 expression, STAT6 is the dominant transcription factor in the combined treatment. ChIP assays consistently demonstrated strong binding of STAT6 to the IL-19 promoter in the treatment of IL-13 with or without IL-17A, whereas no STAT6 binding was observed with IL-17A treatment alone (Fig 6, B and C). However, STAT6 nuclear translocation and IL-19 promoter binding were similar in IL-13– and IL-13/IL-17A–treated cells, despite the fact that IL-17A/IL-13–cotreated cells express much higher IL-19 compared with IL-13 alone. This suggests the possible involvement of other transcriptional coactivators or repressors in IL-13/IL-17A–mediated IL-19 gene expression, which will be a topic of exploration in future studies. Interestingly, IL-13 and IL-17A can individually induce a kinetic translocation of p65 into the nucleus, but this response is slightly diminished in cotreated samples (Fig 4,C), which might affect the kinetics of inducible gene expression at early time points. The diminished p65 translocation in cotreatment still needs to be verified but might account for decreased HBD-2 induction compared with that seen with IL-17A alone (Fig 5, A and B). These hypotheses require further investigation.
In summary, we have identified airway epithelia as a major source of IL-19 expression in airways of asthmatic patients. In airways of asthmatic patients, IL-13 and IL-17A might synergistically activate IL-19 expression. Because IL-19 has been implicated in the regulation of TH2 cytokine production, we hypothesize that this activation might be crucial to the pathogenesis of asthma by maintaining or amplifying a TH2-dominant immune response in the mucosal tissue.
Supplementary Material
Key message.
TH2-dominant mucosal immunity in asthmatic airways is potentially regulated by IL-17A– and IL-13–stimulated IL-19 expression through a STAT6-dependent pathway.
Acknowledgments
Supported in part by grants from the National Institutes of Health (HL077902, HL077315, and ES00628) and the T32 HL07103 (PT).
We thank Sharlene Velichko and Laura Shih for their editing.
Abbreviations used
- CF
Cystic fibrosis
- ChIP
Chromatin immunoprecipitation
- COPD
Chronic obstructive pulmonary disease
- HBD-2
Human β-defensin 2
- NHBE
Normal human bronchial epithelium
- NF-κB
Nuclear factor κB
- STAT
Signal transducer and activator of transcription
Footnotes
Disclosure of potential conflict of interest: The authors have declared that they have no conflict of interest.
REFERENCES
- 1.Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, et al. Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10) Genes Immun. 2000;1:442–450. doi: 10.1038/sj.gene.6363714. [DOI] [PubMed] [Google Scholar]
- 2.Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 3.Conti P, Kempuraj D, Frydas S, Kandere K, Boucher W, Letourneau R, et al. IL-10 subfamily members: IL-19, IL-20, IL-22, IL-24 and IL-26. Immunol Lett. 2003;88:171–174. doi: 10.1016/s0165-2478(03)00087-7. [DOI] [PubMed] [Google Scholar]
- 4.Chang C, Magracheva E, Kozlov S, Fong S, Tobin G, Kotenko S, et al. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J Biol Chem. 2003;278:3308–3313. doi: 10.1074/jbc.M208602200. [DOI] [PubMed] [Google Scholar]
- 5.Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–3702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 6.Sa SM, Valdez PA, Wu J, Jung K, Zhong F, Hall L, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 7.Liao YC, Liang WG, Chen FW, Hsu JH, Yang JJ, Chang MS. IL-19 induces production of IL-6 and TNF-alpha and results in cell apoptosis through TNF-alpha. J Immunol. 2002;169:4288–4297. doi: 10.4049/jimmunol.169.8.4288. [DOI] [PubMed] [Google Scholar]
- 8.Koks S, Kingo K, Vabrit K, Ratsep R, Karelson M, Silm H, et al. Possible relations between the polymorphisms of the cytokines IL-19, IL-20 and IL-24 and plaquetype psoriasis. Genes Immun. 2005;6:407–415. doi: 10.1038/sj.gene.6364216. [DOI] [PubMed] [Google Scholar]
- 9.Koks S, Kingo K, Ratsep R, Karelson M, Silm H, Vasar E. Combined haplotype analysis of the interleukin-19 and-20 genes: relationship to plaque-type psoriasis. Genes Immun. 2004;5:662–667. doi: 10.1038/sj.gene.6364141. [DOI] [PubMed] [Google Scholar]
- 10.Romer J, Hasselager E, Norby PL, Steiniche T, Thorn Clausen J, Kragballe K. Epidermal overexpression of interleukin-19 and-20 mRNA in psoriatic skin disappears after short-term treatment with cyclosporine a or calcipotriol. J Invest Dermatol. 2003;121:1306–1311. doi: 10.1111/j.1523-1747.2003.12626.x. [DOI] [PubMed] [Google Scholar]
- 11.Liao SC, Cheng YC, Wang YC, Wang CW, Yang SM, Yu CK, et al. IL-19 induced Th2 cytokines and was up-regulated in asthma patients. J Immunol. 2004;173:6712–6718. doi: 10.4049/jimmunol.173.11.6712. [DOI] [PubMed] [Google Scholar]
- 12.Hsing CH, Hsu CC, Chen WY, Chang LY, Hwang JC, Chang MS. Expression of IL-19 correlates with Th2 cytokines in uraemic patients. Nephrol Dial Transplant. 2007;22:2230–2238. doi: 10.1093/ndt/gfm179. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher G, Eskdale J, Jordan W, Peat J, Campbell J, Boniotto M, et al. Human interleukin-19 and its receptor: a potential role in the induction of Th2 responses. Int Immunopharmacol. 2004;4:615–626. doi: 10.1016/j.intimp.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Otkjaer K, Kragballe K, Funding AT, Clausen JT, Noerby PL, Steiniche T, et al. The dynamics of gene expression of interleukin-19 and interleukin-20 and their receptors in psoriasis. Br J Dermatol. 2005;153:911–918. doi: 10.1111/j.1365-2133.2005.06800.x. [DOI] [PubMed] [Google Scholar]
- 15.Li HH, Lin YC, Chen PJ, Hsiao CH, Lee JY, Chen WC, et al. Interleukin-19 upregulates keratinocyte growth factor and is associated with psoriasis. Br J Dermatol. 2005;153:591–595. doi: 10.1111/j.1365-2133.2005.06665.x. [DOI] [PubMed] [Google Scholar]
- 16.Boniface K, Lecron JC, Bernard FX, Dagregorio G, Guillet G, Nau F, et al. Keratinocytes as targets for interleukin-10-related cytokines: a putative role in the pathogenesis of psoriasis. Eur Cytokine Netw. 2005;16:309–319. [PubMed] [Google Scholar]
- 17.Zhong H, Wu Y, Belardinelli L, Zeng D. A2B adenosine receptors induce IL-19 from bronchial epithelial cells, resulting in TNF-alpha increase. Am J Respir Cell Mol Biol. 2006;35:587–592. doi: 10.1165/rcmb.2005-0476OC. [DOI] [PubMed] [Google Scholar]
- 18.Molet S, Hamid Q, Davoine F, Nutku E, Taha R, Page N, et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin Immunol. 2001;108:430–438. doi: 10.1067/mai.2001.117929. [DOI] [PubMed] [Google Scholar]
- 19.Huang F, Kao CY, Wachi S, Thai P, Ryu JS, Wu R. Requirement for both JAK-mediated PI3K signaling and ACT1/TRAF6/TAK1-dependent NF-B Activation by IL-17A in enhancing cytokine expression in human airway epithelia cells. J Immunol. 2007;179:6504–6513. doi: 10.4049/jimmunol.179.10.6504. [DOI] [PubMed] [Google Scholar]
- 20.Wu R, Yankaskas J, Cheng E, Knowles MR, Boucher R. Growth and differentiation of human nasal epithelial cells in culture. Serum-free, hormone-supplemented medium and proteoglycan synthesis. Am Rev Respir Dis. 1985;132:311–320. doi: 10.1164/arrd.1985.132.2.311. [DOI] [PubMed] [Google Scholar]
- 21.Wu R, Martin WR, Robinson CB, St George JA, Plopper CG, Kurland G, et al. Expression of mucin synthesis and secretion in human tracheobronchial epithelial cells grown in culture. Am J Respir Cell Mol Biol. 1990;3:467–478. doi: 10.1165/ajrcmb/3.5.467. [DOI] [PubMed] [Google Scholar]
- 22.Kao CY, Huang F, Chen Y, Thai P, Wachi S, Kim C, et al. Up-regulation of CC chemokine ligand 20 expression in human airway epithelium by IL-17 through a JAK-independent but MEK/NF-kappaB-dependent signaling pathway. J Immunol. 2005;175:6676–6685. doi: 10.4049/jimmunol.175.10.6676. [DOI] [PubMed] [Google Scholar]
- 23.Kao CY, Chen Y, Thai P, Wachi S, Huang F, Kim C, et al. IL-17 markedly upregulates beta-defensin-2 expression in human airway epithelium via JAK and NF-kappaB signaling pathways. J Immunol. 2004;173:3482–3491. doi: 10.4049/jimmunol.173.5.3482. [DOI] [PubMed] [Google Scholar]
- 24.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Thai P, Zhao YH, Ho YS, DeSouza MM, Wu R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J Biol Chem. 2003;278:17036–17043. doi: 10.1074/jbc.M210429200. [DOI] [PubMed] [Google Scholar]
- 26.Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-Deficient mice. J Immunol. 1996;157:3220–3222. [PubMed] [Google Scholar]
- 27.McAllister F, Henry A, Kreindler JL, Dubin PJ, Ulrich L, Steele C, et al. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J Immunol. 2005;175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullens DM, Truyen E, Coteur L, Dilissen E, Hellings PW, Dupont LJ, et al. IL-17 mRNA in sputum of asthmatic patients: linking T cell driven inflammation and granulocytic influx? Respir Res. 2006;7:135. doi: 10.1186/1465-9921-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakir J, Shannon J, Molet S, Fukakusa M, Elias J, Laviolette M, et al. Airway remodeling-associated mediators in moderate to severe asthma: effect of steroids on TGF-beta, IL-11, IL-17, and type I and type III collagen expression. J Allergy Clin Immunol. 2003;111:1293–1298. doi: 10.1067/mai.2003.1557. [DOI] [PubMed] [Google Scholar]
- 30.Linden A. Role of interleukin-17 and the neutrophil in asthma. Int Arch Allergy Immunol. 2001;126:179–184. doi: 10.1159/000049511. [DOI] [PubMed] [Google Scholar]
- 31.Linden A, Hoshino H, Laan M. Airway neutrophils and interleukin-17. Eur Respir J. 2000;15:973–977. doi: 10.1034/j.1399-3003.2000.15e28.x. [DOI] [PubMed] [Google Scholar]
- 32.Huang SK, Xiao HQ, Kleine-Tebbe J, Paciotti G, Marsh DG, Lichtenstein LM, et al. IL-13 expression at the sites of allergen challenge in patients with asthma. J Immunol. 1995;155:2688–2694. [PubMed] [Google Scholar]
- 33.Doucet C, Brouty-Boye D, Pottin-Clemenceau C, Canonica GW, Jasmin C, Azzarone B. Interleukin (IL) 4 and IL-13 act on human lung fibroblasts. Implication in asthma. J Clin Invest. 1998;101:2129–2139. doi: 10.1172/JCI741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humbert M, Durham SR, Kimmitt P, Powell N, Assoufi B, Pfister R, et al. Elevated expression of messenger ribonucleic acid encoding IL-13 in the bronchial mucosa of atopic and nonatopic subjects with asthma. J Allergy Clin Immunol. 1997;99:657–665. doi: 10.1016/s0091-6749(97)70028-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Xia Y, Nguyen A, Lai YH, Feng L, Mosmann TR, et al. Effects of Th2 cytokines on chemokine expression in the lung: IL-13 potently induces eotaxin expression by airway epithelial cells. J Immunol. 1999;162:2477–2487. [PubMed] [Google Scholar]
- 37.Wong CK, Ho CY, Ko FW, Chan CH, Ho AS, Hui DS, et al. Proinflammatory cytokines (IL-17, IL-6, IL-18 and IL-12) and Th cytokines (IFN-gamma, IL-4, IL-10 and IL-13) in patients with allergic asthma. Clin Exp Immunol. 2001;125:177–183. doi: 10.1046/j.1365-2249.2001.01602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barczyk A, Pierzchala W, Sozanska E. Interleukin-17 in sputum correlates with airway hyperresponsiveness to methacholine. Respir Med. 2003;97:726–733. doi: 10.1053/rmed.2003.1507. [DOI] [PubMed] [Google Scholar]
- 39.Hellings PW, Kasran A, Liu Z, Vandekerckhove P, Wuyts A, Overbergh L, et al. Interleukin-17 orchestrates the granulocyte influx into airways after allergen inhalation in a mouse model of allergic asthma. Am J Respir Cell Mol Biol. 2003;28:42–50. doi: 10.1165/rcmb.4832. [DOI] [PubMed] [Google Scholar]
- 40.Prause O, Laan M, Lotvall J, Linden A. Pharmacological modulation of interleukin-17-induced GCP-2-, GRO-alpha- and interleukin-8 release in human bronchial epithelial cells. Eur J Pharmacol. 2003;462:193–198. doi: 10.1016/s0014-2999(03)01341-4. [DOI] [PubMed] [Google Scholar]
- 41.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai L, Suboc P, Hogue DA, Fei DT, Filvaroff EH. Interleukin 17 induced nitric oxide suppresses matrix synthesis and protects cartilage from matrix breakdown. J Rheumatol. 2002;29:1725–1736. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.