Abstract
Very young cucumber (Cucumis sativus) fruit are highly susceptible to infection by the oomycete pathogen, Phytophthora capsici. As the fruit complete exponential growth, at approximately 10–12 days post pollination (dpp), they transition to resistance. The development of age-related resistance (ARR) is increasingly recognized as an important defense against pathogens, however, underlying mechanisms are largely unknown. Peel sections from cucumber fruit harvested at 8 dpp (susceptible) and 16 dpp (resistant) showed equivalent responses to inoculation as did whole fruit, indicating that the fruit surface plays an important role in defense against P. capsici. Exocarp from 16 dpp fruit had thicker cuticles, and methanolic extracts of peel tissue inhibited growth of P. capsici in vitro, suggesting physical or chemical components to the ARR. Transcripts specifically expressed in the peel vs. pericarp showed functional differentiation. Transcripts predominantly expressed in the peel were consistent with fruit surface associated functions including photosynthesis, cuticle production, response to the environment, and defense. Peel-specific transcripts that exhibited increased expression in 16 dpp fruit relative to 8 dpp fruit, were highly enriched (P<0.0001) for response to stress, signal transduction, and extracellular and transport functions. Specific transcripts included genes associated with potential physical barriers (i.e., cuticle), chemical defenses (flavonoid biosynthesis), oxidative stress, penetration defense, and molecular pattern (MAMP)-triggered or effector-triggered (R-gene mediated) pathways. The developmentally regulated changes in gene expression between peels from susceptible- and resistant- age fruits suggest programming for increased defense as the organ reaches full size.
Introduction
Fruit development is typified by a progression from fruit set, to exponential fruit growth, maturation, and ripening. Morphological and transcriptomic analyses of early cucumber fruit growth indicate that the period spanning anthesis through the end of exponential expansion is marked by two developmental transitions, one at the onset of exponential growth, the second at the end of exponential growth [1]. The first several days post-pollination (0–4 dpp), prior to exponential growth, are characterized by extensive cell division [1–3]. Transcripts nearly exclusively expressed during this time period include homologs of genes associated with cell cycle and DNA replication [1,4]. The first transition, as is typical of fruit development in general [5], is from cell division to cell expansion. Rapid fruit elongation in cucumber occurs from approximately 4–12 dpp [2,3,6,7]. Genes with peak expression at mid-exponential growth (8 dpp), included genes encoding cytoskeleton, cell wall, cuticle, and phloem-specific proteins [1,6]. A second shift in gene expression occurred at the end of exponential growth, 12–16 dpp, but well before the transition to maturity and ripening that occurs at about 25–30 dpp.
The transition accompanying the late/post-exponential growth stage, which has received little attention in the literature, was marked by strong enrichment for abiotic and biotic-stress related genes and induction of stress-related and development-related transcription factor gene homologs [1]. In certain cultivars, this time period is also associated with transition from susceptibility to resistance to an important cucumber disease, fruit rot caused by Phytophthora capsici. The soil borne oomycete pathogen, P. capsici, causes severe yield and economic losses for a variety of important vegetable crops, including cucumber (Cucumis sativus) [8,9]. The primary infectious agents responsible for spread of disease during the growing season are motile zoospores, which are released from asexual sporangia upon contact with water. For cucumber plants, it is primarily the fruit, rather than the leaves and vines that become infected [10]. Our prior studies showed that that very young cucumber fruit are highly susceptible, but at 10–12 dpp transition to resistance, becoming fully resistant by 16 dpp [10,11]. The transition to resistance was observed in bee-pollinated field-grown fruit, hand-pollinated greenhouse-grown fruit, and fruit that set parthenocarpically in the greenhouse, indicating that the resistance did not depend on the presence or development of seeds within the fruit [10,12]. An age-related reduction in susceptibility to P. capsici also was observed in fruit of several other cucurbit crops, including pumpkin, butternut squash, and acorn squash, although the effect was most pronounced for cucumber [11,13]. Age-related, or ontogenic, resistance also has been observed in strawberry fruit and grape berries in response to powdery mildew (Podosphaera aphanis and Erysiphe nectator, respectively) [14,15] and for apple fruit resistance to scab (Venturia inaequalis) [16].
In addition to the above examples in fruit systems, age-related resistance (ARR) has been observed in a number of other host-pathogen systems in a variety of tissue types, thus ARR is becoming increasingly recognized as an important component of plant defenses against fungal, oomycete, bacterial and viral pathogens [17–20]. However, the specific mechanisms responsible for resistance are just beginning to be explored and appear to vary among systems. By definition, ARR results from developmentally regulated changes that occur prior to contact with the pathogen, possibly by physical or chemical barriers, or by priming the organ for more rapid detection or response to infection. In several cases, ARR coincides with a developmental transition such as transition to flowering or critical leaf number as occurs for resistance of tobacco (Nicotiana tabacum) to P. parasitica, Arabidopsis to Pseudomonas syringa pv. tomato, and maize to Puccinia sorghi [21–24].
As the fruit surface is the first point of contact between the host and pathogen, in this study we sought to examine the role of the cucumber fruit surface in ARR to P. capsici. Tests with cucumber peels at different ages indicated a critical role of the fruit exocarp in expression of ARR, including possible biochemical defenses. Transcriptomic analysis of fruit peels identified developmental changes in gene expression in the surface tissue of resistant-age fruit. Peels from resistant-age fruit exhibited specific increase in transcripts associated with potential physical barriers, chemical defenses, and pathogen recognition responses.
Materials and Methods
Fruit peel experiments
Cucumber plants (pickling type, cv. Vlaspik; Seminis Vegetable Seed Inc, Oxnard, CA) were grown in greenhouse facilities at Michigan State University. No permits were required. Plants were grown in 3.78 L plastic pots filled with BACCTO (Michigan Peat Co., Houston, TX) or Suremix Perlite (Michigan Grower Product, Inc., Galesburg, MI) soil medium and fertilized once per week. Temperature was maintained at 21–25°C; supplemental lights were used to provide an 18 h light period. Pest control was performed according to standard management practices. Sets of 20–25 flowers were hand-pollinated on two dates to provide 15–20 fruit of each age to be harvested on the same day. To avoid competition between fruits, only one fruit was set per plant. The experiment was repeated three times.
The harvested fruit were washed, surface sterilized by brief immersion in a 5% sodium hypochlorite solution, rinsed with water several times, and allowed to air dry. Zoospore suspensions were prepared from 7-day old cultures of P. capsici isolate OP97 [10] grown on diluted V8 media and flooded with 6–10 ml sterile distilled water to release zoospores as described by Gevens et al. [10].
Preparation of zoospores of P. capsici isolate OP97 was performed according to Gevens et al. [10]. Concentration of the zoospore suspensions was determined by hemacytometer and diluted to 1 x 105 per ml. Exocarp sections (3 cm x 3 cm x 1–2 mm) from the middle part of the fruit were excised from both 8 and 15 dpp fruit with petit knife or razor blade without introducing nicks. A sterile plastic tube (0.6 cm height, 0.8 cm diameter) was placed on the exocarp section and anchored to underlying intact 8 or 16 dpp fruit using a strip of 1 cm wide parafilm. A twenty-two gauge sterile needle was used to deliver 30 μl zoospore suspension into the tube by penetrating the parafilm; the needles did not come in contact with the fruit tissue. Inoculated fruit were incubated under constant light at 23–25°C in plastic wrap covered trays lined with moist paper to maintain high humidity. Intact 8 and 15 dpp fruits were similarly inoculated and included in each tray as control. Each tray contained each treatment combination: 8 day peel/8 day fruit; 8 day peel/15 day fruit; 15 day peel/8 day fruit; 15 day peel/15 day fruit; intact 8 day fruit; intact 15 day fruit. The peel sections and underlying fruit were monitored daily for 10 dpi and scored for stage of disease progression (1 –no symptoms, 2 –water soaked, and 3 –sporulation). The experiment was repeated three times. Data were analyzed as a randomized complete block design by ANOVA using the SAS program 9.1 (SAS Institute Inc., Cary, NC) with mixed procedures. Each value is the mean of at least 9 peel sections or fruit ±se. Bars marked with different letters indicate significant difference by LSD, P<0.05.
Peel extract experiments
Fruit exocarp (1-2mm thick) was collected from the middle section of each fruit by razor blade. Frozen peel samples from fruits of the same developmental stage were pooled and used immediately for sequential extraction with water followed by methanol (Fig 1) based on the procedure by Jayaprakasam et al. [25]. Each extract was concentrated by rotary evaporation (BUCHI Rotavapor, BUCHI, Corp., Newcastle, DE) and freeze-dried using Genesis Pilot Freeze Dryer (SP Scientific Industries, Stoneridge, NY). The aqueous and methanolic extracts were redissolved in water and 10% methanol, respectively, to a final concentration of 25 μg ul-1. A 96-well clear (Thermo Fischer Scientific Inc., Waltham MA) or black microtiter plate (Griener Bio-One, Orlando, FL) was prepared with 200 μl clarified V8 media (centrifuged at 10,000 rpm for 10 min) per well. Samples were treated with 10 μl crude extract solution or solvent controls, and inoculated with 20 μl of 1x105 zoospores ml-1 suspension of either P. capsici isolate OP97 or NY0664-1 expressing red fluorescent protein (RFP) ([26]; kindly provided by C. Smart, Cornell University) as described above. The inoculated plates were incubated at 25°C with a 16h light/ 8h dark cycle for 72 hours.
Fig 1. Phytophthora capsici disease development on cucumber fruit and peels.
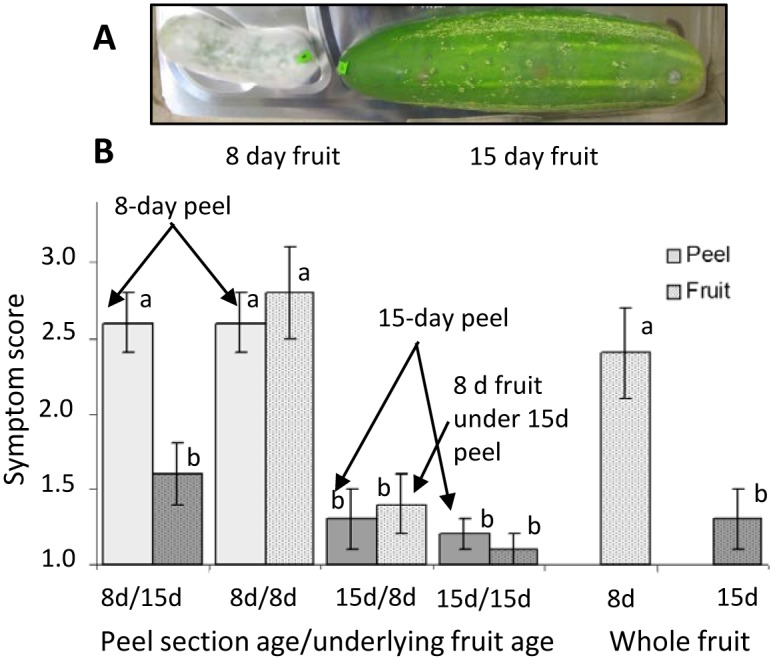
(A) Pathogen growth on whole, unwounded cucumber fruit. Fruit were harvested at 8 or 15 days post pollination (dpp) and inoculated with zoospore suspensions of P. capsisi. The photograph was taken at 5 days post-inoculation. The 8 dpp fruit is covered with mycelial growth. (B) Disease development on peel sections and underlying fruit, or directly inoculated whole fruit as described in methods. x/y indicates age of overlying peel section/age of underlying intact fruit. Symptom score: 1 = no symptoms or localized necrosis; 2 = water soaking; 3 = sporulation. Each value is the mean of at least 9 fruit ±se. Bars marked with different letters indicate significant different, ANOVA, LSD, P<0.05.
Visual ranking was performed on a 1–5 scale at 3 dpi as illustrated in Fig 2A. Fluorescence values of the RFP-expressing cultures were measured at 530nm (excitation) and 590nm (emission) using SpectraMax M2e (Molecular Devices, Sunnyville, CA) at 0, 24, 48 and 72hrs post inoculation. Mean fluorescence measurements from the control (media with aqueous/methanolic extracts) were subtracted from the mean fluorescence values for the corresponding treatments. Samples within the plate were arranged in a randomized complete block design. Data were analyzed by ANOVA, followed by means separation by LSD, P<0.05. Each experiment was repeated two or three times with five replicate samples per treatment.
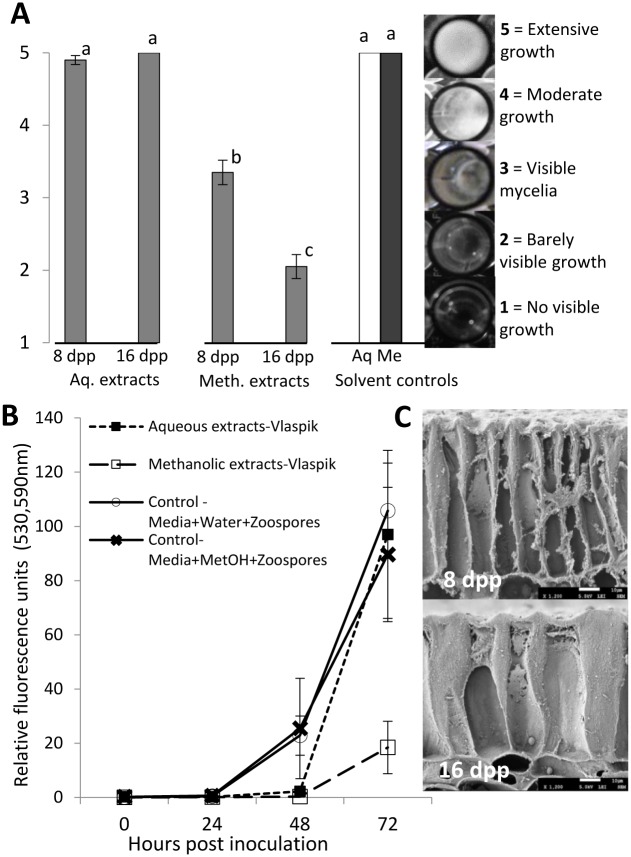
Fig 2. Chemical and physical properties of cucumber fruit peel at 8 and 16 days post pollination (dpp).
(A, B) Effect of aqueous and methanolic extracts from cucumber fruit peel on growth of Phytophthora capsici in vitro. Media in individual wells were treated with 10 μl methanolic or aqueous extracts of cucumber fruit peel or solvent controls prepared as described in methods and inoculated with 20 μl of zoospores at 105 zoospores ml-1. (A) Visual growth rating of isolate OP97 in response to extracts from 8 dpp and 16 dpp fruit peels, 3 days post inoculation. Rating was based on a 1–5 scale as illustrated in the sidebar. (B) Fluorescence emission from isolate NY0664-1RFP in response to aqueous and methanolic extracts. Each value is the mean of 4–5 replicate samples ± S.E. Bars marked with different letters are significantly different (LSD, P<0.05). Each experiment was performed twice with equivalent results. (C) Surface of cucumber fruit at 8 and 16 dpp. Magnification = 1200x; white bar = 10 microns.
Imaging of fruit exocarp by SEM
Sample preparation and imaging of cucumber fruit exocarp sections (2–3 mm) was performed by the Center for Advanced Microscopy of Michigan State University as briefly described here. Exocarp tissues were fixed in glutaraldehyde solution and dried in Balzers Model 010 critical point dryer (Balzers Union Ltd., balzers, Liechtenstein). After drying, the samples were mounted on aluminum stub using high vacuum carbon tabs (SPI supplies, West Chester, PA) and coated with osmium using a NEOC-AT osmium coater (Meiwafosis Co. Ltd., Osaka, Japan. Processed exocarp tissues were examined using a JEOL JSM-7500F scanning electron microscope (JEOL Ltd., Tokyo, Japan).
Sample preparation for pyrosequencing
Cucumber plants were grown in the greenhouse as described above and in Ando et al. [1]. All flowers for each experiment were hand pollinated on a single date (1–2 flowers per plant). The experiment was repeated three times. Randomly assigned groups of twenty fruit were harvested at 8 and 16 dpp and ranked by size; the middle ten fruits were used for RNA extraction. Peel sections (1–2 mm thick) were removed by razor blade, immediately frozen in liquid nitrogen, and stored at– 70°C until RNA was isolated. Each biological replicate consisted of peel sections pooled from ten fruits; two biological replicates were prepared for each age. RNA extraction and oligo(dT)-primed cDNA sample preparation were based on the procedures of Schilmiller et al. [27] and Ando and Grumet [6]. Final concentration was assessed by the nanodrop ND-1000 Final concentration was assessed by the nanodrop ND-1000 (Thermo Scientific, Wilmington, DE) method and subsequent steps for 454 Titanium pyrosequencing analysis were performed by the Michigan State University Research Technology Support Facility (RTSF). Each sample was loaded on a 1/4 plate 454 Pico TiterPlate (454 Life Sciences, a Roche Corporation, CT). Pericarp samples consisting of exocarp, mesocarp, and placenta tissue but not seeds, from fruit grown at the same time in the greenhouse as those used for peel analysis, were sequenced previously [1].
Contig assembly, EST mapping, and gene annotation
Contigs were assembled by the MSU RTSF Bioinformatics Group. Transcript assemblies were created from a collection of EST data sets from Cucumis sativus. An integrated pipeline was used to align individual reads to the C. sativus genome (ICuGI) using BLAT [28] and then to assemble clusters of overlapping alignments. The pipeline, Program to Assemble Spliced Alignments (PASA), is described in Hass et al. [29]. Prior to submitting the EST sequences to PASA they were cleaned using the TIGR SeqClean pipeline [http://compbio.dfci.harvard.edu/tgi/software/]. This was used to remove and vector or primer sequences, poly(A) tails and other low quality or low complexity sequences. Input to the PASA pipeline was comprised of 1.65 million reads generated from the nine libraries of ESTs from fruit at various developmental stages; all libraries were from fruit grown at the same time in the greenhouse. 99.2% of the ESTs were mapped to the genome. PASA assembled 53,677 putative transcripts clustered at ~32,000 loci on the genome. Read data for 8 day post pollination samples is available from the Sequence Read Archive (SRA), accessible through NCBI BioProject ID PRJNA79541. Read data for 0, 4, 12 and 16 dpp samples and the 8 and 16 dpp peel samples in SRA as well as assembled contig sequences deposited as Transcriptome Shotgun Assemblies (TSA) and expression profiling data in the Gene Expression Omnibus (GEO) are available through NCBI BioProject ID PRJNA169904 and DDBJ/EMBL/GenBank under the accession GDIL00000000.
Putative transcripts were annotated by BLAST comparison to both the Arabidopsis proteome (TAIR9) and the NCBI RefSeq Plant database; 37,800 putative transcripts scored a significant (e-value ≤ 10−10) hit to TAIR9 and 40,000 to RefSeq Plant. To estimate relative expression, the number of reads originating from each cDNA library were counted for each contig and reported relative to the total number of reads generated for that library as transcripts per hundred thousand (TPHT).
Transcriptome analysis
The Classification SuperViewer Tool w/Bootstrap web database [30] was used for GO categorization, determination of normalized frequencies relative to Arabidopsis, and calculation of bootstrap standard deviations, and P-values. Princomp procedure SAS 9.1 (SAS Institute, Cary, NC) was used for principal component analysis. The first two principal components, which explain nearly 90% of the total variation were extracted from the covariance matrix. To identify transcripts either preferentially or minimally expressed in peel tissue, the proportion of reads obtained from the peel samples was calculated for each transcript for which there were ≥30 reads [i.e., reads from peel samples at 8 and 16 dpp/total reads (peel + pericarp) at 8 and 16 dpp]. Transcripts with increased expression in 16 dpp peel were identified by the ratio of reads from 16 dpp peel vs. 8 dpp peel. Putative cucumber homologs of the Arabidopsis SYP121/SNP33 regulon [31] were identified within the cucumber fruit transcriptome set and tested for co-expression with cucumber SYP121 and SNP33 by correlation analysis of transcript frequency in peel and pericarp across fruit age.
qRT-PCR
Cucumber fruit used for qRT-PCR analysis were grown as described above for fruit peel experiments and pollinated on two dates, 8 days apart. Five fruits of each age (8 and 16 dpp) were harvested and quickly processed for RNA isolation. Peduncle and blossom ends were removed and peels separated from pericarp of the middle 5–10 cm of fruit tissue using a razor blade. Samples were quickly frozen in liquid nitrogen and stored at -80°C. RNA extraction and oligo(dT)-primed cDNA sample preparation were as described above. qRT-PCR primers were designed using NCBI Primer-BLAST [http://www.ncbi.nlm.nih.gov/tools/primer-blast] and tested for product specificity and reaction efficiency (S1 Table). qRT-PCR reactions were performed with the ABI Prism 7900HT Sequence Detection System (Life Technologies, Inc., Gaithersburg, MD). Samples were prepared using the rEVAlution Master Mix (Syzygy Biotech, Grand Rapids, MI) with ROX reference dye (Syzygy Biotech, Grand Rapids, MI) according to manufacturer’s instructions. Three technical replicates were prepared for each of the five peel and pericarp samples at each age. C. sativus Ubiquitin 3 (CsUBQ3) was used as an endogenous control. For standard curve dilutions a pool of 2 μl from each of the cDNA samples was collected and diluted to 20, 4, 0.8 and 0.16 ng/μl. PCR conditions were 50°C for two minutes, 95°C for 10 minutes enzyme activation, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Samples were quantified using the relative standard curve method for each gene. Relative quantification values (RQ) were normalized to the concentration of CsUBQ3 in each sample.
Results
Cucumber fruit surface is an important determinant of ARR to P. capsici
With the exception of pathogens that enter through wounds or are delivered by a vector, the outer surface of a plant organ is typically the first point of contact with the host. We therefore sought to determine whether the ARR of cucumber fruit to infection by P. capsici was influenced by the fruit surface. Preliminary tests showed that when 16 dpp fruits were peeled prior to inoculation, 100% formed sporulating lesions, whereas none of the intact control fruit at 16 dpp become infected. While these results suggest that the fruit surface plays an important role in the resistance of older fruits to infection by P. capsici, it is also possible that the observed infection was facilitated by wounding as has been observed in other systems.
To eliminate possible confounding effects of injury, exocarp sections from fruits at 8 dpp or 15 dpp were placed on top of a second, intact fruit, and then inoculated with P. capsici. The peel sections responded equivalently to intact fruit (Fig 1). Like whole 8 dpp fruit, the 8 dpp peel pieces exhibited either water-soaking or sporulation, regardless of the age of the fruit underneath. Similarly, peel sections from 15 dpp fruit responded like intact 15 dpp fruit, regardless of fruit age underneath. In addition, even when subjected to the additional disease pressure of contact with an infected 8 dpp fruit peel, the underlying 15 dpp fruit did not typically become infected (8d/15d treatment). Finally, when 15 dpp fruit surface pieces were inoculated, the underlying 8 dpp fruit also maintained a very low disease score (15d/8d treatment), indicating that the 15 dpp fruit surface sections protected the underlying 8 dpp fruit. These results suggest that the surface of 15–16 dpp fruit possesses properties that inhibit P. capsici infection.
Surface factors influencing resistance may include biochemical or structural components. Cucumber leaves have been found to produce methanol-soluble phenolic and flavonoid compounds with antimicrobial properties [32–34]. Therefore we sought to test whether cucumber peels might also produce compounds with antimicrobial activity. Testing of aqueous and methanol extracts from peels of fruit at different ages showed that methanolic extracts from cucumber fruit peels inhibited growth of two P. capsici isolates in vitro (Fig 2A and 2B). Methanolic extracts from 16 dpp peels provided greater inhibition than from 8 dpp peels. Structural changes in the 16 dpp peels included thicker epidermal cell walls, increased cuticle thickness, and increased intercalation of cutin and waxes between adjacent cells in the epidermal layer (Fig 2C).
Gene expression in cucumber fruit peel
To examine changes in gene expression specifically occurring in cucumber peels between fruit at susceptible and resistant ages, cDNA libraries were prepared from peel samples from 8 and 16 dpp fruit. Each biological replicate consisted of peel sections pooled from ten fruits; two biological replicates were prepared for each age. Pyrosequencing analysis yielded 814,250 ESTs from peel samples, which were combined with ESTs obtained from pericarp samples from 0, 4, 8, 12 and 16 dpp fruit that had been grown in the greenhouse at the same time [1], providing a data set of 1.65 million reads. Of those, 99.2% were mapped to the cucumber draft genome [35] at 38,318 loci.
The number of ESTs per assembled transcript ranged from 2–16,817 with a mean of 76 reads/transcript and a median of 9 reads/transcript. As was observed by Ando et al. [1], average assembled transcript length increased with the number of ESTs/transcript until approximately 30 ESTs/transcript (S2 Fig). At ≥30 reads/transcript the average length leveled off at approximately 1.6 kb/transcript. Significant BLAST hits to the NSCI RefSeq Plant gene database (E-value ≤ 10−10) were obtained for 74.5% of the assembled transcripts. Increasing number of ESTs/transcript was also associated with increased identification of putative homologs, again leveling off at approximately 30 ESTs/transcript (S2 Fig). Putative homologs were identified for 97% of the transcripts represented by ≥30 ESTs. Based on the observed relationship between ESTs/transcript, transcript length and identification of putative homologs, all subsequent bioinformatic analysis were performed with the set of transcripts represented by ≥30 ESTs (16,176 transcripts).
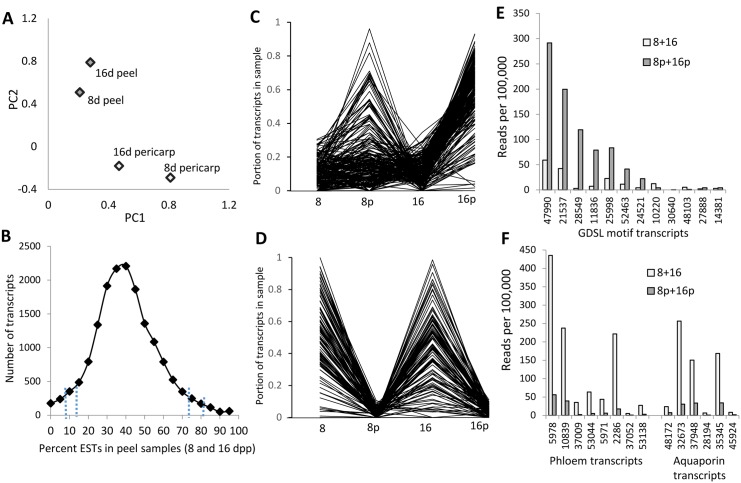
Principal component analysis indicated that pericarp or peel samples at the different ages were more closely associated with each other than were pericarp and peel at the same age, indicating more commonality based on tissue type than fruit age (Fig 3A). To identify transcripts either preferentially or minimally expressed in peel tissue, the proportion of reads obtained from the peel samples relative to pericarp was calculated for each transcript as: reads from peel samples 8 and 16 dpp / total reads (peel + pericarp) at 8 and 16 dpp (Fig 3B). The proportion of reads obtained from the peel samples was approximately normally distributed over the population of transcripts. Genes present in the tails of the distribution, i.e., the top or bottom 5% (Fig 3C and 3D) showed functional differentiation. The 5% of transcripts with highest proportion of expression in the peel relative to the pericarp (a set of 813 transcripts for which >72% of the reads for each transcript were obtained from one or both of the peel samples), were specifically significantly enriched for cellular component categories of extracellular, ER, cell wall and plastid-related genes (S2 Fig). For example, cuticle-related transcripts, such as GDSL motif lipase gene homologs, were primarily expressed in the peel while phloem and aquaporin related gene homologs had minimal expression in the peel (Fig 3E and 3F)
Fig 3. Gene expression in cucumber fruit peel and pericarp samples.
(A) Principal component analysis of transcript expression levels for 8 and 16 days post pollination (dpp) peel and pericarp samples for all transcripts with ≥30 reads. The first two principal components account for 88% of the total variation. (B) Distribution of the portion of gene expression observed in the peel of 8 and 16 dpp fruit for all transcripts with ≥30 reads. The area to the right or left of the dotted vertical lines demarks those transcripts most strongly or most weakly expressed in the peel (top and bottom 2.5% and 5% respectively). (C, D) Expression patterns relative to fruit age and tissue type of genes preferentially expressed in (C) or excluded from (D) the peel at 8 and/or 16 dpp (8 = 8 dpp pericarp; 8p = 8 dpp peel; 16 = 16 dpp pericarp; 16p = 16 dpp peel). (E, F) Examples of transcripts differentially expressed between the peel (8p, 16p) and pericarp (8, 16) samples. (E) GDSL-motif transcripts. (F) Phloem and aquaporin transcripts. Gene labels refer to cucumber transcript assembly numbers (Sup. File 1).
Putative transcription factor genes that were primarily expressed in the peel (>80% reads from peel samples) included many that have been annotated to be involved in biotic and abiotic stress responses, including: R2R3 MYB domain proteins 30 and 96; WRKY 15 and 40; ethylene response factors (ERF6); heat shock factors (HSFA3); and salt tolerance zinc finger (STZ) factors (Table 1). The peel-expressed MYB factor genes all belong to R2R3 subgroup 1, which in Arabidopsis, is involved in drought stress and disease resistance functions including the ABA signal cascade, regulation of stomatal movement, and hypersensitive cell death response [36]. In contrast, expression of many development-related transcription factors observed in the pericarp, such as homologs of AGAMOUS, Auxin response factor 4, FLOWERING LOCUS T, SEEDSTICK, SEPALLATA, and SHATTERPROOF 1 and 2, were largely excluded from the peel samples.
Table 1. Putative transcription factors enriched or reduced in cucumber peel at 8 and 16 days post-pollination.
| Transcript Number | Cucumber CDS | ESTs/100,000 | % reads in peel | Hit ID Arabidopsis | Hit Description | E value | |
|---|---|---|---|---|---|---|---|
| Pericarp | Peel | ||||||
| 14161 | 3M710870.1 | 0.00 | 5.99 | 100.0 | At1g80840 | WRKY40 transcription factor | 1.0E-77 |
| 9034 | 3M809420.1 | 1.07 | 7.95 | 88.2 | At2g42200 | SPL9 (SQUAMOSA Promoter binding protein-like 9) | 1.0E-25 |
| 4603 | 5M603910.1 | 1.03 | 6.81 | 86.8 | At1g10470 | ARR4 (RESPONSE REGULATOR 4) | 1.0E-62 |
| 29023 | 2M301540.1 | 1.07 | 6.77 | 86.4 | At5g01880 | Zn finger (C3HC4-type RING finger) | 4.0E-33 |
| 9225 | 3M816030.1 | 8.96 | 52.03 | 85.1 | At1g08810 | MYB60 (myb domain protein 60) | 4.0E-76 |
| 29496 | 2M354820.1 | 2.09 | 10.54 | 83.5 | At1g27730 | STZ (salt tolerance zinc finger) | 2.0E-53 |
| 22834 | 6M094760.1 | 2.65 | 11.47 | 83.5 | At5g50570 | Squamosa promoter binding protein, putative | 5.0E-43 |
| 15026 | 3M750350.1 | 1.03 | 5.17 | 83.3 | At5g03720 | At-HSFA3 (heat shock transcription factor A3) | 2.0E-74 |
| 5285 | 2M428380.1 | 1.55 | 7.24 | 82.4 | At2g23320 | WRKY15; calmodulin binding | 1.0E-69 |
| 42396 | 3M018320.1 | 4.17 | 19.11 | 82.1 | At4g17490 | AtERF6 (ethylene response element binding factor 6) | 1.0E-52 |
| 43150 | 3M826690.1 | 2.65 | 11.40 | 81.1 | At3g28910 | MYB30 (myb domain protein 30) | 9.0E-81 |
| 13524 | 1M033200.1 | 4.14 | 17.34 | 80.7 | At5g62470 | MYB96 (myb domain protein 96) | 2.0E-89 |
| 7109 | 7M429520.1 | 2.59 | 10.41 | 80.1 | At5g06710 | HAT14 (Homeobox from Arabidopsis thaliana) | 9.0E-79 |
| 14554 | 3M733980.1 | 7.36 | 0.00 | 0.0 | At2g37740 | ZFP10 (Zinc finger protein 10) | 6.0E-19 |
| 41443 | 7M378520.1 | 29.80 | 0.00 | 0.0 | At3g04730 | IAA16; early auxin induced, transcription factor | 1.0E-72 |
| 25851 | Chrom5NA a | 7.86 | 0.00 | 0.0 | At4g09960 | STK (SEEDSTICK) | 4.0E-54 |
| 2743 | 6M520410.3 | 10.46 | 0.00 | 0.0 | At4g18960 | AG (AGAMOUS) | 5.0E-45 |
| 42893 | 1M467100.1 | 84.30 | 0.55 | 0.7 | At2g42830 | SHP2 (SHATTERPROOF 2) | 4.0E-74 |
| 38366 | 5M146260.1 | 9.46 | 0.25 | 2.6 | At5g47610 | Zn finger (C3HC4-type RING finger) | 8.0E-35 |
| 50868 | 3M016400.1 | 7.78 | 0.21 | 2.7 | At4g18020 | APRR2 (pseudo response regulator 2) | 1.0E-101 |
| 22831 | 6M095270.1 | 5.80 | 0.21 | 3.6 | At1g24260 | SEP3 (SEPALATTA3) | 2.0E-65 |
| 48984 | 6M291920.1 | 17.25 | 0.78 | 4.3 | At5g60450 | ARF4 (Auxin response factor 4) | 1.0E-129 |
| 22337 | 5M270900.1 | 16.64 | 0.75 | 4.3 | At2g02070 | AtIDD5 (Arabidopsis thaliana Intermediate domain 5) | 1.0E-150 |
| 18472 | 1M651710.1 | 8.44 | 0.50 | 5.6 | At1g65480 | FT (FLOWERING LOCUS T) | 2.0E-73 |
| 2433 | 6M526230.1 | 10.51 | 0.71 | 6.4 | At2g34830 | WRKY35 (WRKY DNA binding protein 35) | 2.0E-71 |
| 25082 | 4M645240.1 | 10.07 | 0.96 | 8.7 | At1g10120 | DNA binding transcription factor | 2.0E-63 |
| 24179 | 2M000630.1 | 28.67 | 2.78 | 8.8 | At2g19810 | Zn finger (CCCH-type family protein) | 1.0E-101 |
| 51052 | 5M198240.1 | 18.85 | 1.99 | 9.6 | At5g05790 | MYB family transcription factor | 3.0E-71 |
| 48460 | 3M073900.1 | 37.82 | 4.09 | 9.8 | At1g50640 | ERF3 (ethylene response element binding factor 3) | 8.0E-61 |
a Not annotated—located in cucumber genome, but not currently annotated in Chinese Long v.2 draft
Collectively, these observations reflect functional differentiation between the peel and pericarp. Transcripts predominantly expressed in the peel were consistent with fruit surface associated functions including photosynthesis, cuticle production, response to the environment, and defense.
Transcripts expressed specifically in peel from 16 dpp fruit
We next sought to identify transcripts that were primarily expressed in the peel, and also showed increased expression in 16 dpp fruit relative to 8 dpp fruit, i.e., specifically expressed at 16 dpp rather than 8 dpp. The 105 transcripts that met both criteria (top 5% for expression in peel, and top 5% for increase in 16 dpp peel vs. 8 dpp peel) exhibited 8–800 fold enrichment in 16 dpp peel relative to 8 dpp peel (S2 Table). Of those transcripts, 9 did not have putative homologs in the NSCI RefSeq Plant gene database, and an additional 12 with homologs did not have functional annotation. Those with annotation showed strong enrichment for GO categories of response to stress, extracellular, response to abiotic or biotic stimulus, signal transduction, and transport functions (Table 2). The greatest reductions in expression in 16 dpp peel relative to 8 dpp peel were observed for the categories of chloroplast, and plastids. This observation is developmentally consistent with the reduced chlorophyll content observed at these ages [1].
Table 2. Functional enrichment of genes preferentially expressed in peels at 16 days post pollination (dpp).
Transcripts were selected based on criteria of top 5% for expression in peel and top 5% for increase in 16 dpp peel vs. 8 dpp peel. The resulting 105 transcripts had >72% of reads in peel samples and >8-fold increase above 8 dpp peel samples.
| Classification a | Normalized frequency | Bootstrap Std Dev | P value |
|---|---|---|---|
| Other biological processes | 3.09 | 0.357 | 1.13 E-11 |
| Response to stress | 2.70 | 0.334 | 3.89 E-10 |
| Other enzyme activity | 2.43 | 0.300 | 4.18 E-07 |
| Extracellular | 2.40 | 0.444 | 1.74 E-05 |
| Response to abiotic of biotic stimulus | 2.32 | 0.361 | 1.73 E-06 |
| Signal transduction | 2.29 | 0.456 | 6.81 E-04 |
| Transport | 1.99 | 0.328 | 2.28 E-04 |
| Other membranes | 1.64 | 0.295 | 5.23 E-03 |
| Other metabolic processes | 1.50 | 0.080 | 4.37 E-07 |
| Other cytoplasmic components | 1.40 | 0.19 | 9.28 E-03 |
| Other cellular processes | 1.36 | 0.083 | 6.77 E-05 |
a Functional distribution, normalized frequency, and bootstrap standard deviation (SD) of contigs with putative Arabidopsis homologs was determined using the categories classification from the Classification SuperViewer from Bio-Array Resource for Arabidopsis Functional Genomics for Gene Ontology [http://compbio.dfci.harvard.edu/tgi/982 software96/].
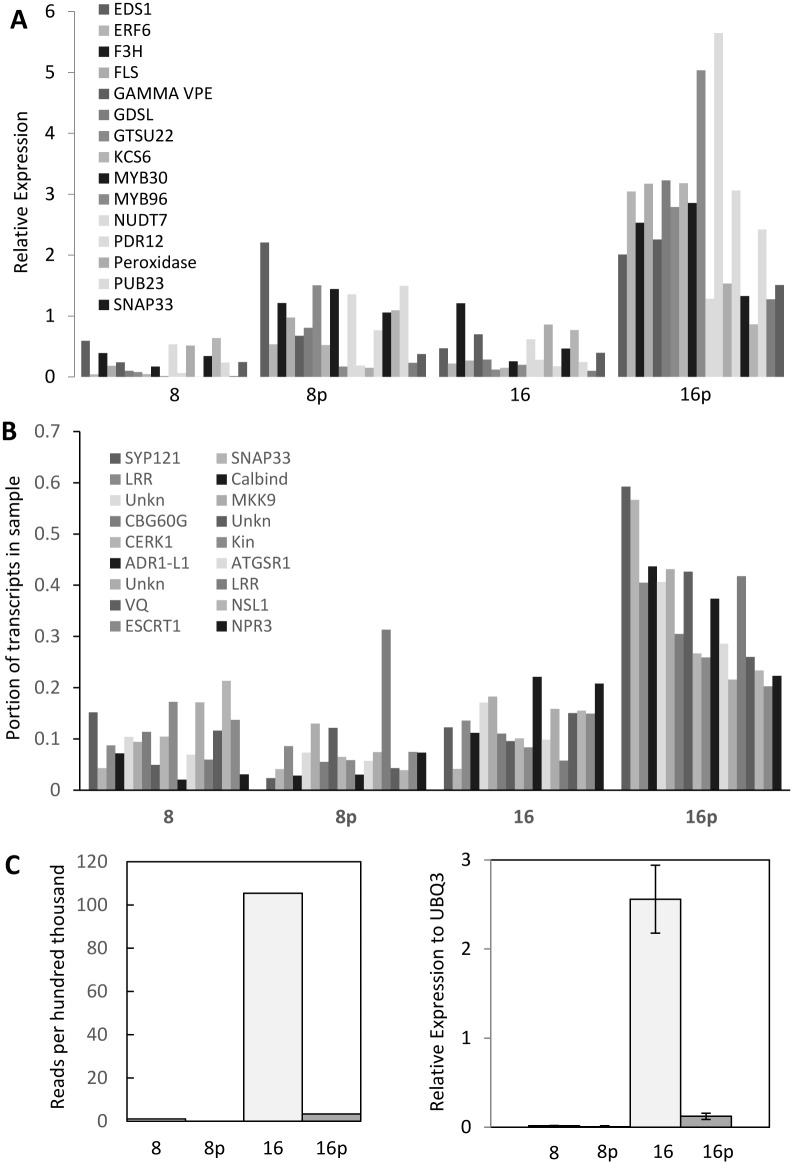
Greater than 40% of the 16 dpp peel-enriched genes were potentially associated with pathogen defense based on annotations from other systems (Table 3). A subset of 20 putative defense-related genes whose transcriptome patterns showed elevated expression in 16 dpp peel samples was selected for verification of expression by qRT-PCR analysis of pericarp and peel samples from 8 dpp and 16 dpp fruit (Fig 4A). The qRT-PCR results mirrored those of the 454 analysis, substantiating the 454 analysis, and also demonstrating reproducibility of gene expression patterns across experiments grown at different times in the greenhouse. Predominant expression was observed in 16 dpp peel tissue, although for a few genes (EDS1, NUDT7, SYP121), the difference between 8 dpp peel and 16 dpp peel was less pronounced in the qRT-PCR analysis than from the transcriptome data.
Table 3. Putative pathogen defense-associated transcripts preferentially expressed in cucumber fruit peel at 16 days post pollination.
| Transcript Number | Cucumber CsaCDS | ESTs/105 16 dpp peel | % reads in peel | 16dpp peel/8dpp peel | Hit ID Arabidopsis | Hit Description | E value |
|---|---|---|---|---|---|---|---|
| 42396 | 3M018320.1 | 18.33 | 82.1 | 21.0 | At4617490 | ATERF6 (ethylene responsive element binding factor 6) | 1E-52 |
| 53532 | 1M033160.1 | 17.01 | 87.0 | 27.3 | At5g62480 | ATGSTU9 (glutathione-S-transferase TAU 9) | 1E-64 |
| 43980 | 4M064630.1 | 59.49 | 87.5 | 8.9 | At1g78340 | ATGSTU22 (glutathione-S-transferase TAU 22) | 2E-42 |
| 43984 | 4M064650.1 | 33.46 | 73.9 | 16.7 | At1g17180 | ATGSTU25 (glutathione-S-transferase TAU 25) | 1e-91 |
| 3557 | 6M507520.1 | 59.35 | 80.5 | 157.7 | At3g12500 | ATHCHIB (Arabidopsis thaliana basic chitinase) | 1E-128 |
| 34558 | 7M031650.1 | 9.45 | 80.2 | 9.4 | At4g12720 | ATNUDT7 (nudix hydrolase homolog 7) | 3E-86 |
| 24234 | 2M000460.1 | 2.85 | 72.8 | 29.5 | At4g29720 | ATPA05 (Polyamine oxidase 5) | 1E-170 |
| 38780 | 4M622230.1 | 17.98 | 75.6 | 11.1 | At3g14690 | CYP72A15; electron carrier | 1E-161 |
| 15574 | 1M006320.1 | 3.46 | 87.0 | 35.6 | At3g48090 | EDS1 (enhanced disease susceptibility 1) | 1E-117 |
| 3105 | 6M516960.1 | 7/56 | 80.1 | 8.7 | At4g21510 | F-box family protein | 2E-30 |
| 3642 | 6M507140.1 | 52.72 | 87.3 | 46.8 | At1g75900 | Family II extracellular lipase (EXL3) | 1E-100 |
| 22665 | 6M108510.1 | 8.10 | 80.2 | 21.8 | At3g51240 | F3H (flavonone-3-hydroxylase) | 1E-120 |
| 29623 | 2M354820.1 | 36.49 | 76.9 | 10.0 | At4g32940 | GAMMA-VPE (Gamma vacuolar processing enzyme) | 0 |
| 21123 | 6M117710.1 | 6.71 | 86.9 | 18.1 | At1g76490 | HMG1 (hydroxy methylglutaryl CoA reductase 1) | 0 |
| 4009 | 5M609650.1 | 71.39 | 83.6 | 38.5 | At1g68530 | KCS6 (3-ketoacyl-CoA synthase 6) | 0 |
| 1636 | 3M144140.1 | 14.91 | 91.1 | 13.6 | At5g43760 | KCS20 (3-ketoacyl-CoA synthase 20); fatty acid elongase | 0 |
| 13524 | 1M033200.1 | 16.51 | 80.7 | 17.9 | At5g62470 | MYB96 (MYB domain protein 96) | 2E-89 |
| 10009 | 4M008780.1 | 8.67 | 1.00 | 13.4 | At3g53260 | PAL; phenylalanine ammonia-lyase | 0 |
| 44827 | 2M433930.1 | 7.53 | 87.9 | 76.3 | At1g59870 | PEN3; PDR12 (PLEIOTROPIC DRUG RESISTANCE 12) | 0 |
| 27490 | 4M285730.1 | 53.46 | 88.2 | 85.4 | At5g06720 | Peroxidase 2 | 7E-95 |
| 53444 | 4M285760.1 | 33.59 | 78.6 | 23.5 | At5g06730 | Peroxidase superfamily protein | 3E-98 |
| 52067 | 6M213910.1 | 20.47 | 76.7 | 205.7 | At5g05340 | Peroxidase superfamily protein | 1E-116 |
| 8758 | 3M782630.1 | 9.45 | 90.4 | 27.3 | At2g35930 | PUB23 (PLANT U-BOX 23) ubiquitin protein ligase | 4E-123 |
| 13606 | 1M039020.1 | 14.12 | 87.8 | 12.6 | At5g61210 | SNAP33 (soluble N-ethylmaleimide-sensitive factor adaptor protein) | 3E-91 |
| 50760 | 2M251460.3 | 3.85 | 79.4 | 11.3 | At4g34640 | SQS1 (squalene synthase 1) | 0 |
| 25013 | 4M642460.1 | 41.05 | 77.5 | 21.9 | At1g27730 | STZ (salt tolerance zinc finger) | 2E-58 |
| 29496 | 2M354820.1 | 9.98 | 83.5 | 15.4 | At1g27730 | STZ (salt tolerance zinc finger) | 2E-53 |
| 8776 | 3M782680.1 | 6.95 | 92.9 | 70.5 | At3g11820 | SYP121 (SYNTAXIN OF PLANTS 121); PEN1 (penetration 1) | 1E-69 |
| 34134 | 3M643770.1 | 8.74 | 90.1 | 10.1 | At5g07990 | TT7 (Transparent testa 7) flavonoid 3’-monoxygenase | 1E-114 |
| 14161 | 3M710870.1 | 5.49 | 100.0 | 9.3 | At1g80840 | WRKY40 transcription factor | 1E-77 |
| 47100 | 7M390100.1 | 7.92 | 77.2 | 8.6 | At1g48910 | YUC10; FAD binding/monooxygenase | 7E-98 |
| 3738 | 6M505230.1 | 4.21 | 89.1 | 43.1 | At2g21320 | Zinc finger (B box type) family protein | 2E-51 |
Fig 4. Expression analysis of putative defense related genes.
(A) qRT-PCR verification of potential pathogen defense-related genes with elevated expression in 16 days post pollination (dpp) peels. (B) Relative expression of SYP121/SNAP33 co-expressed genes in 8 and 16 dpp pericarp and peel samples as assessed by 454 pyrosequencing. Genes shown are in the order listed in Table 4. (C) Expression of CsFM01 as assessed by 454 pyrosequencing (left) and qRT-PCR analysis (right). 8 = 8 dpp pericarp; 8p = 8 dpp peel; 16 = 16 dpp pericarp; 16p = 16 dpp peel.
Age-related, peel-specific, gene expression potentially associated with biochemical and structural defenses
The 16 dpp peel samples were specifically enriched for expression of several groups of genes potentially associated with biochemical or structural defenses. These include putative homologs of genes encoding enzymes associated with flavonoid biosynthesis such as: phenylalanine amonia-lyase (PAL), flavanone-3-hydroxylase (F3H) and flavonoid 3'-monooxygenase (also flavonoid 3' hydroxylase; TT7) (Table 3). A putative homolog of flavonol synthase At5g08640 (1.0E-120) was also highly expressed in the peel (92% of transcript reads) with 4.7-fold higher expression in 16 dpp peel than 8 dpp peel.
Another group of genes with strong expression in the 16 dpp peel samples included members of the glutathione S-transferase (GST) gene family (Table 3). GSTs, which are highly expressed in plants and can comprise 1–2% of the soluble proteins, have been associated with a range of biotic stress response related functions, including increased resistance to several fungal or oomycete pathogens [37]. The GST family is classified into six groups (phi, tau, theta, zeta, lambda, and dehydroascorbate reductase) that exhibit tissue- and developmental- specific expression. The cucumber fruit transcriptome included twelve putative GST family members representing five of the six groups (Additional File 4). Three showed strong expression specifically in the 16 dpp peel samples (Table 3); all three were members of the tau group, which has been associated with resistance to biotic stresses [38, 39].
One of the genes with the strongest 16 dpp peel-specific expression was a putative member of the fungal and plant-specific, pleiotropic drug resistance (PDR) ATP-binding cassette (ABC) transporter sub-family. CsPDR12 (PEN3) showed greater than 70-fold elevated expression in 16 dpp peel relative to 8 dpp peels, and essentially exclusive expression in 16 dpp peel as assessed by qRT-PCR analysis (Table 3, Fig 4A). ABC transporters have been implicated in transport of a wide variety of structurally unrelated molecules, including flavonoid related compounds [40–42].
The 16 dpp peel sample also showed peak expression of putative homologs of several cuticle associated genes [43, 44] including: two GDSL motif genes; two GDSL-like extracellular lipases; a long chain fatty acid- CoA ligase family protein gene; and two very long chain fatty acid synthesis related genes [KCS6 and 20 (3-ketoacyl-CoA synthase 6 and 20)] (Table 3; S2 Table; Fig 4A).
Age-related, peel-specific, gene expression of defense pathway associated genes
Several of the genes strongly represented in the 16 dpp peel samples are putative homologs of genes that have been specifically associated with microbial associated molecular pattern (MAMP)-triggered defense, suggesting possible priming for defense (Table 3). A central feature of MAMP defense is an oxidative burst resulting from rapid production of reactive oxygen species (ROS), including peroxidase-mediated production of hydrogen peroxide [45,46]. The oxidative burst can inhibit pathogen growth, signal induction of host defense responses, and promote hypersensitive response. The genes showing increased expression in the 16 dpp peel included putative homologs of three members of the peroxidase gene family with 20-200- fold up-regulation relative to 8 dpp peel. Peroxidases also have been implicated in additional roles that may contribute to plant defense, including strengthening of cell walls (e.g., protein cross linking, lignification), potentially inhibiting pathogen penetration [46].
Notably, two of the genes specifically highly expressed in the 16 dpp peel were putative homologs of genes that confer resistance to penetration of fungal and oomycete pathogens in Arabidopsis and barley, SYP121 (SYNTAXIN OF PLANTS)/PEN1(PENETRATION1) and SNAP33 [47]. SYN121/PEN1 function is associated with more rapid formation of penetration-resistant papillae structures containing callose, phenolic compounds, lignin, and reactive oxygen species [48]. As was observed for Arabidopsis and barley, the cucumber SYP121 and SNAP33 homologs were highly co-expressed (R = 0.976, P<0.001). In silico analysis of Arabidopsis genes identified a set of 107 genes co-expressed with PEN1, SNAP33 and MLO2 [31]. Putative homologs for 37 of these genes were observed in the cucumber fruit data set, of which 17 showed patterns of expression correlated with expression of the cucumber SYP121/PEN1 and SNAP33 homologs (Table 4; Fig 4B). These genes included putative homologs of the elicitor response genes, CERK1 (chitin elicitor receptor kinase 1), and MKK9 (MAP kinase kinase 9) [48]; SA-mediated and hypersenstitive response genes, SARD1 (SAR deficient 1) and CPB60G [49]; the SA receptor, NPR3 (NPR1-like gene 3; non-expressor of PR genes 1-like protein 3) [50], and a defense response regulator, NSL1 (necrotic spotted lesions 1) [51].
Table 4. Co-expression analysis of putative cucumber homologs of MLO2/SYP121/SNAP33- co-expressed genes from Arabidopsis.
Putative homologs of MLO2/SYP121/SNAP33- co-expressed genes from Arabidopsis (as identified by Humphrey et al.[31] were tested for co-expression with cucumber SYP121 and SNAP33 genes in cucumber pericarp (0, 4, 8, 12, and 16 dpp) and peel (8 and 16 dpp) samples.
| TranscriptNumber | Cucumber Csa CDS | ESTs/105 16 dpp peel | Corr. With SYP121/SNAP33 | P-value | Hit ID Arabidopsis | Hit Description | E value |
|---|---|---|---|---|---|---|---|
| 8776 | 3M782680.1 | 12.5 | At3g11820 | SYP121 (Syntaxin of plants 121);PEN1 (penetration1)b | 1.0E-124 | ||
| 13606 | 1M039020.1 | 14.1 | At5g61210 | SNAP33 (soluble N-ethylmaleimide-senstive factor adaptor protein 33) | 3.0E-91 | ||
| 33242 | 1M601530.1 | 94.1 | 0.984 | <0.001 | At5g21090 | Leucine-rich repeat protein, putative | 1.0E-100 |
| 13843 | 1M042460.1 | 16.2 | 0.975 | <0.001 | At1g73805 | Calmodulin binding; SARD1 (systemic acquired resistance deficient 1) | 1.0E-94 |
| 14259 | 3M730710.1 | 27.1 | 0.963 | <0.001 | At5g01750 | Unknown protein | 9.0E-60 |
| 13997 | 1M042980.1 | 9.8 | 0.925 | 0.002 | At1g73500 | MKK9 (MAP kinase kinase 9) | 1.0E-114 |
| 6185 | 1M569130.1 | 4.3 | 0.897 | 0.004 | At5g26920 | CBP60G (CAM-binding protein 60-like G); calmodulin binding | 1.0E-51 |
| 8202 | 3M889130.1 | 9.2 | 0.876 | 0.005 | At2g25737 | Unknown protein | 0.0 |
| 34393 | 7M041930.1 | 5.5 | 0.856 | 0.007 | At3g21630 | CERK1 (Chitin elicitor receptor kinase 1) | 0.0 |
| 46112 | 3M651840.1 | 6.2 | 0.847 | 0.008 | At3g14050 | RSH2 (RelA SpoT homolog 2) | 0.0 |
| 31565 | 2M070870.1 | 12.8 | 0.835 | 0.010 | At2g24360 | Serine/threonine/tyrosine kinase, putative | 1.0E-117 |
| 24820 | 4M638480.1 | 9.6 | 0.833 | 0.010 | At4g33300 | ADR1-L1 (Activated disease resistance1-like 1); NB-LRR family protein | 0.0 |
| 38247 | 3M304140.1 | 33.0 | 0.794 | 0.018 | At5g37600 | ATGSR1 (copper ion binding / glutamate-ammonia ligase) | 0.0 |
| 38772 | 4M621210.1 | 14.8 | 0.793 | 0.018 | At1g17080 | Unknown protein | 1.0e-44 |
| 47275 | 7M373520.1 | 3.7 | 0.734 | 0.031 | At5g48380 | Leucine-rich repeat family protein | 4.0E-58 |
| 40525 | 1M074920.1 | 10.7 | 0.718 | 0.036 | At1g28280 | VQ motif containing protein | 4.0E-66 |
| 40561 | 1M075570.1 | 4.7 | 0.689 | 0.044 | At1g28380 | NSL1 (necrotic spotted lesions 1) | 2.0E-63 |
| 14951 | 3M746590.1 | 6.3 | 0.626 | 0.068 | At2g36680 | Located in ESCRT 1 complex | 8.0E-71 |
| 43939 | 4M063470.1 | 3.9 | 0.614 | 0.073 | At5g45110 | NPR3 (NPR1-like protein 3), non-expressor of PR genes1-like protein3 | 1.0E-152 |
Putative homologs of genes associated with effector triggered-defense such as EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1) and NUDT7 (NUDIX HYDROLASE HOMOLOG 7) (Table 3) were also enriched in the 16 dpp peel samples. EDS1 in Arabidopsis regulates R gene-mediated and systemic resistance, acting in combination with several other factors including AtNUDT7 and the flavin-dependent monooxygenase, FMO1, to regulate cell death responses [52,53]. A homolog of FMO1 (At1g19250, 1.0E-116) was minimally expressed in cucumber fruit samples at 0, 4 or 8 dpp, but then increased 50- and 100-fold at 12 and 16 dpp, respectively. The transcripts, however, were almost exclusively located in the pericarp (97%) samples, rather than peel (Fig 4C), suggesting that if FMO1 plays a role in resistance to P. capsici, it likely occurs at an infection step post-penetration.
The set of 16 dpp peel-enriched genes also included putative homologs of R2R3 subgroup 1 MYB transcription factor MYB 96 and a vacuolar processing cysteine protease enzyme, Gamma-VPE, which is a critical component of the hypersensitive programmed cell death response (Table 3) [36,54]. The peel-expressed putative homologs of abiotic and biotic stress MYB factor 96 was 18 fold up-regulated in the 16 dpp peel relative to 8 dpp peel (Table 3), while MYB30, which in Arabidopsis has been found to encode an activator of the hypersensitive cell death response via regulation of 402 biosynthesis of very long chain fatty acids [55], exhibited 4.1-fold increase in 16 dpp peel relative to 8 dpp peel (Table 1, S2 Table).
Discussion
These studies showed that ARR of cucumber fruit to infection by P. capsici is associated with the fruit surface. Peel sections from 16 dpp fruit exhibited resistance, while peel sections from 8 dpp fruit were highly susceptible. We therefore sought to examine changes in the fruit peel that might contribute to increased resistance. Peel sections from 16 dpp had obvious differences in surface morphology and produced increased levels and/or types of methanol-soluble compounds capable of inhibiting growth of P. capsici in vitro. Transcriptome analysis reflected functional differentiation for gene expression between the peel and pericarp with increased expression of fruit surface associated functions such as photosynthesis, cuticle production, response to the environment, and defense in the peel tissue. Gene expression that was specifically associated with peel sections from resistant age fruit showed strong enrichment for transcripts annotated to be associated with response to stress or abiotic or biotic stimuli, signal transduction, and transport and extracellular functions. Greater than 40% of transcripts of the 16 dpp peel-enriched genes were potentially associated with pathogen defense.
Consistent with methanol-soluble compounds capable of inhibiting growth of P. capsici, was increased expression of homologs of several genes associated with flavonoid and phenylpropanoid biosynthesis, PAL, F3H, TT7 and FLS. Previous studies have observed phenylpropanoid-derived phenolics, C-glycosyl flavonoids, and aglycones associated with resistance to powdery mildew (Podosphaera xanthii) in cucumber leaves [32–34] and inhibitory glycoside-linked phenolic compounds that increase with leaf age, have been found to localize to cucumber leaf cells beneath penetrating appressoria of Colletotrichum orbiculare [56]. Such defensive compounds may serve roles as phytoanticipins, accumulating prior to infection, or may exhibit increased resistance in response to infection. The developmental regulation of expression of phenylpropanoid-associated genes, and the presence of pathogen-inhibitory, methanol-soluble compounds in the 16 dpp peel, suggests synthesis of preformed chemical barriers. However, induced resistance in cucumber leaves to powdery mildew has been found to be dependent on elevated activity of flavonoid pathway enzymes [32], suggesting potential for induced response as well. Increased expression of PAL, a key upstream enzyme for flavonoids, as well as salicylic acid and lignin biosynthesis, has been observed in a wide range of systems, including cucumber and melon, in response to treatment with pathogens or chemical inducers (e.g., [57,58]).
The 16 dpp fruit peel sections also had specific elevation of expression of several GST genes and the pleiotropic drug resistance gene family member homolog, CsPDR12. Members of both GST and PDR gene families can facilitate export of flavonoid and terpenoid related compounds to the cell wall where they can accumulate in the cuticle [59,60]. The PDR12 homolog from Nicotiana plumbaginifolia (NpPDR1), shows age-related and epidermal-specific transcription and exports an anti-fungal/oomycete terpenoid to the leaf surface [41,61]. Several members of the Arabidopsis PDR family show age-specific expression in developing seedlings [62].
Other types of defense related genes that were specifically, highly expressed in the peel of 16 dpp fruit were homologs of genes that have been associated with resistance to fungal and oomycete penetration by more rapid formation of papillae, as well as numerous putative elicitor-, effector-, and SA- response genes which may play roles in MAMP or R-gene mediated resistance. While at this time, we cannot determine whether these genes are specifically associated with ARR to P. capsici, the transcriptomic analysis indicating increased expression of a large number of putative-defense related genes, raises the possibility that ARR results from systematic, developmental reprogramming for defense.
In summary, analysis of cucumber fruit indicated importance of the fruit surface for ARR to P. capsici and a potential role for methanol-soluble inhibitory compounds. Transcriptomic studies of the fruit peel suggest developmentally-regulated expression of defense genes potentially associated with structural, biochemical (flavonoid pathway and transporters), MAMP response, and effector-triggered or R-gene mediated resistances.
Supporting Information
(PPTX)
(PPTX)
(DOCX)
(XLSX)
Acknowledgments
We thank Sue Hammar and Elizabeth Straley for greenhouse assistance, Carol Flegler of the Center for Advanced Microscopy, Shari Tjugum-Holland of the Michigan State University Research Technology Support Facility for DNA pyrosequencing, Chris Smart (Cornell University) for providing the fluorescent P. capsici, and Cornelius Barry and Brad Day for critical reading of the manuscript.
Data Availability
This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GDIL00000000. The version described in this paper is the first version, GDIL01000000.
Funding Statement
This work was supported by USDA-NIFA-SCRI (United States Department of Agriculture-National Institute for Food and Agriculture-Specialty Crops Research Initiative) 2011-51181-30661, http://nifa.usda.gov/funding-opportunity/specialty-crop-research-initiative-scri, to RG and Pickle Packers International - Agriculture Research Fund, https://www.ilovepickles.org/, to RG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ando K, Carr KM, Grumet R. Transcriptome analysis of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genomics 2012; 13: 518 10.1186/1471-2164-13-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Colle M, Hodges-King C, Grumet R. Factors influencing fruit size and shape differences in cucumber In: Sari N, Solmaz I, Aras V (eds). Cucurbitaceae 2012. Cukurova University Press; 2012. pp. 585–589. [Google Scholar]
- 3. Fu FQ, Mao WH, Shi K, Zhou YH, Asami T, Yu JQ. A role of brassinosteroids in early fruit development in cucumber. J Exp Bot 2008; 9: 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang XY, Wang Y, Jiang WJ, Liu XL, Zhang XM, Yu HJ et al. Characterization and expression profiling of cucumber kinesin genes during early fruit development: revealing the roles of kinesins in exponential cell production and enlargement of cucumber fruit. J Exp Bot 2013; 64: 4541–4557. 10.1093/jxb/ert269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillaspy G, Ben-David H, Gruissem W. Fruits: A developmental perspective. Plant Cell 1993; 5: 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ando K, Grumet R. Transcriptional profiling of rapidly growing cucumber fruit by 454-pyrosequencing analysis. J Amer Soc Hortic Sci 2010; 135: 291–302. [Google Scholar]
- 7. Marcelis LFM, Hofman-Eijer LRB. Cell division and expansion in the cucumber fruit. J Hort Sci 1993; 68: 665–671. [Google Scholar]
- 8. Hausbeck M, Lamour K. Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis 2004; 88: 1292–1302. [DOI] [PubMed] [Google Scholar]
- 9. Sonogo S, Ji PS. Integrated management of Phytophthora capsici on solanaceous and cucurbitaceous crops: current status, gaps in knowledge and research needs. Can J Plant Pathol 2012; 34: 479–492. [Google Scholar]
- 10. Gevens AJ, Ando K, Lamour KH, Grumet R, Hausbeck MK. A detached cucumber fruit method to screen for resistance to Phytophthora capsici and effect of fruit age on susceptibility to infection. Plant Dis 2006; 90: 1276–1282. [DOI] [PubMed] [Google Scholar]
- 11. Ando K, Hammar SA, Grumet R. Age-related resistance of diverse cucurbit fruit to infection by Phytophthora capsici . J Amer Soc Hort Sci 2009; 134: 176–182. [Google Scholar]
- 12.Ando K. Evaluation of the role of plant architecture and cucumber fruit development in Phytophthora capsici disease development. Ph.D. Thesis, Michigan State University, Graduate Program in Plant Breeding, Genetics, and Biotechnology; 2009.
- 13. Meyer MD, Hausbeck MK. Age-related resistance to Phytophthora fruit rot in ‘Dickenson Field’ processing pumpkin and ‘Golden Delicious’ winter squash fruit. Plant Dis 2013; 97: 446–452. [DOI] [PubMed] [Google Scholar]
- 14. Asalf B, Gadoury DM, Tronsmo AM, Seem RC, Dobson D, Peres NA, et al. Ontogenic resistance of leaves and fruits, and how leaf folding influences the distribution of powdery mildew on strawberry plants colonized by Podosphaera aphanis . Phytopathology 2014; 104:954–963. 10.1094/PHYTO-12-13-0345-R [DOI] [PubMed] [Google Scholar]
- 15. Gee CT, Gadoury DM, Cadle-Davidson L. Ontogenic resistance to Uncinula necator varies by genotype and tissue type in a diverse collection of Vitis spp. Plant Dis 2008; 92:1067–1073. [DOI] [PubMed] [Google Scholar]
- 16. Gusberti M., Gessler C, Broggini GAL. RNA-seq analysis reveals candidate genes for ontogenic resistance in Malus-Venturia pathosystem. PLoS One 2013; 8: e78457 10.1371/journal.pone.0078457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alcazar R, Reymond M, Schmitz G, De Meaux J. Genetic and evolutionary perspectives on the interplay between plant immunity and development. Curr Opin Plant Biol 2012; 14: 378–384. [DOI] [PubMed] [Google Scholar]
- 18. Develey-Rivière MP, Galiana E. Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytologist 2007; 175: 405–416. [DOI] [PubMed] [Google Scholar]
- 19. Panter SN, Jones DA. Age-related resistance to plant pathogens. Adv Bot Res 2002; 38: 251–280. [Google Scholar]
- 20. Whallen MC. Host defense in a developmental context. Mol Plant Pathol 2005; 6: 347–360. 10.1111/j.1364-3703.2005.00286.x [DOI] [PubMed] [Google Scholar]
- 21. Abedon BG, Tracy WF. Corngrass1 of maize (Zea mays L) delays development of adult plant resistance to common rust (Puccinia sorghi Schw) and European corn borer (Ostrinia nubilalis hubner). J Hered 1996; 87: 219–223. [Google Scholar]
- 22. Hugot K, Riviere MP, Moreilhon C, Dayem MA, Cozzitorto J, Arbiol G et al. Coordinated regulation of genes for secretion in tobacco at late developmental stages: Association with resistance against oomycetes. Plant Physiol 2004; 134: 858–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rusterucci C, Zhao Z, Haines K, Mellersh D, Neumann A, Cameron RK. Age-related resistance to Pseudomonas syringae pv. tomato is associated with the transition to flowering in Arabidopsis and is effective against Peronospora parasitica . Physiol Molec Plant Pathol 2005; 66: 222–231. [Google Scholar]
- 24. Wilson DC, Carella P, Isaacs M, Cameron RK. The floral transition is not the developmental switch that confers competence for the Arabidopsis age-related resistance response to Pseudomonas syringae pv. tomato. Plant Molec Biol 2013; 83: 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jayaprakasam B, Seeram NP, Nair MG. Anticancer and anti-inflammatory activities of cucurbitacins from Cucurbita andreana . Cancer Lett 2003; 189: 11–16. [DOI] [PubMed] [Google Scholar]
- 26. Dunn AR, Fry BA, Lee TY, Conley KD, Balaji V, Fry WE et al. Transformation of Phytophthora capsici with genes for green and red fluorescent protein for use in visualizing plant-pathogen interactions. Austral Plant Pathol 2013; 42: 583–593. [Google Scholar]
- 27. Schilmiller AL, Schauvinhold I, Larson M, Xu M, Charbonneaua AL, Schmidt A et al. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Nat Acad Sci USA 2009; 106: 0865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res 2002; 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hass BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 2003; 31: 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Provart N, Zhu T. A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr Computat Mol Biol 2003: 271–272. [Google Scholar]
- 31. Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Gobel U et al. A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Nat Acad Sci USA 2010; 107: 21896–21901. 10.1073/pnas.1003619107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fofana B, Benhamou N, McNally DJ, Labbe C, Seguin A, Belanger RR. Suppression of induced resistance in cucumber through disruption of the flavonoid pathway. Phytophathology 2005; 95: 114–123. [DOI] [PubMed] [Google Scholar]
- 33. McNally DJ, Wurms KV, Labbe C, Belanger RR. Synthesis of C-glycosyl flavonoid phytoalexins as a site-specific response to fungal penetration in cucumber. Physiol Molec Plant Pathol 2003; 63: 293–303. [Google Scholar]
- 34. Ongena M, Daayf F, Jacques P, Thonart P, Benhamou N, Paulitz TC, et al. Systemic induction of phytoalexins in cucumber in response to treatments with fluorescent pseudomonads. Plant Pathol 2000; 49: 523–530. [Google Scholar]
- 35. Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., Lucas W.J., Wang X., et al. The genome of the cucumber, Cucumis sativus L. Nature Genet 2009. November 1, 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- 36. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Cell 2010; 15: 573–581. [DOI] [PubMed] [Google Scholar]
- 37. Oztetik E. A tale of plant glutathione S-transferases: Since 1970. Botanical Rev 2008; 74: 419–437. [Google Scholar]
- 38. Sappl PG, Onate-Sanchez L, Singh KB, Millar AH. Proteomic analysis of glutathione S-transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes. Plant Molec Biol 2004; 54: 205–219. [DOI] [PubMed] [Google Scholar]
- 39. Wisser RJ, Kolkman JM, Patzoldt ME, Holland JB, Yu JM, Krakowsky M et al. Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc Natl Acad Sci USA 2011; 108: 7339–7344. 10.1073/pnas.1011739108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Banasiak J, Biala W, Staszkow A, Swarcewicz B, Kepoczynska E, Figlerowicz, et al. A Medicago trucatula ABC transporter belonging to subfamily G modulates the level of isoflavonoids. J Exp Bot 2013; 64: 1005–1015. 10.1093/jxb/ers380 [DOI] [PubMed] [Google Scholar]
- 41. Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 2001; 13: 1095–1107. [PMC free article] [PubMed] [Google Scholar]
- 42. Verrier PJ, Bird D, Buria B, Dassa E, Forestier C, Geisler M et al. Plant ABC protiens—a unified nomenclature and updated inventory. Trends Plant Sci. 2008; 13: 151–159. 10.1016/j.tplants.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 43. Dominguez E, Heredia-Guerrero JA, Heredia A. Plant cutin genesis: unanswered questions. Trends Plant Sci 2015; 20: 551–558. 10.1016/j.tplants.2015.05.009 [DOI] [PubMed] [Google Scholar]
- 44. Yeats TH, Rose JKC. The formation and function of plant cuticles. Plant Physiol 2013; 163:5–20. 10.1104/pp.113.222737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI). Molec Plant 2015; 8:521–539. [DOI] [PubMed] [Google Scholar]
- 46. O-Brien JA, Daudi A, Butt VS, Bolwell GP. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 2012; 236: 765–779. 10.1007/s00425-012-1696-9 [DOI] [PubMed] [Google Scholar]
- 47. Underwood W, Somerville SC. Focal accumulation of defenses at sites of fungal penetration attack. J Exp Bot 2008; 59: 3501–3508. 10.1093/jxb/ern205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pel MJC, Pieterse CMJ. Microbial recognition and evasion of host immunity. J Exp Bot 2013; 64: 1237–1248. 10.1093/jxb/ers262 [DOI] [PubMed] [Google Scholar]
- 49. Zhang YX, Xu SH, Ding PT, Wang DM, Cheng YT, He J et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 2010; 107: 18220–18225. 10.1073/pnas.1005225107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fu ZQ, Yan SP, Saleh A, Wang W, Ruble J, Oka N et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012; 486: 228–232. 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noutoshi Y, Kuromori T, Wada T, Hirayama T, Kamiya A, Imura Y et al. Loss of necrotic spotted lesions 1 associates with cell death and defense responses in Arabidopsis thaliana . Plant Molec Biol 2006; 62: 29–42. [DOI] [PubMed] [Google Scholar]
- 52. Koch M, Vorwerk S, Masure C, Sharifi-Sirchi G, Olivieri N, Schlaich NL. A role for flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J 2006; 47: 629–239. [DOI] [PubMed] [Google Scholar]
- 53. Strauss MR, Rietz S, van Thaemaat EVL, Bartsch M, Parker JE. Salicylic acid antagonism of EDS-1 driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 2010; 62: 628–640. 10.1111/j.1365-313X.2010.04178.x [DOI] [PubMed] [Google Scholar]
- 54. Rojo E, Martin R, Carter C, Zouhar J, Pan SQ, Plotnikova J et al. VPE gamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 2004; 14: 1897–1906. [DOI] [PubMed] [Google Scholar]
- 55. Raffaele S, Vailleau F, Leger A, Joube J, Miersch O, Huard C, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008; 20: 752–767. 10.1105/tpc.107.054858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin TC, Ishizaka M, Ishii H. Acibenzolar-S-methyl-induced systemic resistance against anthracnose and powdery mildew diseases on cucumber plants without accumulation of phytoalexins. J Phytopathol 2009; 157: 40–45. [Google Scholar]
- 57. Ge YH, Guest KI, Bi Y. Differences in the induction of defense responses in resistant and susceptible muskmelon plants infected with Colletotrichum lagenarium . J Phytophathol 2014; 162: 48–54. [Google Scholar]
- 58. Kuzniak E, Wielanek M, Chwatko G, Glowacki R, Libik-Konieczny M, PIatek M, et al. Salicylic acid and cysteine contribute to arbutin-induced alleviation of angular leaf spot disease development in cucumber. J Plant Physiol 2015; 181: 9–13. 10.1016/j.jplph.2015.03.017 [DOI] [PubMed] [Google Scholar]
- 59. Agati G, Azzarello E, Pollastri S, Tattini M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci 2012; 196: 67–76. 10.1016/j.plantsci.2012.07.014 [DOI] [PubMed] [Google Scholar]
- 60. Zhao J, Dixon RA. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci 2009; 15: 72–80. 10.1016/j.tplants.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 61. Stukkens Y, Bultreys A, Grec S, Trombik T, Vanham D, Boutry M. NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transported from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol 2005; 139: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Badri DV, Loyola-Vargas VM, Broeckling CD, Dela-Pena C, Jasinski M, Santelia D, et al. Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol 2008; 146: 762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX)
(PPTX)
(DOCX)
(XLSX)
Data Availability Statement
This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GDIL00000000. The version described in this paper is the first version, GDIL01000000.