Abstract
DNA methylation in the form of 5-methylcytosine (5mC) is essential for normal development in mammals and influences a variety of biological processes including transcriptional regulation, imprinting and the maintenance of genomic stability. The recent discovery of TET proteins, which oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), has changed our understanding of the process of DNA demethylation. Here, we summarize our current knowledge of the roles of DNA methylation and TET proteins in cell differentiation and function. The intensive research on this subject has so far focused primarily on ES cells and neurons. Here we summarize what is known about DNA methylation in T cell function.
Methylation of cytosines in metazoan genomes adds epigenetic information onto DNA without changing the genetic information encoded in the DNA sequence. Until recently, the only known modified base in DNA was 5-methylcytosine (5mC), an epigenetic mark established by DNA methyltransferases (DNMTs) (Ooi et al. 2009). In somatic cells, 5mC is almost exclusively found in the CpG sequence context, although non-CpG methylation has been documented in embryonic stem (ES) cells and in neurons (Lister et al. 2009; Lister et al. 2013). The promoters of the most highly expressed genes show the lowest levels of CpG methylation; conversely, dense CpG methylation of promoters is generally associated with decreased gene expression (Suzuki and Bird 2008; Laurent et al. 2010; Deaton and Bird 2011). There is also dense DNA methylation in gene bodies (Lister et al. 2009; Laurent et al. 2010), but the association of gene body CpG methylation with transcriptional regulation is less clear.
Two classes of DNMTs are involved in DNA methylation. The de novo DNA methyltransferases DNMT3A and 3B are required to establish DNA methylation patterns, while the maintenance DNA methyltransferase DNMT1 reestablishes DNA methylation patterns following DNA replication (Klose and Bird 2006) (Ooi et al. 2009). DNMT1 acts with its cofactor UHRF1, which binds hemimethylated DNA (Avvakumov et al. 2008) (Hashimoto et al. 2008), to reestablish symmetrical CpG methylation on the newly synthesized DNA strand, thus maintaining DNA methylation patterns during replication (Bestor et al. 1988; Ooi et al. 2009).
The distribution of 5mC has been mapped at single nucleotide resolution in human and mouse ES cells, ES cells differentiated to distinct lineages, somatic tissues, cultured cell lines, and various cancer cells (Hansen et al. 2011; Stadler et al. 2011; Kulis et al. 2012; Gifford et al. 2013; Lister et al. 2013; Xie et al. 2013; Ziller et al. 2013). These studies showed that most of the genome is highly methylated (~80–90% of CpGs with >50% methylation), with the remainder subdivided into unmethylated regions (UMRs) with less than 10% methylation, and low-methylated regions (LMRs) with 10–50% methylation (Stadler et al. 2011). UMRs correspond largely to unmethylated CpG islands (CGIs), many of which are located at transcription start sites (TSSs), whereas LMRs often coincide with promoter-distal gene regulatory elements enriched for transcription factor binding sites (Stadler et al. 2011).
TET proteins are 5mC oxidases
The recent discovery that TET (Ten-Eleven Translocation) proteins are 5-methylcytosine oxidases added an additional layer of complexity to our understanding of the biological role of DNA methylation (Iyer et al. 2009; Tahiliani et al. 2009). TET proteins are named after the rare ten-eleven translocation (t(10;11)(q22;q23) observed in cases of acute myeloid and lymphocytic leukemia, in which the MLL1 (mixed-lineage leukemia 1) gene located on human chromosome 10 is fused with the TET1 gene located on chromosome 11 (Ono et al. 2002; Lorsbach et al. 2003). The three TET proteins in mammals, TET1, TET2 and TET3 (Fig. 1A) were identified by homology with the J-binding proteins of trypanosomes (Iyer et al., 2008), and are known to be members of the larger family of 2-oxoglutarate- and Fe(II)-dependent dioxygenases (Iyer et al., 2009; Tahiliani 2009) (Loenarz and Schofield 2008; Loenarz and Schofield 2011). JBP1 and JBP2 oxidize the methyl group of thymine; the resulting 5-hydroxymethyluracil is then glycosylated to generate Base J (Iyer et al. 2009; Pastor et al. 2013), whereas the mammalian TET proteins are 2-oxoglutarate- and Fe(II)-dependent dioxygenases oxidize 5mC to generate 5hmC, 5fC and 5caC (Tahiliani et al. 2009) (Ito et al. 2011), (He et al. 2011) (Fig. 1B; reviewed in Pastor et al., 2013). Representatives of the TET/JBP superfamily are found in every metazoan organism that uses DNA methylation (Iyer et al. 2009), suggesting a major role for TET proteins in regulating DNA methylation status through production of oxi-mC (Kohli and Zhang 2013; Pastor et al. 2013).
Figure 1.
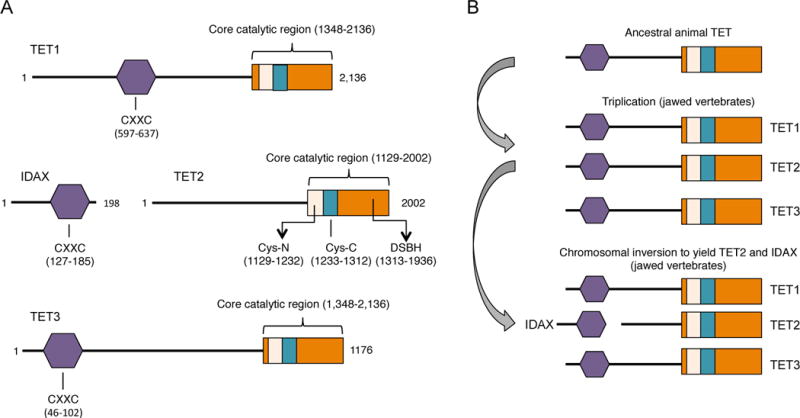
A, Schematic representation of the predicted functional domains of mammalian TET proteins. Depicted is the CXXC domain that is present in TET1 and TET3, the C-terminal catalytic domain that contains a cysteine-rich (Cys-rich) region and the double-stranded β-helix (DSBH) region that are common features in all three members of the TET family. The crystal structure of human TET2 reveals that the Cys-rich region is divided into N- and C-terminal regions that flank the DSBH domain (Hu et al. 2013). Also depicted is IDAX, a CXXC-domain protein that was part of TET2 prior to chromosomal inversion ((Pastor et al. 2013), see B). The numbers indicate aminoacids and they correspond to human TET proteins. B, Evolutionary history of TET proteins. The original gene encoding TET underwent triplication in jawed vertebrates, giving rise to the three members of the TET family. A subsequent chromosomal inversion resulted in the separation of the TET2 catalytic domain from the CXXC domain, which became a separate gene, IDAX/CXXC4.
The prediction that the core catalytic domain of the TET proteins is a double-stranded beta-helix (DSBH) adjacent to a unique cysteine-rich domain involved in DNA recognition (Iyer 2009) was confirmed experimentally by studies on recombinant human Tet1 expressed in insect cells, which showed that the DSBH region was catalytically inactive unless the Cys-rich region was present (Tahiliani 2009), and subsequently by determination of the crystal structures of the human TET2 catalytic domain (Hu et al., Cell 2013) and a TET-like dioxygenase from Naegleria gluberi (Hashimoto et al., Nature 2013). In the human TET2 structure, the DSBH region forms the active site with the signature His-Xaa-Asp motif (where Xaa is any amino acid) motif and a conserved Arg residue that binds 2-oxoglutarate, whereas the Cys-rich region wraps around the DSBH core and stabilizes DNA binding (Hu et al. 2013). Notably, the TET2 catalytic domain binds CpG-containing and methyl-CpG-containing DNA with similar affinity; the fact that the methyl group itself does not contact TET2 presumably allows the catalytic cavity to accommodate the partly oxidized 5mC derivative 5hmC and 5fC for further oxidation to 5caC (Hu et al. 2013).
TET1 and TET3 contain N-terminal CXXC domains, whereas TET2 became separated from its CXXC domain as a result of a chromosomal inversion (Iyer et al. 2009); Pastor review (Fig. 1B). The separated CXXC domain is now a distinct gene, IDAX (inhibition of the Dvl and axin complex; also known as CXXC4), whose product, when DNA-bound, recruits TET2 and regulates TET2 protein levels through caspase activation (Ko et al. 2013). CXXC domains typically bind unmethylated CpGs and are found in many chromatin- associated proteins including the maintenance methyltransferase DNMT1 and MLL1, a component of the SET/COMPASS complex.
TET proteins and oxi-mC in the pathway to DNA demethylation
The oxidative derivatives of 5mC that are generated by TET proteins are likely to serve as intermediates in DNA demethylation (Kohli and Zhang 2013; Pastor et al. 2013). Two main pathways for demethylation have been proposed: passive dilution of the oxidized base during DNA replication, and active DNA demethylation through DNA repair; the latter pathway is shown in Fig. 2. We note, however, that 5hmC is abundant in postmitotic neurons – in Purkinje neurons where it comprises ~40% of the level of 5mC (Kriaucionis and Heintz 2009), as well as in other neurons in the brain (Szulwach et al. 2011b; Mellen et al. 2012; Hahn et al. 2013) – indicating that like 5mC itself, oxidized methylcytosines are likely to function as epigenetic marks. Indeed, many oxi-mC ‘readers’ have been identified by mass spectrometry (Spruijt et al. 2013); these include diverse transcription factors and chromatin regulatory protein, but the importance of oxi-mC recognition for gene regulation remains to be defined.
Figure 2.
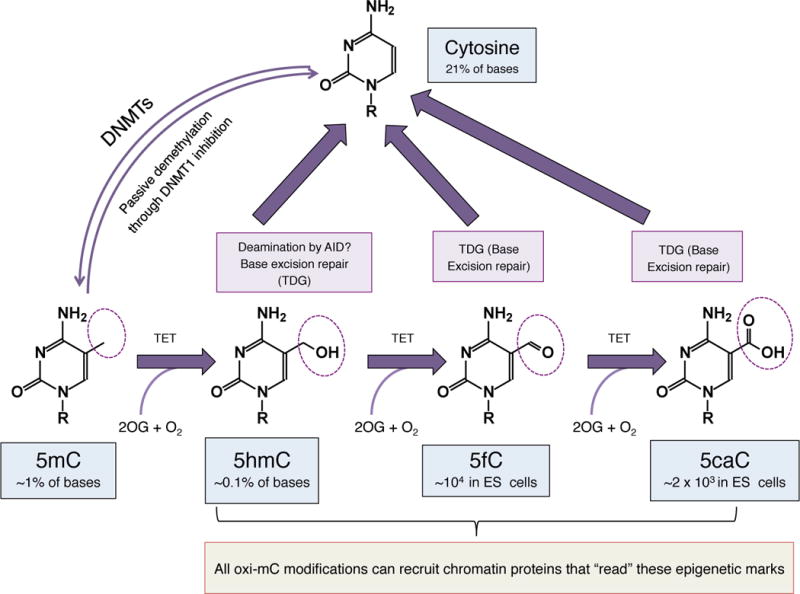
The cycle of DNA demethylation by TET proteins and the generation of oxi-mCs. DNA methyltransferases (DNMTs) add a methyl group to the 5 position of cytosine, and thus are responsible for DNA methylation. DNA demethylation can occur passively by inhibition of the maintenance methyltransferase DNMT1, especially in the presence of oxi-mCs which inhibit DNMT1, resulting in dilution of the methyl mark during replication. TET proteins in the presence of 2-oxoglutarate (OG) can oxidize mC to 5hmC, which is further oxidized by the TET proteins to 5fC and 5caC, which are much less abundant than 5hmC in the genome. 5fC and 5caC are recognized and excised by thymine DNA glycosylase (TDG) and subsequently replaced through base excision repair with unmethylated C. 5hmC has also been suggested to be directly demethylated by deamination through AID, followed by base excision repair mediated by TDG. Notably, the oxi-mCs (5hmC, 5fC and 5caC) can exert functions beyond mediating DNA demethylation, acting as marks that recruit chromatin-bound regulatory proteins.
1. Passive dilution of the oxidized base during replication
Blocking the action of the maintenance DNA methyltransferases would result in loss of DNA methylation during replication, a process known as passive DNA demethylation (Fig. 2). Conversion of 5mC to 5hmC could promote passive demethylation as a consequence of replication by inhibiting the ability of DNMT1 to recognize 5hmCpGs as observed in vitro (Valinluck and Sowers 2007). However the obligate DNMT1 partner UHRF1 binds 5mC and 5hmC with similar affinities (Frauer et al. 2011), indicating that further investigation is needed. Current in vivo evidence from pre-implantation embryos suggests that 5hmC is passively removed through replication (Inoue and Zhang 2011). Passive dilution of 5hmC has also been reported in primordial germ cells (PGCs) during the second wave of DNA demethylation observed in these cells (Hackett et al. 2013). Notably, 5hmC levels are dramatically reduced during in vitro expansion of activated mouse CD4+ and CD8+ T cells (our unpublished data), as well as in many cancers (Ko et al. 2010; Pfeifer et al. 2013) and normal proliferating tissues (Pfeifer et al. 2013) suggesting that passive 5hmC dilution occurs in these cases as well.
2. Active DNA demethylation through DNA repair
The DNA repair enzyme TDG (thymine DNA glycosylase) was originally identified as recognizing and repairing T:G mismatches in DNA. However, TDG also efficiently excises 5fC and 5caC that are properly paired to G in double stranded DNA (He et al. 2011) (Fig. 2), and depletion of TDG in ES cells results in increased levels of 5fC and 5caC at specific genomic locations (Shen et al. 2013; Song et al. 2013). Consistent with the hypothesis that TDG-mediated excision of 5fC and 5caC constitutes the last step of a pathway of TET-mediated active DNA demethylation, TDG is required for embryonic development and TDG-null embryos as well as embryos that bear a catalytically inactive mutation of TDG exhibit epigenetic abnormalities, with predominant reduction in the expression of hox genes due to aberrant methylation observed in their regulatory sequences (Cortazar et al. 2011; Cortellino et al. 2011).
Biological functions of TET proteins and 5hmC
TET enzymes are widely expressed, with at least one member being represented in every cell type examined. Tet1 and Tet2 are the two major TET proteins expressed in mouse embryonic stem (ES) cells whereas Tet3 is barely expressed (Fig. 3); Tet1 and Tet2 are also high in primordial germ cells and in the inner cell mass of the mouse embryo from which ES cells are derived. Tet1 mRNA expression drops rapidly in differentiating mouse ES cells (Koh et al. 2011), whereas Tet2 protein levels are regulated by Idax during ES cell differentiation (Ko et al., Nature 2013). Tet2 and Tet3 are the major TET proteins expressed in differentiated tissues and cell types (Fig. 3). Tet1-deficient mice on a pure C57BL/6 genetic background are born at less than Mendelian ratios and display female sterility (unpublished data), Tet2-deficient mice are viable and fertile (Ko et al. 2011) with a mild haematopoietic phenotype, whereas Tet3-deficient mice are perinatally lethal (our unpublished data, (Gu et al. 2011)).
Figure 3.
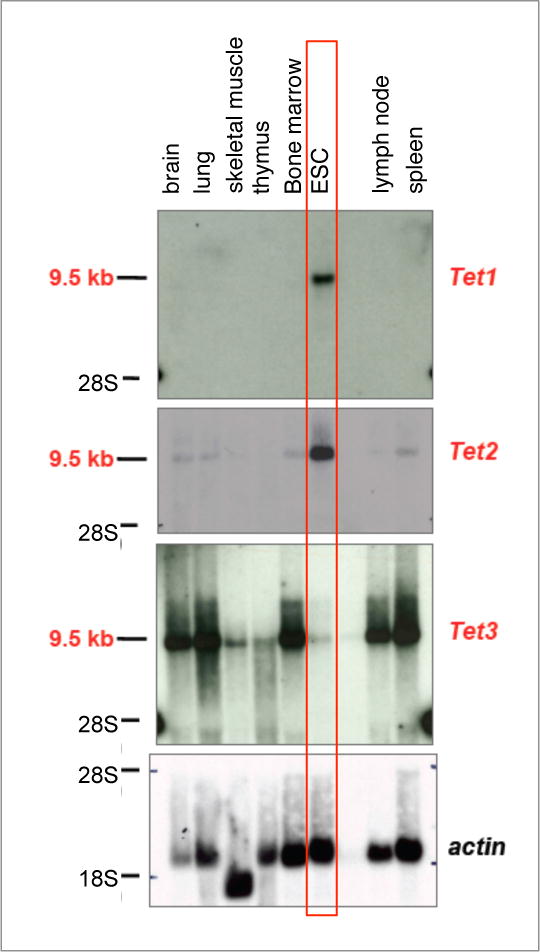
Northern blot analysis of RNA expression of all TET members in different murine tissues. Probes for actin were used for normalization. Mouse embryonic stem cells (ESC), which express primarily Tet1 and Tet2, are highlighted. Tet3 is not expressed in ESC but is abundantly expressed in all the differentiated cell types.
There have been extensive studies of 5hmC distribution in ES cells and in brain. 5hmC is found predominantly in euchromatic regions, suggesting that it is associated with gene activity (Ficz et al. 2011). In mouse ES cells, Tet1 and 5hmC are enriched at gene bodies, transcription start sites, promoters and genomic regions with moderate CpG density (Ficz et al. 2011; Pastor et al. 2011; Williams et al. 2011; Wu et al. 2011a; Wu et al. 2011b; Xu et al. 2011). CpG islands (CGIs) that bind Tet1 in undifferentiated ES cells show an increase in 5mC upon Tet1 deletion (Wu et al. 2011a), implicating Tet1 in maintaining CGI hypomethylation (Williams et al. 2012) either by blocking the DNA methylation machinery or by promoting DNA demethylation. There is also evidence for specific enrichment of 5hmC at ‘bivalent’ gene promoters marked with H3K4me3 and H3K27me3 histone modifications, associated with transcriptionally active and silenced genes respectively (Gu et al. 2011; Pastor et al. 2011; Williams et al. 2011; Wu et al. 2011a). This dual modification is thought to mark genes that are poised for activation in response to an external signal (Bernstein et al. 2006). Finally, 5hmC is located at many intergenic cis-regulatory elements such as active enhancers, pluripotent transcription factor-binding sites, and insulator-binding sites (Ficz et al. 2011; Pastor et al. 2011; Wu et al. 2011a; Yu et al. 2012); indeed low-methylated regions (LMRs), which correspond generally to gene-distal regulatory elements, show enrichment for TET1 binding and for 5hmC (Stadler et al. 2011).
The presence of 5hmC in gene bodies correlates with gene expression in mouse and human ES cells (Ficz et al. 2011; Pastor et al. 2011; Stroud et al. 2011; Szulwach et al. 2011a; Wu et al. 2011a; Xu et al. 2011), neural cells (Mellen et al. 2012), during neurogenesis, where the gain of 5hmC is accompanied by loss of H3K27me3 at promoters and gene bodies (Hahn et al. 2013) and in diverse other cell types and tissues including thymocytes and peripheral T cells (our unpublished data), mouse liver and human melanomas(Lian et al. 2012). Based on our studies in undifferentiated mouse ES cells (Koh et al. 2011; Huang Y 2014), we propose that Tet2 is primarily responsible for depositing gene body 5hmC. Briefly, the use of ES cells depleted of either Tet1 or Tet2 showed that Tet1 primarily regulated 5hmC levels at promoter/TSS regions whereas Tet2 primarily regulated 5hmC in the gene bodies of highly-expressed genes (Huang Y 2014). This difference may reflect the fact that Tet1 possesses a CXXC domain which would be expected to bind unmethylated CpGs and therefore would be targeted to CpG islands and high CpG promoters as is in fact observed (Wu and Zhang 2011; Williams et al. 2012); in contrast Tet2 would not be expected to be targeted preferentially to promoters since IDAX, the separated CXXC domain of Tet2, is not expressed in undifferentiated ES cells (Ko et al. 2013) (Fig. 1B). The ability of Tet2 to control 5hmC deposition at gene bodies may reflect its reported association with the SET1/COMPASS complex (Deplus et al. 2013), which travels with RNA polymerase II (Shilatifard 2012). Overall, Tet1 and Tet2 have distinct as well as redundant functions in mouse ES cells; similarly, Tet2 and Tet3 may have some overlapping as well as distinct functions in differentiated cell types including T cells and haematopoietic precursor cells (our unpublished data).
DNA methylation, TET proteins and 5hmC in T cells
It is well established that gene expression in T cells, as in many other cell types, is influenced by DNA methylation. (i) Genes such as Cd4 are sequentially activated and repressed during different stages of T cell development, in a manner thought to be epigenetically controlled (Taniuchi et al. 2002). (ii) The upregulation of Lck gene transcription from DN1 to DN3 stage of thymic development is correlated with progressively demethylation of CpGs in Lck exon 1 through intron 2 (Ji et al. 2010). (iii) Naïve CD4 T cells exhibit an impressive plasticity and readily differentiate to Th1, Th2 and Th17 cells depending on the cytokine milieu and the environmental cues (inflammatory stimuli, microorganisms) that they receive (Ansel et al. 2003; Zhou et al. 2009; Mukasa et al. 2010). Demethylation within the transcribed sequences of Il4 and Ifng correlates with high expression levels of these cytokines in TH2 and TH1 cells respectively (Ansel et al. 2006). (iv) A small region in the promoter-enhancer of the interleukin-2 (Il2) gene is demethylated in T cells following activation (Bruniquel and Schwartz 2003), in response to OCT1 binding. Demethylation occurs closely after transcriptional activation, suggesting that it may involve an active process independent of DNA replication (Bruniquel and Schwartz 2003), (Murayama et al. 2006). Il2 promoter demethylation stabilizes OCT1 binding and ensures that secondary activation in ensuing cell progeny is more rapid and more intense (Murayama et al. 2006). (v) Naïve T cells that lack expression of the DNA methyltransferase Dnmt1 upregulate sets of cytokines that are normally silenced and methylated (Lee et al. 2001), whereas Dnmt1-null CD8 T cells aberrantly upregulate CD4 T cell cytokines (Makar and Wilson 2004). (vi)The methylation status of conserved non-coding sequence 2 (CNS2) in the first intron of the Foxp3 gene influences the stability of Foxp3 expression in the regulatory T cell (Treg) lineage. The transcription factor RUNX1 and its obligate partner, core binding factor beta (CBFβ), bind to this locus in conjunction with rapid demethylation, providing a window for Foxp3 binding; this in turn stabilizes lineage progression over ensuing cell divisions in an autoregulatory loop (Zheng et al. 2010). The conclusion is that localized loss of DNA methylation at promoter/enhancer regions is associated not only with transcription factor binding but also with stabilization of DNA-protein interactions to ensure robust expression of target genes after their activation.
We previously showed that all three Tet proteins are expressed in CD4+ CD8+ double positive (DP) thymocytes as well as CD4 single positive (SP) and CD8 SP thymocytes (Ko et al, 2010). 5hmC levels in DP cells are lower than in CD4 and CD8 SP cells (0.7 compared to 2.5–3 pmol 5hmC/μg DNA respectively); Th1 and Th2 cells derived from naïve CD4 T cells, and effector cytolytic T cells derived from naïve CD8 T cells, have even lower levels of 5hmC (unpublished data). Global or conditional deletion of individual Tet proteins in T cells (using Tet2−/− mice and Tet3fl/fl CD4Cre mice) resulted in relatively normal T cell development and function, but T cells that lacked Tet3 showed compensatory upregulation of Tet2 mRNA (unpublished data). Thus for a full understanding of T function in T cells, it will be necessary to analyse the phenotypes of Tet2−/− Tet3 flx/flx CD4cre mice, which lack both Tet2 and Tet3 in T cells.
Methods for detection and mapping of 5mC and oxi-mC
Several methods have been developed to map modified cytosine species in the genome (summarized in table 1). They can be roughly divided into affinity-based approaches for enriching genomic regions that contain the modified cytosines, and methods for mapping the modified bases at single-nucleotide resolution (Song et al. 2012; Pastor et al. 2013; Wu and Zhang 2014). We describe some of the most useful methods here; commercial kits have been developed for hME-Seal, TAB-seq and oxBS.
Table 1.
Summary of the available methods used to detect 5mC and oxi-mCs
| Base | Affinity based Approach | Single nucleotide resolution |
|---|---|---|
| 5mC | MeDIP | WGBS, RRBS |
| 5hmC | CMS-IP, GLIB, 5hmC-IP, hMe-Seal, JBP1 IP | TAB-seq, oxBS-seq, oxRRBS |
| 5fC | 5fC-IP, 5fC-pulldown, 5fC-Seal | fCAB-seq |
| 5caC | 5caC IP | caCAB-seq |
Abbreviations: MeDIP: methylated DNA immunoprecipitation (IP); WGBS: whole-genome bisulfite sequencing; RRBS: reduced-representation bisulfite sequencing; CMS: cytosine methylenesulfonate; GLIB: glucosylation, periodate oxidation, biotinylation; hMe-Seal: 5-hmC selective chemical labeling; TAB-seq: TET assisted bisulfite sequencing; oxBS-seq: Oxidative bisulfite sequencing; 5fC-Seal: 5-fC selective chemical labeling; CAB-seq: Chemically assisted bisulfite sequencing
A. Affinity approaches
1. Immunoprecipitation of modified cytosines
The development of specific antibodies for each of the known modified cytosines – 5mC, 5hmC, 5caC and 5fC – enabled the immunoprecipitation and sequencing of genomic regions enriched for these modifications (Ficz et al. 2011; Williams et al. 2011; Wu et al. 2011a; Shen et al. 2013). The methylated DNA-IP (MeDIP) approach allows enrichment of methylated DNA by using an antibody that specifically recognizes methylated cytosines, whether or not they are in a CpG context (Weber et al. 2005). The DNA is denatured in order to convert it to a single-stranded form, thus exposing the 5mCs to permit efficient immunoprecipitation of genomic fragments enriched for 5mC (Weber et al. 2005). In all these cases the efficiency of immunoprecipitation is markedly density-dependent, and the methods described below are preferred for 5hmC mapping (Pastor et al. 2013).
2. Biotin modification of 5hmC followed by streptavidin pulldown
The enzyme β-glucosyl-transferase (BGT) from phage T4 very efficiently attaches glucose to the hydroxyl group of 5hmC, after which the glucose can be modified by oxidation with sodium periodate followed by treatment with an aldehyde-reactive probe in a method termed GLIB (glucosylation, periodate oxidation, biotinylation) (Pastor et al. 2011; Pastor et al. 2012). A similar method that uses click chemistry to attach the biotin has been developed (5hmC-selective chemical labeling, hMe-Seal (Song et al. 2011) (Song et al. 2012).
3. Immunoprecipitation of the 5hmC derivative CMS (CMS-IP)
This method takes advantage of the fact that treatment of DNA with sodium bisulfite efficiently converts 5hmC in single-stranded DNA to a stable and highly antigenic derivative, cytosine 5-methylenesulfonate (CMS)(Ko et al. 2010). Antibodies to CMS can thus be used to immunoprecipitate and sequence 5hmC containing fragments of genomic DNA with high specificity and low background; the method is much less density-dependent than immunoprecipitation (IP) with antibodies to 5hmC itself (Pastor et al. 2011; Huang et al. 2012). Numerous studies have used this method to map 5hmC efficiently in different systems (Pastor et al. 2011; Jeong et al. 2013; Lister et al. 2013; Vincent et al. 2013; Huang Y 2014).
B. Single nucleotide resolution approaches
Most single base-resolution methods used to map oxi-mC species involve a step of treatment of the DNA with sodium bisulfite. Bisulfite treatment has long been used to distinguish unmodified cytosine from 5mC, since it results in the deamination of unmodified cytosine to uracil, which is subsequently read as thymine after PCR amplification. In contrast methylated cytosines (and CMS) are resistant to bisulfite conversion and are read as cytosines, enabling quantitative discrimination between unmodified cytosines and 5mC (Frommer et al. 1992). It is now recognized, however, that bisulfite treatment cannot distinguish 5mC from 5hmC, or C from 5fC and 5caC (Fig. 4A,table 2).
Figure 4.
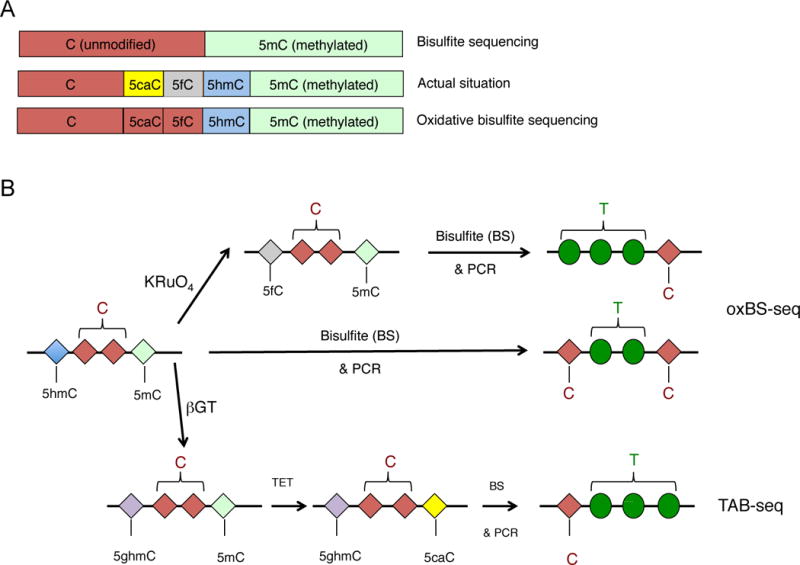
A. Top, Bisulfite sequencing (BS-seq) cannot discriminate methylated C from 5hmC, since both are converted after bisulfite treatment and PCR amplification to C. Middle, The actual state of C in the genome can be unmethylated C, 5caC, 5fC, 5hmC or 5mC. Bottom, The advent of oxidative bisulfite (oxBS) sequencing, which introduces chemical treatment of DNA prior to the bisulfite treatment, allows discrimination of 5hmC from 5mC, however the unmodified C is still not distinguished from 5cac or 5fC. B. Schematic representation of oxBS-seq and TAB-seq, two of the most useful methods that enable detection of 5hmC at single nucleotide resolution when combined with bisulfite sequencing. In the oxBS approach (top), DNA is treated with potassium perruthenate (KRuO4) that oxidizes 5hmC to 5fC. Subsequent bisulfite treatment converts 5fC (as well as 5caC and C) to U that will be read as T after PCR amplification, so only 5mC stays unconverted and is thus read as C after PCR amplification. In contrast in DNA samples that have been subjected exclusively to bisulfite treatment, both 5hmC and 5mC are read as C after PCR amplification, so 5mC, 5hmC and “unmodified” C can be discriminated by subtracting the results of BS-seq from those of oxBS. In TAB-seq (TET-assisted bisulfite sequencing), the DNA is treated with T2 phage beta-glucosyltransferase (βGT) that specifically recognizes and glycosylates 5hmC, generating glucosylated 5hmC (5ghmC). Subsequent treatment with purified and active TET1 leads to oxidization of 5mC to 5caC, and finally treatment of the sample with bisulfite results in complete conversion of all the bases to U, read as T after PCR amplification, with the exception of 5ghmC that stays unconverted and thus is read as C
Table 2.
Behavior of 5mC and its oxi-mCs in bisulfite conversion and the following PCR step
| Base | Bisulfite conversion | BS Sequencing result |
|---|---|---|
| 5mC | 5mC | C |
| 5hmC | CMS | C |
| 5gmC | 5gmC | C |
| 5fC | U | T |
| 5caC | U | T |
| C | U | T |
Abbreviations: CMS: cytosine methylenesulfonate; 5gmC: β-glucosyl-5-hydroxymethylcytosine
1. TAB-seq
TET-assisted BS-seq has been used to map 5hmC quantitatively at single-base resolution (Yu et al. 2012). In this method, treatment with βGT blocks 5hmC, and subsequently, recombinant mouse TET1 is used to convert 5mC to oxi-mC – ideally, all the way to 5caC (Figure 4B). 5caC (originally 5mC) and unmodified C are both converted by bisulfite treatment to uracil; thus the only base that is read as cytosine is the glucosylated 5hmC (Figure 4B). The main caveat of this approach is that highly active TET enzyme is required for efficient conversion of 5mC to 5caC: if the conversion is inefficient such that there is no conversion of the 5mC or the conversion does not proceed beyond 5hmC, there will be false-positive base calling of 5hmC at sites that were actually 5mC. Bisulfite treatment can also be inefficient: even with a high efficiency of 5mC oxidation by TET1 at sites with 80–100% 5mC, if only 95% of the unmodified C is converted to uracil, the remaining 5% of unconverted 5mCs will be falsely identified as 5hmC (Yu et al. 2012).
2. Oxidative Bisulfite (OxBs) sequencing
This is a chemical conversion method that also allows the quantitative mapping of 5hmC in genomic DNA at single-nucleotide resolution. Potassium perruthenate selectively oxidizes 5hmC to 5fC, and subsequent bisulfite treatment of the DNA converts 5fC to U (Figure 4B). By comparing the data from oxidized and bisulfite-treated DNA to those from only bisulfite-treated DNA, it is possible to distinguish 5mC, 5hmC and unmodified C from one another. However, bases that are read as T can be derived from unmodified C, 5fC or 5caC (Booth et al. 2012). The method is more cost-effective than TAB-seq, however one needs to be extremely cautious since the method is very sensitive to the purity of the isolated genomic DNA (Booth et al. 2013) and also tends to damage DNA, resulting in biases due to overamplification.
3. CMS-IP
An advantage of the CMS-IP method is that the modification status of cytosines in the IP’d DNA fragments can be estimated by determining whether they are read as T (i.e were originally C, 5fC or 5caC) or remain as C (in which case they were originally 5mC or 5hmC, converted to CMS). Moreover the input DNA can be analysed by BS-seq to yield the methylation status of all cytosines in the genome.
C. The effect of sequencing depth on mapping performance
Because of the relatively low levels of 5hmC in the genome, high coverage is needed to sequence this base quantitatively. Furthermore, for the oxBS and TAB-seq single-base resolution methods, a subtraction is needed in order to obtain the actual levels of 5hmC. As a result, there is an increase in the “noise level”, thus further increasing the need to increase both coverage and the number of biological replicates.
In our experience and at current sequencing costs, a range of ~30 million reads per sample for MeDIP-seq, CMS-IP and hMe-Seal constitute a viable compromise between breadth and depth of sequencing. In contrast, whole-genome bisulfite sequencing and oxBS provide comprehensive genomic coverage at the cost of having to sequence over a billion reads per sample, which increases the cost of the experiment tremendously. An alternative approach would be to combine TAB-seq and oxBS-seq with Reduced representation bisulfite sequencing (RRBS) which allows selective, but deep, sequencing of a fraction of the genome that is highly enriched for CpG islands (Meissner et al. 2008); in this case only ~25 million 40-bp short reads are required to achieve an average sequencing depth of ~120× by combining oxBS-Seq with RRBS (oxRRBS) (Booth et al. 2012), making this method cost-efficient and suitable for some applications although it captures only a fraction of 5mC and thus 5hmC sites in the genome (less than 10% of the 28 million CpGs (Bock et al. 2010) and ~11.5% of the dynamically active CpGs that overlap with differentially methylated and hydroxymethylated regions eg during development or during different time points of differentiation/treatment with different substances (Ziller et al. 2013).
Overall, in our opinion, an approach that is cost-efficient without compromising the quality of the data and thus the confidence of the conclusions deduced, would be to combine affinity-based approaches such as CMS-IP to assess 5mC and oxi-mC at a genome-wide level, then apply single-nucleotide resolution approaches to specific loci of interest.
Conclusions
The oxi-mC species 5hmC, 5fC and 5caC are now established as important intermediates in DNA demethylation, as well as potential epigenetic marks that regulate chromatin structure by recruiting interacting proteins. Despite extensive research in numerous biological systems, the physiological functions of these modifications remain elusive. For instance, what targets TET proteins to specific regions of DNA? Is the correlation of high gene-body 5hmC with high gene expression merely a byproduct of gene transcription or does the presence of 5hmC actually facilitate transcription? The large datasets that have been generated provide stimulating ideas but are far from providing conclusive information. The advent of new sequencing approaches, the increase in depth of sequencing coverage, the study of more biological systems in combination with detailed experimental strategies for monitoring gene expression, the use of incisive mouse models, and a strong focus on individual genes will allow us to gain a better understanding of the biological functions of TET proteins and the oxi-mC modifications that they generate, in the context of physiological development as well as disease.
Acknowledgments
We apologize to those whose work could not be cited due to space limitations. We thank Drs. Will Pastor and Elizabeth Thompson for the Northern analysis shown in Figure 2, and Drs. Tarmo Äijö, Nancy Huang and Lukas Chavez for fruitful discussions. The work was supported by NIH grants AI44432 and a grant from the California Institute of Regenerative Medicine (RM1-01729) to A.R. A.T. is the recipient of an Irvington Postdoctoral Fellowship from the Cancer Research Institute.
References
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annual review of immunology. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nature immunology. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. Journal of molecular biology. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nature biotechnology. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nature protocols. 2013;8:1841–1851. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nature immunology. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013 doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PloS one. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell reports. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nature genetics. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Huang YCL, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, Rao A. Proceedings of the National Academy of Sciences of the United States of America. 2014. Distinct roles of Tet1 and Tet2 in mouse embryonic stem cells. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nature genetics. 2013 doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends in biochemical sciences. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nature genetics. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome research. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nature chemical biology. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends in biochemical sciences. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. Journal of immunology. 2004;173:4402–4406. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. The EMBO journal. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer research. 2002;62:4075–4080. [PubMed] [Google Scholar]
- Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. Journal of cell science. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Huang Y, Henderson HR, Agarwal S, Rao A. The GLIB technique for genome-wide mapping of 5-hydroxymethylcytosine. Nature protocols. 2012;7:1909–1917. doi: 10.1038/nprot.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics & chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature biotechnology. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nature biotechnology. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome biology. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS genetics. 2011a;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011b;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer research. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, et al. Stage-Specific Roles for Tet1 and Tet2 in DNA Demethylation in Primordial Germ Cells. Cell stem cell. 2013 doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO reports. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes & development. 2011a;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011b;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells. Cell cycle. 2011;10:2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]


