The Dengue virus NS5 proteins from all four serotypes are elongated and flexible in solution, with DENV-4 NS5 being more compact than the others. The methyltransferase domain, along with the ten-residue linker, contributes to the flexibility and is positioned at the top of the RNA-dependent RNA polymerase domain in solution.
Keywords: flavivirus, dengue, nonstructural proteins, viral polymerase, methyltransferase, small-angle X-ray scattering, protein flexibility
Abstract
Infection by the four serotypes of Dengue virus (DENV-1 to DENV-4) causes an important arthropod-borne viral disease in humans. The multifunctional DENV nonstructural protein 5 (NS5) is essential for capping and replication of the viral RNA and harbours a methyltransferase (MTase) domain and an RNA-dependent RNA polymerase (RdRp) domain. In this study, insights into the overall structure and flexibility of the entire NS5 of all four Dengue virus serotypes in solution are presented for the first time. The solution models derived revealed an arrangement of the full-length NS5 (NS5FL) proteins with the MTase domain positioned at the top of the RdRP domain. The DENV-1 to DENV-4 NS5 forms are elongated and flexible in solution, with DENV-4 NS5 being more compact relative to NS5 from DENV-1, DENV-2 and DENV-3. Solution studies of the individual MTase and RdRp domains show the compactness of the RdRp domain as well as the contribution of the MTase domain and the ten-residue linker region to the flexibility of the entire NS5. Swapping the ten-residue linker between DENV-4 NS5FL and DENV-3 NS5FL demonstrated its importance in MTase–RdRp communication and in concerted interaction with viral and host proteins, as probed by amide hydrogen/deuterium mass spectrometry. Conformational alterations owing to RNA binding are presented.
1. Introduction
Dengue virus (DENV), a mosquito-transmitted member of the Flaviviridae family, consists of four antigenically distinct serotypes (DENV-1 to DENV-4) and is an infection threat to nearly 40% of the world’s population (World Health Organization, 2009 ▸). The Dengue virus genome is an ∼11 kb positive-strand RNA comprising a single open reading frame (ORF). This encodes a single polyprotein that is co-translationally and post-translationally processed by both host cellular proteases and the DENV protease (NS2B/NS3) into three structural proteins (capsid, membrane and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) (Lindenbach et al., 2007 ▸).
The replication of the Dengue virus genome takes place in the ER membrane-associated replication complexes (RCs), which include NS and host proteins (Mackenzie, 2005 ▸; Fernandez-Garcia et al., 2011 ▸; Welsch et al., 2009 ▸), with NS3 and NS5 residing in the functional centre. NS3 possesses protease activity (with the cofactor NS2B) for polyprotein cleavage, helicase activity for unwinding dsRNA in the process of replication and NTPase/RTPase activities that are essential for genome capping (Luo et al., 2008 ▸). The 900-amino-acid full-length NS5 protein (NS5FL) harbours multiple enzymatic activities: an RNA-dependent RNA polymerase (RdRp) activity that is required for genome replication and methyltransferase (MTase)/guanylyltransferase (GTase) activities for genome capping (Egloff et al., 2002 ▸; Issur et al., 2009 ▸). The cap, a 7-methylguanosine (m7G) moiety linked to the first nucleotide of the transcript via a 5′–5′ triphosphate bridge, has roles in protecting viral mRNA from degradation by 5′-exoribonucleases and confers stability to mRNAs and ensures their efficient recognition by eukaryotic translation initiation factor 4E (eIF4E) for translation (Decroly et al., 2012 ▸; Liu & Kiledjian, 2006 ▸). The MTase of flaviviral NS5 has been shown to contain both N-7 and 2′-O methylation activities to form the type 1 cap (Dong et al., 2010 ▸). The RNA guanylyltransferase (GTase) function is attributed to the N-terminal segment of the MTase domain (Fig. 1 ▸) and NS3 has been described to stimulate this activity (Issur et al., 2009 ▸).
Figure 1.
Multiple sequence alignment of full-length NS5 proteins from DENV-1 (GenBank EU081230), DENV-2 (GenBank M29095.1), DENV-3 (GenBank AY662691) and DENV-4 (GenBank GQ398256) with secondary structure derived from the crystallographic structure of DENV-3 NS56–895 (PDB entry 4v0r; Zhao et al., 2015 ▸). The secondary structures are indicated by boxes (α-helices), lines (loops) and arrows (β-sheets), and are coloured blue for the methyltransferase domain (dark blue, N-terminal GTP-binding subdomain; blue, core subdomain; light blue, C-terminal subdomain) and magenta for the RdRp domain (magenta, fingers subdomain; dark magenta, palm subdomain; light magenta, thumb subdomain) (Davidson et al., 2009 ▸). Despite high sequence conservation in the methyltransferase and RdRp domains, low sequence similarity was identified in the GTP-binding site, ten-residue linker region and a/b NLS region (red dashed box) across the four Dengue virus serotypes, as well as in the C-terminal region of the RdRp domain. The proposed NS3-binding site is also highlighted.
The MTase domain forms the N-terminal domain (residues 1–262) of NS5FL and contains an N-terminal subdomain, a core MTase subdomain and an C-terminal subdomain. The N-terminal subdomain includes the GTP-binding site, while the core MTase subdomain is responsible for S-adenosylmethionine (AdoMet) binding and catalytic activity (Egloff et al., 2002 ▸; Yap et al., 2010 ▸; Zhao et al., 2015 ▸). The MTase domain is connected via a ten-residue linker to the C-terminal RdRp domain (Fig. 1 ▸) composed of residues 273–900, forming the canonical polymerase subdomains called the fingers, palm and thumb subdomains (Yap et al., 2007 ▸; Davidson, 2009 ▸; Zhao et al., 2015 ▸). The fingers and thumb subdomains interconnect via the N-terminal region of the RdRp domain and, through loops protruding from the fingers subdomain, encircle the active site on the palm subdomain, forming a closed structure (Yap et al., 2007 ▸). Two tunnels that run perpendicular to each other allow access of the single-stranded RNA template to the active site on the palm subdomain and diffusion of dNTPs to the active site.
The recently determined crystallographic structure of DENV-3 NS5, including residues 6–895 (NS56–895), adopts a compact shape, with the MTase domain, which is located above the fingers subdomain of the RdRp domain, positioned such that its GTP-binding and SAH-binding sites are positioned away from the interdomain interface (Zhao et al., 2015 ▸). This assembly arrangement of the MTase and RdRp domains, which differs from that in NS5 from Japanese encephalitis virus (JEV; Lu & Gong, 2013 ▸), is proposed to be established by four residues (263–266 of DENV-3) forming a short 310-helix (Zhao et al., 2015 ▸). However, the mechanism of how the two domains of NS5FL communicate and how the catalytic entities are arranged relative to each other in solution remains elusive. Solution X-ray scattering experiments on DENV-3 NS51–878 (residues 1–878) revealed that the protein adopts multiple conformations, including compact and extended forms (Bussetta & Choi, 2012 ▸).
The NS5 proteins from the distinct serotypes DENV-1 to DENV-4 are not identical in sequence, in particular with regard to the N-terminal GTP-binding site, the ten-residue linker region, the a/b NLS region, which is important for nuclear-targeting ability and importin-α/β-binding activity (Davidson, 2009 ▸), and the fingers and catalytic palm subdomains as well as the C-terminal region of the RdRp domain (Fig. 1 ▸). Differences in the a/b NLS region are related to differences in the cellular localization of the four DENV NS5 proteins. Whereas the DENV-2 and DENV-3 NS5 proteins accumulate in the nuclei of infected cells, the DENV-1 and DENV-4 NS5 proteins are predominantly, if not exclusively, localized to the cytoplasm (Hannemann et al., 2013 ▸; Tay et al., 2013 ▸). In addition, NS5 exists in differentially phosphorylated states, with the hyperphosphorylated form predominantly in the nucleus (Kapoor et al., 1995 ▸). Nuclear transport of DENV-2 and DENV-3 NS5 via the importin-α/β heterodimer to the nucleus of infected cells enables the NS5 proteins of both serotypes to interact with nuclear host proteins and to perturb the host innate immune-system response. Shuttling of both proteins between the cytoplasm and nucleus may also reflect differences in the concerted action of RNA replication among the NS5 proteins from the four serotypes.
The key processes in viral replication performed by NS5 require intramolecular changes to the conformation of NS5 itself, in addition to the concerted interactions with viral proteins, viral RNA in the replication complex and host proteins. Here, we have studied the structural traits and flexibility of the full-length DENV-3 NS5 protein (900 residues) by solution X-ray scattering (SAXS), showing that the MTase and RdRp domain ensemble is more extended and less rigid in solution compared with the crystallographic structure of NS56–895. Solution studies of the individual DENV-3 MTase and RdRp domains reveal that the MTase domain contributes to the flexibility of DENV-3 NS5FL and reflects its importance in drug design. The low-resolution solution structures of all four (DENV-1 to DENV-4) recombinant NS5FL proteins are presented for the first time, indicating a more compact shape profile of DENV-4 NS5FL compared with the NS5FLs from the other serotypes. Inserting the genetically engineered linker from DENV-4 NS5FL into DENV-3 NS5FL and vice versa demonstrated the importance of the ten-residue linker of NS5FL in flexibility. These results, together with hydrogen/deuterium-exchange mass spectrometry (HDXMS) of the wild-type NS5FL proteins from DENV-3 and DENV-4, highlight the inherent differences in flexibility between the two linker regions. Finally, the effect of RNA binding (GpppAGUUGUU or m7GpppAGUUGUU) on the overall dimensions of NS5FL is presented.
2. Experimental procedures
2.1. Purification of full-length NS5 and single domains of NS5 from DENV
The plasmids carrying the full-length NS5 genes for all four serotypes are described in Table 1 ▸. In order to obtain the methyltransferase (MTase1–272) and RdRp (RdRp263–900) domains with the linker (residues 263–272) of DENV-3 NS5, the following primers were designed. The forward and reverse primers for MTase1–272 were 5′-ATG GGA ATT CAC GGA ACA GGC TCA-3′ and 5′-CCA TAA GCT TCT AGT TGG GTG TTT CTG G-3′, respectively. RdRp263–900 of DENV-3 NS5 was created using the forward primer 5′-GAC TGA ATT CAT GTT AAT GCG GAA-3′ and the reverse primer 5′-CAC ATA AAG CTT TCA TAC CAA TAT GTG CTA CC-3′. The plasmid DNA of DENV-3 NS5FL was used as the template (Tay et al., 2013 ▸). EcoRI and HindIII restriction sites (underlined) were incorporated in all of the constructs. Following digestion with EcoRI and HindIII, the PCR products were ligated into a modified pET-24b vector. The modification of the plasmid enables the introduction of a 6×His tag at the N-terminus.
Table 1. List of proteins used in SAXS experiments.
| Strain | Plasmid | Residue Nos. | GenBank | |
|---|---|---|---|---|
| DENV-1 NS5FL | Singapore | pProEx HTb (Life Technologies) | 1899 | EU081230 |
| DENV-2 NS5FL | New Guinea-C | pET-15b (Novagen) | 1900 | M29095.1 |
| DENV-3 NS5FL | Singapore | pProEx HTb (Life Technologies) | 1900 | AY662691 |
| DENV-3 NS5FL | Singapore | pProEx HTb (Life Technologies) | 1900 | GQ398256 |
| DENV-3 RdRp263900 | Singapore | pET-24b | 263900 | AY662691 |
| DENV-3 MTase1272 | Singapore | pET-24b | 1272 | AY662691 |
| d3-NS5-d4linker | Singapore | pProEx HTb (Life Technologies) | 1900 | AY662691 |
| d4-NS5-d3linker | Singapore | pProEx HTb (Life Technologies) | 1900 | GQ398256 |
The DENV-3 NS5 linker mutant and the DENV-4 NS5 linker mutant were generated by swapping the NS5 linker regions of DENV-3 and DENV-4 NS5 as shown in Fig. 8(a). Linker swapping was performed using KAPA HiFi DNA polymerase and amplification of the whole plasmid. For this purpose, the following primers were designed for the DENV-3 NS5 with the DENV-4 linker: forward primer 5′-ACT CGA AGT GTC TCC ACT GAA ACA GAA AAA CCA GAC ATG GAT GTC ATT GGG GAA AGA-3′ and reverse primer 5′-TGT CAT GTC TGG TTT TTC TGT TTC AGT GGA GAC ACT TCG AGT TCC TGC TCC TAA ATC-3′. In the case of mutant DENV-4 NS5 with the DENV-3 linker, the forward primer 5′-ACG AGA CAT GTT AAT GCG GAA CCA GAA ACA CCC AAC ATG ACA GTT ATT GGG AGA AGG-3′ and the reverse primer 5′-TGT CAT GTT GGG TGT TTC TGG TTC CGC ATT AAC ATG TCT CGT TCC TGC CCC AAG-3′ were used. The linker region is underlined in both mutants.
All plasmids for MTase1–272, RdRp263–900 and the DENV-3 and DENV-4 linker mutants were transformed into Escherichia coli BL21 CodonPlus (DE3)-RIL cells (Stratagene, USA) for protein expression and purification. The cells were lysed on ice by sonication for 3 × 1 min in buffer A [50 mM Tris–HCl pH 7.5, 500 mM NaCl, 5% glycerol, 0.8 mM DTT, 2 mM PefablocSC (Biomol)] for full-length NS5 from all four serotypes, NS5 linker mutants and RdRp263–900 or buffer B (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 0.8 mM DTT, 2 mM PefablocSC) for MTase1–272. The cell lysate was centrifuged at 12 500g for 25 min and the supernatant was then filtered (0.45 µm; Millipore). The filtered supernatant was incubated with Ni–NTA agarose (Qiagen) for 1.5 h at 4°C and the His6-tagged protein was eluted with an imidazole gradient from 20 to 500 mM in buffer A or buffer B. Fractions containing the proteins of interest were pooled and applied onto a gel-filtration column (Superdex 200 HR 10/300, GE Healthcare) in buffer C (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5% glycerol, 1 mM DTT) for full-length NS5 from all four serotypes and NS5 mutants or buffer D (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM DTT) for MTase1–272 and RdRp263–900. Fractions containing the proteins of interest were pooled and concentrated using Amicon Ultra-4 Centrifugal Units (50 and 10 kDa molecular-mass cutoff; Millipore).
2.2. Small-angle X-ray scattering data collection
Small-angle X-ray scattering (SAXS) data for the four serotypes of NS5, NS5 mutants and single domains of DENV-3 NS5 were measured with a Bruker NANOSTAR SAXS instrument equipped with a MetaJet X-ray source (Excillum, Germany) and a VÅNTEC 2000 detector system. The X-ray radiation was generated from a liquid gallium alloy with a microfocus X-ray source (λ = 0.13414 nm, with a potential of 70 kV and a current of 2.857 mA). The X-rays were filtered through Montel mirrors and collimated by a two-pinhole system. The sample-to-detector distance was set to 0.67 m and the sample chamber and X-ray paths were evacuated (Balakrishna et al., 2014 ▸; Dip et al., 2014 ▸; Tay et al., 2014 ▸). This setup covers a range of momentum transfer of 0.16 < q < 4 nm−1 [q = 4πsin(θ)/λ, where 2θ is the scattering angle]. SAXS experiments were carried out at 15°C in a concentration series ranging from 0.45 to 6.86 mg ml−1 in buffer C (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5% glycerol, 1 mM DTT) for full-length NS5 and NS5 mutants or in buffer D (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM DTT) for single domains of DENV-3 NS5, with a sample volume of 40 µl in a vacuum-tight quartz capillary. The data were collected for 30 min and for each measurement a total of six frames were recorded at 5 min intervals. Using the built-in SAXS software (Bruker AXS, Germany), the data were flood-field corrected to produce a smooth detector data image and spatially corrected to correct the inherent geometrical pincushion distortion, followed by the integration of two-dimensional images to one-dimensional data. The data were tested for possible radiation damage by comparing the six data frames, and no changes were detected. The scattering of the buffer was subtracted from the scattering of the sample, and all of the scattering data were normalized by the concentration as well as the incoming intensity.
SAXS data for DENV-2 NS5FL in the absence or presence of the substrates guanosine triphosphate (GTP), guanosine diphosphate (GDP), S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), GpppAGUUGUU (RNA1) or m7GpppAGUUGUU (RNA2) were collected on the Bio-SAXS beamline BL4-2 at Stanford Synchrotron Radiation Lightsource (SSRL; Smolsky et al., 2007 ▸). Data were collected using a Rayonix MX225-HE CCD detector (Rayonix, Evanston, Illinois, USA) with a 1.7 m sample-to-detector distance and a beam energy of 11 keV (λ = 0.1127 nm), covering a range of momentum transfer of 0.068 < q < 6.7 nm−1 [q = 4πsin(θ)/λ, where 2θ is the scattering angle]. SAXS experiments were performed using the BL4-2 autosampler (Martel et al., 2012 ▸). An aliquot of 30 µl of buffer and sample was subsequently exposed to the X-ray beam via a 1.5 mm quartz capillary cell and moved during exposure to minimize radiation damage.
For each sample, two protein concentrations, 1.25 and 2.5 mg ml−1, were measured in buffer C (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5% glycerol, 1 mM DTT). 5 mM DTT, which acts as radical scavenger, was freshly added to all samples prior to data collection. The data for each sample were recorded as a total of 15 frames with 1 s exposure. The data were tested for possible radiation damage by comparing all 15 data frames. Only data without radiation damage were subsequently averaged and the buffer-scattering intensity was subtracted using SasTool (http://ssrl.slac.stanford.edu/~saxs/analysis/sastool.htm).
2.3. Small-angle X-ray scattering data analysis
All of the data-processing steps were performed using the PRIMUS program package (Konarev et al., 2003 ▸, 2006 ▸). The experimental data obtained for all of the protein samples were analyzed for aggregation using the Guinier region (Guinier, 1939 ▸). The forward scattering I(0) and the radius of gyration R g were computed using the Guinier approximation, assuming that at very small angles (q < 1.3/R g) the intensity is represented as I(q) = I(0)exp[−(qR g)2/3] (Guinier, 1939 ▸; Glatter & Kratky, 1982 ▸). These parameters were also computed from the extended scattering patterns using the indirect transform package GNOM (Svergun, 1992 ▸), which provides the distance distribution function P(r) and hence the maximum particle dimension D max, as well as the radius of gyration R g. Qualitative particle motion was inferred by plotting the scattering patterns in the so-called normalized Kratky plot (Durand et al., 2010 ▸). The Porod–Debye plot was generated by transforming scattering data as q 4 I(q) versus q 4, and the Porod–Debye law approximates the scattering for a well folded compact particle decay as q −4 scaled by the particle’s surface area, concentration and electron-density contrast within a limited angle range. The compact particle will produce a sharp scattering contrast and subsequently display a plateau in a Porod–Debye plot. This plateau will disappear with increasing flexibility of the particle owing to the diffuse scattering contrast (Rambo & Tainer, 2011 ▸). The hydrated volume V p, which is used to estimate the molecular mass of the protein, was computed using the Porod invariant. Ab initio low-resolution models of the proteins were built by DAMMIF (Svergun, 1999 ▸; Franke & Svergun, 2009 ▸) considering low-angle data (q < 0.2 nm−1). The algorithm constructs bead models yielding a scattering profile with the lowest possible discrepancy (χ2) to the experimental data while keeping the beads interconnected and the model compact. 20 independent ab initio reconstructions were performed for NS5 from each serotype and were averaged using DAMAVER (Volkov & Svergun, 2003 ▸). The average excluded volume V Ex computed using DAMAVER was further used to estimate the molecular mass of the protein (Petoukhov et al., 2012 ▸).
Rigid-body modelling was performed for NS5FL from all four serotypes using BUNCH (Petoukhov & Svergun, 2005 ▸) by docking the individual domains of the available high-resolution structures [PDB entries 3p8z (MTase domain; Lim et al., 2010 ▸) and 4c11 (RdRp domain; Lim et al., 2013 ▸)] against the experimental data. The theoretical scattering curves from the atomic structures were generated and evaluated against the experimental scattering curves using CRYSOL (Svergun et al., 1995 ▸). Superimposition between the ab initio reconstruction and the atomic model was performed using SUPCOMB (Kozin & Svergun, 2001 ▸). Quantitative assessment of the flexibility was performed using the Ensemble Optimization Method (EOM) 2.0 (Tria et al., 2015 ▸), which assumes the coexistence of a range of conformations in solution for which an average scattering intensity fits the experimental data (Bernadó et al., 2007 ▸; Mertens & Svergun, 2010 ▸; Svergun et al., 2013 ▸; Tria et al., 2015 ▸). In EOM 2.0, a random pool of 10 000 independent models is created as a first step, with the aim of approximating the conformational space for a protein exhibiting considerable motion. For each model in the pool the theoretical scattering curve is automatically computed using CRYSOL (Svergun et al., 1995 ▸). Subsequently, a genetic algorithm (GA) is used to select ensembles with varying numbers of conformers by calculating the average theoretical profile and fitting it to the experimental SAXS data. For each NS5 serotype, the GA was repeated 100 independent times and the ensemble with the lowest discrepancy value (χ2) was reported as the best solution out of 100 final ensembles. Independent repetitions of the GA allow the computation of R g and D max distributions, from which structural information about the motion of the particle can be extracted. Distributions with average R g values that are above the average R g value from the random pool are considered as extended, whereas models with values below the average are considered as compact. Quantification of the flexibility is computed as the Shannon information entropy of the R g distributions and is reported as R flex. Ensemble R flex values close to that extracted from the random pool (pool R flex) are considered as an indication of random motion. Potential artifactual solutions are identified by using the complementary metric R σ, which represents the ratio of the standard deviation of the solution distribution against that of the pool distribution. Values of R σ lower than 1.0 are expected when the ensemble R flex is smaller than the pool R flex and values of greater than 1.0 otherwise.
2.4. Amide hydrogen/deuterium-exchange mass spectrometry (HDX)
The NS5FL samples from DENV-3 and DENV-4 were concentrated to ∼20 µM in buffer C. Amide deuterium exchange was carried out by diluting 3 µl of the protein sample in buffer C with 22 µl deuterated buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5% glycerol, 1 mM DTT reconstituted in 99.9% D2O), resulting in a final D2O concentration of 88%. This reaction was incubated for deuterium-labelling time points of 1 and 10 min. The exchange reaction was quenched by the addition of 25 µl 0.1%(v/v) trifluoroacetic acid at 0°C to obtain a final pHread of 2.5. 50 µl of the quenched sample was injected onto a chilled nano-ACQUITY UPLC sample manager (Waters, Milford, Massachusetts, USA) as described previously (Wales et al., 2008 ▸). The sample was trapped and digested with an immobilized pepsin column [Enzymate BEH Pepsin Column (2.1 × 30 mm, 5 µm), Waters] with 0.1%(v/v) formic acid in water at 100 µl min−1. The pepsin-digested peptides were trapped on a trap column (ACQUITY UPLC BEH C-18 VanGuard Pre-column, 2.1 × 5 mm, Waters ) and eluted using a gradient of 8–40% acetonitrile in 0.1%(v/v) formic acid at 40 µl min−1 using a nano-ACQUITY Binary solvent manager (Waters) in a reverse-phase C18 column (ACQUITY UPLC BEH C18 reverse-phase column, Waters). The peptides were detected and their masses were measured using a SYNAPT G2-Si mass spectrometer (Waters ). The data were acquired in MSE mode with continuous calibration with Glu-fibrinogen peptide at 200 fmol µl−1.
The peptides were identified from undeuterated samples (3 µl protein sample + 22 µl buffer C in H2O) using ProteinLynx Global Server (PLGS v.2.4, Waters) and searched against the primary sequences of NS5FL from DENV-3 and DENV-4 with a mass tolerance of less than 5 p.p.m. The deuteration profiles for the peptides identified by PLGS were extracted and analyzed using DynamX 2.0 (Waters). Heat maps for fractional deuterium uptake (the fraction of deuterons taken up over the total exchangeable amides) for every residue across the complete protein were also generated using DynamX 2.0 and were aligned and mapped onto the structure of DENV-3 NS56–895 (PDB entry 4v0q; Zhao et al., 2015 ▸). The heat map is colour-coded as a gradient from yellow to red representing increasing deuterium uptake, with yellow and red representing 0 and 100% deuterium uptake, respectively (Fig. 6d). Regions in grey represent data that are absent owing to lack of peptide coverage.
2.5. Sequence-alignment analysis
The amino-acid sequences of the NS5 proteins from DENV-1, DENV-2, DENV-3 and DENV-4 were obtained from the National Center for Biotechnology Information database (Altschul et al., 1997 ▸). The selected sequences were then aligned using Clustal Omega (Sievers et al., 2011 ▸) as included in JalView 2.8.2 (Waterhouse et al., 2009 ▸). The secondary structure of the crystallographic structure DENV-3 NS56–895 (PDB entry 4v0r; Zhao et al., 2015 ▸) is included in Fig. 1 ▸.
3. Results
3.1. Purification of full-length NS5 from DENV-3
In order to prepare highly pure and monodisperse DENV-3 NS5FL, Ni–NTA affinity chromatography was employed as the first step of purification, as described in §2. The fractions containing DENV-3 NS5FL were eluted at an imidazole concentration of 50–150 mM using an imidazole gradient from 20 to 500 mM. These fractions were pooled together and concentrated before application onto a gel-filtration column (Superdex 200 HR 10/300). 15% of the peak fractions which were identified by SDS–PAGE were pooled (Supplementary Figs. S1a and S1b) and concentrated to the respective protein concentrations for small-angle X-ray scattering (SAXS) experiments.
3.2. SAXS study of NS5FL from DENV-3 in solution
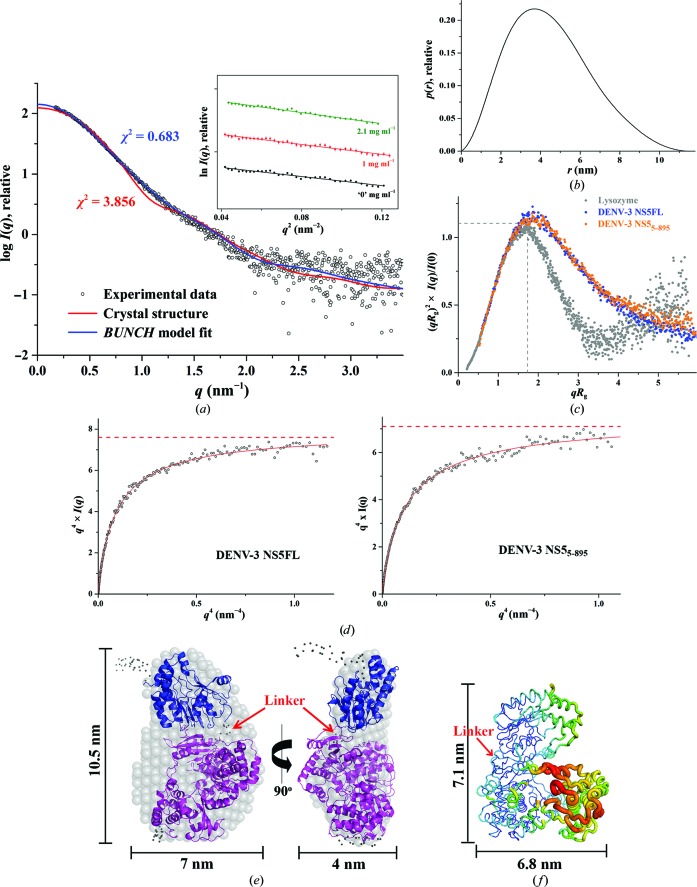
In order to characterize DENV-3 NS5FL in solution and to understand the orientation as well as the flexibility of the two domains, methyltransferase and RdRp, of NS5FL in solution, SAXS experiments were performed with freshly purified proteins. SAXS patterns of full-length DENV-3 were recorded at 1.0, 2.1 and 4.1 mg ml−1. Extrapolation to theoretical infinite dilution was used for the analysis (Fig. 2 ▸ a, Table 2 ▸). The Guinier plot at low angles for different concentrations appeared to be linear and confirmed the good data quality and monodispersity of DENV-3 NS5FL, with no indication of protein aggregation (Fig. 2 ▸ a, inset). From the Guinier approximation, a radius of gyration of 3.37 ± 0.04 nm was derived. The distance distribution [P(r)] function showed a trend that is usually associated with a slightly elongated shape conformation with a small tail, pointing to a maximum particle dimension, D max, of 11.5 ± 1 nm (Fig. 2 ▸ b). The R g value of 3.47 ± 0.02 nm extracted from the P(r) function, which takes the whole scattering curve into consideration, is in agreement with that derived from the Guinier region. Accordingly, the low-resolution shape of DENV-3 NS5FL was reconstructed ab initio and the averaged model (DAMMIF) is shown in Fig. 2 ▸(e) with a normalized spatial discrepancy (NSD) value of 0.717 ± 0.08. A model that is more elongated when compared with the atomic crystal structure ensued, with dimensions of 10.5 × 7.0 × 4.0 nm compared with 8.7 × 7.2 × 5.5 nm for the crystallographic structure of NS56–895 (PDB entry 4v0r; Zhao et al., 2015 ▸; Figs. 2 ▸ e and 2 ▸ f).
Figure 2.
Solution X-ray scattering studies of DENV-3 NS5FL. (a) Small-angle X-ray scattering pattern (black circles) and calculated scattering profiles (red lines, crystal structure, PDB entry 4v0r; blue lines, BUNCH model). Inset: Guinier plots show linearity at the different concentrations used and indicate no aggregation of the protein. (b) Pair-distance distribution function P(r) of DENV-3 NS5FL. (c) Normalized Kratky plot of DENV-3 NS5FL (blue filled circles) compared with the DENV-3 NS56–895 protein (orange filled circles) and the compact globular lysozyme (grey filled circles) with a peak (grey dashed lines) representing the theoretical peak and assuming an ideal Guinier region of a globular particle. The scattering pattern of DENV-3 NS5FL exhibits a broad bell-shaped profile shifted to the right with respect to standard globular proteins, indicating the presence of motion in the protein. (d) Porod–Debye plot (black circles) and fit line (red) of DENV-3 NS5FL (left) and DENV-3 NS56–895 (right). The absence of a Porod–Debye plateau in the plot suggests that the DENV-3 NS5FL (Porod exponent = 3.7) is flexible in solution and that this flexibility is not owing to a flexible tag. (e) Ab initio DAMMIF envelopes superimposed onto the cartoon representation of the BUNCH model for DENV-3 NS5FL, with the RdRp domain coloured magenta and the methyltransferase domain blue. Regions that are not resolved in the crystal structures are presented as dots. Front (left) and side (right) views are displayed. (f) Temperature-factor (B-factor) putty representation of the crystal structure of DENV-3 NS56–895 (PDB entry 4v0r) coloured by B factor from low (blue) to high (red).
Table 2. Data-collection and scattering-derived parameters for NS5FL from all serotypes.
| Protein sample | DENV-1 NS5FL | DENV-2 NS5FL | DENV-3 NS5FL | DENV-4 NS5FL |
|---|---|---|---|---|
| Data-collection parameters | ||||
| Instrument (source and detector) | Bruker NANOSTAR equipped with MetalJet Excillum X-ray source and VNTEC-2000 detector | |||
| Beam geometry | 100m slit | |||
| Wavelength (nm) | 0.134 | |||
| q range (nm1) | 0.164 | |||
| Exposure time (min) | 30 (6 frames 5min) | |||
| Concentration range (mgml1) | 1.174.56 | 1.004.40 | 1.004.10 | 0.456.86 |
| Temperature (K) | 288.15 | 288.15 | 288.15 | 288.15 |
| Structural parameters | ||||
| I(0) from P(r) (cm1) | 140.3 0.9 | 148.9 0.8 | 137.6 0.7 | 114.7 0.5 |
| R g from P(r) (nm) | 3.51 0.02 | 3.57 0.02 | 3.47 0.02 | 3.36 0.02 |
| I(0) from Guinier (cm1) | 141.9 2.3 | 149.5 1.5 | 135.5 1.0 | 113.0 0.7 |
| R g from Guinier (nm) | 3.55 0.1 | 3.56 0.05 | 3.37 0.04 | 3.27 0.03 |
| D max (nm) | 11 1 | 11.5 1 | 11.5 1 | 11.5 1 |
| Porod volume estimate (nm3) | 140 | 150 | 140 | 140 |
| DAMMIF excluded volume (nm3) | 197 | 192 | 185 | 183 |
| Dry volume from sequence† (nm3) | 131 | 128 | 131 | 131 |
| Molecular-mass determination | ||||
| Calculated monomeric molecular mass from sequence‡ (kDa) | 107.9 | 105.3 | 107.9 | 107.9 |
| Molecular mass from MALDI§ (kDa) | 108 | 105 | 108 | 108 |
| Molecular mass from Porod invariant (kDa) | 85 10 | 91 10 | 87 10 | 87 10 |
| Molecular mass from excluded volume (kDa) | 98 10 | 96 10 | 93 10 | 92 10 |
| Software employed | ||||
| Primary data reduction | BRUKER SAS | |||
| Data processing | PRIMUS | |||
| Ab initio analysis | DAMMIF | |||
| Validation and averaging | DAMAVER | |||
| Computation of model intensities | CRYSOL | |||
| Rigid-body modelling | BUNCH | |||
| Flexibility | EOM 2.0 | |||
| Three-dimensional graphical representations | PyMOL | |||
Calculated with Peptide Property Calculator (http://www.basic.northwestern.edu/biotools/proteincalc.html).
Calculated with Compute pI/Mw (http://web.expasy.org/compute_pi/).
MALDI-TOF/TOF service provided by the Proteomic Core Facility of the School of Biological Sciences at Nanyang Technological University.
Considering that NS5FL from DENV-3 adopted an elongated conformation in solution compared with a compact conformation in the DENV-3 NS56–895 crystal structure (Zhao et al., 2015 ▸), computational docking of the single domains against the SAXS pattern was performed using the rigid-body modelling software BUNCH (Petoukhov & Svergun, 2005 ▸). The BUNCH models revealed an arrangement of the full-length DENV-3 NS5FL with the methyltransferase domain positioned at the top of the RdRP domain rather than at the side as observed in the NS56–895 crystal structure (Zhao et al., 2015 ▸; PDB entry 4v0r). The fittings of the theoretical scattering curves derived from the BUNCH model and the NS56–895 crystal structure against the experimental scattering curve are shown in Fig. 2 ▸(a), resulting in χ2 values of 0.683 and 3.856, respectively, and indicating that DENV-3 NS5FL adopts an elongated conformation in solution. Furthermore, the ab initio envelope (DAMMIF) and the BUNCH atomic model superimpose well with an NSD of 0.9402 for DENV-3 NS5FL, as shown in Fig. 2 ▸(e).
3.3. Ensemble formation of DENV-3 NS5FL
Since the methyltransferase and RdRp domains of DENV-3 NS5FL were arranged differently in solution to in the NS56–895 crystal structure, domain movement within NS5 was taken into consideration. The normalized Kratky plot {(qR g)2[I(q)/I(0)] versus qR g} for DENV-3 NS5FL (Fig. 2 ▸ c) was slightly shifted from a well defined bell-curve profile, as is usually seen for a globular protein such as lysozyme, indicating that DENV-3 NS5FL exhibits a slightly extended conformation (Durand et al., 2010 ▸; Receveur-Brechot & Durand, 2012 ▸).
In order to determine (i) whether the phenomena of a more extended conformation and less rigidity can be observed for DENV-3 NS56–895 in solution and (ii) whether the peak shifts observed in the normalized Kratky plot are not owing to the flexible tags in the protein, DENV-3 NS56–895 without any tag was produced, purified (Supplementary Fig. S1c) and analyzed by SAXS. The normalized Kratky plot of DENV-3 NS56–895 showed a similar shift as DENV-3 NS5FL, indicating that DENV-3 NS56–895 in solution is also more extended and less rigid compared with the crystallographic structure and that the changes observed in the normalized Kratky plot of DENV-3 NS5FL are not owing to the flexible tag in the protein. To assess the potential flexibility of DENV-3 NS5FL, a Porod–Debye plot was generated with a Porod exponent of 3.7 (Fig. 2 ▸ d). The shift in the normalized Kratky plot and the absence of a Porod plateau in the Porod–Debye plot (Fig. 2 ▸ d) suggest that DENV-3 NS5FL as well as DENV3 NS56–895 are flexible in solution.
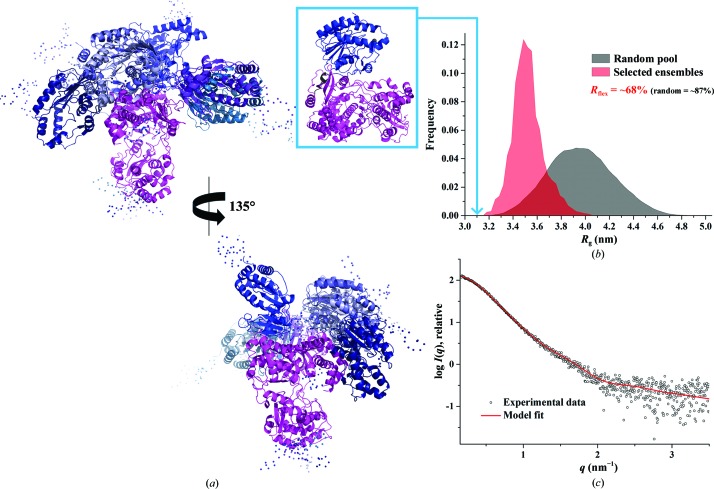
Previous SAXS studies of DENV-3 NS51–878 showed a bimodal R g distribution with values of between 3.4 and 4.1 nm using the Ensemble Optimization Method (EOM; Bussetta & Choi, 2012 ▸). In order to study the R g distribution of the freshly purified DENV-3 NS5FL and to further characterize the dynamic behaviour of NS5FL, in particular with respect to the potential flexibility of the methyltransferase domain relative to the RdRP domain, EOM (Tria et al., 2015 ▸) was used. As a result, the ensemble solution selected by EOM 2.0 provided a discrepancy χ2 of 0.542 by selecting an ensemble of six models with different conformations of the methyltransferase domain (Fig. 3 ▸ a). The quantification of the flexibility (ensemble R flex = ∼68%) suggests the presence of a nonrandom motion for the methyltransferase domain, when compared with the threshold of randomness as derived from the original random pool (pool R flex = ∼87%; Fig. 3 ▸ b). The quality of the ensemble solution was further confirmed by the control value R σ = 0.46 (expected to be lower than 1.0 when the ensemble R flex < the pool R flex). In addition, a monomodal R g distribution was revealed for DENV-3 NS5FL, with a major peak between 3.4 and 3.5 nm (Fig. 3 ▸ b).
Figure 3.
Flexibility characterization of DENV-3 NS5FL showing an ensemble of six possible positions for the methyltransferase domain based on analysis with EOM 2.0. (a) Different views of the ensemble models in cartoon representation with the RdRp domain of DENV-3 NS5FL in magenta and the flexible methyltransferase domain in blue, together with their conformational contribution (deep blue, 40%; blue, 10%; slate blue, 10%; sky blue, 10%; purple blue, 20%; light blue, 10%; the connecting linkers are rendered semi-transparent). (b) Comparison of the R g distributions (pool, black; ensemble, red) shows a nonrandom flexibility as confirmed by R flex (∼68 versus ∼87%, with 87% being the threshold value for randomness) and R σ = 0.46. The R g corresponding to the NS56–895 crystal structure (PDB entry 4v0r) is indicated (light blue arrow). (c) Small-angle X-ray scattering pattern (black circles) and calculated ensemble scattering profiles (red lines) with discrepancy χ2 = 0.542.
In comparison, the R g of the NS56–895 crystal structure (PDB entry 4v0r) was determined to be 3.1 nm using CRYSOL, being smaller than the R g distribution of selected ensembles of DENV-3 NS5FL in solution (Fig. 3 ▸ b). This confirms the normalized Kratky and the Porod–Debye plot data (Figs. 2 ▸ c and 2 ▸ d indicating that DENV-3 NS5FL and DENV3 NS56–895 are extended and less rigid in solution.
3.4. Solution X-ray studies of single domains of DENV-3 NS5FL
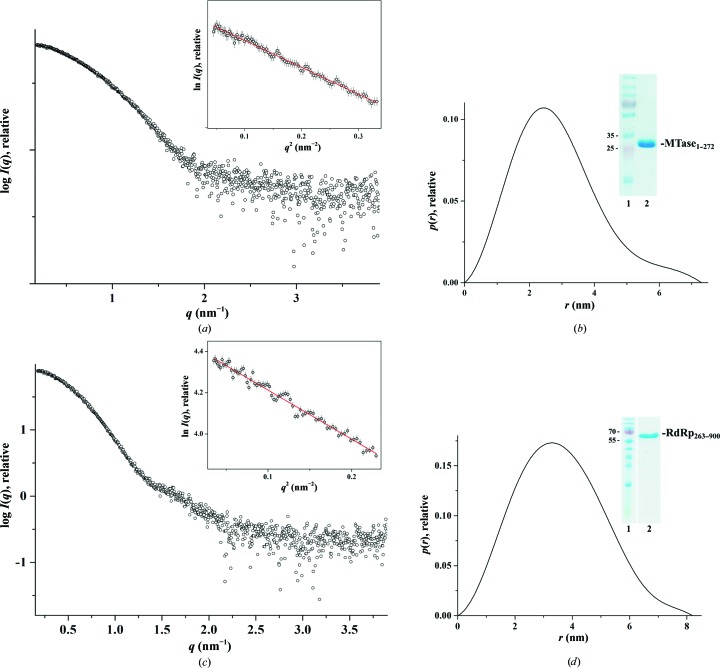
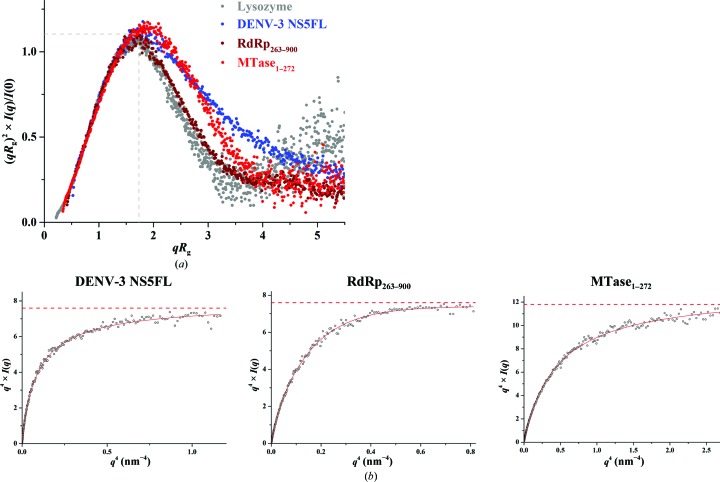
In order to understand whether the methyltransferase and/or RdRp domain makes the main contribution to the flexibility seen in DENV-3 NS5FL, the methyltransferase (MTase1–272) and RdRp (RdRp263–900) domains, both including the linker residues 263–272, were generated and purified (Figs. 4 ▸ b and 4 ▸ d, inset). SAXS patterns for DENV-3 MTase1–272 (1.5, 3.5 and 5.0 mg ml−1) and DENV-3 RdRp263–900 (1.1, 2.1 and 5.6 mg ml−1) were recorded at different protein concentrations to yield the final composite scattering curves shown in Figs. 4 ▸(a) and 4 ▸(c), respectively. The Guinier plots at low angles are linear and revealed good data quality, with no indication of protein aggregation (Figs. 4 ▸ a and 4 ▸ c). The overall structural parameters for MTase1–272 and RdRp263–900 are given in Table 3 ▸. MTase1–272 and RdRp263–900 have R g values of 2.2 and 2.68 nm, respectively, and their D max values were determined to be 7.3 and 8.2 nm, respectively. By overlapping the normalized Kratky plots of DENV-3 NS5FL and single domains of DENV-3 NS5 (Fig. 5 ▸ a), a similar shift of the bell-curve profile was seen for DENV-3 MTase1–272. In comparison, DENV-3 RdRp263–900 reveals a well defined bell-curve profile, as found for lysozyme, which suggests that the changes observed in the full-length NS5 from DENV-3 were driven by the methyltransferase domain and its linker. RdRp263–900 showed a Porod–Debye plateau with a Porod exponent of 4, consistent with the Porod exponent derived from a globular molecule without flexibility (Rambo & Tainer, 2011 ▸; Burke et al., 2012 ▸; Stiegler et al., 2013 ▸), while the Porod–Debye decay of MTase1–272 was best fitted by a Porod exponent of 3.5 (Fig. 5 ▸ b), validating that the MTase1–272 domain with the linker region contributes to the flexibility observed for full-length DENV-3 NS5.
Figure 4.
Solution X-ray scattering studies of individual domains of DENV-3 NS5FL. (a) Small-angle X-ray scattering pattern of MTase1–272 (circles). Inset: the Guinier plot shows linearity for MTase1–272, indicating no aggregation. (b) Pair-distance distribution function P(r) of MTase1–272. Inset: a 17% SDS gel reveals the high purity of the recombinant MTase1–272 (lane 2); lane 1 contains molecular-mass protein marker. (c) Small-angle X-ray scattering pattern of RdRp263–900 (circles). Inset: the Guinier plot shows linearity, indicating no aggregation of the protein. (d) Pair-distance distribution function P(r) of RdRp263–900. Inset: a 17% SDS gel reveals the high purity of the recombinant RdRp263–900 (lane 2); lane 1 contains molecular-mass protein marker.
Table 3. Data-collection and scattering-derived parameters for the methyltransferase (MTase) and RNA-dependent RNA polymerase (RdRp) domains of DENV-3 N5.
| Protein sample | MTase1272 | RdRp263900 |
|---|---|---|
| Data-collection parameters | ||
| Instrument (source and detector) | Bruker NANOSTAR equipped with MetalJet Excillum X-ray source and VNTEC-2000 detector | |
| Beam geometry | 100m slit | |
| Wavelength (nm) | 0.134 | |
| q range (nm1) | 0.164 | |
| Exposure time (min) | 30 (6 frames 5min) | |
| Concentration range (mgml1) | 1.505.00 | 1.105.56 |
| Temperature (K) | 288.15 | 288.15 |
| Structural parameters | ||
| I(0) from P(r) (cm1) | 40.8 0.2 | 86.8 0.3 |
| R g from P(r) (nm) | 2.21 0.01 | 2.73 0.01 |
| I(0) from Guinier (cm1) | 40.9 0.3 | 85.9 0.4 |
| R g from Guinier (nm) | 2.20 0.02 | 2.68 0.02 |
| D max (nm) | 7.3 0.7 | 8.2 0.8 |
| Porod volume estimate (nm3) | 40.2 | 98.5 |
| DAMMIF excluded volume (nm3) | 54.2 | 131 |
| Dry volume from sequence† (nm3) | 39 | 91 |
| Molecular-mass determination | ||
| Calculated monomeric molecular mass from sequence‡ (kDa) | 32.2 | 75.3 |
| Molecular mass from Porod invariant (kDa) | 24 2.4 | 60 10 |
| Molecular mass from excluded volume (kDa) | 27 2.7 | 65 6.5 |
| Software employed | ||
| Primary data reduction | BRUKER SAS | |
| Data processing | PRIMUS | |
Calculated with Peptide Property Calculator (http://www.basic.northwestern.edu/biotools/proteincalc.html).
Calculated with Compute pI/Mw (http://web.expasy.org/compute_pi/).
Figure 5.
(a) Normalized Kratky plot of DENV-3 NS5FL (blue filled circles), MTase1–272 (red filled circles) and RdRp263–900 (brown filled circles) compared with the compact globular lysozyme (grey filled circles), with the expected peak (grey dashed line) representing the theoretical peak assuming an ideal Guinier region of a globular particle. Similar to DENV-3 NS5FL, MTase1–272 adopted a similar peak to DENV-3 NS5L, while RdRp263–900 had a peak at the theoretical peak for a globular protein, suggesting that the RdRp domain is slightly more rigid in solution when compared with the methyltransferase domain and full-length NS5 of DENV-3. (b) Porod–Debye plots (black circles) and fit lines (red) of DENV-3 NS5FL, MTase1–272 and RdRp263–900. The data for RdRp263–900 show a Porod–Debye plateau (Porod exponent = 4.0), while this plateau was absent for MTase1–272 (Porod exponent = 3.5), showing that the flexibility seen in DENV-3 NS5FL is partially driven by the methyltransferase domain and its linker.
3.5. Solution X-ray studies of NS5 from serotypes DENV-1 to DENV-4
The key processes in viral replication performed by NS5 require intramolecular changes to the conformation of NS5 itself, in addition to interactions with viral proteins, viral RNA in the replication complex and host proteins. A multiple sequence alignment of the NS5 proteins from all four serotypes showed high conservation in the methyltrasferase and polymerase domains, with lower sequence similarity in the N-terminal GTP-binding site of the methyltransferase domain, the ten-residue linker region, the a/b NLS region important for β-Gal nuclear-targeting ability and importin-α/β-binding activity (Davidson, 2009 ▸), and the fingers and catalytic palm subdomains, as well as the C-terminal region of the RdRp domain (Fig. 1 ▸). SAXS experiments were conducted for the remaining three (DENV-1, DENV-2 and DENV-4) NS5FL proteins to investigate whether differences in NS5FL in solution can be observed among the four serotypes. Firstly, pure NS5FL proteins from DENV-1 to DENV-4 were produced and purified according to the protocol described in §2 and shown in Supplementary Fig. S1(b). The molecular masses of all four NS5FL proteins from the four serotypes were confirmed by MALDI-TOF spectrometry, as reported in Table 2 ▸. The freshly prepared samples were used for SAXS experiments using three different protein concentrations per protein, resulting in the final composite scattering curves shown in Fig. 6 ▸(a). The Guinier plots at low angles are linear and revealed good data quality, with no indication of protein aggregation (Fig. 6 ▸ a, inset). The overall parameters for all of the serotypes are listed in Table 2 ▸. From the Guinier approximation, R g values of 3.55 ± 0.09, 3.56 ± 0.05, 3.37 ± 0.02 and 3.27 ± 0.02 nm were calculated for NS5FL from DENV-1, DENV-2, DENV-3 and DENV-4, respectively, confirming the values derived from the P(r) function of 3.51 ± 0.02, 3.57 ± 0.02, 3.47 ± 0.02 and 3.36 ± 0.02 nm, respectively, and indicating the similar R g values for NS5 from DENV-1 and DENV-2 and the slight and more prominent decrease in the R g values for DENV-3 and DENV-4, respectively. As described for DENV-3 NS5FL, the distance distribution [P(r)] function of all four serotype NS5 proteins showed a trend towards a slightly elongated shape conformation with a small tail. As indicated by the R g value, a more compact shape profile for DENV-4 NS5FL was revealed by the P(r) function in the range 6–11 nm (Fig. 6 ▸ b). The maximum particle dimension D max of NS5FL of DENV-1 to DENV-4 was determined to be 11 ± 1 to 11.5 ± 1 nm (Table 2 ▸).
Figure 6.
Solution X-ray scattering studies of NS5FL from the four DENV serotypes. (a) Small-angle X-ray scattering patterns (circles) for DENV-1 NS5FL (red), DENV-2 NS5FL (green), DENV-3 NS5FL (blue) and DENV-4 NS5FL (magenta). Inset, Guinier plots show linearity for the NS5 from all four serotypes, indicating no aggregation. (b) Pair-distance distribution function P(r) for DENV-1 NS5FL (red line), DENV-2 NS5FL (green line), DENV-3 NS5FL (blue line) and DENV-4 NS5FL (magenta line). (c) Normalized Krakty plots for lysozyme (grey filled circles), DENV-1 NS5FL (red filled circles), DENV-2 NS5FL (green filled circles), DENV-3 NS5FL (blue filled circles) and DENV-4 NS5FL (magenta filled circles). The NS5FLs from all four serotypes reveal a shifted bell-curve profile in their normalized Kratky plots, indicating that the proteins exhibited a less rigid conformation. The normalized Kratky plot of DENV-4 NS5FL was shifted inwards (light blue arrow) compared with the plots for serotypes DENV-1 to DENV-3, suggesting that DENV-4 NS5FL adopts a more compact conformation. (d) Ab initio envelopes (grey) superimposed onto the cartoon representation of the BUNCH models for all four DENV serotypes, with the RdRp domain coloured magenta and the methyltransferase domain coloured blue. Regions that are not resolved in the crystal structures are presented as grey dots. Front (top) and side (bottom) views are displayed.
Ab initio envelope reconstructions superimposed onto rigid-body BUNCH models showed a similar arrangement of the methyltransferase and RdRP domains for all four serotypes (Fig. 6 ▸ d). In the normalized Kratky plots, similar to that for DENV-3 NS5FL, the NS5s from other three serotypes showed slightly shifted bell-curve profiles from the bell-curve profile observed for a globular protein such as lysozyme (Fig. 6 ▸ c), indicating that NS5 from all four serotypes exhibited an elongated conformation and less rigidity in solution. When comparing the normalized Kratky plots among all four serotypes, the curve for DENV-4 NS5 was shifted inwards (indicated by the blue arrow in Fig. 6 ▸ c) compared with the curves derived for the NS5FLs from DENV-1 to DENV-3, suggesting that the DENV-4 NS5FL has a more compact conformation compared with those from the other three serotypes. This phenomenon was also seen in the pair-distance distribution function P(r) of DENV-4 NS5FL (indicated by the blue arrow in Fig. 6 ▸ b).
Quantitative characterization of the flexibility of NS5FL from DENV-1, DENV-2 and DENV-4 using EOM showed the presence of motion for NS5FL from all of the remaining serotypes of Dengue virus (Supplementary Figs. S2, S3 and S4). As for DENV-3 NS5FL, we compared the R g distribution of the selected ensemble against the random ensemble formation. In comparison, the random R g distributions of the DENV-2, DENV-3 and DENV-4 NS5FLs mainly have monomodal distributions, covering the R g range 3.2–3.9 nm for DENV-2 and DENV-3 NS5FL and a major R g distribution between 3.2 and 3.6 nm for DENV-4 NS5FL, confirming the smaller R g value previously determined for this protein (Table 2 ▸). In the case of DENV-1 NS5FL, besides a maximum R g distribution between 3.2 and 4.0 nm, an R g distribution of the selected ensemble with low frequency was obtained. As in the comparison with DENV-3 NS5FL shown above, the R g value (3.1 nm) determined from the NS56–895 crystal structure (PDB entry 4v0r) is smaller than the R g distribution of selected ensembles of all four DENV NS5FLs in solution (Fig. 3 ▸ b, Supplementary Figs. S2, S3 and S4). The quantification of the flexibility (ensemble R flex) resulted in R flex values of 81, 67, 68 (see above) and ∼74% for DENV-1, DENV-2, DENV-3 and DENV-4, respectively. In all cases the threshold of randomness was ∼87% (Supplementary Figs. S2, S3 and S4).
3.6. Differences in the conformational flexibility of DENV-3 and DENV-4 NS5FL in solution
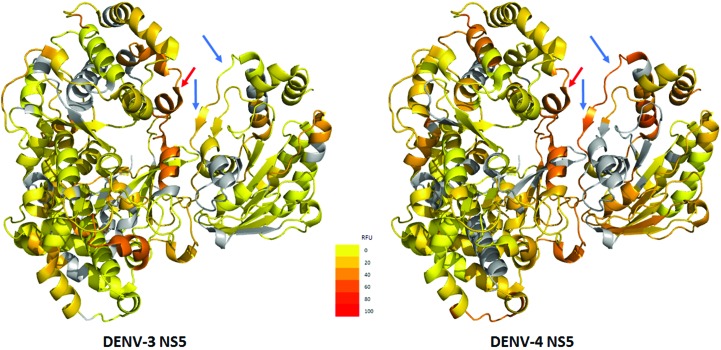
In order to localize the differences in flexibility observed for DENV-3 and DENV-4 NS5FL, we performed amide hydrogen/deuterium-exchange mass spectrometry (HDX-MS) experiments using wild-type DENV-3 and DENV-4 NS5FL proteins. The results show that DENV-4 NS5FL is more flexible overall, as inferred by HDX-MS, than the DENV-3 protein (Fig. 7 ▸). In order to understand the differences in linker-region dynamics between the proteins from the two serotypes, we examined deuterium exchange of the linker and flanking regions. The linker helix in DENV-4 NS5FL showed greater deuterium exchange (45% residue-wise fractional deuterium uptake; RFU) compared with the DENV-3 NS5FL linker (27% RFU). The regions flanking the linker also showed interesting differences between the two serotypes. DENV-3 NS5FL has a more stable structure in the region surrounding the linker, while the same regions in DENV-4 NS5FL showed greater flexibility, with close to 60% relative deuterium exchange. The region TMTHRRPTEK (269–279) showed only 27% deuteration in DENV-3 NS5FL, while the equivalent region in the DENV-4 protein (FTTRHRKPTYEKD; 269–281) showed twice the exchange at 53%. The high-exchanging locus in DENV-4 NS5FL spans the helical segment (relative fractional uptake of 56%). Higher deuterium exchange suggests that this helix is less stable or is transiently unfolded (Wang et al., 2012 ▸) and suggests a role in the compaction of the DENV-4 NS5FL. Contrastingly, the regions surrounding the linker in DENV-3 NS5FL are more likely to resist compaction owing to the presence of more ordered stable structures.
Figure 7.
Comparison of the flexibility of DENV-3 and DENV-4 NS5 using HDX-MS. Residue-wise fractional deuterium uptake (RFU) for DENV-3 (left) and DENV-4 (right) are mapped onto the available NS5 structure (PDB entry 4v0r; Zhao et al., 2015 ▸). Blue arrows indicate regions surrounding the linker that show significant differences in deuterium uptake between the serotypes. The sheet–turn–helix region in DENV-4 is more flexible, while that in DENV-3 is more stable and unyielding. This is indicated by the 53% RFU observed for DENV-4 in contrast to the 27% RFU for DENV-3. Interestingly, these differences are only observed in the region adjacent to the linker in the MTase domain (blue arrows). Similar regions in the RdRp domain adjacent to the linker show very similar deuterium uptake and indicate that the differences in compaction are primarily from the MTase domain. Residue-wise RFU is calculated by normalizing the deuterons exchanged to the number of maximum exchangeable amide H atoms for each peptide. If a residue position is covered by multiple peptides, then the RFU is an average over all of these peptides. The heat map is drawn with a continuous gradient scale (inset) from yellow to red representing increasing RFU. Yellow represents 0% RFU, while red represents 100% deuterium exchange.
3.7. Solution X-ray studies of the DENV-3 NS5FL and DENV-4 NS5FL mutants
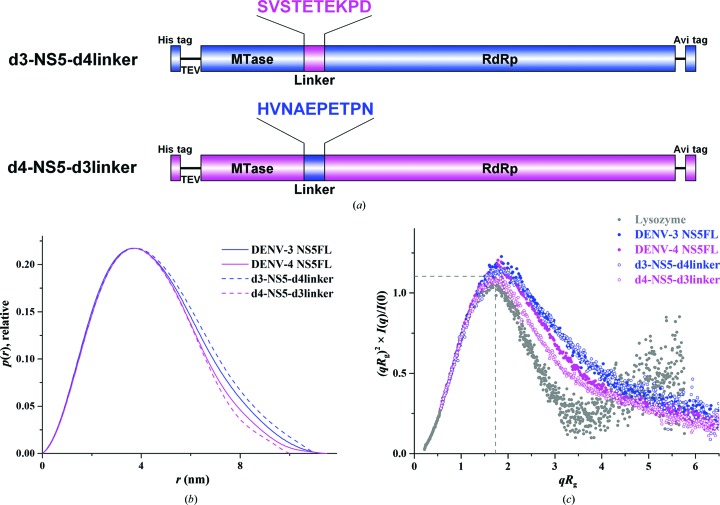
Considering that DENV-4 NS5 has a more compact conformation in solution and that low sequence conservation of the ten-residue linker region across the four NS5 serotypes has been revealed, two NS5FL mutants (d3-NS5-d4linker and d4-NS5-d3linker) were created by mutating the linker region of DENV-3 NS5FL (263-HVNAEPETPN-272) to the linker of DENV-4 NS5FL (264-SVSTETEKPD-273) and vice versa (Fig. 8 ▸ a) in order to understand the importance of the linker to the compactness of NS5FL. Both mutant proteins, d3-NS5-d4linker and d4-NS5-d3linker, have been produced in high amounts and with high purity (Supplementary Fig. S5a). As shown from the scattering profile and the Guinier plot, no protein aggregation was observed for either of the mutant proteins (Supplementary Figs. S5b and S5c). The derived R g and D max values were 3.58 ± 0.02 and 11.0 ± 1 nm for the d3-NS5-d4linker protein and 3.26 ± 0.01 and 10.0 ± 1 nm for the d4-NS5-d3linker protein, respectively (Table 4 ▸). The R g value of d3-NS5-d4linker increased slightly, and its P(r) function (Fig. 8 ▸ b) shifted moderately outwards compared with the wild-type DENV-3 NS5FL, indicating a small enhancement in the size of the protein. However, no significant differences can be seen in the normalized Kratky plot (Fig. 8 ▸ c) between d3-NS5-d4linker and DENV-3 NS5FL, implying that genetically engineering the linker from DENV-4 into DENV-3 NS5FL has no major effect on the flexibility of the d3-NS5-d4linker mutant protein. In comparison, introduction of the linker from DENV-3 NS5FL into the DENV-4 NS5FL protein induced a more compact structure, as verified by a decreased R g value as well as the overall size of the d4-NS5-d3linker mutant compared with wild-type DENV-4 NS5FL. These findings are well documented by the normalized Kratky plot, in which the bell-shaped curve of the d4-NS5-d3linker mutant is shifted inwards compared with that of wild-type DENV-4 NS5FL (Fig. 8 ▸ c). The data suggested that the linker swapping affects the dynamics of DENV-4 NS5FL and that its linker plays an important role in maintaining the proper compact conformation of this protein.
Figure 8.
Solution X-ray scattering studies of the NS5FL mutants d3-NS5-d4linker and d4-NS5-d3linker. (a) Schematic diagrams of the d3-NS5-d4linker and d4-NS5-d3linker mutant constructs. The linker of DENV-3 NS5 (263-HVNAEPETPN-272) was mutated to the linker of DENV-4 NS5 (264-SVSTETEKPD-273) and vice versa. (b) Pair-distance distribution functions P(r) for DENV-3 NS5FL (blue unbroken line), DENV-4 NS5FL (magenta unbroken line), d3-NS5-d4linker (blue dashed line) and d4-NS5-d3linker (magenta dashed line). (c) Normalized Krakty plots for lysozyme (grey filled circles), DENV-3 NS5FL (blue filled circles), DENV-4 NS5FL (magenta filled circles), d3-NS5-d4linker (blue circles) and d4-NS5-d3linker (magenta circles). The d3-NS5-d4linker mutant protein shows a bell-curve profile with a slight difference from that of wild-type DENV-3 NS5FL in the normalized Kratky plots, while the more bell-curved profile of d4-NS5-d3linker shifts inwards in the normalized Kratky plots when compared with those for wild-type DENV-3 NS5FL and DENV-4 NS5FL. This indicates that the linker of DENV-4 NS5FL is essential to preserve its compactness.
Table 4. Data-collection and scattering-derived parameters for d3-NS5-d4linker and d4-NS5-d3linker.
| Protein sample | d3-NS5-d4linker | d4-NS5-d3linker |
|---|---|---|
| Data-collection parameters | ||
| Instrument (source and detector) | Bruker NANOSTAR equipped with MetalJet Excillum X-ray source and VNTEC-2000 detector | |
| Beam geometry | 100m slit | |
| Wavelength (nm) | 0.134 | |
| q range (nm1) | 0.164 | |
| Exposure time (min) | 30 (6 frames 5min) | |
| Concentration range (mgml1) | 1.003.10 | 1.906.30 |
| Temperature (K) | 288.15 | 288.15 |
| Structural parameters | ||
| I(0) from P(r) (cm1) | 143.4 0.8 | 104.7 0.4 |
| R g from P(r) (nm) | 3.58 0.02 | 3.26 0.01 |
| I(0) from Guinier (cm1) | 144.2 1.2 | 102.8 0.3 |
| R g from Guinier (nm) | 3.57 0.05 | 3.17 0.03 |
| D max (nm) | 11.0 1 | 10.0 1 |
| Porod volume estimate (nm3) | 147 | 130 |
| DAMMIF excluded volume (nm3) | 197 | 179 |
| Dry volume from sequence† (nm3) | 131 | 131 |
| Molecular-mass determination | ||
| Calculated monomeric molecular mass from sequence‡ (kDa) | 107.9 | 107.9 |
| Molecular mass from Porod invariant (kDa) | 89 10 | 79 10 |
| Molecular mass from excluded volume (kDa) | 99 10 | 90 10 |
| Software employed | ||
| Primary data reduction | BRUKER SAS | |
| Data processing | PRIMUS | |
Calculated with Peptide Property Calculator (http://www.basic.northwestern.edu/biotools/proteincalc.html).
Calculated with Compute pI/Mw (http://web.expasy.org/compute_pi/).
Furthermore, since the linker of DENV-3 NS5 leads to a more compact form of the d3-NS5-d4linker mutant protein instead of the expected extended form, the data also demonstrate the importance of the regions surrounding the linker in DENV-3 and DENV-4 NS5FL as shown by the HDX-MS data above. Specifically, the substitution of nonpolar residues (Ala266 and Pro268) and polar residues (Thr270 and Asn272) in the DENV-3 linker by polar residues (Thr267 and Thr269) and charged residues (Lys271 and Asp273) in the DENV-4 linker might have generated additional interdomain interactions either with the MTase and/or RdRp domains, which may help in the stabilization of the d3-NS5-d4linker protein in the native conformation. Similarly, for the d4-NS5-d3linker mutant protein, because of the substitution of polar by nonpolar and charged by polar residues, the probable interdomain interactions between the linker residues with the MTase and/or RdRp domain may be weakened, causing a more unstable and dynamic linker region and leading to a more compact conformation. This is in line with the HDX-MS data described above, in which the unstable regions surrounding the linker in DENV-4 NS5FL provide a more dynamic environment and slight compactness of the protein.
3.8. Solution X-ray studies of DENV-2 NS5FL with substrates
In order to understand the effect of substrates of NS5 on the flexibility of this protein, DENV-2 NS5FL, which like the DENV-3 form is located in the cytoplasm and the nucleus (Hannemann et al., 2013 ▸; Tay et al., 2013 ▸), and which is one of the more predominant serotypes in Southeast Asia, was studied in the presence of GTP, GDP, S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), RNA1 (GpppAGUUGUU) and RNA2 (m7GpppAGUUGUU), respectively. To observe any changes in the NS5 after adding substrate, high-resolution SAXS data were collected using a synchrotron source (§2). SAXS data for DENV-2 NS5FL were collected in the absence (apo form) or the presence of substrates. For each sample measurement, the data were collected at protein concentrations of 1.25 and 2.5 mg ml−1 and the two scattering profiles were merged to obtain higher resolution data. The molar ratio of protein to substrate for GTP, GDP, SAM and SAH was 1:3, whereas equimolar ratios were used for RNA1 and RNA2. The merged scattering profiles of the DENV-2 NS5FL apo form and the presence of different substrates are shown in Supplementary Fig. S6. In general, no protein aggregation was observed in all of the data collected. As shown in Table 5 ▸, no significant change in the R g or D max values could be observed for the apo form of DENV-2 NS5FL compared with the protein in the presence of GTP, GDP, SAM and SAH. However, a reproducible increase in the R g value and D max were determined for the RNA1-bound (R g = 3.71 ± 0.01 nm and D max = 13.0 nm) and RNA2-bound (R g = 3.73 ± 0.01 nm and D max = 13.0 nm) forms when compared with the apo form (R g = 3.62 ± 0.01 nm and D max = 12.3 nm) of DENV-2 NS5FL. The normalized Kratky plots of the RNA1-bound and RNA2-bound forms revealed a slight change in the flexibility compared with the apoprotein, indicating conformational alterations owing to the binding of the small RNA.
Table 5. Scattering-derived parameters for DENV-2 NS5FL in the absence or presence of substrates.
| DENV-2 NS5FL | I(0) (Guinier) (cm1) | R g (Guinier) (nm) | I(0) [P(r)] (cm1) | R g [P(r)] (nm) | D max (nm) |
|---|---|---|---|---|---|
| Apo form | 1714.2 1.7 | 3.62 0.01 | 1718.0 1.1 | 3.69 0.01 | 12.3 |
| + Guanosine triphosphate (GTP) | 1722.8 1.8 | 3.64 0.01 | 1719.0 1.3 | 3.68 0.01 | 12.2 |
| + Guanosine diphosphate (GDP) | 1746.2 1.8 | 3.65 0.01 | 1742.0 1.4 | 3.68 0.01 | 12.3 |
| + RNA1 (GpppAGUUGUU) | 2323.9 1.9 | 3.71 0.01 | 2327.0 1.4 | 3.78 0.01 | 13.0 |
| + RNA2 (m7GpppAGUUGUU) | 2343.0 2.1 | 3.73 0.01 | 2347.0 1.5 | 3.79 0.01 | 13.0 |
| + S-Adenosylmethionine (SAM) | 1730.9 1.8 | 3.63 0.01 | 1727.0 1.5 | 3.66 0.01 | 12.1 |
| + S-Adenosylhomocysteine (SAH) | 1729.3 2.0 | 3.57 0.01 | 1732.0 1.4 | 3.62 0.01 | 11.5 |
| + GDP and SAM | 1728.8 2.1 | 3.60 0.01 | 1726.0 1.7 | 3.64 0.01 | 12.0 |
| + GDP and SAH | 1742.6 2.2 | 3.61 0.01 | 1741.0 1.8 | 3.66 0.01 | 12.1 |
4. Discussion
4.1. Arrangements of the two-enzyme ensemble NS5
The DENV two-enzyme complex NS5 is multifunctional and interacts with a variety of viral and host proteins to enable replication of the viral genome and to perturb the host innate immune response. The inter-ensemble and intra-ensemble formation and cross-talk require structural alterations and dynamic properties of the enzymes involved. The core enzymes of the Dengue virus replication machinery, NS3 and NS5, with its helicase, MTase and RdRp domains, perform the essential steps of dsRNA unwinding, NTPase/RTPase activity, genome replication and methyltransferase/guanylyltransferase activity (Egloff et al., 2002 ▸; Tay et al., 2014 ▸). Therefore, the catalytic centres of these enzymes have to come into proximity during the enzymatic events.
The very recently determined DENV-3 NS56–895 crystal structure reveals a compact shape with a total length of 8.7 nm, in which the MTase domain is oriented above the fingers subdomain of the RdRp domain such that its GTP-binding and SAH-binding sites are positioned away from the interdomain interface (Zhao et al., 2015 ▸). In comparison, the related flavivirus NS5 structure from Japanese encephalitis virus (JEV) is 10 nm in length and its MTase domain is attached to the back side of the RdRp domain through key hydrophobic interactions, shielding the rim of the NTP entry channel (Fig. 9 ▸; Lu & Gong, 2013 ▸). When the RdRp domain structures of JEV NS5 and DENV-3 NS56–895 are superimposed, the MTase domain has to be rotated by 105° to bring them into coincidence. In comparison, a structural model of NS5 derived from combining structures of the MTase and RdRp domains of the West Nile virus (WNV) NS5 and reverse genetics data from DENV show the MTase domain to be off the front channel, indicating a cooperative model (Malet et al., 2007 ▸; Fig. 9 ▸). The position of the MTase domain differs in each of the models, indicating the need to identify the most stable conformation in solution that is essential for the function. Furthermore, although both the MTase and RdRp domains are functionally independent (Selisko et al., 2006 ▸), the MTase domain in NS5FL enhances the RNA template affinity of the RdRp domain either by completing a high-affinity RNA-binding site and/or by promoting the correct formation of the template tunnel, for which the MTase domain orientation is important (Potisopon et al., 2014 ▸). The solution model derived from the presented SAXS studies of DENV-3 revealed an arrangement of the full-length NS5 proteins as well of DENV-3 NS56–895 with the methyltransferase domain positioned at the top of the RdRP domain, resulting in a total length of about 11.5 nm. Importantly, the overall arrangement of the MTase and RdRp domains, as well as the maximum particle dimension (11.0–11.5 nm), of all four serotypes of NS5FL is similar in solution (Table 2 ▸). The slightly extended and less rigid behaviour of DENV-3 NS5FL and DENV-3 NS56–895 in solution compared with the crystallographic structure is demonstrated (Fig. 2 ▸ d). The crystallographic units of the DENV-3 NS56–895 structure show a tight packing compared with the JEV NS5 protein (Supplementary Fig. S7). Because of this lattice arrangement, the interstitial spaces between the JEV molecules are larger compared with DENV-3 NS56–895, which may partially explain the lower R g value and overall dimensions of the crystallographic DENV-3 NS56–895 structure compared with the solution and DENV-3 NS5FL forms as well as with the JEV NS5 structure (Lu & Gong, 2013 ▸).
Figure 9.
Possible arrangements of the methyltransferase domain (blue) with respect to the RdRp domain (magenta) of NS5, connected by the linker (brown). The fingers subdomain is labelled ‘F’ and the thumb subdomain is labelled ‘T’. The yellow arrow indicates the RNA-exit site. (a) The methyltransferase domain in the DENV-3 NS56–895 crystallographic structure (PDB entry 4v0r) is positioned above the fingers subdomain of the RdRp domain (Zhao et al., 2015 ▸). (b) The BUNCH model derived from the SAXS data for DENV-3 NS5FL. The methyltransferase domain is located slightly away from the back side of RdRp and the NTP-entry channel is exposed. (c) The highly extended conformation of DENV-3 NS51–878 derived from a SAXS experiment (Bussetta & Choi, 2012 ▸). The methyltransferase is arranged side-by-side with respect to RdRp. (d) Similar to the DENV-3 NS56–895 crystallographic structure, the methyltransferase in the JEV NS5FL crystallographic structure (PDB entry 4k6m) is located at the back side of RdRp, with a 105° rotation of the methyltransferase domain when the structures are superimposed (Lu & Gong, 2013 ▸). (e) The methyltransferase domain in the cooperative model is positioned closer to the RNA-exit site (Malet et al., 2007 ▸).
Interestingly, the MTase domain is shown to be more flexible in solution compared with the compact RdRp domain (Fig. 5 ▸). Such flexibility may be essential for the multifunctional and sequential steps of capping mRNA inside the MTase. In addition, the R g and D max values of DENV-2 NS5FL bound to GpppAGUUGUU (R g = 3.71 ± 0.01 nm and D max = 13.0 nm) or m7GpppAGUUGUU (R g = 3.73 ± 0.01 nm and D max = 13.0 nm) compared with the apo form of DENV-2 NS5FL confirm that it is undergoing structural alterations owing to substrate binding, leading to a more extended enzyme complex. The overall increase in R g and D max of the capped RNA-bound form of DENV-2 NS5FL suggests that either the MTase domain and/or the RdRp domain might have undergone conformational changes allowing the NS5FL to adopt a more open or extended conformation necessary for the elongation step after the initiation/transition phase. Such conformational changes are inherent to the NS5FL during RNA synthesis. These alterations would also provide the conformations needed for binding and/or dissociation of the helicase domain of NS3 during the sequential steps of RNA replication. The present SAXS data for DENV-2 NS5FL also enable it to be determined that no significant changes occur owing to the binding of the smaller substrates GTP, GDP, S-adenosylmethionine and S-adenosylhomocysteine, and confirm the recent crystallographic structures of individual domains (Egloff et al., 2002 ▸; Yap et al., 2010 ▸) or DENV-3 NS56–895 (Zhao et al., 2015 ▸) in the presence of these substrates.
In order to ascertain the predominant conformations of DENV-3 NS5FL in solution, EOM was used, in which the flexibility of the MTase domain with respect to the RdRp domain is considered. The calculations revealed that the protein has an ensemble of bent and elongated conformations (monomodal R g distribution mainly between 3.4 and 3.5 nm). These studies reveal the flexible nature of DENV-3 NS5FL, with the MTase domain existing in several different conformations in solution, which is vital for its catalytic function and also for the preferential interaction with the RdRp and helicase domains of NS3 during replication, as described in the presence of capped RNA above. In comparison, previous SAXS studies of DENV-3 NS51–878 showed a bimodal R g distribution with values between 3.4 and 4.1 nm in EOM, with the latter showing a highly extended form of 12.5 nm in dimension and a side-by-side model of the MTase–RdRP assembly (Bussetta & Choi, 2012 ▸). However, such a bimodal R g distribution could not be observed for freshly purified NS5FL proteins from all four serotypes in solution.
4.2. Similarity and diversity of the DENV-1 to DENV-4 NS5FLs
The determined R g values for the NS5FLs of DENV-1, DENV-2, DENV-3 and DENV-4 of 3.55 ± 0.09, 3.56 ± 0.05, 3.37 ± 0.02 and 3.27 ± 0.02 nm, respectively, indicate a similarity in the overall dimensions of the DENV-1 and DENV-2 NS5FLs as well as a slight and reproducible decrease for the DENV-3 and in particular the DENV-4 NS5FLs. The solution data for the DENV-4 NS5FL reflect the differences in flexibility and indicate a more compact shape profile for the DENV-4 NS5FL. Comparison of the normalized Kratky and Porod–Debye plots of the individual MTase and RdRp domains of DENV-3 and the full-length NS5 of all four serotypes show an increase in the flexibility and extension of the NS5FL, indicating that the linker which connects the two enzymatic domains provides an ideal element to allow inter-dynamic and intra-dynamic processes to occur. HDX-MS displays high deuterium exchange for the ten-residue linker region of DENV-3 and DENV-4 NS5FL, supporting the concept of flexibility between the MTase and RdRp, which is in line with the high B factors of the partially determined linker region in the JEV NS5 structure (Lu & Gong, 2013 ▸). The higher deuterium exchange seen in the linker region of the DENV-4 NS5FL compared with the DENV-3 NS5FL may reflect a degree of dynamism, which enables DENV-4 to become slightly more compact in the conformational interplay of complex and extended formation during catalysis. The changes seen after swapping the ten-residue linker between the DENV-4 and DENV-3 NS5FLs in the elution profiles (Supplementary Fig. S5), overall dimensions and flexibility (Fig. 8 ▸) confirm the importance of the linker for MTase–RdRp communication and dynamics. Interestingly, substitution of the ten-residue linker from the DENV-4 NS5FL into the DENV-3 NS5FL (Fig. 8 ▸) shows a change in the R g value but no major effect on the overall size or the compactness of the d3-NS5-d4linker protein, whereas substitution of the DENV-3 NS5FL linker into DENV-4 NS5FL (Fig. 8 ▸) leads to a protein with a smaller D max value and a greater compactness compared with the wild-type protein. This phenomenon may be caused by the different amino-acid partners in the linker region and the residues of the MTase and RdRp domains of the d4-NS5-d3linker mutant, which are supposed to form interdomain interactions via charged side chains and water-mediated contacts, as described for DENV-3 NS56–895 (Zhao et al., 2015 ▸). In particular, the substitution of nonpolar residues (Ala266 and Pro268) and polar residues (Thr270 and Asn272) in the DENV-3 linker by polar residues (Thr267 and Thr269) and charged residues (Lys271 and Asp273) in the DENV-4 linker might induce additional interdomain interactions that might help in the stabilization of the preferred conformation. The importance of the linker is confirmed by mutagenesis studies in which Arg263 and Glu270 of DENV-4 NS5FL were substituted by an alanine, leading to significant inhibition of MTase and polymerase activities, and neither viral RNA nor infectious virus could be observed (Zhao et al., 2015 ▸).
Furthermore, comparison of the HDX-MS heat maps of the DENV-3 and DENV-4 NS5FLs yield the differences in deuterium exchange previously observed for the linker as well as for the different regions of the MTase and RdRp domains. The results of HDX-MS point towards differences in dynamics between the linker and linker-flanking segments, which in turn promote the greater compaction of DENV-4 compared with DENV-3. The helix–turn–sheet motif in DENV-4 adjacent to the linker (residues 255–266 and 270–281) shows 50% relative fractional uptake, indicative of less stable hydrogen bonding and consequently suggesting local unfolding within the locus. There are equivalent regions in the RdRp domain on the other side of the linker that also show higher flexibility common to both the DENV-3 and DENV-4 NS5 proteins. We infer therefore that the differences in R g mainly arise from the dissimilarity in compaction of the MTase domain between the two serotypes owing to differences in linker stabilities between the two serotype proteins. Such local unfolding, as inferred from the higher deuterium exchange (50% RFU), has also previously been observed in the osmosensing protein EnvZ, where osmolyte-mediated modulation of the local unfolding in a critical helical subdomain allows the protein to function as an osmosensor (Wang et al., 2012 ▸). Although this region in the MTase domain is expected to be a helix–turn–sheet based on the DENV-3 NS5 structure, the deuterium exchange indicates high flexibility in the region in solution, which drives the compaction observed by SAXS for DENV-4 NS5FL. This is consistent with our results from single-domain studies with SAXS, in which the compaction is shown to be driven by the MTase and the linker. Using HDX-MS, we have localized the region responsible for compaction to residues 255–281.
In conclusion, the studies here provide a detailed insight into the overall dimensions and flexibility of the NS5FL complex as well as its individual MTase and RdRP domains. Considering the available structures and solution studies of apo and substrate-bound NS5FL, implications and hypothesis concerning functional conformations were discussed. The first comparison of all four DENV NS5FLs shines a light onto the similarities and diversity of the DENV-1 to DENV-4 proteins in compactness and dynamics, which may also partially influence the localization of the four DENV NS5FL forms inside the cytoplasm and/or nucleus and finally their interaction with nuclear host proteins to perturb the host innate immune-system response. Swapped linkers of DENV-3 and DENV-4 reveal the importance of this region for the cross-talk of the MTase and RdRp domains and possibly the contact with NS3 as well as for the differences seen in compactness and dynamics, in particular of the DENV-4 NS5, making this epitope and serotype a valuable drug target. As drug-binding sites are composed of residues located on loops, or from two or more domains that are highly flexible, the inherent flexibility of the proteins is often ignored in drug design, leading to failures in docking (Teague, 2003 ▸). However, understanding the protein flexibility makes it possible to exploit high-affinity interactions and thereby obtain a high-affinity drug (Teague, 2003 ▸). Ongoing NMR solutions and future in vivo studies will add to the understanding of the connection between these dynamics and the pathogenesis of the virus, providing a valuable drug target for antiviral drug development.
Acknowledgments
We thank Mr Abhijeet Ghode (Department of Biological Sciences, NUS, Singapore) for his help in HDX-MS experiments. WGS thanks the authority of Nanyang Technological University for awarding a research scholarship. This work was supported by the Ministry of Education (MOE) Tier 3 (MOE2012-T3-1-008), Singapore to GG and GSA and Duke–NUS Signature Research Program (funded by the Ministry of Health, Singapore), the National Medical Research Council, Singapore (NMRC/1315/2011) to SGV. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH. We thank Drs Feng Gu, Pei Yong Shi and Siew Pheng Lim (Novartis Institute for Tropical Diseases) for providing GpppAGUUGUU and m7GpppAGUUGUU.
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Balakrishna, A. M., Basak, S., Manimekalai, M. S. S. & Grüber, G. (2015). J. Biol. Chem. 290, 3183–3196. [DOI] [PMC free article] [PubMed]
- Bernadó, P., Mylonas, E., Petoukhov, M. V., Blackledge, M. & Svergun, D. I. (2007). J. Am. Chem. Soc. 129, 5656–5664. [DOI] [PubMed]
- Burke, J. R., Hura, G. L. & Rubin, S. M. (2012). Genes Dev. 26, 1156–1166. [DOI] [PMC free article] [PubMed]
- Bussetta, C. & Choi, K. H. (2012). Biochemistry, 51, 5921–5931. [DOI] [PMC free article] [PubMed]
- Davidson, A. D. (2009). Adv. Virus Res. 74, 41–101. [DOI] [PubMed]
- Decroly, E., Ferron, F., Lescar, J. & Canard, B. (2012). Nature Rev. Microbiol. 10, 51–65. [DOI] [PMC free article] [PubMed]
- Dip, P. V., Kamariah, N., Subramanian Manimekalai, M. S., Nartey, W., Balakrishna, A. M., Eisenhaber, F., Eisenhaber, B. & Grüber, G. (2014). Acta Cryst. D70, 2848–2862. [DOI] [PubMed]
- Dong, H., Chang, D. C., Xie, X., Toh, Y. X., Chung, K. Y., Zou, G., Lescar, J., Lim, S. P. & Shi, P.-Y. (2010). Virology, 405, 568–578. [DOI] [PubMed]
- Durand, D., Vivès, C., Cannella, D., Pérez, J., Pebay-Peyroula, E., Vachette, P. & Fieschi, F. (2010). J. Struct. Biol. 169, 45–53. [DOI] [PubMed]
- Egloff, M.-P., Benarroch, D., Selisko, B., Romette, J.-L. & Canard, B. (2002). EMBO J. 21, 2757–2768. [DOI] [PMC free article] [PubMed]
- Fernandez-Garcia, M. D., Meertens, L., Bonazzi, M., Cossart, P., Arenzana-Seisdedos, F. & Amara, A. (2011). J. Virol. 85, 2980–2989. [DOI] [PMC free article] [PubMed]
- Franke, D. & Svergun, D. I. (2009). J. Appl. Cryst. 42, 342–346. [DOI] [PMC free article] [PubMed]
- Glatter, O. & Kratky, O. (1982). Small-angle X-ray Scattering. London: Academic Press.
- Guinier, A. (1939). Ann. Phys. (Paris), 12, 161–237.
- Hannemann, H., Sung, P.-Y., Chiu, H.-C., Yousuf, A., Bird, J., Lim, S. P. & Davidson, A. D. (2013). J. Biol. Chem. 288, 22621–22635. [DOI] [PMC free article] [PubMed]
- Issur, M., Geiss, B. J., Bougie, I., Picard-Jean, F., Despins, S., Mayette, J., Hobdey, S. E. & Bisaillon, M. (2009). RNA, 15, 2340–2350. [DOI] [PMC free article] [PubMed]
- Kapoor, M., Zhang, L., Ramachandra, M., Kusukawa, J., Ebner, K. E. & Padmanabhan, R. (1995). J. Biol. Chem. 270, 19100–19106. [DOI] [PubMed]
- Konarev, P. V., Petoukhov, M. V., Volkov, V. V. & Svergun, D. I. (2006). J. Appl. Cryst. 39, 277–286.
- Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. J. & Svergun, D. I. (2003). J. Appl. Cryst. 36, 1277–1282.
- Kozin, M. B. & Svergun, D. I. (2001). J. Appl. Cryst. 34, 33–41.
- Lim, S. P., Koh, J. H. K., Seh, C. C., Liew, C. W., Davidson, A. D., Chua, L. S., Chandrasekaran, R., Cornvik, T. C., Shi, P.-Y. & Lescar, J. (2013). J. Biol. Chem. 288, 31105–31114. [DOI] [PMC free article] [PubMed]
- Lim, S. P. et al. (2010). J. Biol. Chem. 286, 6233–6240.
- Lindenbach, B. D., Thiel, H. & Rice, C. M. (2007). Fields Virology, 5th ed., edited by D. M. Knipe & P. M. Howley, pp. 1101–1152. Philadelphia: Lippincott Williams & Wilkins.
- Liu, H. & Kiledjian, M. (2006). Biochem. Soc. Trans. 34, 35–38. [DOI] [PubMed]
- Lu, G. & Gong, P. (2013). PLoS Pathog. 9, e1003549. [DOI] [PMC free article] [PubMed]
- Luo, D., Xu, T., Hunke, C., Grüber, G., Vasudevan, S. G. & Lescar, J. (2008). J. Virol. 82, 173–183. [DOI] [PMC free article] [PubMed]
- Mackenzie, J. (2005). Traffic, 6, 967–977. [DOI] [PMC free article] [PubMed]
- Malet, H., Egloff, M. P., Selisko, B., Butcher, R. E., Wright, P. J., Roberts, M., Gruez, A., Sulzenbacher, G., Vonrhein, C., Bricogne, G., Mackenzie, J. M., Khromykh, A. A., Davidson, A. D. & Canard, B. (2007). J. Biol. Chem. 282, 10678–10689. [DOI] [PubMed]
- Martel, A., Liu, P., Weiss, T. M., Niebuhr, M. & Tsuruta, H. (2012). J. Synchrotron Rad. 19, 431–434. [DOI] [PMC free article] [PubMed]
- Mertens, H. D. & Svergun, D. I. (2010). J. Struct. Biol. 172, 128–141. [DOI] [PubMed]
- Petoukhov, M. V., Franke, D., Shkumatov, A. V., Tria, G., Kikhney, A. G., Gajda, M., Gorba, C., Mertens, H. D. T., Konarev, P. V. & Svergun, D. I. (2012). J. Appl. Cryst. 45, 342–350. [DOI] [PMC free article] [PubMed]
- Petoukhov, M. V. & Svergun, D. I. (2005). Biophys. J. 89, 1237–1250. [DOI] [PMC free article] [PubMed]
- Potisopon, S., Priet, S., Collet, A., Decroly, E., Canard, B. & Selisko, B. (2014). Nucleic Acids Res. 42, 11642–11656. [DOI] [PMC free article] [PubMed]
- Rambo, R. P. & Tainer, J. A. (2011). Biopolymers, 95, 559–571. [DOI] [PMC free article] [PubMed]
- Receveur-Brechot, V. & Durand, D. (2012). Curr. Protein Pept. Sci. 13, 55–75. [DOI] [PMC free article] [PubMed]
- Selisko, B., Dutartre, H., Guillemot, J.-C., Debarnot, C., Benarroch, D., Khromykh, A., Desprès, P., Egloff, M.-P. & Canard, B. (2006). Virology, 351, 145–158. [DOI] [PubMed]
- Sievers, F., Wilm, A., Dineen, D. G., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. (2011). Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed]
- Smolsky, I. L., Liu, P., Niebuhr, M., Ito, K., Weiss, T. M. & Tsuruta, H. (2007). J. Appl. Cryst. 40, s453–s458.
- Stiegler, A. L., Grant, T. D., Luft, J. R., Calderwood, D. A., Snell, E. H. & Boggon, T. J. (2013). PLoS One, 8, e55591. [DOI] [PMC free article] [PubMed]
- Svergun, D. I. (1992). J. Appl. Cryst. 25, 495–503.
- Svergun, D. I. (1999). Biophys. J. 76, 2879–2886. [DOI] [PMC free article] [PubMed]
- Svergun, D., Barberato, C. & Koch, M. H. J. (1995). J. Appl. Cryst. 28, 768–773.
- Svergun, D. I., Koch, M. H. J., Timmins, P. A. & May, R. P. (2013). Small Angle X-ray Scattering from Solutions of Biological Macromolecules. Oxford University Press.
- Tay, M. Y. F., Fraser, J. E., Chan, W. K. K., Moreland, N. J., Rathore, A. P., Wang, C., Vasudevan, S. G. & Jans, D. A. (2013). Antiviral Res. 99, 301–306. [DOI] [PubMed]
- Tay, M. Y. F., Saw, W. G., Zhao, Y., Chan, W. K. K., Singh, D., Chong, Y., Forwood, J. K., Ooi, E. E., Grüber, G., Lescar, J., Luo, D. & Vasudevan, S. G. (2014). J. Biol. Chem. 290, 2379–2394. [DOI] [PMC free article] [PubMed]
- Teague, S. J. (2003). Nature Rev. Drug Discov. 2, 527–541. [DOI] [PubMed]
- Tria, G., Mertens, H. D. T., Kachala, M. & Svergun, D. I. (2015). IUCrJ, 2, 207–217. [DOI] [PMC free article] [PubMed]
- Volkov, V. V. & Svergun, D. I. (2003). J. Appl. Cryst. 36, 860–864.
- Wales, T. E., Fadgen, K. E., Gerhardt, G. C. & Engen, J. R. (2008). Anal. Chem. 80, 6815–6820. [DOI] [PMC free article] [PubMed]
- Wang, L. C., Morgan, L. K., Godakumbura, P., Kenney, L. J. & Anand, G. S. (2012). EMBO J. 31, 2648–2659. [DOI] [PMC free article] [PubMed]
- Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. (2009). Bioinformatics, 25, 1189–1191. [DOI] [PMC free article] [PubMed]
- Welsch, S., Miller, S., Romero-Brey, I., Merz, A., Bleck, C. K., Walther, P., Fuller, S. D., Antony, C., Krijnse-Locker, J. & Bartenschläger, R. (2009). Cell Host Microbe, 5, 365–375. [DOI] [PMC free article] [PubMed]
- World Health Organization (2009). Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. Geneva: World Health Organization. [PubMed]
- Yap, L. J., Luo, D., Chung, K. Y., Lim, S. P., Bodenreider, C., Noble, C., Shi, P.-Y. & Lescar, J. (2010). PLoS One, 5, e12836. [DOI] [PMC free article] [PubMed]
- Yap, T. L., Xu, T., Chen, Y.-L., Malet, H., Egloff, M.-P., Canard, B., Vasudevan, S. G. & Lescar, J. (2007). J. Virol. 81, 4753–4765. [DOI] [PMC free article] [PubMed]
- Zhao, Y., Soh, T. S., Zheng, J., Chan, K. W. K., Phoo, W. W., Lee, C. C., Tay, M. Y. F., Swaminathan, K., Cornvik, T. C., Lim, S. P., Shi, P.-Y., Lescar, J., Vasudevan, S. G. & Luo, D. (2015). PLoS Pathog. 11, e1004682. [DOI] [PMC free article] [PubMed]