Abstract
Acidithiobacillus ferrooxidans is a chemolithoautotrophic acidophile capable of obtaining energy by oxidizing ferrous iron or sulfur compounds such as metal sulfides. Some of the proteins involved in these oxidations have been described as forming part of the periplasm of this extremophile. The detailed study of the periplasmic components constitutes an important area to understand the physiology and environmental interactions of microorganisms. Proteomics analysis of the periplasmic fraction of A. ferrooxidans ATCC 23270 was performed by using high resolution linear ion trap-FT MS. We identified a total of 131 proteins in the periplasm of the microorganism grown in thiosulfate. When possible, functional categories were assigned to the proteins: 13.8% were transport and binding proteins, 14.6% were several kinds of cell envelope proteins, 10.8% were involved in energy metabolism, 10% were related to protein fate and folding, 10% were proteins with unknown functions, and 26.1% were proteins without homologues in databases. These last proteins are most likely characteristic of A. ferrooxidans and may have important roles yet to be assigned. The majority of the periplasmic proteins from A. ferrooxidans were very basic compared with those of neutrophilic microorganisms such as Escherichia coli, suggesting a special adaptation of the chemolithoautotrophic bacterium to its very acidic environment. The high throughput proteomics approach used here not only helps to understand the physiology of this extreme acidophile but also offers an important contribution to the functional annotation for the available genomes of biomining microorganisms such as A. ferrooxidans for which no efficient genetic systems are available to disrupt genes by procedures such as homologous recombination.
Acidithiobacillus ferrooxidans is generally found in acidic environments such as mining dumps and acid mine drainages. It is a chemolithoautotrophic Gram-negative γ-proteobacterium that obtains its energy from the oxidation of ferrous iron, elemental sulfur, or partially oxidized sulfur compounds (Refs. 1–3 and references therein). The ability of this and other microorganisms present in their habitat to solubilize metal sulfides has been successfully applied in biomining operations (1, 4).
The reactions involved in ferrous iron oxidation have been studied in detail (1, 5). The terminal electron acceptor is assumed to be a cytochrome oxidase anchored to the cytoplasmic membrane. The transfer of electrons would occur through several periplasmic carriers, including at least the blue copper protein rusticyanin and a cytochrome c552. A high molecular weight c-type cytochrome, Cyc2, which is located in the outer membrane of A. ferrooxidans, has been suggested to be the prime candidate for the initial electron acceptor in the respiratory pathway between ferrous iron and oxygen (6). This pathway would be Cyc2 → rusticyanin→ Cyc1 (c552) → aa3 cytochrome oxidase (1).
The aerobic oxidation of elemental sulfur by A. ferrooxidans and other microorganisms is carried out by a sulfur dioxygenase (2, 7). On the other hand, thiosulfate has been postulated as a key intermediate compound in the oxidation of the sulfur moiety of pyrite (thiosulfate mechanism) (2). Sulfur compound-oxidizing enzymes such as thiosulfate-oxidizing enzyme in A. ferrooxidans (8) or tetrathionate hydrolase in Acidithiobacillus thiooxidans or Thiobacillus ferrooxidans (9, 10) may be involved in the process. Also a rhodanese activity has been described previously in A. ferrooxidans (11). This enzyme is a thiosulfate sulfurtransferase (TST),1 which breaks the S–S bond present in thiosulfate, generating sulfur and sulfite. We have recently described in A. ferrooxidans several predicted cytoplasmic TST-like proteins and an exported TST-like protein (P21) that is highly expressed when the bacterium is grown in pyrite and sulfur but not in ferrous iron (12). The genomic contexts of some of the rhodanese-like genes suggested their implication in sulfur oxidation and metabolism, formation of Fe-S clusters, or detoxification mechanisms (13).
The most relevant reactions for ferrous iron and sulfur oxidation take place at the periplasmic space of A. ferrooxidans (1, 3). However, little is known about the periplasm of this acidophile and its components. To further study the proteins that may be involved in the oxidation of sulfur and metal sulfides, we analyzed and characterized by high throughput expression proteomics the proteins present in the periplasmic fraction of this bacterium.
We found 131 proteins in the periplasmic fraction of the acidophilic A. ferrooxidans grown in thiosulfate. These include many transport and binding proteins, cell envelope proteins, energy metabolism proteins, and proteins involved in fate and folding. In addition, several proteins were identified as having unknown functions, and around 26.1% of them were proteins with no homologues in databases, many of which may be characteristic of this microorganism. The results presented not only contribute to understand the physiology of A. ferrooxidans but are also important for the genomic annotation of the new periplasmic proteins from this extremophile.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
A. ferrooxidans strain ATCC 23270 was grown in thiosulfate by using DSMZ medium 71 containing 20 mM thiosulfate and the following components: 3.0 g liter−1 KH2PO4, 0.5 g liter−1 MgSO4·7H2O, 3.0 g liter−1 (NH4)2SO4, 0.25 g liter−1 CaCl2·2H2O. The pH was adjusted to about 4.6 by addition of 1 M NaOH.
Preparation of Periplasmic Fractions
To determine the most appropriate and efficient method to obtain the periplasmic proteins from A. ferrooxidans, we tested both the most commonly used osmotic shock procedure of Laundenbach et al. (14) and the chloroform-based method of Ames et al. (15). Possible cell lysis during the liberation of periplasmic proteins was controlled by determining the presence of an abundant cytoplasmic protein such as elongation factor (EF)-Tu by using Western blotting analysis with an antiserum against EF-Tu from Escherichia coli.
We found that the method of Laundenbach et al. (14) as applied to A. ferrooxidans released a high concentration of EF-Tu together with the periplasmic fraction, clearly indicating some cell disruption. On the other hand, when the chloroform method was used, no detectable EF-Tu was identified in the periplasmic preparations by using the immunological method (not shown). Furthermore the chloroform method gave a higher enrichment of rusticyanin, a well characterized periplasmic protein from A. ferrooxidans. Consequently the method of choice was that from Ames et al. (15) with minor modifications.
Briefly the harvested cell pellet (from a 200-ml culture at ~1 × 108 cells/ml) was washed twice with a basal salt solution (0.77 mM (NH4)2SO4, 1.63 mM MgSO4, 0.175 mM K2HPO4, pH 2.5) and was resuspended in 200 μl of the same acidic solution followed by the addition of 20 μl of chloroform. After briefly vortexing, the tubes were maintained at room temperature for about 15 min and adjusted to pH 2.5 by adding 10 mM Tris-HCl, pH 8. The cells were separated by centrifugation at 6,000 × g for 20 min, and the supernatant fraction containing the periplasmic proteins was withdrawn and brought to 100 μg/ml phenylmethylsulfonyl fluoride. Finally the proteins present in the periplasmic fraction were concentrated by vacuum to dryness in a vacuum concentrator (UniVapo 100H, Uniequip).
Two-dimensional (2-D) Non-equilibrium pH Polyacrylamide Gel Electrophoresis (NEPHGE) and SDS-PAGE
Total cell proteins were separated by 2-D NEPHGE (16) performed as described before for A. ferrooxidans (12, 17) using ampholytes (pH 3–10) from Bio-Rad. Cell samples (4 mg wet weight of cells) to be analyzed were resuspended in 80 μl of sonication buffer (10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 50 μg of pancreatic RNase/ml). Cell disruption was done by using a Misonix XL2020 sonicator with a microtip (six times during 30 s) followed by treatment with DNase (50 μg/ml final concentration). The mixture was then lyophilized. Total cell proteins obtained or the proteins from the periplasmic fraction were dissolved in lysis buffer and were analyzed by 12.5% SDS-PAGE, and the protein arrays obtained were visualized by silver staining as described before (17).
In-gel Trypsin Digestion
Lyophilized periplasmic protein pellets were solubilized directly in sample loading buffer, and an aliquot of the sample (~20 μg of proteins/lane) was separated on a 12.5% SDS-polyacrylamide gel. The gels were stained with Coomassie Blue R-250 for 18 h. The lanes containing the total periplasmic samples were cut evenly into five slices from the top to the bottom of the gel. In-gel trypsin digestion was performed using the procedure of Shevchenko et al. (18).
Liquid Chromatography-coupled Mass Spectrometric Analysis
An aliquot of the periplasmic protein digest was loaded onto a fused silica precolumn (outer diameter × inner diameter, 360 × 75 μm) packed with irregular C18 beads (5–20 μm, ODS-AQ, YMC, Waters, Milford, MA) and washed with 0.1% acetic acid. The precolumn was then Teflon sleeve-connected (outer diameter × inner diameter, 0.012 × 0.060 inch; Zeus, Orangerburg, SC) to a fused silica analytical column (outer diameter × inner diameter, 360 × 50 μm) packed with 5 cm of regular C18 beads (5 μm, ODS-AQ, YMC). An electrospray emitter tip (2–4 μm in diameter) was pulled with a laser on one end of this analytical column as described previously (19). An Agilent 1100 Series binary HPLC system (Palo Alto, CA) was interfaced with a hybrid linear ion trap (LTQ)-FT mass spectrometer (ThermoElectron, San Jose, CA) equipped with a microelectrospray source for on-line peptide separations. The instrument was operated in the data-dependent mode using Xcalibur software (ThermoElectron). It cycled through a single MS experiment using the FT-ICR cell as the mass analyzer (resolution, R = 25,000 at m/z 400; target value, 1 × 106) followed by 10 MS/MS experiments using the linear ion trap as the mass analyzer. The dynamic exclusion duration was set to be 45 s. The HPLC gradient used for the sample analysis was: 0–7% B in 5 min, 7–45% B in 70 min, 45–100% in 15 min, and 100–0% B in 5 min (A = 0.1 M acetic acid (Sigma-Aldrich), B = 70% acetonitrile (Mallinckrodt, Paris, KY) in 0.1 M acetic acid).
Data Analysis
When we started this study, the available A. ferrooxidans genome (The Institute for Genomic Research (TIGR), www.tigr.org) was not yet annotated. Therefore, we used the GLIMMER (Gene Locator and Interpolated Markov Modeler, version 2.10) software tools to predict protein-encoding ORFs within the available A. ferrooxidans genomic sequence and hence constructed our local putative A. ferrooxidans protein database to be used in the MS/MS proteomics analysis (20, 21). Briefly long ORFs were extracted from the genomic contigs with “long-orfs(1)” and used to train an interpolated Markov model with “build-icm.” The resulting model was used to scan the genomic contigs for high scoring ORFs with “glimmer2(1),” yielding 3,727 non-overlapping ORF predictions. Each ORF or putative protein was provisionally annotated by identifying the highest scoring homologous protein from a Smith-Waterman search of the National Center for Biotechnology Information (NCBI) nonredundant protein database (downloaded July 27, 2003) (22), requiring a statistical expectation of 1e–6 or better. The acquired MS/MS data were then searched against the putative A. ferrooxidans protein database using SEQUEST (23). SEQUEST search parameters were conducted with “no enzyme” specificity and a static modification of 57 amu on cysteine, representing alkylation with iodoacetamide, and a differential modification of 16 amu on methionine, representing the possibility of oxidation. The parent ion mass window was set as ±0.05 Da (monoisotopic mass), and the fragment ion mass tolerance was set as ±0.35 Da (monoisotopic mass). The parameters DelMass < 1.0, Xcorr > 2.4, DelCn > 0.1, Sp > 500, RSp < 10, and Ion Ratio > 0.6 were used to evaluate database search results, and the reported peptide sequences were manually validated (24). Detailed additional information for all annotated proteins with predicted export signals identified in the periplasmic fraction can be seen in Supplemental Table S1. For protein identifications based on single peptide assignments, the sequences of the peptides used to make each assignment, the precursor masses and charges observed, and the scores for each of these peptides are shown in Supplemental Table S2, and their corresponding spectra are shown in Supplemental Fig. S1.
Sequence Analysis
Identity/similarity and conserved domain searching in databases was done by using the BlastP program (25) and Conserved Domain Database from NCBI (www.ncbi.nlm.nih.gov). Because we started using the finished non-annotated available A. ferrooxidans ATCC 23270 genomic sequence (www.tigr.org), we determined the ORFs for all the periplasmic proteins found. Now that the TIGR final annotation is available, we compared both databases and have indicated the minor discrepancies when pertinent. Molecular masses and isoelectric points of ORFs were obtained by using Compute pI/Mw tool (www.expasy.org/tools/pi_tool.html). The presence of export signals for each protein found in the periplasmic fraction was predicted by using SignalP (www.cbs.dtu.dk/services/SignalP), TatP (www.cbs.dtu.dk/services/TatP-1.0), SecretomeP (www.cbs.dtu.dk/services/SecretomeP), and LipoP (www.cbs.dtu.dk/services/LipoP). The predicted subcellular locations of the proteins coded by the different ORFs analyzed were obtained using PENCE Proteome Analyst (www.cs.ualberta.ca/%7Ebioinfo/PA/Sub/index.html) and CELLO (cello.life.nctu.edu.tw) programs. The functional categories for the different identified proteins were obtained from the genomic sequence of A. ferrooxidans (www.tigr.org).
RESULTS AND DISCUSSION
General Characteristics of the Periplasmic Proteins from A. ferrooxidans
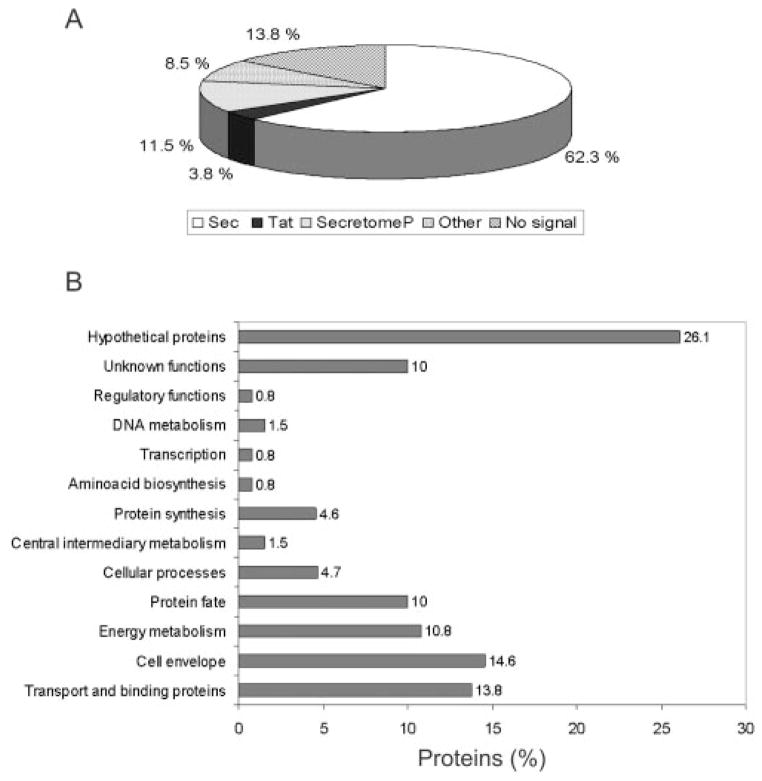
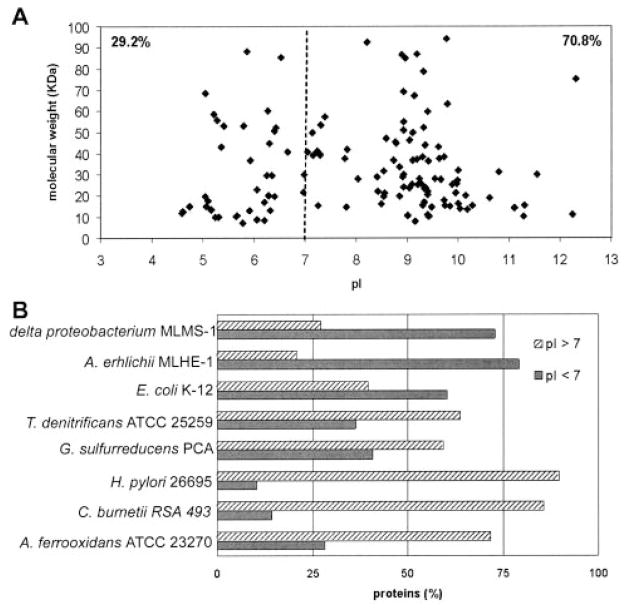
A. ferrooxidans cells were obtained by growing the bacteria in thiosulfate as described under “Experimental Procedures.” The periplasmic fraction was prepared and checked by SDS-PAGE and 2-D PAGE as shown in Fig. 1. As expected, the periplasmic fraction (Fig. 1B) contained a reduced number of proteins compared with the total cellular proteins (Fig. 1A). Furthermore many of the most abundant proteins present in the total fraction were absent in the periplasmic preparation. These results together with the absence of EF-Tu in the periplasmic fraction (see “Experimental Procedures”) indicate a low amount of cell lysis during the preparation of this fraction. Fig. 1C shows the periplasmic fraction proteins separated in a monodimensional SDS gel from which the five sections from top to bottom of the gel were obtained (see “Experimental Procedures”). A total of 131 proteins were identified from the A. ferrooxidans periplasmic sample using tandem mass spectrometry. The proteins identified were first categorized according to their predicted export signals as illustrated in Fig. 2A. Most of the proteins (62.3%) possessed export signals of the Sec type. Five of them (3.8%) showed a twin arginine translocation (Tat) signal, suggesting that they would be proteins exported in a folded form across the cytoplasmic membrane by the Tat pathway. Of the remaining proteins, 11.5% could be considered as possibly periplasmic according to the SecretomeP program, 8.5% could be exported according to their subcellular location prediction (CELLO and PENCE Proteome Analyst), and 13.8% of the proteins found in the periplasmic fraction did not contain any predicted export signal. These last putative proteins were expected to have a cytoplasmic localization. However, it is known that some proteins are exported without having typical export signals (26). Therefore, some of them could be exported, although others could be the result of some degree of cell lysis during the preparation of the periplasmic fraction. The identified 131 periplasmic proteins with typical export signals were further grouped according to functional categories as shown in Fig. 2B and Table I. Fig. 3A shows the distribution of the A. ferrooxidans periplasmic proteins according to their molecular weights and their corresponding calculated isoelectric points. The majority (70.8%) of the periplasmic proteins of this acidophilic microorganism showed pI values higher than 7, and only 29.2% showed pI values below pH 7. When the results obtained for the periplasmic proteins of A. ferrooxidans were compared with those obtained experimentally for the periplasmic proteins from E. coli (27) and Synechocystis sp. (28) a different distribution of the pI values for the proteins was observed. As predicted in Fig. 3B using SignalP or TatP, pI distribution of proteins in neutrophiles E. coli, Thiobacillus denitrificans, and Geobacter sulfurreducens are comparable. They differ drastically from the pI distributions of the acidophilic A. ferrooxidans and Coxiella burnetii or the acid tolerant Helicobacter pylori. These acid-resistant microorganisms have much higher percentages of basic proteins in their periplasms. On the contrary, the bioinformatics analysis of alkalophiles such as the δ-proteobacterium MLMS-1 and Alkalilimnicola ehrlichii showed a much higher percentage of acidic proteins. The great amount of highly basic proteins in the periplasm of A. ferrooxidans (16.8% of the proteins with pI higher than 10) revealed in this study a special adaptation of this acidophile to its acidic environment. It has been reported that the pH of its periplasm is in the 2.5–3.0 range (5). Most of these proteins are therefore likely acid-tolerant and/or acid-stable. One example is rusticyanin, an abundant redox blue copper protein with a pI of 8.85 that is typically purified by acidifying the A. ferrooxidans extract to pH 2–4 (29).
Fig. 1. 2-D PAGE of the periplasmic fraction of A. ferrooxidans grown on thiosulfate.
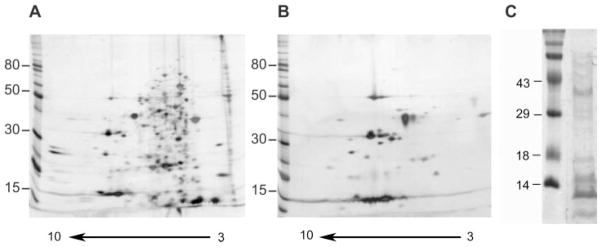
Total proteins (A) or periplasmic fraction (B and C) were separated by 2-D NEPHGE (A and B) with a pH gradient between 3.0 (right side of the gel) and 10.0 (left side of the gel) or by monodimensional SDS-PAGE (C). Spots were detected by silver stain (A and B) or by colloidal Coomassie Blue (C). Molecular mass standards (in kilodaltons) are given on the left of the gels.
Fig. 2. Global analysis of periplasmic proteins from A. ferrooxidans.
A, percent distribution of proteins according to their export signals. B, distribution of periplasmic proteins in functional categories according to TIGR annotation.
Table I. Proteins with predicted export signals identified in the periplasmic fraction of A. ferrooxidans ATCC 23270 grown in thiosulfate.
AFE, predicted protein-coding genes from TIGR-Comprehensive Microbial Resource. Names of proteins correspond to those assigned by TIGR. Sec, SignalP prediction; Tat, TatP prediction; SecP, SecretomeP prediction. Molecular weight, pI, and export signal prediction was done with sequences from a local database.
| AFE | Function/similarity | Molecular weight/pI | Export signal | No. unique peptides | Sequence coverage |
|---|---|---|---|---|---|
|
| |||||
| % | |||||
| Transport and binding proteins | |||||
| 1151 | Phosphate ABC transporter, periplasmic phosphate- binding protein PstS2 | 38.4/9.3 | Sec | 16 | 47 |
| 1648 | Phosphate ABC transporter, periplasmic phosphate-binding protein PstS1 | 36.2/9.41 | Sec | 11 | 45 |
| 174 | TonB-dependent receptor | 86.9/9.19 | Sec | 6 | 10 |
| 1054 | TonB-dependent receptor | 85.3/6.53 | Sec | 20 | 37 |
| 1590 | Iron compound ABC transporter, periplasmic iron-binding protein, putative | 38.4/9.72 | Sec | 3 | 10 |
| 2438 | Cation ABC transporter, periplasmic cation-binding protein, putative | 33.5/8.85 | Sec | 6 | 38 |
| 2957 | Tol-Pal system β propeller repeat protein TolB | 45.5/8.76 | Sec | 14 | 54 |
| 3103 | TonB-dependent receptor | 84.8/8.96 | Sec | 7 | 9 |
| 848 | Carbohydrate-selective porin, OprB family | 40.7/7.05 | Sec | 5 | 15 |
| 1256 | Toluene tolerance protein, putative | 25.2/9.08 | Sec | 2 | 9 |
| 2865 | Toluene tolerance protein, putative | 23.5/9.33 | Sec | 8 | 62 |
| 1586 | ABC transporter, periplasmic substrate-binding protein, putative | 37.3/9.18 | Sec | 13 | 53 |
| 2975 | Periplasmic solute-binding protein, putative | 36.6/8.73 | Sec | 17 | 73 |
| 2981 | Periplasmic solute-binding protein, putative | 36.7/9.1 | Sec | 21 | 59 |
| Cell envelope | |||||
| 2087 | Periplasmic glucan biosynthesis protein, MdoG | 57.4/7.38 | Sec | 2 | 7 |
| 2621 | Pilin, putative | 17.7/5.09 | SecP | 1 | 5 |
| 3018 | Pilus chaperone protein, putative | 30/11.54 | Sec | 5 | 27 |
| 158 | Lipoprotein, putative | 15.5/11.3 | Sec, LipoP | 2 | 20 |
| 297 | Lipoprotein, putative | 10.2/9.42 | Sec, LipoP | 3 | 49 |
| 560 | Outer membrane protein, OMPP1/FadL/TodX family | 47/8.59 | Sec | 2 | 9 |
| 745 | Lipoprotein, putative | 43.7/9.35 | SecP | 14 | 45 |
| 783 | Outer membrane lipoprotein Slp, putative | 20.1/6.28 | SecP, LipoP | 2 | 18 |
| 850 | Lipoprotein, putative | 16.2/8.49 | Sec, LipoP | 6 | 47 |
| 982 | VacJ lipoprotein | 30.2/6.97 | Sec, LipoP | 1 | 3 |
| 1632 | Outer membrane protein, OMP85 family | 86.6/8.9 | Sec | 7 | 12 |
| 2445 | Lipoprotein, putative | 13.8/10.03 | Sec, LipoP | 5 | 41 |
| 2579 | Rare lipoprotein B, putative | 18.9/10.61 | Sec LipoP | 1 | 5 |
| 2654 | Outer membrane protein transport protein, OMPP1/FadL/TodX family | 36.9/5.92 | Sec | 8 | 28 |
| 2722 | OmpA family protein | 22.3/9.4 | Sec | 5 | 24 |
| 2748 | Lipoprotein, SmpA-OmlA family | 7.9/9.16 | SecP | 2 | 22 |
| 2953 | Peptidoglycan-associated lipoprotein, putative | 19.6/6.4 | Sec, LipoP | 2 | 13 |
| 2996 | Outer membrane protein | 55/8.93 | Sec | 24 | 55 |
| Energy metabolism | |||||
| 555 | Glycine cleavage system H protein, putative | 15.2/4.74 | SecP | 3 | 22 |
| 8 | Cytochrome c4 (CycA-1) | 27.5/9.65 | Sec | 1 | 3 |
| 373 | Iron oxidase (Iro) | 10.8/9 | Tat | 3 | 41 |
| 378 | Cytochrome c4 (CycA-2) | 26.2/9.95 | Sec | 4 | 13 |
| 3179 | Cytochrome c (Cyc2) | 52.8/5.4 | Sec | 5 | 15 |
| 3180 | Cytochrome c552 (Cyc1) | 25.2/9.2 | Sec | 6 | 37 |
| 3186 | Rusticyanin (Rus) | 19.9/8.85 | Sec | 15 | 94 |
| Protein fate | |||||
| 2463 | Type I secretion outer membrane protein, TolC family | 50.4/6.4 | Sec | 1 | 2 |
| 259 | Serine protease, DO-DeqQ family | 53.4/7.3 | Sec | 8 | 21 |
| 1685 | Serine protease, DO-DeqQ family | 49.7/7.14 | Sec | 3 | 9 |
| 2514 | C-terminal peptidase (CtpA) | 50.8/8.93 | Sec | 2 | 5 |
| 75 | Survival protein SurA (SurA) | 52.3/6.42 | Sec | 11 | 29 |
| 162 | Thiol:disulfide interchange protein DsbG, putative | 31.8/8.54 | Sec | 3 | 16 |
| 2516 | Peptidyl-prolyl cis-trans isomerase, PPIC-type | 28/9.24 | Sec | 6 | 25 |
| Cellular processes | |||||
| 1399 | Carboxysome peptide B | 92.4/8.21 | SecP | 1 | 10 |
| 2607 | Competence lipoprotein ComL, putative | 30.2/8.93 | Sec, LipoP | 7 | 21 |
| Central intermediary metabolism | |||||
| 2979 | Sulfur-pyrite-thiosulfate-sulfide-induced protein (P21) | 23.8/9.36 | Sec | 5 | 32 |
| 225 | Choloylglycine hydrolase, putative | 39.3/7.15 | Sec | 4 | 14 |
| Protein synthesis | |||||
| 491 | Ribosomal protein L35 | 75/12.31 | SecP | 1 | 22 |
| 2701 | Ribosomal protein S19 | 10.3/11.28 | SecP | 1 | 9 |
| 2717 | Ribosomal protein L11 | 14.9/9.84 | SecP | 1 | 8 |
| Unknown function | |||||
| 831 | Phosphoesterase family protein | 60/6.26 | Tat | 14 | 33 |
| 2266 | Phosphorylase, putative | 39.5/7.28 | SecP | 4 | 13 |
| 169 | TonB-dependent receptor domain protein | 28.9/8.41 | SecP | 2 | 15 |
| 847 | PQQ enzyme repeat domain protein | 43.2/9.61 | Sec | 19 | 48 |
| 851 | DsbG domain protein | 22.3/8.42 | Sec | 3 | 19 |
| 1154 | PQQ enzyme repeat domain protein | 41.7/7.82 | Sec | 4 | 14 |
| 1396 | Tat pathway signal sequence domain protein (carboxysome structural protein CsoS2) | 78.7/9.32 | Tat | 3 | 5 |
| 2576 | YceI-like family protein | 25.2/9.28 | Sec | 10 | 42 |
| 2650 | ErfK-YbiS-YcfS-YnhG family protein | 37.6/7.77 | Sec | 1 | 3 |
| 2753 | Thiol:disulfide interchange domain protein | 21.9/6.96 | Sec | 5 | 30 |
| 2974 | Tat pathway signal sequence domain protein | 29.5/6.24 | Tat | 7 | 44 |
| 2982 | Tat pathway signal sequence domain protein | 29.7/6.35 | Tat | 7 | 47 |
| 3006 | LysM domain protein | 37.6/9.62 | Sec | 7 | 22 |
Fig. 3. pI distribution among periplasmic proteins found in A. ferrooxidans compared with some other Gram-negative micro-organisms.
A, distribution of all proteins found in the periplasmic fraction from A. ferrooxidans ATCC 23270 grown in thiosulfate according to their theoretical molecular masses and isoelectric points. B, percentage of SignalP or TatP proteins with pI values above and below 7.0 predicted in complete genomes of Gram-negative bacteria. Alkalophiles, δ-proteobacterium MLMS-1 and A. ehrlichii MLHE-1; neutrophiles, E. coli K-12, T. denitrificans ATCC 25259, and G. sulfurreducens PCA; acidophiles, C. burnetii RSA 493 and A. ferrooxidans ATCC 23270. Also included in this group for comparison is the acid-tolerant H. pylori 26695.
In a bacterium such as H. pylori, both the inner and outer membrane of the organism can be exposed to high acidity. It has been argued that because most of these proteins have an isoelectric point significantly more alkaline than that of neutrophiles (30), they would retard the flux of protons into the periplasmic space. A similar situation may take place for several of the A. ferrooxidans exposed proteins. Accordingly we estimated before that all the external putative loops of the major outer membrane protein Omp40 from A. ferrooxidans would have a positive net charge at pH 2.5 (31). Being positively charged, the pore would restrict the diffusion of protons from both outside and the periplasmic space toward the environment. These modifications may be a general functional adaptation of acidophiles that provides them with a degree of resistance to the acid present in their environment.
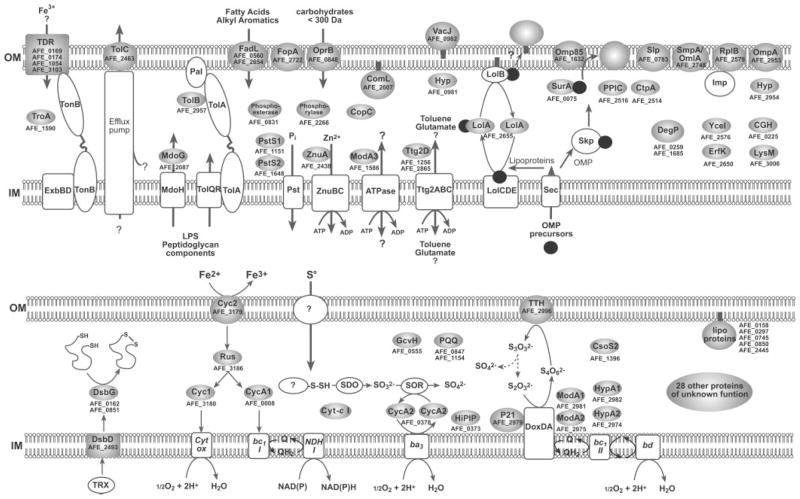
As shown in Fig. 2B and Table I, periplasmic proteins identified were further grouped according to functional categories. Almost 50% of the periplasmic proteins belonged to the cell envelope, protein fate, transport and binding proteins, and energy metabolism categories. On the other hand, about 36% of the proteins comprised proteins with no homologues in databases, being proteins with unknown functions that most likely are unique to A. ferrooxidans. Based on what is known or has been proposed for other microorganisms, we show schematically in Fig. 4 the possible locations and in some cases the suggested functions of the proteins present in the periphery of A. ferrooxidans.
Fig. 4. Schematic location and suggested putative functions of the majority of the proteins identified in the periplasmic fraction from A. ferrooxidans.
Most of the proposed localizations and possible roles of the indicated proteins are based on previously reported studies in E. coli and other microorganisms (1, 33, 40, 43, 73). The proteins shaded in gray were identified experimentally by proteomics analysis of the periplasmic fraction. Transport functions are highlighted in the upper part, and energy metabolism is in the lower portion of the figure. The OMPs are shown in their final destination into the outer membrane. Black circles represent OMP precursors in transit to the outer membrane. The order in which the different proteins are included in this schematic is arbitrary.
Transport and Binding Proteins
A. ferrooxidans contains two pstS genes, both of which form part of an apparent Pho regulon (32) because both are induced when A. ferrooxidans is grown in the absence of phosphate and may represent an adaptation of the microorganism for the survival under scarce phosphate availability. AFE_1151 and AFE_1648 in Table I corresponded to the genes coding for phosphate-binding proteins PstS2 and PstS1, respectively (Fig. 4), which may form part of ABC-type periplasmic phosphate transporters in A. ferrooxidans.
TonB-dependent receptors are outer membrane proteins that form channels permeable to large solutes such as vitamin B12 and siderophores. These receptors bind ligands with high affinity and require the interaction with the TonB protein (Ref. 33 and references therein). In A. ferrooxidans, AFE_0174, AFE_1054, and AFE_3103 are putative TonB-dependent receptors (Fig. 4) and may have a function equivalent to those from E. coli. We also identified in the periplasm another protein (AFE_0169) that showed only a conserved TonB-dependent receptor domain.
AFE_1590 has been annotated as a putative periplasmic iron compound-binding protein that belongs to the TroA superfamily. Bioinformatics analysis of the A. ferrooxidans genome showed 11 putative genes coding for siderophore outer membrane receptors, all grouped in seven gene clusters (34). According to these authors, AFE_1590 is located at “cluster B,” which potentially encodes for a complete suite of proteins needed for Fe(III) uptake and also has an upstream putative fur box. The identification and presence of the periplasmic binding protein TroA (AFE_1590) (Fig. 4) in the periplasmic fraction from A. ferrooxidans grown in thiosulfate strongly suggests that this gene cluster is used in A. ferrooxidans for Fe(III) uptake under this growth condition.
AFE_2463 is a putative protein with similarity to TolC, a common component of a wide variety of efflux pumps (see Fig. 4). TolC and similar proteins are involved in the export of chemically diverse molecules ranging from large protein toxins such as α-hemolysin to small toxic compounds such as antibiotics (33).
The Tol system is involved in outer membrane stability by establishing the structural network linking outer membrane to the peptidoglycan, driving newly synthesized outer membrane components across the periplasm, and the surface expression of O-lipopolysaccharide (35). AFE_2957 was found to be similar to TolB, the periplasmic component of the Tol biopolymer transport system. Candidate genes coding for the other components of the Tol-Pal system (TolAQR) were also present in the genetic context of AFE_2957, but we did not find them in our periplasmic fraction.
Fig. 4 also shows the location for AFE_2438, which is a putative periplasmic cation-binding protein of the TroA family similar to the periplasmic zinc-binding protein ZnuA. Downstream in the genetic context of AFE_2438 there are genes coding for proteins with similarities to the high affinity zinc ABC-type transporter: the ATP-binding protein (ZnuC) (AFE_2437) and a duplicated permease (ZnuB) (AFE_2436 and AFE_2435) that could form part of one transcriptional unit (36).
AFE_2865 and AFE_1256 are periplasmic putative toluene tolerance proteins (Ttg2D) (Fig. 4). The ttg2D gene encodes a periplasmic component of an ABC-type transport system involved in resistance to organic solvents (37). The surrounding three ORFs in this A. ferrooxidans putative cluster (AFE_2866, AFE_2867, and AFE_2868) have similarity to genes ttg2ABC, which are part of the Pseudomonas putida operon involved in toluene resistance (37). It is expected that toluene resistance would be related to extrusion of toluene; however, the operon in P. putida has been shown to be a four-member system typical of the ABC uptake family systems. In addition, the whole-genome analysis of transporters in the plant pathogen Xylella fastidiosa suggests that a ttg putative operon may be related to uptake of glutamate and other polar amino acids instead of being a system for toluene resistance (38). In fact, proteins with homology to Ttg proteins of P. putida were described in Neisseria meningitidis, and their participation in the uptake of L-glutamate was verified (39). Interestingly it has recently been found that despite being an obligate autotroph A. ferrooxidans can use exogenous glutamate2 suggesting the existence of a possibly functional glutamate transporter in this microorganism.
AFE_1586 (ModA3) is the candidate gene for the periplasmic component of an ABC-type molybdate transport system (Fig. 4) that belongs to the ModA family (40). Upstream of the genetic context of AFE_1586 from A. ferrooxidans there is a hypothetical protein (AFE_1587) and a cation diffusion facilitator family transporter (AFE_1588) suggesting a role for AFE_1586 in cation transport. Two other putative periplasmic components of an ABC-type molybdate transporter system were found in the periplasmic fraction: AFE_2981 or ModA1 and AFE_2975 or ModA2 (Fig. 2). These transport systems allow both uptake and efflux and have different ion specificities. Structural modeling of ModA proteins with crystal structures of known similar proteins strongly suggests a functional and conserved mechanism for the transport of thiosulfate/sulfate or molybdate in A. ferrooxidans (41). By expression proteomics we found previously that ModA1 and ModA2 are synthesized in much higher amounts in cells grown in sulfur or thiosulfate as compared with those grown in ferrous iron (17, 41). The almost absent expression of these putative binding proteins in ferrous iron suggests that these proteins are related with sulfur compound metabolism in this acidophile (Fig. 4). Recently in a proteomics study of A. ferrooxidans total proteins, Bouchal et al. (42) described a sulfate/molybdate-binding protein expressed in high abundance during sulfur growth condition. These authors suggested that in all probability this protein is the same gene product we previously annotated as ModA1 (13, 17).
In the “central intermediary metabolism: sulfur metabolism category” we found that AFE_2979 (Table I) corresponded to the periplasmic sulfur-pyrite-thiosulfate-sulfide-induced protein P21 that we have described before (12). The genomic context of the rhodanese-like gene p21 contains the gene modA1 and modA2 and the two putative thiosulfate quinone oxidoreductases (doxDA1 and doxDA2) (3, 13). These genes were all highly expressed as determined by real time PCR and DNA macroarray studies in cells grown in thiosulfate compared with the levels seen in ferrous iron (12, 13, 17). Furthermore we have described in A. ferrooxidans that the transcription levels of doxDA1 and doxDA2 genes are induced when the microorganism was grown in pyrite compared with ferrous iron growth.3
Cell Envelope
Outer membrane proteins (OMPs) have been identified as adhesins, architectural proteins, passive diffusion pores, siderophore receptors, efflux channels, protein translocation pores, and enzymes (e.g. lipases, proteases, and palmitoyltransferases) (33). OMPs are synthesized in the cytoplasm and are translocated across the inner membrane. Passage through the periplasm presents a number of challenges due to the hydrophobic nature of the OMPs, the acidic pH of the periplasm in the case of A. ferrooxidans, and the choice of membranes into which they can insert (43). Most likely, the periplasmic intermediates of the OMPs diffuse through the periplasm before they are inserted into the outer membrane (44). In this regard, Eppens et al. (45) demonstrated that PhoE passes through the periplasm on its way to the outer membrane. It is also known that misfolded OMPs are present in the periplasm of E. coli under stressing conditions (46). Several possible outer membrane precursor proteins were found in the periplasmic fraction from A. ferrooxidans analyzed in Table I. It is possible that these intermediates or possibly misfolded envelope proteins were detected in our case due to the high dynamic range and specificity of the FT MS that allows their recognition.
AFE_0848 showed identity to a carbohydrate-selective porin of the OprB family (Fig. 4). The OprB porin from P. aeruginosa is permeable to solutes of <300 kDa but shows specificity for monosaccharides (33).
AFE_2654 showed similarity to an outer membrane protein of the OMPP1/FadL/TodX family involved in translocation of long-chain fatty acids across the outer membrane and in alkyl-aromatic compound catabolism (47). Downstream of the genomic context of AFE_2654 is AFE_2655, a candidate gene for a LolA protein that was detected in the periplasmic fraction (Fig. 4). AFE_2655 did not show a typical export signal but was predicted to be located in the periplasm by the PENCE Proteome Analyst and CELLO programs. LolA is a periplasmic chaperone involved in lipoprotein transport through the bacterial cell envelope (48). After transport via the Sec system, lipoproteins bind to the ABC transporter LolCDE. Energy from ATP hydrolysis by LolD is transferred to LolC and LolD and then used to open the hydrophobic LolA cavity to accommodate the lipoprotein. When the LolA-lipoprotein complex interacts with the outer membrane receptor LolB, the lipoprotein is transferred to LolB and then inserted into the outer membrane (48) (Fig. 4). Candidate genes for LolCD (AFE_2555 and AFE_2556) and LolB (AFE_0343) were also present in the genome sequence from A. ferrooxidans ATCC 23270 (www.tigr.org).
Fig. 4 indicates a possible location for the protein coded by AFE_0560 that also belongs to the OMPP1/FadL/TodX outer membrane protein transport protein family. This protein was identified previously in our laboratory by 2-D PAGE as a sulfur-induced protein called Omp44 (17). A. ferrooxidans contained in its periplasmic fraction proteins identified as candidates SurA (AFE_0075) and Omp85 (AFE_1632) (Table I). Members of the Omp85 family are involved in the assembly of outer membrane proteins, LPS, glycolipids, and phospholipids (49). Most likely then outer membrane proteins from A. ferrooxidans are assembled and inserted into the outer membrane according to the currently proposed model for Gram-negative bacteria. The presence of LolA in the periplasmic fraction and of genes coding for putative LolB and LolCD in A. ferrooxidans strongly suggests that in this acidophilic microorganism the mechanism for the transport of lipoproteins from the inner membrane to the outer membrane is similar to that proposed for E. coli (48) (Fig. 4).
AFE_2579 showed similarity to a rare lipoprotein homologue of RlpB from E. coli. Recently it was reported that r1pB is an essential gene in E. coli that forms a complex with Imp, an important protein for outer membrane biogenesis, and functions in LPS assembly (50). In the genome of A. ferrooxidans there is a candidate gene for an Imp protein codified in a different genetic region (AFE_0076) that could function together with RlpB in LPS assembly in A. ferrooxidans (Fig. 4).
Fig. 4 also shows the location for the proteins coded in the AFE_0783 and AFE_2748. AFE_0783 is a candidate gene for an outer membrane lipoprotein of the Slp family that in E. coli is known to be an outer membrane lipoprotein (43). AFE_2748 is a putative gene for an outer membrane lipoprotein of the SmpA/OmlA family that is possibly involved in maintaining the structural integrity of the cell envelope.
AFE_2953 and AFE_2722 are peptidoglycan-associated lipoproteins of the OmpA family (Fig. 4). As in the case of OmpA, they could have a role in maintaining the bacterial surface integrity acting as a physical linkage between the outer membrane and the peptidoglycan (51). AFE_2722 showed similarity to an OmpA-like outer membrane protein, FopA (Fig. 4), which was identified previously as an NaCl-regulated major outer membrane proteins of A. ferrooxidans strain NASF-1.4
We also found in the periplasmic fraction AFE_2087 that is similar to the osmoregulated periplasmic glucan biosynthesis protein MdoG (52) and AFE_0158, AFE_0297, AFE_0745, AFE_0850, and AFE_2445 that were identified as putative lipoproteins of unknown function (Fig. 4).
AFE_2621 (not shown in Fig. 4) has a short N-terminal sequence that directs the methylation of a conserved phenylalanine residue (Pfam N-methyl, PF07963) often found at the N terminus of pilins and other proteins involved in secretion. However, the periplasmic protein encoded by AFE_2621 appears to be unique for A. ferrooxidans. AFE_3018 (not included in Fig. 4) was found to be similar to a putative pili assembly chaperone, transmembrane protein of the FimC and PapD family (Table I). PapD-like chaperones bind, stabilize, and cap interactive surfaces of subunits until they are assembled into the pilus (53). The presence of fimbria and pili-like appendages has been reported previously for A. ferrooxidans (54).
Protein Fate
A range of periplasmic proteins were identified and implicated in the targeting and assembly of extracytoplasmic proteins and as such are potential candidates as facilitators for OMP biogenesis. These folding factors include molecular chaperones, protein folding catalysts, and proteases. Two types of folding catalysts are found in the periplasm, namely protein-disulfide isomerases and peptidyl-prolyl cis-trans isomerases (PPIases) (43). We found two putative PPIases in the periplasm of A. ferrooxidans (AFE_0075 and AFE_2516) (Table I and Fig. 4). PPIases have been shown to facilitate the cis-trans isomerization of proline residues both in vitro and in vivo. AFE_0075 is a protein with similarity to E. coli SurA, a periplasmic protein that possesses both chaperone and PPIase activity. AFE_2516 is a protein with a PPIase domain, but it did not show similarity to known proteins. For this putative protein we can infer a function similar to that of SurA and other PPIases (55).
Periplasmic and outer membrane proteins often contain disulfide bonds. Their formation is facilitated in vivo by a number of specialized thiol-disulfide exchange enzymes, which are involved in the formation, isomerization, and reduction of disulfide bonds. DsbC and DsbG are homodimeric disulfide isomerases that resolve incorrectly formed disulfide bonds. They are maintained in a reduced state by DsbD, which is regenerated by the cytoplasmic thioredoxin reductase system (56). We found one DsbD homologue (AFE_2493) (Fig. 4), a protein without a predicted export signal (not shown in Table I), and one DsbG homologue (AFE_162) in the periplasm, but we did not find proteins with similarities to DsbC protein in the complete genome of A. ferrooxidans. However, we found another protein in the periplasmic fraction (AFE_0851) with a DsbG domain that possesses an active site similar to the active site of SoxW, a thioredoxin involved in the transfer of electrons to an unknown periplasmic target of the thiosulfate-oxidizing enzyme during lithotrophic growth of Paracoccus pantotrophus in thiosulfate (57).
AFE_0259 and AFE_1685 (Table I) were found to be similar to two putative periplasmic serine proteases of the DO-DeqQ family. They are trypsin-like serine proteases, typically periplasmic, that contain a C-terminal PDZ domain. These serine proteases belong to the HtrA family of proteins (Fig. 4). In E. coli, DegP, DegQ, and DegS comprise the HtrA family of proteins. These three proteins exhibit a high degree of sequence similarity in their protease domains. However, DegP and DegQ harbor two PDZ domains (like AFE_0259 and AFE_1685), whereas DegS contains only one (58). In contrast to the high sequence similarity found in HtrA homologues of various species, one specific region in the protease domain exhibits only minimal sequence identity. This region of DegP is called the Q-linker as many Gln residues have been discovered in DegP of E. coli (59). Upon comparison of the sequences found near the Q-linker in AFE_0259 and AFE_1685, it was clear that both exhibited sequence similarity to the Q-linker of E. coli DegP, which is of a similar length. Therefore, we propose that the HtrA-like proteins annotated in A. ferrooxidans (www.tigr.org) should be renamed as DegP-like proteins (Fig. 4).
Energy Metabolism
In the electron transfer subgroup of the “energy metabolism functional category” (Table I), which is a key subgroup for the substrate oxidation ability of A. ferrooxidans, we detected and identified six proteins. All of them showed signal peptides, and the only protein showing a Tat signal in this group was the putative high redox potential iron-sulfur protein (HiPIP) or iron oxidase (Iro, AFE_0373). The HiPIP protein has been characterized previously in A. ferrooxidans (60, 61). This protein was proposed to be the first electron acceptor in several alternative models of the electron transfer chain between Fe(II) and oxygen (60, 62). However, recently Bruscella et al. (63) suggested that Iro is involved in an electron transfer chain between a cytochrome bc1 complex functioning in the forward direction and a terminal oxidase as it occurs in other known HiPIPs (64). Bruscella et al. (63) cloned the gene coding for A. ferrooxidans HiPIP and overproduced it in the periplasm of E. coli. Translocation of this protein in the periplasm of E. coli was dependent on the tatC gene, indicating that the A. ferrooxidans HiPIP protein is dependent on the Tat system. Our results demonstrate indeed that this protein is present in the periplasm of A. ferrooxidans (Fig. 4).
The other five proteins with Sec export signals were cytochromes c4 (CycA-1, AFE_0008 and CycA-2, AFE_0378), cytochrome c (Cyc2, AFE_3179), cytochrome c552 (Cyc1, AFE_3180), and rusticyanin (AFE_3186). All these redox proteins have been described to be located in the periplasm of A. ferrooxidans (1, 5, 17), and it has been confirmed here by our mass spectrometric analysis (Fig. 4). It is known that cycA1 is codified by the petI operon and the cycA2 is in petII operon (65). Very recently, a model has been presented in which petI was proposed to encode the bc1 complex, functioning in the uphill flow of electrons from iron to NAD(P), whereas petII was suggested to be involved in electron transfer from sulfur to oxygen (66). The participation of these two c4 cytochromes in thiosulfate oxidation has not been reported previously. However, considering thiosulfate is a principal intermediary in pyrite (FeS2) oxidation, these two c4 cytochromes most likely have an important function during the mineral oxidation.
AFE_2996 has a pyrroloquinoline quinone (PQQ) enzyme repeat that has been found in several enzymes that utilize pyrroloquinoline quinone as a prosthetic group (67). This protein was previously reported as a sulfur-induced outer membrane protein in A. ferrooxidans MSR, and the corresponding gene was cloned and sequenced (68). This sulfur-induced outer membrane protein was recently annotated (EMBL-EBI accession number AB259312) as a tetrathionate hydrolase, and we have tentatively included it in Fig. 4. AFE_0847 and AFE_1154 (Table I and Fig. 4) were also annotated as PQQ enzyme repeat domain proteins.
AFE_0555 was a candidate gene for the lipoate-binding protein H of the glycine cleavage system. Lipoate acts as a swinging arm in the glycine cleavage system and 2-oxo acid dehydrogenase complex moving the substrate to the different active sites of the complex (69). Putative genes nearby are similar to genes related to lipoate metabolism and a putative heterodisulfide reductase complex. The function of these genes is entirely unknown in A. ferrooxidans.
Cellular Processes and Unknown Proteins with General Putative Functions
AFE_2607 (Table I) is a candidate gene for ComL similar to that of Neisseria gonorrhoeae (70). Although no information regarding this putative protein is currently available in A. ferrooxidans, we have tentatively included it in Fig. 4. AFE_0831 showed similarity to an acid phosphatase from Methylococcus capsulatus, and AFE_2266 has an incomplete purine nucleoside permease (NUP) domain, which is present in both bacteria and fungi (71). The function and specificity of both putative proteins (Fig. 4) are unknown in A. ferrooxidans.
HypA1 (AFE_2974) and HypA2 (AFE_2982) are export proteins, with no homologues in databases, that contain a Tat signal peptide (Fig. 4). Downstream of AFE_2982 in the genome of A. ferrooxidans are codified P21 and ModA1 proteins (12, 13). AFE_1396 is the carboxysome structural protein CsoS2 of A. ferrooxidans. This protein also contains a Tat signal peptide and forms a putative operon with the second copy of the Rubisco genes (cbbL2 and cbbS2) (72).
Ribosomal Proteins
Table I shows the presence of three putative ribosomal proteins (S19, L11, and L35). These proteins showed a predicted SecretomeP export signal. This could be an indication that the predicting program gives some false positive hits in its predictions because ribosomal proteins would be expected to be located at the cytoplasm. Alternatively the exportation of these proteins may have an interesting but not yet defined role in this microorganism. However, it is also possible that these proteins were present in the periplasmic fraction due to cell lysis contamination.
Unknown Proteins
We found 28 proteins with export signals that were currently annotated as conserved hypothetical proteins, proteins containing domains of hypothetical proteins, or hypothetical proteins (Table II).
Table II. New periplasmic conserved proteins, unique periplasmic proteins, and periplasmic proteins not previously annotated in the genome of A. ferrooxidans ATCC 23270.
Conditions and sequence analysis were the same as in Table I.
| AFE | Function/similarity | Score | E value | Molecular weight/pI | Export signal | No. unique peptides | Sequence coverage |
|---|---|---|---|---|---|---|---|
|
| |||||||
| % | |||||||
| Hypothetical proteins | |||||||
| 136 | Conserved hypothetical protein | 63.2/9.78 | Sec | 4 | 8 | ||
| 163 | Conserved hypothetical protein | 59.6/9.4 | Sec | 3 | 7 | ||
| 278 | Conserved hypothetical protein | 23.9/9.04 | Sec | 1 | 4 | ||
| 981 | Conserved hypothetical protein | 24.2/8.92 | Sec | 6 | 29 | ||
| 2570 | Conserved hypothetical protein | 27.4/9.98 | SecP | 2 | 11 | ||
| 2954 | Conserved hypothetical protein | 28.9/8.91 | Sec | 3 | 14 | ||
| 3181 | Conserved hypothetical protein | 19.9/8.54 | Sec | 6 | 42 | ||
| 1060 | Conserved domain protein | 39.3/7.29 | Sec | 12 | 47 | ||
| 1061 | Conserved domain protein | 10.8/5.66 | Sec | 7 | 63 | ||
| 2507 | Conserved domain protein | 28/8.04 | SecP | 1 | 4 | ||
| 2910 | Conserved domain protein | 52.2/9.31 | Sec | 2 | 6 | ||
| 157 | Hypothetical protein | 31.3/10.79 | Sec | 3 | 11 | ||
| 292 | Hypothetical protein | 15.5/9.3 | Sec | 8 | 54 | ||
| 457 | Hypothetical protein | 15.4/9.74 | Sec | 6 | 52 | ||
| 610 | Hypothetical protein | 10.9/12.24 | Sec | 3 | 31 | ||
| 826 | Hypothetical protein | 17.8/9.72 | Sec | 3 | 25 | ||
| 1110 | Hypothetical protein | 16.1/10 | Sec | 4 | 20 | ||
| 2111 | Hypothetical protein | 21.4/9.94 | Sec | 4 | 25 | ||
| 2167 | Hypothetical protein | 25.4/9.88 | SecP | 3 | 15 | ||
| 2418 | Hypothetical protein | 27.9/9.55 | Sec | 1 | 4 | ||
| 2429 | Hypothetical protein | 46.3/9.04 | Sec | 7 | 21 | ||
| 2434 | Hypothetical protein | 44.5/8.78 | Sec | 2 | 7 | ||
| 2770 | Hypothetical protein | 19.9/5.04 | Sec | 2 | 14 | ||
| 2928 | Hypothetical protein | 14.8/7.8 | Sec | 3 | 32 | ||
| 2994 | Hypothetical protein | 49.8/9.1 | Sec | 13 | 44 | ||
| 3060 | Hypothetical protein | 20.3/10.14 | Sec | 3 | 25 | ||
| 3099 | Hypothetical protein | 21.3/8.56 | Sec | 3 | 18 | ||
| 3197 | Hypothetical protein | 14.5/9.48 | Sec | 8 | 50 | ||
| ORFs not annotated by TIGR | |||||||
| Copper resistance protein CopC (Burkholderia sp. 383) | 85.9 | 4e–16 | 13.5/10.17 | Sec | 4 | 45 | |
| Putative sterol carrier protein (T. denitrificans ATCC 25259) | 120 | 3e–26 | 20.5/9.4 | Sec | 1 | 10 | |
| Cytochrome c, class I (Anaeromyxobacter dehalogenans 2CP-C) | 74.3 | 5e–12 | 32.2/9.99 | SecP | 12 | 31 | |
| Hypothetical protein | 15.4/7.25 | Sec | 3 | 33 | |||
| Hypothetical protein | 10.4/9.43 | Sec | 1 | 11 | |||
| Hypothetical protein | 15.3/10.28 | Sec | 4 | 30 | |||
| Hypothetical protein | 16.8/9.33 | Sec | 9 | 69 | |||
The proteins in Table II are most likely characteristic of A. ferrooxidans and may have important functions yet to be defined. Therefore, they should be reannotated as periplasmic proteins with unknown functions. The high throughput proteomics approach used here offers an important contribution to the functional annotation for the available genomes of acidophilic biomining microorganisms such as A. ferrooxidans for which no genetic systems are available to perform functional analysis by homologous recombination and genetic disruption of the genes of interest.
Non-annotated Proteins
All ORFs coding for the proteins shown in Tables I and II were searched against local synthetic databases before the A. ferrooxidans genome annotation was made available (www.tigr.org). The majority of them coincided with the TIGR annotation. However, we did identify a number of proteins in the periplasmic fraction that are not currently included in the TIGR database. This includes four proteins with no homologues in the databases, a putative sterol carrier protein, and a class 1 cytochrome c. We also experimentally confirmed a putative periplasmic copper resistance protein, CopC. The transcription of its gene has been validated by Northern electrophoresis,5 thus supporting a possible functional role for it in A. ferrooxidans.
Proteins without Predicted Export Signals and Present in the Periplasmic Fraction
We also detected 30 proteins without export signals in the periplasmic fraction of A. ferrooxidans (not shown). Most of these proteins could be attributed to cell lysis during the preparation of the periplasm due to the detection of some abundant cytoplasmic proteins such as EF-Tu, carboxysome shell components, subunits of ribulose-bisphosphate carboxylase, and some ribosomal proteins (L17 and L7–L12). However, it is not clear why only some of the ribosomal proteins were present. Interestingly chaperone DnaK, the chaperonin GroEl, superoxide dismutase, EF-Tu, and some ribosomal proteins have also been found as non-classically secreted proteins mainly in Gram-positive bacteria (26). Therefore, their cellular locations would have to be further verified. Another protein without a typical export signal but found in the periplasm was AFE_2655, a protein with high similarity to LolA (Fig. 4). LolA was predicted to be periplasmically located when using PENCE Proteome Analyst and CELLO programs. It is therefore possible that in A. ferrooxidans its LolA-like protein is exported to the periplasm in a non-classical form.
Final Remarks
Reported here is the first detailed proteomics analysis of periplasmic proteins of A. ferrooxidans. The data gathered from this important biomining microorganism will (i) be a valuable contribution to complete the annotation of A. ferrooxidans and future microbial genomes; (ii) aid in understanding the physiology of this acidophilic bacterium and its adaptation to its very acidic environment with high toxic metals concentrations; (iii) provide information on several unknown periplasmic proteins, most likely characteristic of A. ferrooxidans and that may have important roles in its functioning; and (iv) be the basis for further differential proteomics studies to determine the interactions of A. ferrooxidans peripheric components with its oxidizable substrates.
Supplementary Material
Fig. S1. MS/MS spectra of single peptides matching to proteins.
Table S2. Protein identifications based on single-peptide assignments.
Footnotes
This work was supported in part by Project Fondo de Ciencia y Tecnología 1030767, by Iniciativa Científica Milenio Project P-05-001-F, and by National Institutes of Health Grant GM37537 (to D. F. H.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The abbreviations used are: TST, thiosulfate sulfurtransferase; ABC, ATP-binding cassette; OMP, outer membrane protein; LPS, lipopolysaccharide; PPIase, peptidyl-prolyl cis-trans isomerase; HiPIP, high redox potential iron-sulfur protein; Tat, twin arginine translocation; PQQ, pyrroloquinoline quinone; 2-D, two-dimensional; NEPHGE, non-equilibrium pH polyacrylamide gel electrophoresis; TIGR, The Institute for Genomic Research; CELLO, subcellular localization predictor; PENCE, Protein Engineering Network of Centres of Excellence; Rubisco, ribulose-bisphosphate carboxylase/oxygenase; EF, elongation factor.
O. Orellana, personal communication.
Valenzuela, L., Chi, A., Beard, S., Shabanowitz, J., Hunt, D. F., and Jerez, C. A. (2007) Differential-expression proteomics for the study of sulfur metabolism in the chemolithoautophic Acidithiobacillus ferrooxidans in Microbial Sulfur Metabolism (Friedrich, C. G., and Dahl, C., eds), pp. 77–86, Springer Press, Berlin.
K. Kamikura, unpublished results.
C. Navarro and C. A. Jerez, unpublished results.
References
- 1.Rawlings DE. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb Cell Fact. 2005;4:13. doi: 10.1186/1475-2859-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohwerder T, Gehrke T, Sand W. Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl Microbiol Biotechnol. 2003;63:239–248. doi: 10.1007/s00253-003-1448-7. [DOI] [PubMed] [Google Scholar]
- 3.Valenzuela L, Chi A, Beard S, Orell A, Guiliani N, Shabanowitz J, Hunt DF, Jerez CA. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnol Adv. 2006;24:197–211. doi: 10.1016/j.biotechadv.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Olson JG, Brierley JA, Brierley CL. Bioleaching review part B: progress in bioleaching: applications of microbial processes by the mineral industries. Appl Microbiol Biotechnol. 2003;63:249–257. doi: 10.1007/s00253-003-1404-6. [DOI] [PubMed] [Google Scholar]
- 5.Ingledew WJ. Thiobacillus ferrooxidans. The bioenergetics of an acidophilic chemolithotroph. Biochim Biophys Acta. 1982;683:89–117. doi: 10.1016/0304-4173(82)90007-6. [DOI] [PubMed] [Google Scholar]
- 6.Yarzábal A, Brasseur G, Ratouchniak J, Lund K, Lemesle-Meunier D, DeMoss JA, Bonnefoy V. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J Bacteriol. 2002;184:313–317. doi: 10.1128/JB.184.1.313-317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silver M, Lundgren DG. Sulfur-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans) Can J Biochem. 1968a;46:457–461. doi: 10.1139/o68-069. [DOI] [PubMed] [Google Scholar]
- 8.Silver M, Lundgren DG. The thiosulfate-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans) Can J Biochem. 1968b;46:1215–1220. doi: 10.1139/o68-181. [DOI] [PubMed] [Google Scholar]
- 9.De Jong GAH, Hazeu W, Bos P, Kuenen G. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology. 1997;143:499–504. doi: 10.1099/00221287-143-2-499. [DOI] [PubMed] [Google Scholar]
- 10.Kelly DP, Shergill JK, Lu WP, Wood AP. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leewenhoek. 1997;71:95–107. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 11.Tabita R, Silver M, Lundgren DG. The rhodanese enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans) Can J Biochem. 1969;47:1141–1145. doi: 10.1139/o69-184. [DOI] [PubMed] [Google Scholar]
- 12.Ramírez P, Toledo H, Guiliani N, Jerez CA. An exported rhodanese-like protein is induced during growth of Acidithiobacillus ferrooxidans in metal sulfides and different sulfur compounds. Appl Environ Microbiol. 2002;68:1837–1845. doi: 10.1128/AEM.68.4.1837-1845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta M, Beard S, Ponce J, Vera M, Mobarec JC, Jerez CA. Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins. OMICS. 2005;9:13–29. doi: 10.1089/omi.2005.9.13. [DOI] [PubMed] [Google Scholar]
- 14.Laundenbach DE, Ehrhardt D, Green L, Grossman A. Isolation and characterization of a sulfur-regulated gene encoding a periplasmically localized protein with sequence similarity to rhodanese. J Bacteriol. 1991;173:2751–2760. doi: 10.1128/jb.173.9.2751-2760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ames GFL, Prody C, Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Farrel PZ, Goodman HM, O’Farrel PH. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 17.Ramírez P, Guiliani N, Valenzuela L, Beard S, Jerez CA. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds or metal sulfides. Appl Environ Microbiol. 2004;70:4491–4498. doi: 10.1128/AEM.70.8.4491-4498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevchenko A, Mann M, Wilm M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 19.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Sub-femtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 20.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Interpolated Markov models for eukaryotic gene finding. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 22.Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 23.Eng J, McCormack A, Yates J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Corbin RW, Paliy O, Yang F, Shabanowitz J, Platt M, Lyons CE, Root K, McAuliffe J, Jordan MO, Kustu S, Soupene E, Hunt DF. Toward a protein profile of Escherichia coli: comparison to its transcription profile. Proc Natl Acad Sci U S A. 2003;100:9232–9237. doi: 10.1073/pnas.1533294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DL. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rey S, Acab M, Gardy JL, Laird MR, deFays K, Lambert C, Brinkman FS. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;1:D164–D168. doi: 10.1093/nar/gki027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulda S, Huang F, Nilsson F, Hagemann M, Norling B. Proteomics of Synechocystis sp. Strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur J Biochem. 2000;267:5900–5907. doi: 10.1046/j.1432-1327.2000.01642.x. [DOI] [PubMed] [Google Scholar]
- 29.Blake RC, II, Shute EA. Respiratory enzymes of Thiobacillus ferrooxidans: kinetic properties of an acid-stable iron:rusticyanin oxidoreductase. Biochemistry. 1994;33:9220–9228. doi: 10.1021/bi00197a025. [DOI] [PubMed] [Google Scholar]
- 30.Scott DR, Weeks D, Melchers K, Sachs G. The life and death of Helicobacter pylori. Gut Suppl. 1998;1:S56–S60. doi: 10.1136/gut.43.2008.s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guiliani N, Jerez CA. Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl Environ Microbiol. 2000;60:2318–2324. doi: 10.1128/aem.66.6.2318-2324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vera M, Guiliani N, Jerez CA. Proteomic and genomic analysis of the phosphate starvation response of Acidithiobacillus ferrooxidans. Hydrometallurgy. 2003;71:125–132. [Google Scholar]
- 33.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quatrini R, Jedlicki E, Holmes DS. Genomic insights into the iron uptake mechanisms of the biomining microorganism Acidithiobacillus ferrooxidans. J Ind Microbiol Biotechnol. 2005;32:606–614. doi: 10.1007/s10295-005-0233-2. [DOI] [PubMed] [Google Scholar]
- 35.Lloubès R, Cascales E, Walburger A, Bouveret E, Lazdunski C, Bernadac A, Journet L. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res Microbiol. 2001;152:523–529. doi: 10.1016/s0923-2508(01)01226-8. [DOI] [PubMed] [Google Scholar]
- 36.Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim K, Lee S, Lee K, Lim D. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J Bacteriol. 1998;180:3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meidanis J, Braga MD, Verjovski-Almeida S. Whole-genome analysis of transporters in the plant pathogen Xylella fastidiosa. Microbiol Mol Biol Rev. 2002;66:272–299. doi: 10.1128/MMBR.66.2.272-299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monaco C, Tala A, Spinosa MR, Progida C, De Nitto E, Gaballo A, Bruni CB, Bucci C, Alifano P. Identification of a meningococcal L-glutamate ABC transporter operon essential for growth in low-sodium environments. Infect Immun. 2006;74:1725–1740. doi: 10.1128/IAI.74.3.1725-1740.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Self WT, Grunden AM, Hasona A, Shanmugam KT. Molybdate transport. Res Microbiol. 2001;152:311–321. doi: 10.1016/s0923-2508(01)01202-5. [DOI] [PubMed] [Google Scholar]
- 41.Valenzuela L, Beard S, Guiliani N, Jerez CA. In: Harrison STL, Rawlings DE, Petersen J, editors. 16th International Biohydrometallurgy Symposium, Compress.Proceedings of the 16th International Biohydrometallurgy Symposium; Cape Town, South Africa. September 25–29, 2005; 2005. pp. 773–780. [Google Scholar]
- 42.Bouchal P, Zdráhal Z, Helánová S, Janiczek O, Hallberg K, Mandl M. Proteomic and bioinformatic analysis of iron- and sulfur-oxidizing Acidithiobacillus ferrooxidans using immobilized pH gradients and mass spectrometry. Proteomics. 2006;6:4278–4285. doi: 10.1002/pmic.200500719. [DOI] [PubMed] [Google Scholar]
- 43.Mogensen J, Otzen D. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 44.Ureta AR, Endres RG, Wingreen NS, Silhavy T. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2006;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eppens EF, Nouwen N, Tommassen J. Folding of bacterial outer membrane protein during passage through the periplasm. EMBO J. 1997;16:4295–4301. doi: 10.1093/emboj/16.14.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- 47.Black PN, DiRusso CC. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol Mol Biol Rev. 2003;67:454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bos MP, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Curr Opin Microbiol. 2004;7:610–616. doi: 10.1016/j.mib.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Gentle LE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 50.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 52.Bohin JP. Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol Lett. 2000;186:11–19. doi: 10.1111/j.1574-6968.2000.tb09075.x. [DOI] [PubMed] [Google Scholar]
- 53.Sauer FG, Knight SD, Waksman GJ, Hultgren SJ. PapD-like chaperones and pilus biogenesis. Semin Cell Dev Biol. 2000;11:27–34. doi: 10.1006/scdb.1999.0348. [DOI] [PubMed] [Google Scholar]
- 54.Dispirito A, Silver M, Voss L, Tuovinen O. Flagella and pili of iron-oxidizing Thiobacilli isolated from an uranium mine in Northern Ontario, Canada. Appl Environ Microbiol. 1982;46:1196–1200. doi: 10.1128/aem.43.5.1196-1200.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer G, Tradler T, Zarnt T. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett. 1998;426:17–20. doi: 10.1016/s0014-5793(98)00242-7. [DOI] [PubMed] [Google Scholar]
- 56.Bessette PH, Cotto JJ, Gilbert HF, Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 57.Bardischewsky F, Fischer J, Höller B, Friedrich CG. SoxV transfers electrons to the periplasm of Paracoccus pantotrophus—an essential reaction for chemotrophic sulfur oxidation. Microbiology. 2006;152:465–472. doi: 10.1099/mic.0.28523-0. [DOI] [PubMed] [Google Scholar]
- 58.Kim DY, Kim KK. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol. 2005;38:266–274. doi: 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- 59.Wootton JC, Drummond MH. The Q-linker—a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 60.Fukumori Y, Yano T, Sato A, Yamanaka T. Fe(II)-oxidizing enzyme purified from Thiobacillus ferrooxidans. FEMS Microbiol Lett. 1988;50:169–172. [Google Scholar]
- 61.Kusano T, Takeshima T, Sugarawa K, Inoue C, Shiratori T, Yano T, Fukumori Y, Yamanaka T. Molecular cloning of the gene encoding Thiobacillus ferrooxidans Fe(II) oxidase. High homology of the gene product with HiPIP. J Biol Chem. 1992;267:11242–11247. [PubMed] [Google Scholar]
- 62.Yamanaka T, Fukumori Y. Molecular aspects of the electron transfer system which participates in the oxidation of ferrous ion by Thiobacillus ferrooxidans. FEMS Microbiol Lett. 1995;17:401–413. doi: 10.1111/j.1574-6976.1995.tb00222.x. [DOI] [PubMed] [Google Scholar]
- 63.Bruscella P, Cassagnaud L, Ratouchniak J, Brasseur G, Lojou E, Amils R, Bonnefoy V. The HiPIP from the acidophilic Acidithiobacillus ferrooxidans is correctly processed and translocated in Escherichia coli, in spite of the periplasm pH difference between these two micro-organisms. Microbiology. 2005;151:1421–1431. doi: 10.1099/mic.0.27476-0. [DOI] [PubMed] [Google Scholar]
- 64.Bonora P, Principi I, Monti B, Ciurli S, Zannoni D, Hochkoeppler A. On the role of high-potential iron sulfur proteins and cytochromes in the respiratory chain of two facultative phototrophs. Biochim Biophys Acta. 1999;1410:51–60. doi: 10.1016/s0005-2728(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 65.Levican G, Bruscella P, Guacunano M, Inostroza C, Bonnefoy V, Holmes DS, Jedlicki E. Characterization of the petI and res operons of Acidithiobacillus ferrooxidans. J Bacteriol. 2002;184:1498–1501. doi: 10.1128/JB.184.5.1498-1501.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bruscella P, Appia-Ayme C, Levicán G, Ratouchniak J, Jedlicki E, Holmes DS, Bonnefoy V. (2007) Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiol. 2007;153:102–110. doi: 10.1099/mic.0.2006/000067-0. [DOI] [PubMed] [Google Scholar]
- 67.Oubrie A, Dijkstra BW. Structural requirements of pyrrolo-quinoline quinone dependent enzymatic reactions. Protein Sci. 2000;9:1265–1273. doi: 10.1110/ps.9.7.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buonfiglio V, Polidoro M, Soyer F, Valenti P, Shively J. A novel gene encoding a sulfur-regulated outer membrane protein in Thiobacillus ferrooxidans. J Biotechnol. 1999;72:85–93. doi: 10.1016/s0168-1656(99)00097-8. [DOI] [PubMed] [Google Scholar]
- 69.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 70.Fussenegger M, Facius D, Meier J, Meyer TF. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol. 1996;5:1095–1105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- 71.Detke S. Cloning of the Candida albicans nucleoside transporter by complementation of nucleoside transport-deficient Saccharomyces. Yeast. 1998;14:1257–1265. doi: 10.1002/(sici)1097-0061(1998100)14:14<1257::aid-yea326>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- 72.Cannon GC, Baker SH, Soyer F, Johnson DR, Bradburne CE, Mehlman JL, Davies PS, Jiang QL, Heinhorst S, Shively JM. Organization of carboxysome genes in the thiobacilli. Curr Microbiol. 2003;46:115–119. doi: 10.1007/s00284-002-3825-3. [DOI] [PubMed] [Google Scholar]
- 73.Collet JF, Bardwell JC. Oxidative protein folding in bacteria. Mol Microbiol. 2002;44:1–8. doi: 10.1046/j.1365-2958.2002.02851.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. MS/MS spectra of single peptides matching to proteins.
Table S2. Protein identifications based on single-peptide assignments.