Abstract
Neurotrophins regulate diverse aspects of neuronal development and plasticity, but their precise in vivo functions during central brain neural circuit assembly remain unclear. We show that the neurotrophin receptor tropomyosin-related kinase C (TrkC) is required for dendritic growth and branching of mouse cerebellar Purkinje cells. Sparse TrkC knockout reduced dendrite complexity, but global Purkinje cell knockout had no effect. Removal of the TrkC ligand Neurotrophin-3 (NT-3) from cerebellar granule cells, which provide major afferent input to developing Purkinje cell dendrites, rescued the dendrite defects caused by sparse TrkC disruption in Purkinje cells. Our data demonstrate that NT-3 from presynaptic neurons—granule cells—is required for TrkC-dependent competitive dendrite morphogenesis in postsynaptic neurons—Purkinje cells—a novel mechanism of neural circuit development.
Main Text
Neurotrophins regulate the survival, differentiation, and plasticity of peripheral and central neurons (1–3). The mammalian neurotrophin family signals through three tropomyosin-related kinase (Trk) receptors as well as the p75 neurotrophin receptor (p75NTR). While brain-derived neurotrophic factor (BDNF) has been intensely studied, much less is known about Neurotrophin-3 (NT-3) and its receptor TrkC, despite their widespread expression in the developing and adult brain (4, 5). The lack of central brain cell death in mice lacking NT-3 and TrkC contrasts starkly with the severe reductions in sensory and sympathetic neurons, and suggests survival-independent functions (6, 7). NT-3 functions in dendrite morphogenesis in brain slice culture and in proprioceptive axon patterning (8–10). However, evaluating the roles of NT-3/TrkC signaling in the central brain in vivo has been hindered by the early postnatal lethality of NT-3 or TrkC knockout mice and the limited cellular resolution of phenotypic analyses (7, 11).
To study the cell-autonomous function of TrkC in mouse neural development, we employed Mosaic Analysis with Double Markers (MADM) (12, 13) with a null allele of trkC that removes all isoforms of the receptor (14), and a pan-neural Nestin-Cre line (15) to drive recombination throughout the brain. Thus, in an otherwise heterozygous animal (trkC+/−), Cre/loxP-mediated mitotic recombination between homologous chromosomes sparsely labels wild-type (trkC+/+) and homozygous mutant (trkC−/−) cells in distinct colors (fig. S1A, S2). In these animals, sparse trkC−/− cells were labeled with GFP (green), trkC+/+ cells with tdTomato (red), and trkC+/− cells with both GFP and tdTomato (yellow). Cells without recombination, which remained colorless, were all trkC+−. Although survival of central brain cells was not apparently affected by sparse trkC removal (fig. S2, S3A), consistent with previous observations (6, 7), we observed a striking Purkinje cell dendrite phenotype (Fig. 1).
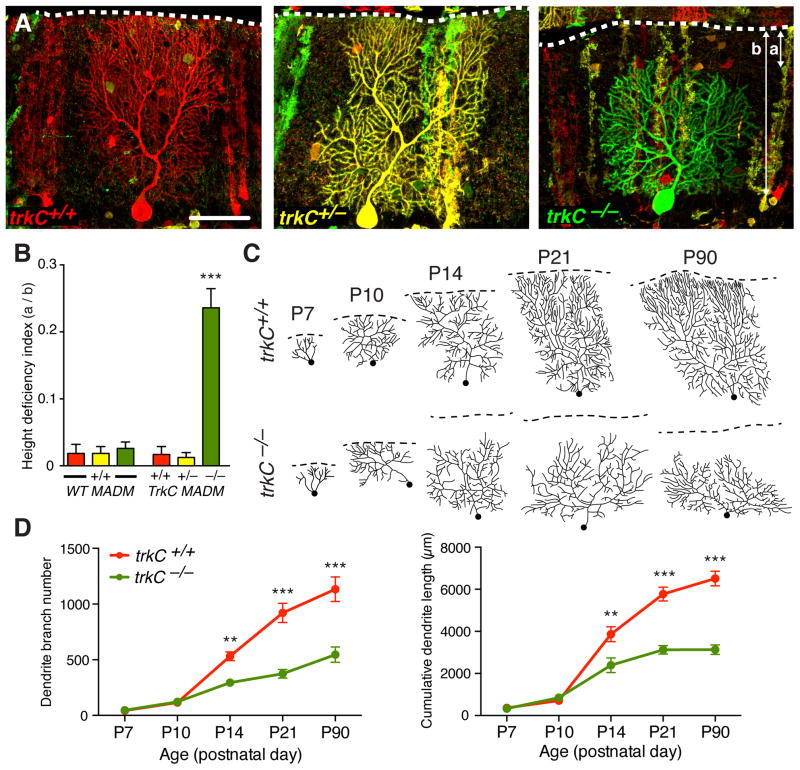
Fig. 1. TrkC is cell-autonomously required for Purkine cell dendrite arborization.
(A) Postnatal day 21 (P21) red trkC+/+ and yellow trkC+/− Purkinje cell dendritic arbors span the entire molecular layer of the cerebellar cortex. In contrast, trkC−/− Purkinje cell dendritic arbors are shorter and fail to reach the pial surface (dashed white lines). Short (a) and long (b) arrows indicate distance from pial surface to top of arbor and molecular layer span, respectively. Scale bar, 50μm. (B) Quantifications of height deficiency index (a/b) for WT MADM and trkC MADM. Here and in subsequent figures, all values are means ± SEM. *** p<0.001, one-way ANOVA with Tukey’s multiple comparisons test. (C) Traces of Purkinje cell dendritic arbors between P7 and P90. Dashed lines indicate the external granule layer margin in P7-P10, or the pial surface in P14–90. Dots indicate primary branch start point. (D) Quantification of dendrite branch number (left) and cumulative dendrite length (right). Red, trkC+/+; green, trkC−/−. **p<0.01, ***p<0.001, two-way ANOVA with Bonferroni’s multiple comparisons test. See Table S1 for additional information including N for each experiment.
Both trkC+/+ and trkC+/− Purkinje cells extended complex dendritic arbors that spanned the entire molecular layer of the cerebellum (Fig. 1A, left and middle). In contrast, arbors from trkC−/− cells failed to reach the pial surface (65/72 cells; Fig. 1A, right). These stunted arbors exhibited normal dendritic spine density (fig. S3B), but spanned only ~75% of the molecular layer (Fig. 1B). To determine when the trkC−/− dendrite phenotype emerges, we compared trkC+/+ and trkC−/− cells between postnatal days 7 and 21, when Purkinje cells normally elaborate their dendrites and begin to form synapses (Fig. 1C; fig. S4). While indistinguishable from control cells at postnatal day 7 and 10, trkC−/− cells exhibited reduced dendrite arbor height, branch number, and total dendritic length compared to trkC+/+ cells by day 14 (Fig. 1C–D; fig. S4). Growth and branching phenotypes persisted in 3-month old animals (Fig. 1C–D; fig. S4), indicating that loss of TrkC caused a morphogenesis defect rather than a developmental delay. Furthermore, the most distal dendrite branches were preferentially lost in trkC−/− cells (fig. S3C). Uniparental disomies (13; fig. S5) and fluorescent markers (fig. S6) did not affect Purkinje cell dendrite phenotypes. Thus, our mosaic analysis suggested that TrkC is required cell-autonomously for proper Purkinje cell dendrite growth and branching.
Although Trk signaling normally requires the tyrosine kinase domain (16), TrkC also has a kinase-independent role in synaptogenesis (17). To test whether dendrite arborization relies on kinase activity, we examined MADM mice harboring a conditional allele in which loxP sites flank an exon encoding part of the TrkC kinase domain (18). Here, Nestin-Cre mediated inter-chromosomal recombination within the MADM cassettes, and also excised this exon to generate a “kinase-dead” allele (trkCKD; fig. S1B). At P21, Purkinje cells homozygous for this allele (trkCKD/KD) exhibited dendrite height, branch number, and total dendritic length phenotypes comparable to those of trkC−/− cells (Fig. 2). Thus, proper dendrite arborization requires TrkC kinase activity.
Fig. 2. Dendrite arborization requires TrkC kinase activity.
(A) Analogous to trkC−/− arbors (Fig. 1), trkCKD/KD arbors are shorter than those of trkC+/+ and trkCKD/+ cells at P21, and fail to reach the pial surface. (B) Quantifications of height deficiency index (*** p<0.001, one-way ANOVA with Tukey’s multiple comparisons test), branch number, and cumulative dendrite length (** p<0.01, *** p<0.001, unpaired t-test) as in Fig. 1. Also see Table S1.
Since perturbing TrkC in a sparse population (0.5–1%) of Purkinje cells disrupted their dendrite arbors, we examined how TrkC ablation from all Purkinje cells affects dendrite morphology. Calbindin immunostaining labels Purkinje cells and their dendrites, which normally span the entire molecular layer of the cerebellum (Fig. 3A). trkC conditional knockout using Purkinje-cell specific pcp2-Cre (19) (fig. S1C) did not reduce Purkinje cell dendrite height compared to controls (Fig. 3A–B). To analyze dendrite branching in more detail, we used MADM cassettes to sparsely label Purkinje cells in the Purkinje-cell specific trkC conditional knockout background. In this context, pcp2-Cre removed trkC from all Purkinje cells, but GFP and tdTomato were reconstituted only in a sparse subset (5–10%) of Purkinje cells through inter-chromosomal recombination (fig. S1D). Such individually labeled trkCKD/− Purkinje cells exhibited normal dendrite height, branching, and length (Fig. 3C–D). Thus, in contrast to sparse MADM-based knockout of TrkC, TrkC ablation from all Purkinje cells did not disrupt dendrite arborization.
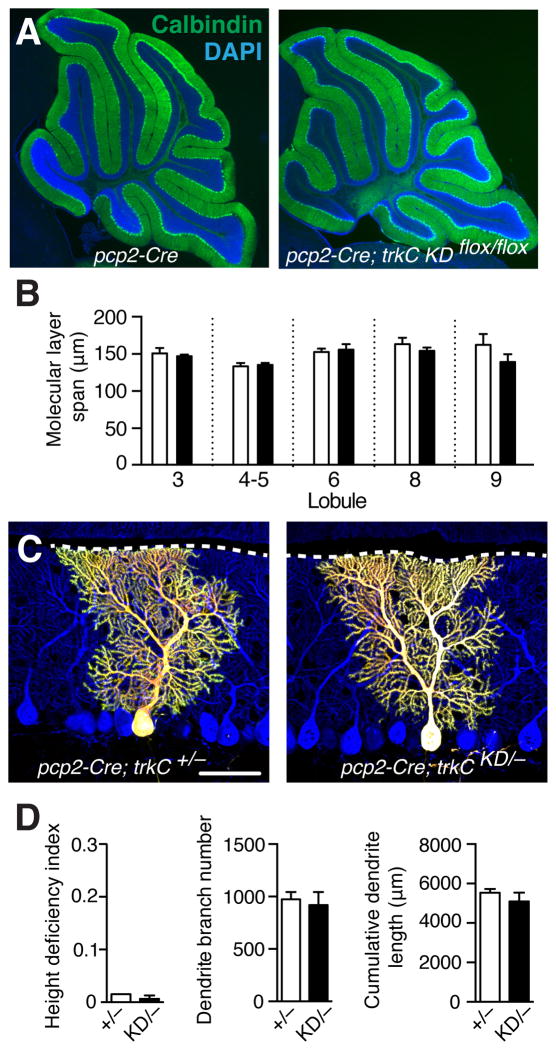
Fig. 3. Normal dendrite morphogenesis when all Purkinje cells lack TrkC.
(A) Calbindin staining labels Purkinje cells and their dendrites (green). Removing trkC from all Purkinje cells (right panel) does not cause global reductions in dendrite height relative to controls (left panel) at P21. (B) Molecular layer thickness across multiple lobules were similar for control pcp2-Cre+/− (white) and pcp2-Cre+/−;trkC KDflox/flox (black) mice. No significant differences, two-way ANOVA with Tukey’s multiple comparisons test. (C) trkCKD/− Purkinje cells do not exhibit dendrite height or branching defects relative to trkC+/− cells. (D) Quantifications of height deficiency index, branch number and cumulative length as in Fig. 1 (no significant differences, unpaired t-test). Also see Table S1.
Differences in the timing of trkC removal are unlikely to account for the different outcomes of global and sparse knockout (Fig. 1–3). pcp2-Cre mediated recombination in nearly all Purkinje cells by P7, and markedly reduced trkC mRNA levels in Purkinje cells by P10 (fig. S7), well before dendrite phenotypes emerge at P14. A more likely interpretation is that the observed dendrite defects depend on sparseness of trkC deletion. This raised the possibility of a competitive mechanism (20), in which dendrite morphogenesis depends on relative differences in TrkC signaling between neighboring Purkinje cells.
We next investigated the expression pattern of NT-3, the ligand for TrkC (16), using a lacZ knock-in reporter in the NT-3 locus (21). The lacZ product β-Gal was transiently expressed at P7 (but not at P14) in a small subset of inferior olive neurons, which extend climbing fibers to Purkinje cell dendrites (Fig. 4A, left). β-Gal was also expressed in cerebellar granule cells, which send parallel fibers to provide major inputs to Purkinje cell dendrites, but was undetectable in the external granular layer, which contains granule cell progenitors (Fig. 4A, middle and right). This suggests that postmitotic granule cells express NT-3 after migrating to the internal granular layer. Although restricted to granule cells in posterior folia at P7 (Fig. 4A, middle), β-Gal was expressed in all folia by P10 (Fig. 4A, right), coinciding with the time window of TrkC-dependent dendritic development (Fig. 1C–D). Purkinje cell dendrite morphogenesis likely relies on NT-3 produced around or after P10, as dendritic phenotypes of sparse trkC−/− Purkinje cells were equally severe in anterior and posterior folia (fig. S8). Given that β-Gal was undetectable in the deep cerebellar nuclei, the postsynaptic targets of Purkinje cells (Fig. 4A), presynaptic granule cells are the best candidate cellular source of NT-3.
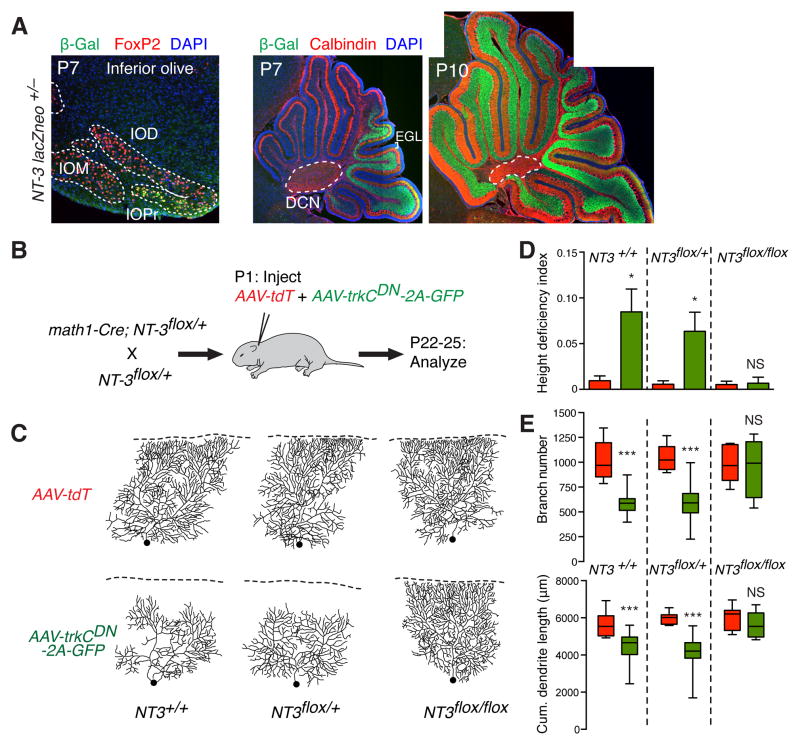
Fig. 4. Testing NT-3/TrkC-dependent competition.
(A) β-Gal staining reports NT-3 expression patterns in a NT-3lacZneo/+ knock-in mouse (green).
Left: coronal section of the mouse medulla at postnatal day 7. FoxP2 staining labels inferior olive neurons (red). β-Gal is present in a subset of neurons in the inferior olive principal nucleus
(IOPr), but not in the medial or dorsal nuclei (IOM, IOD). Middle and right: At P7, β-Gal is enriched in granule cells of posterior folia. By P10, β-Gal is present in all folia. No signal was detectable in the external granular layer (EGL, bracket), which contains granule cell precursors. Likewise, no signal was detectable in Purkinje cells or the deep cerebellar nuclei (DCN, dashed line), the postsynaptic targets of Purkinje cells. (B) Schematic of a viral approach to test relative NT-3/TrkC signaling in dendrite morphogenesis. Viruses encoding either tdTomato (AAV-tdT) or a dominant negative TrkC together with GFP (AAV-trkCDN-2A-GFP) were co-injected into math1-Cre;NT-3flox/flox, math1-Cre;NT-3flox/+, or control math1-Cre mice at P1. tdT or GFP/TrkCDN-expressing Purkinje cells were analyzed 21–24 days post-injection. (C) Dendrite arbors of tdT or GFP/TrkCDN-expressing Purkinje cells. In math1-Cre;NT-3+/+ or math1-Cre;NT-3flox/+ mice, TrkCDN-expressing cells exhibited decreased arbor complexity much like MADM trkC−/− cells (Fig. 1). However, in math1-Cre;NT-3flox/flox animals, dendrite defects were rescued to WT levels. (D) Quantification of height deficiency index. * p<0.05, one-way ANOVA with Tukey’s multiple comparisons test. (E) Quantification of tdT and GFP/TrkCDN dendritic branch number (top) and cumulative length (bottom). Boxes indicate the mean (middle line) and 25–75% range, while whiskers indicate maximum and minimum values. ***p<0.001, two-way ANOVA with Sidak’s multiple comparisons test. Also see Table S1.
Conditional knockout of NT-3 from granule cell progenitors using math1-Cre (22) did not affect Purkinje cell dendrite height (fig. S9), branch number, or total length (Fig. 4B–E). This is consistent with the absence of dendrite phenotypes in Purkinje cell-specific trkC knockout (Fig. 3). We next devised a method to investigate NT-3/TrkC signaling in a competitive context (Fig. 4B). Adeno-associated virus serotype 8 (AAV8) preferentially transduces Purkinje cells when injected into neonatal mice (23). We exploited this tropism to co-transduce two AAV vectors, the first expressing tdTomato as a control, and the second expressing a dominant negative TrkC construct (fig. S10) together with GFP (TrkCDN-2A-GFP). We achieved sparse labeling (~0.5%) by controlling the titer and volume of neonatal injections (fig. S11). In P21 mice, TrkCDN-expressing GFP-positive Purkinje cells exhibited reduced dendrite height, branch number and total length relative to control, tdTomato-expressing Purkinje cells (Fig. 4C–E, left columns). This viral approach thus corroborated results from MADM-based sparse knockout.
To test sparse TrkC loss-of-function in the absence of NT-3, we neonatally transduced AAV viruses into conditional knockout mice in which math1-Cre removes NT-3 from all granule cells (Fig. 4B). In heterozygous (NT-3flox/+) conditional knockout animals, TrkCDN-expressing GFP-positive Purkinje cells still exhibited reduced branching number and length compared to tdTomato-positive control cells (Fig. 4C–E, middle columns). However, in homozygous (NT-3flox/flox) conditional knockout animals, the dendrite phenotypes of TrkCDN-expressing cells were completely suppressed (Fig. 4C–E, right columns). Thus, granule cell-derived NT-3 is necessary for TrkC-dependent competitive dendrite morphogenesis in Purkinje cells.
In summary, while Purkinje cell dendrite development can proceed in the absence of NT-3 or TrkC, our data indicate that relative intercellular differences in NT-3/TrkC signaling can profoundly modulate dendrite morphogenesis. Specifically, Purkinje cells with lower TrkC levels relative to their neighbors exhibit reduced dendritic arbor complexity. Future studies must elucidate how Purkinje cells compare TrkC levels, and how such intercellular comparisons contribute to normal circuit function. For example, heterogeneous TrkC activation during development may diversify Purkinje cell dendrite complexity, or homeostatically adjust dendrite branching rates to ultimately equalize Purkinje cell participation in the cerebellar circuit. Furthermore, NT-3/TrkC may cooperate with other signals to regulate dendrite development. Although p75NTR likely acts as a receptor for paracrine signals during competitive axon stabilization and pruning (24, 25), we found that sparse trkC knockout phenotypes persisted in p75−/− mice (fig. S12), suggesting alternative mechanisms. Indeed, TrkC/NT-3 signaling may mediate competition by enhancing neuronal activity or synaptogenesis, both of which are known to modulate dendrite arborization (26, 27). In one scenario, differential TrkC/NT-3 signaling may drive competitive parallel fiber synapse formation and locally stabilize Purkinje cell dendritic branches. Consistent with this idea, each parallel fiber forms synapses with only a subset of Purkinje cells along its trajectory (28), while TrkC can mediate postsynaptic differentiation (17) and NT-3 can be anterogradely transported and released from central neuron presynaptic terminals (29).
According to the classic neurotrophic theory, developing axons compete for limiting amounts of neurotrophic factors from their target tissues, which signal retrogradely to support neuronal survival and axon growth (30, 31). Our findings expand this paradigm and suggest that growing dendrites analogously require anterograde NT-3 from their presynaptic partners during competitive dendrite growth.
Supplementary Material
Acknowledgments
We thank D. Ginty, L. Tessarollo, and D. Rowitch for mouse lines, the Stanford Gene Vector and Virus Core for AAV production, P. Mehlen and S. Tauszig-Delamasure for advice on cell culture experiments, B. Weissbourd for assistance with in situ hybridization, and T. Clandinin and members of the Luo Lab for helpful discussions and comments on the manuscript. W.J.J. was supported by the Stanford Neurosciences Graduate Program, the Lorraine Kendall Predoctoral Fellowship Fund, and an NIH NRSA Predoctoral Fellowship (5 F31 NS071697). This research was also supported by an NIH grant (R01-NS050835). L.L. is an investigator of the Howard Hughes Medical Institute. Supplement contains additional data.
References and Notes
- 1.Huang EJ, Reichardt LF. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park H, Poo M. Nature Rev Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- 3.Nikoletopoulou V, et al. Nature. 2010;467:59–63. doi: 10.1038/nature09336. [DOI] [PubMed] [Google Scholar]
- 4.Maisonpierre PC, et al. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- 5.Tessarollo L, et al. Development. 1993;118:463–475. doi: 10.1242/dev.118.2.463. [DOI] [PubMed] [Google Scholar]
- 6.Minichiello L, Klein R. Genes Dev. 1996;10:2849–2858. doi: 10.1101/gad.10.22.2849. [DOI] [PubMed] [Google Scholar]
- 7.Silos-Santiago I, Fagan AM, Garber M, Fritzsch B, Barbacid M. Eur J Neurosci. 1997;9:2045–2056. doi: 10.1111/j.1460-9568.1997.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 8.McAllister AK, Lo DC, Katz LC. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 9.McAllister AK, Katz LC, Lo DC. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 10.Patel TD, et al. Neuron. 2003;38:403–416. doi: 10.1016/s0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- 11.Ernfors P, Lee K, Kucera J, Jaenisch R. Cell. 1994;77:503–512. doi: 10.1016/0092-8674(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 12.Zong H, Espinosa JS, Suh HH, Muzumdar MD, Luo L. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Hippenmeyer S, Johnson RL, Luo L. Cell Rep. 2013;3:960–967. doi: 10.1016/j.celrep.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tessarollo L, et al. Proc Natl Acad Sci USA. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- 16.Huang EJ, Reichardt LF. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi H, et al. Neuron. 2011;69:287–303. doi: 10.1016/j.neuron.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, et al. Neuron. 2005;46:13–21. doi: 10.1016/j.neuron.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XM, et al. Genesis. 2004;40:45–51. doi: 10.1002/gene.20062. [DOI] [PubMed] [Google Scholar]
- 20.English CN, et al. PNAS. 2012;109:19456–19461. doi: 10.1073/pnas.1206492109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farinas I, Jones KR, Backus C, Wang XY, Reichardt LF. Nature. 369:658–661. doi: 10.1038/369658a0. [DOI] [PubMed] [Google Scholar]
- 22.Matei V, et al. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson, et al. Neuron. 2014;81:1040–1056. doi: 10.1016/j.neuron.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao L, et al. Curr Biol. 2007;17:911–921. doi: 10.1016/j.cub.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh KK, et al. Nature Neurosci. 2008;11:649–658. doi: 10.1038/nn.2114. [DOI] [PubMed] [Google Scholar]
- 26.Niell CM, Meyer MP, Smith SJ. Nat Neurosci. 2004;7:254–259. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- 27.Cline H, Haas K. J Physiol. 2008;586:1509–1517. doi: 10.1113/jphysiol.2007.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. Plos ONE. 2010;5:e11503. doi: 10.1371/journal.pone.0011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altar CA, DiStefano PS. Trends in Neurosci. 1998;21:433–437. doi: 10.1016/s0166-2236(98)01273-9. [DOI] [PubMed] [Google Scholar]
- 30.Levi-Montalcini R. Science. 237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 31.Zweifel LS, Kuruvilla R, Ginty D. Nature Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.