Abstract
New genes in human genomes have been found relevant in evolution and biology of humans. It was conservatively estimated that the human genome encodes more than 300 human-specific genes and 1,000 primate-specific genes. These new arrivals appear to be implicated in brain function and male reproduction. Surprisingly, increasing evidence indicates that they may also bring negative pleiotropic effects, while assuming various possible biological functions as sources of phenotypic novelties, suggesting a non-progressive route for functional evolution. Similar to these fixed new genes, polymorphic new genes were found to contribute to functional evolution within species, e.g. with respect to digestion or disease resistance, revealing that new genes can acquire new or diverged functions in its initial stage as prototypic genes. These progresses have provided new opportunity to explore the genetic basis of human biology and human evolutionary history in a new dimension.
Introduction
Evolutionarily new genes, referred to genes emerged in recent evolution [1], have attracted a broad interest, since the first mechanistic model was proposed in the 1930s [2]. Thanks to extensive studies of molecular evolution and genomic biology in the last decade, a dozen of distinct molecular mechanisms to generate new genes were found, including the most frequently investigated DNA- or RNA-based duplication mechanisms and a recent additional hot topic of de novo origination [1,3]. These mechanisms lead to pervasive new gene origination, which in turn participated in lineage- or species-specific phenotypic evolution [4]. For human biology, tremendous efforts have been dedicated to study human-specific genes absent in other primates or polymorphic genes within human species, which leads to a significant progress in understanding how often these new genes contributed to phenotypic evolution and how they are implicated in disease [5–7].
To discuss the progress in technical and conceptual investigation of new human genes, we provide here a concise and updated overview. We first focus on the rate and describe a few efforts in identifying primate-specific or even human-specific new genes encoded by the human genome. We describe the emerging themes in the functionality of these recently evolved genes and highlight their significance for brain and testis evolution. Then, we discuss a new hypothesis regarding the phenotypic evolution by new genes in the light of recent functional data indicating that new genes can promote tumorigenesis, while evolving toward advantageous functions. We further discuss the initial stage of new gene evolution when a new gene is polymorphic in a species population and discuss how these genes contribute to phenotypic difference between individuals or populations. We end the review with a summary of potentially important directions.
The human genome gains a high flux of new genes
The pioneering effort via cDNA array-based comparative genomic hybridization (aCGH) identified 134 genes showing copy number expansion after the split of human and great apes [8]. Further genomic analysis including three additional mammalian species identified 689 human-specific genes, i.e. the ones not shared by chimpanzees, and 870 hominoid genes shared by human and chimpanzee but absent in mouse and dog [9]. A third analysis of 18 vertebrate genomes detected 389 human-specific genes and 1,828 primate-specific genes [10]. Besides different identification strategies, the changing number could also result from ever-changing annotation. For new genes, this issue became more serious due to their poor conservation and narrow expression [11]. For example, out of 1,828 primate-specific genes, more than half of them were revised by later Ensembl updates as pseudogenes or noncoding transcripts, or unduly removed from the annotation [11]. In other words, the annotation database is getting more conservative when including entries of new gene. Such issue should be cautioned when studying new gene evolution.
The difficulty that the unstable and insufficient annotation brought to the study of new gene evolution was demonstrated by the contrasting number of human-specific de novo genes across different studies. Comparative analyses across multiple primate genomes in Ensembl v47 revealed three human-specific de novo genes supported by both transcription and proteomics data [12]. Pooling of multiple Ensembl versions (v40~v56) led to an exciting discovery of 60 human-specific de novo genes [13]. A third analysis based on Ensembl v51 pooled out 11 human-specific de novo genes [14]. All these efforts are similar technically: 1) to call proteins with the corresponding orthologous region in outgroups incapable of coding the open reading frame in the genomes of recent human ancestors; 2) to ensure that candidate de novo genes are supported by peptide databases. However, as pointed out in [15], the difficulty roots in the lability of human annotation of new genes and the arbitrariness of bioinformatic parameters. Nevertheless, combining complementary efforts on both duplicated new genes and de novo new genes, it seems prudent to conclude that substantial changes occurred in the human gene reservoir with about 300 human-specific genes and 1,000 primate-specific genes added.
Primate- or human-specific new genes are often implicated in brain development and male reproduction
Whether or not a new gene contributes a critical phenotypic effect in evolution is an interesting problem. As one of the early reported primate-specific gene families, morpheus was found to encode nuclear pore complex interacting protein (NPIP) with wide transcription in numerous tissues and organs [16] and its specific function has been known more for its activity involved in the HIV replication [17]. Recently, more cases of new genes were reported related to various molecular functions or phenotypic effects (more examples can be seen in Table 1). Quite a few cases appear to be related with brain functions such as the glutamate dehydrogenase 2 (GLUD2) [18,19] or neuroblastoma breakpoint (DUF1220) family [20,21]. A recently well characterized case in support of the significance of new gene emergence for human brain evolution is Slit-Robo Rho GTPase-activating protein 2 or SRGAP2C, which is a DNA-level duplicate originated around 2 million years ago [22]. As a partial copy, SRGAP2C inhibits the function of its parental gene SRGAP2A and induces neoteny during dendric spine maturation [23].
Table 1.
Examples of new genes evolved after the split of primate from other mammals. Human-specific new genes refer to those genes absent in the other primates including chimpanzee. Homininae-specific genes refer to those shared by human, chimpanzee and gorilla. Hominoid-specific genes refer to those evolved recently in the lineages of apes but absent in rhesus monkey and other primates. Primate-specific genes refer to those absent in non-primate mammals.
| Gene | Origination Mechanism | Age | Function | Citation |
|---|---|---|---|---|
| Brain-related new genes
| ||||
| GLUD2 | Retroposition | Hominoid-specific | Glutamate metabolism in brain | [18,19] |
| DUF1220 family | DNA-based duplication | Primate-specific | Transcribed in brain | [20,21] |
| SRGAP2C | DNA-based duplication | Human-specific | Dendric spine maturation | [22,23] |
|
| ||||
| Testis related
| ||||
| SPANXA/D | DNA-based duplication | Homininae-specific | Spermatid morphogenesis | [24] |
|
| ||||
| Cancer related
| ||||
| CT45A1 | DNA-based duplication | Primate-specific | Upregulate oncogenic and metastatic genes | [25] |
| TBC1D3 | DNA-based duplication | Hominoid-specific | Modulator of epidermal growth factor receptor signaling pathway | [26,27] |
| NCYM | De novo | Homininae-specific | Stabilize the oncogene MYCN | [28,29] |
| PBOV1 | De novo | Human-specific | Possibly repress tumorigenesis | [30] |
|
| ||||
| Other
| ||||
| morpheus family | DNA-based duplication | Primate-specific | Broadly transcribed | [16,17] |
The enriched recruitment of new genes into brain expression is not only detected by these case studies, but also strongly supported by genome-wide studies. Comparative transcriptome profiling across major organs revealed that the proportion of brain transcriptome contributed by primate-specific genes in human is significantly higher than that contributed by rodent-specific genes in mouse [31].
Analogously, transcriptome profiling of hominoid- and human- specific de novo genes also showed that these genes tend to be transcribed in brain and testis [13,14]. Primate-specific genes transcribed in brain is enriched for zinc finger (ZNF) genes [31], which appear to be mainly contributed by the Kruppel-type or KRAB family [32]. Interestingly, about 40% of primate-specific KRAB-ZNF genes are differentially transcribed between human and chimpanzee prefrontal cortex, which may lead to extensive gene expression difference between the two species [33]. Why brain acts like an evolutionary hotbed in recruiting new genes likely roots in the complexity of its molecular network. Before the genomic era, it was already known that the amount of RNAs transcribed in brain was two or three fold higher than that in tissues such as liver or kidney [34]. The updated transcriptome data via RNA-sequencing confirmed that there were more genes with highest expression in brain compared to liver and kidney [35]. Thus, the complex nature of the brain provides more interaction partners for a preexisting old gene of interest that might compromise its evolvability [36]. In contrast, a new gene has much less pleotropic constraint and it can be integrated into this network under positive natural selection [37].
However, it is important to note that the transcription of new genes is often not limited to brain. Actually, the out-of-testis hypothesis stated that new genes tend to gain their functionality initially in testis possibly due to its permissive transcriptional regulation and then extend expression into other tissues [38,39], which was supported by two separate genome-wide analyses showing that primate-specific genes, especially X-linked ones, are often predominantly or specifically transcribed in testis [10,38]. A recent case study provided the valuable insight into the biological role of testis-biased expressed new genes. A better-characterized case is SPANXA/D family, which consists of three X-linked sperm proteins associated with nucleus, SPANXA1, SPANXA2 and SPANXD present only in human and great ape [24]. It was found that during spermatid morphogenesis, the SPANXA/D protein migrated into the base of the sperm head [24].
New genes appear to promote tumorigenesis while evolving new advantageous functions, which supports a mode of non-progressive functional evolution
It was not expected that the newly evolved testis genes could be implicated in tumorigenesis until the SPANX actually as a Cancer/Testis (CT) antigen was found [40]. Despite the poorly characterized function, most CT genes display a characteristic transcription pattern only in testis and somatic cancer tissues possibly due to functional similarity between gametogenesis and tumorigenesis [41]. Although CTs were known for decades, recent evidence began to indicate that these proteins can facilitate tumorigenesis [42]. For example, primate-specific X-linked CT45A1 upregulated various oncogenic and metastatic genes in breast tumor [25] (Table 1). For some CTs, hypomethylation appears to be correlated with their misexpression in tumors, but whether or not copy number increase occurs is barely known [25].
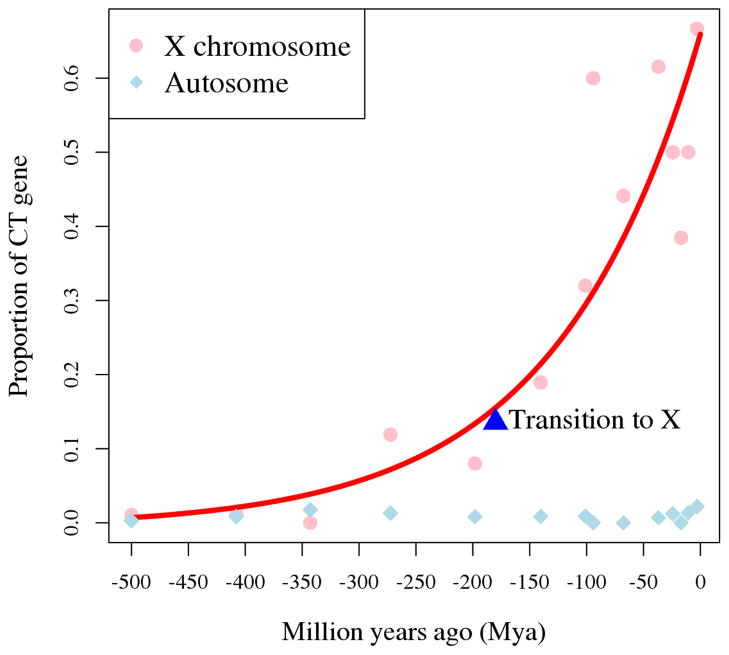
Like SPANX or CT45A1, CT genes overall tend to arise during or after the origin of placental mammals as found in recent comparative genomic studies [43]. Furthermore, there is a significant correlation between the age of X-linked CTs (or CT-Xs) and their proportion out of corresponding age groups (Figure 1): the proportion increases when CT-Xs become younger and about 50% of X-linked primate-specific genes are CTs. By contrast, the proportion of autosomal CTs remains almost constant across various periods. This pattern indicates that the previously reported increase of X-linked testis-biased genes in human [10] is largely contributed by CT-Xs.
Figure 1.
Distribution of new genes with CT expression with respect to their evolutionary ages. For the X chromosome, the proportion is defined as the number of CT-X genes divided by the number of all X-linked genes in the same age group. It was analogously defined for autosomes. The age class was computationally generated on Ensembl v69 by using the pipelines developed in [10] with the time information in TimeTree [44], while the CT gene list was downloaded from CTpedia database in July 2014 [40]. We fitted the observed frequencies of CT-Xs in various stages of human genome evolution to an exponential decay formula (f(t) ~ ert). We mark the time with a blue triangle when the mammalian X chromosome was originated, i.e., before the split of human and opossum [10,45].
More than that, the young ages of the CT genes show the intriguing connection between new gene origination and tumorigenesis. As a matter of fact, new genes could drive tumorigenesis even they are not categorized as CTs as shown in following cases. One of the earliest case is the hominoid-specific oncogene TBC1 domain family, member 3 (TBC1D3), which modulated epidermal growth factor receptor signaling pathway [26]. In breast cancer, TBC1D3 was found in recurrent amplicons which were associated with lower survival span [46]. This line of facts suggests that TBC1D3 situates in a genomic region which is prone to duplication. In other words, such a mutagenic nature not only increases the duplicability of TBC1D3 in evolution, but creates more copies in tumor and supports tumorigenesis. Compared to TBC1D3, NCYM emerged de novo in the ancestor of human and chimpanzee as an antisense transcript of the well characterized oncogene, MYCN [28]. Given such a topology, NCYM is always co-amplified with MYCN in neuroblastomas [28]. More than that, the NYCM protein stabilized MYCN, by repressing GSK3β, which promoted the degradation of MYCN [28]. Meanwhile, it was found that new genes could also repress tumorigenesis. For instance, prostate and breast cancer overexpressed gene 1 (PBOV1) evolved as a human specific de novo gene, whose expression is associated positively with the survival possibility of patients [30].
Different from new genes categorized as CTs (e.g. CT45A1), TBC1D3 is widely transcribed [27] and NCYM is expressed in fetal development [29]. Supposedly, CT type new genes emerged in the ancestral genomes of humans to aid the male functions in testis while non-CT type new genes play some other or general functionality. However, CT45A1, TBC1D3 and NCYM act like oncogenes while they may assume various biological functions as their expression patterns suggested, which bear a previously unexpected theoretical significance in understanding evolutionary process of new gene functions. Specifically, while new genes evolved advantageous functions as is expected, they might also bring negative effects for the survival of organisms, which may be viewed as pleiotropic consequence. This has been predicted by recently proposed the selection, pleiotropy and compensation hypothesis (SPC) for adaptive evolution [47]. Based on the SPC hypothesis, widely observed adaptive evolution of new genes (e.g. [1,48,49]) might be a consequence of further evolution to “solve” a new negative problem(s) brought by the fixation of new genes as compensatory changes. This is a derivation from the SPC hypothesis to new gene evolution, different from the notion of progressively adaptive improvement of new gene function, awaiting further test in the future.
Evolutionarily underexplored polymorphic new genes contribute to within-species phenotypic variation
Almost all the above studies are performed to understand how human differ compared to other primates or other mammals under the conventional framework of comparative genomics. The revolution of the 2nd generation sequencing techniques rapidly promotes the field from between-species level to within-species level. From this angle, we now have the opportunity to understand the early picture on new gene origination. For DNA- or RNA-level duplicates, they should initially arise as copy number variation (CNV). Since CNVs tend to be deleterious [50], human CNVs have revealed the important phenotypic consequence in the context of disease [51]. Progress has also been made in understanding the evolutionary consequence of CNVs, as revealed in numerous case studies that detect their adaptive functional evolution [6]. Among these cases, salivary amylase gene (AMY1) and CC chemokine ligand 3-like 1 (CCL3L1) gene copy number gains have been relatively better characterized, which enables adaptation to a high-starch diet and is linked with lower susceptibility to HIV infection, respectively [52,53]. Genome-wide association studies further revealed that low copy number of AMY1 predisposes the carrier with high possibility of obesity [54] suggesting that a single polymorphic duplicated gene locus may induce multifold phenotypic difference between individuals.
Regardless of the prevalence of CNVs [55], their evolutionary consequence is less known, except for a handful cases such as AMY1 or CCL3L1. The difficulty partially roots in that the exact structure and sequence of CNVs could not be readily inferred based on the short reads (~100 bp) provided by 2nd generation sequencing [56] because a significant proportion of CNVs in human is much bigger than 1 kb [57]. Clearly, such information is helpful or even essential for studying the function of these loci. Fortunately, technical advancement based on the 3rd generation sequencing (e.g. PacBio) enables the full assembly of complex duplicate [56]. As shown in [58], a 100 kb region enriched with repeats was fully assembled with high accuracy (>99.9%) based on targeted sequencing on the PacBio platform.
Compared to polymorphic new genes generated through duplication-based mechanisms, polymorphic de novo genes just began to be appreciated for its scale in the standing genetic variation. Limited evidence indicates that there may be more pervasive de novo gene origination than currently appreciated. As shown by a Drosophila survey, 144 testis-expressed de novo genes emerged recently, which were subject to adaptive selection as shown by the valley of the nucleotide diversity [3]. By contrast, the de novo genes in human genomes may be even much more abundant, as implicated by a recent test that detected 5,737 polymorphic open reading frames [59].
Conclusion and the prospective
Tremendous efforts in recent years have revealed a high rate of new gene origination in the human genome and their significant roles in evolution towards versatile functionality in human biology. The sequencing data accumulated rapidly in astronomical scale have unveiled the evolutionary processes in which new genes emerged and evolved; previous studies also raised new and interesting conceptual and technical problems to solve. These progresses have provided an unprecedented opportunity to explore the genetic basis of human biology and human evolutionary history. The literatures we reviewed above can be taken as starting points to further detect underlying mechanistic processes and evolutionary forces. Deciphering the new genes-related gene-gene interaction networks would help understand how a new gene gets integrated into an ancestral gene network and examining the effects of new genes on the human phenotypes would help reconstruct the phenotypic evolution that our ancestors might have experienced.
The role of new genes in functional evolution will continue to be an enthusiastic topic for research. It should be noted here that the new gene studies have been almost all focused on protein-coding genes with a few exceptions (e.g. [60]). However, non-coding genes may represent an underexplored but potentially valuable field, especially considering their rapid turnover rates [61]. Actually, primate-specific miRNAs may account for 19% of the whole annotated miRNA pool in human, which is much higher than the proportion (9%) of protein-coding genes [10]. More strikingly, a transcriptome survey identified 14,682 long noncoding RNAs in human with 70% (10,359) of them being primate-specific [35]. These short or long noncoding genes present an exciting challenge to understanding their roles in evolution of the human genome.
Finally, the improvement in gene annotation can be expected if the technical endeavor accounting for the observed serious bias against young genes [11] is made. The integration of rapidly developing functional genomics techniques such as ribosome-profiling or proteogenomics and computational methods to detect evolutionary constraint [62,63] can increase reliability in detecting de novo genes from genome sequences and in discerning pseudogenes from functional genes.
Acknowledgments
We appreciate two anonymous referees’ insightful comments. We thank Zhang Lab members including Yi Shao, Chenyu Ma and Hangxing Jia for helpful comments. We are grateful for discussion with Mihaela Pavlicev and Gunter Wagner about the SPC model they proposed. YEZ is supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010400), the National Key Basic Research Program of China (2013CB531200) and the National Natural Science Foundation of China (91331114, 31322050). ML is supported by a NIH R01GM100768-01A1 grant and a NSF 1051826 grant from National Institutes of Health and National Science Foundation in USA, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Long MY, VanKuren NW, Chen SD, Vibranovski MD. New Gene Evolution: Little Did We Know. Annu Rev Genet. 2013;47:307–333. doi: 10.1146/annurev-genet-111212-133301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller HJ. Bar Duplication. Science. 1936;83:528–530. doi: 10.1126/science.83.2161.528-a. [DOI] [PubMed] [Google Scholar]
- 3.Zhao L, Saelao P, Jones CD, Begun DJ. Origin and Spread of de Novo Genes in Drosophila melanogaster Populations. Science. 2014;343:769–772. doi: 10.1126/science.1248286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen SD, Krinsky BH, Long MY. New genes as drivers of phenotypic evolution. Nat Rev Genet. 2013;14:645–660. doi: 10.1038/nrg3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper DN, Kehrer-Sawatzki H. Exploring the potential relevance of human-specific genes to complex disease. Hum Genomics. 2011;5:99–107. doi: 10.1186/1479-7364-5-2-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Iskow RC, Gokcumen O, Lee C. Exploring the role of copy number variants in human adaptation. Trends Genet. 2012;28:245–257. doi: 10.1016/j.tig.2012.03.002. This review summerized CNVs which appears to show signals of adaptive evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet. 2012;13:853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:e207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang YE, Vibranovski MD, Landback P, Marais GAB, Long M. Chromosomal Redistribution of Male-Biased Genes in Mammalian Evolution with Two Bursts of Gene Gain on the X Chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Zhang YE, Landback P, Vibranovski M, Long M. New genes expressed in human brains: implications for annotating evolving genomes. Bioessays. 2012;34:982–991. doi: 10.1002/bies.201200008. This paper reported an unexpected correlation between the ages of genes and the proportion of the genes that have been functionally investigated as recorded with the Gene Ontology terms, revealing that the current gene annotation is dominantly biased toward old or ancient genes. [DOI] [PubMed] [Google Scholar]
- 12.Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu DD, Irwin DM, Zhang YP. De novo origin of human protein-coding genes. PLoS Genet. 2011;7:e1002379. doi: 10.1371/journal.pgen.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang RL, Wei LP, Li CY. Hominoid-Specific De Novo Protein-Coding Genes Originating from Long Non-Coding RNAs. PLoS Genet. 2012:8. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerzoni D, McLysaght A. De novo origins of human genes. PLoS Genet. 2011;7:e1002381. doi: 10.1371/journal.pgen.1002381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Oliveira NM, Cheney KM, Pade C, Dreja H, Bergin AM, Borgdorff V, Beach DH, Bishop CL, Dittmar MT, et al. A whole genome screen for HIV restriction factors. Retrovirology. 2011;8:94. doi: 10.1186/1742-4690-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- 19.Rosso L, Marques AC, Reichert AS, Kaessmann H. Mitochondrial Targeting Adaptation of the Hominoid-Specific Glutamate Dehydrogenase Driven by Positive Darwinian Selection. PLoS Genet. 2008:4. doi: 10.1371/journal.pgen.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 21.Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mosse YP, Wood A, Lynch JE, et al. Copy number variation at 1q21. 1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Dennis Megan Y, Nuttle X, Sudmant Peter H, Antonacci F, Graves Tina A, Nefedov M, Rosenfeld Jill A, Sajjadian S, Malig M, Kotkiewicz H, et al. Evolution of Human-Specific Neural SRGAP2 Genes by Incomplete Segmental Duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. Together with the reference 23, this two papers identified a human-specific partial duplicate which contributed to brain evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, et al. Inhibition of SRGAP2 Function by Its Human-Specific Paralogs Induces Neoteny during Spine Maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westbrook VA, Schoppee PD, Vanage GR, Klotz KL, Diekman AB, Flickinger CJ, Coppola MA, Herr JC. Hominoid-specific SPANXA/D genes demonstrate differential expression in individuals and protein localization to a distinct nuclear envelope domain during spermatid morphogenesis. Mol Hum Reprod. 2006;12:703–716. doi: 10.1093/molehr/gal079. [DOI] [PubMed] [Google Scholar]
- 25.Shang B, Gao A, Pan Y, Zhang G, Tu J, Zhou Y, Yang P, Cao Z, Wei Q, Ding Y. CT45A1 acts as a new proto-oncogene to trigger tumorigenesis and cancer metastasis. Cell Death Dis. 2014;5:e1285. doi: 10.1038/cddis.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wainszelbaum MJ, Charron AJ, Kong C, Kirkpatrick DS, Srikanth P, Barbieri MA, Gygi SP, Stahl PD. The hominoid-specific oncogene TBC1D3 activates ras and modulates epidermal growth factor receptor signaling and trafficking. J Biol Chem. 2008;283:13233–13242. doi: 10.1074/jbc.M800234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodzic D, Kong C, Wainszelbaum MJ, Charron AJ, Su XO, Stahl PD. TBC1D3, a hominoid oncoprotein, is encoded by a cluster of paralogues located on chromosome 17q12. Genomics. 2006;88:731–736. doi: 10.1016/j.ygeno.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 28••.Suenaga Y, Islam SMR, Alagu J, Kaneko Y, Kato M, Tanaka Y, Kawana H, Hossain S, Matsumoto D, Yamamoto M, et al. NCYM, a Cis-Antisense Gene of MYCN, Encodes a De Novo Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas. PLoS Genet. 2014;10:e1003996. doi: 10.1371/journal.pgen.1003996. This paper functionally characterized a human-specific de novo gene on how it promoted tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong BC, Krystal GW. Isolation and Characterization of Complementary-DNA for N-Cym, a Gene Encoded by the DNA Strand Opposite to N-Myc. Cell Growth Differ. 1992;3:385–390. [PubMed] [Google Scholar]
- 30.Samusik N, Krukovskaya L, Meln I, Shilov E, Kozlov AP. PBOV1 Is a Human De Novo Gene with Tumor-Specific Expression That Is Associated with a Positive Clinical Outcome of Cancer. PLoS ONE. 2013;8:e56162. doi: 10.1371/journal.pone.0056162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang YE, Landback P, Vibranovski MD, Long M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Chang LH, Sun Y, Lu X, Stubbs L. Deep vertebrate roots for mammalian zinc finger transcription factor subfamilies. Genome Biol Evol. 2014;6:510–525. doi: 10.1093/gbe/evu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A. 2009;106:22358–22363. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutcliffe JG. mRNA in the mammalian central nervous system. Annu Rev Neurosci. 1988;11:157–198. doi: 10.1146/annurev.ne.11.030188.001105. [DOI] [PubMed] [Google Scholar]
- 35.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014 doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 36.Wang HY, Chien HC, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou CK, Tsai SF, Wu CI, Shen CK. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 38.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. This paper describe the transcriptome complexity of testis which further faciliate new gene origination in this organ. [DOI] [PubMed] [Google Scholar]
- 40.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T, Chen YT, Chua R, Gurung S, Gnjatic S, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 42.Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014;54:251–272. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- 43.Dobrynin P, Matyunina E, Malov SV, Kozlov AP. The novelty of human cancer/testis antigen encoding genes in evolution. Int J Genomics. 2013;2013:105108. doi: 10.1155/2013/105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 45.Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W-L, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 47••.Pavlicev M, Wagner GP. A model of developmental evolution: selection, pleiotropy and compensation. Trends Ecol Evol. 2012;27:316–322. doi: 10.1016/j.tree.2012.01.016. This paper presented a general model that adaptive mutation may be often fixed with negative pleiotropy, which can be resolved by subsequent compensatory mutation. [DOI] [PubMed] [Google Scholar]
- 48.Han M, Demuth J, McGrath C, Casola C, Hahn M. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Su B. Evolutionary Origin and Human-Specific Expansion of a Cancer/Testis Antigen Gene Family. Mol Biol Evol. 2014 doi: 10.1093/molbev/msu188. [DOI] [PubMed]
- 50.Katju V, Bergthorsson U. Copy-number changes in evolution: rates, fitness effects and adaptive significance. Front Genet. 2013:4. doi: 10.3389/fgene.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 54•.Falchi M, El-Sayed Moustafa JS, Takousis P, Pesce F, Bonnefond A, Andersson-Assarsson JC, Sudmant PH, Dorajoo R, Al-Shafai MN, Bottolo L, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet. 2014;46:492–497. doi: 10.1038/ng.2939. This paper provided the evidence that amylase copy number gain is associated with low body mass index. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L, Li YH, Li SL, Hu N, He YM, Pong R, Lin DN, Lu LH, Law M. Comparison of Next-Generation Sequencing Systems. J Biomed Biotechnol. 2012 doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, Hon L, Sudmant PH, Graves TA, Alkan C, Dennis MY, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014;24:688–696. doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wijaya E, Frith MC, Horton P, Asai K. Finding Protein-Coding Genes through Human Polymorphisms. PLoS ONE. 2013;8:e54210. doi: 10.1371/journal.pone.0054210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W, Brunet FG, Nevo E, Long M. Origin of sphinx, a young chimeric RNA gene in Drosophilamelanogaster. Proc Natl Acad Sci USA. 2002;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Marques AC, Ponting CP. Intergenic lncRNAs and the evolution of gene expression. Curr Opin Genet Dev. 2014;27C:48–53. doi: 10.1016/j.gde.2014.03.009. This timely updated review summerized the fast turnover of long noncoding genes and provided a valuable biological base in further investigating biology and evolution of noncoding genes. [DOI] [PubMed] [Google Scholar]
- 62.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. This paper reannotated 140 pseudogenes and 9 noncoding genes as protein-coding genes by mapping mass-spectra peptide against the human genome directly. [DOI] [PMC free article] [PubMed] [Google Scholar]