A recombinant version of the lectin CGL from the sea mussel C. grayanus was expressed, purified and crystallized. The structure determined at 2.12 Å resolution indicated that this lectin is a member of the β-trefoil family.
Keywords: lectin, molecular replacement, carbohydrate binding, structure comparison, β-trefoil
Abstract
CGL is a 150 amino-acid residue lectin that was originally isolated from the sea mussel Crenomytilus grayanus. It is specific for binding GalNAc/Gal-containing carbohydrate moieties and in general does not share sequence homology with other known galectins or lectins. Since CGL displays antibacterial, antifungal and antiviral activities, and interacts with high affinity with mucin-type receptors, which are abundant on some cancer cells, knowledge of its structure is of significant interest. Conditions have been established for the expression, purification and crystallization of a recombinant variant of CGL. The crystal structure of recombinant CGL was determined and refined at a resolution of 2.12 Å. The amino-acid sequence of CGL contains three homologous regions (73% similarity) and the folded protein has a β-trefoil topology. Structural comparison of CGL with the closely related lectin MytiLec allowed description of the glycan-binding pockets.
1. Introduction
Lectins are proteins with the ability to bind specifically and reversibly to carbohydrate moieties, often through multivalent interactions (Sharon & Lis, 2004 ▸). Such a mode of interaction gives some lectins their well known ability to agglutinate cells. Owing to their highly specific interactions with carbohydrates, lectins are used for the purification of polysaccharides and glycoproteins and in a variety of biological applications. The latter include cell separation, cellular localization of glycoconjugates, identification of blood groups and microorganisms, and monitoring alterations on the surface of normal and neoplastic cells (Lowry et al., 1951 ▸; Ozeki et al., 1991 ▸). Many lectins also exhibit intrinsic antimicrobial and/or antiviral properties (Koharudin & Gronenborn, 2014 ▸; Iordache et al., 2015 ▸). Because of these versatile properties, lectins are actively sought proteins with great potential therapeutic and biotechnological utility (Varrot et al., 2013 ▸; Yau et al., 2015 ▸; Zarogoulidis et al., 2015 ▸). Lectins are generally classified based on either their origin, their glycan specificity or their molecular topology. One such group consists of lectins from marine invertebrates, which currently encompasses examples from several hundred species (Luk’yanov et al., 2007 ▸).
CGL is a lectin isolated from the sea mussel Crenomytilus grayanus (Belogortseva et al., 1998 ▸) with specificity for N-acetyl-2-deoxy-2-amino-galactose (GalNAc/Gal). Therefore, this 18 kDa protein may be classified as a member of the family of galectins. CGL interacts strongly with the surfaces of both Gram-positive and Gram-negative bacteria. However, its inhibition of bacterial growth and agglutination was only detected for a few bacterial strains, including Escherichia coli, Bacillus subtilis and Staphylococcus aureus (Kovalchuk et al., 2013 ▸). CGL is expressed in various tissues of a healthy mussel and it is able to inhibit growth of the fungi often associated with this mollusk (Chikalovets et al., 2015 ▸). Such antimicrobial activities suggest that the physiological role of CGL may involve combating bacterial and fungal infections in mollusks. CGL binds with high affinity to mucin-type receptors, which are characteristic for human colon adenocarcinoma, thus opening the possibility of using CGL as a marker of neoplastic transformation of these cancers (Furtak et al., 1999 ▸). Additionally, anti-HIV activity of CGL has also been reported (Luk’yanov et al., 2007 ▸).
The cDNA sequence of CGL became available only recently and bioinformatics analysis showed that the amino-acid sequence of CGL (150 amino acids) does not share significant similarity with other galectins or with other lectins in general (Kovalchuk et al., 2013 ▸). The only exception is another mollusk lectin from the mussel Mytilus galloprovincialis (MytiLec), which was also characterized quite recently (Fujii et al., 2012 ▸) and shares 83% amino-acid sequence identity with CGL. Based on analysis of the amino-acid sequence and the appearance of CD spectra, Kovalchuk et al. (2013 ▸) have shown that the CGL molecule predominantly assumes a β-structure, that its sequence is a repeat of three highly homologous regions (73% similarity) and that its overall fold is expected to be the β-trefoil, as observed previously for B-type lectins such as ricin. It is worth noting that the β-trefoil fold is not unique to B-like lectins but is shared by several other families of proteins including cytokines, agglutinins, actin-cross-linking proteins etc., which share little to no sequence similarity and have distinct functions (Broom et al., 2012 ▸; Renko et al., 2012 ▸).
In this report, we describe the protocols used for the expression, production and purification of the recombinant variant of CGL. The protein was subsequently crystallized and the crystal structure, described here, was determined and refined at a resolution of 2.12 Å.
2. Materials and methods
2.1. Cloning, expression and purification
The synthetic gene encoding the amino-acid sequence of CGL, with codons optimized for expression in E. coli, was purchased from Bio Basic Inc. (http://store.biobasic.com/; Markham, Ontario, Canada). The gene was cloned into a pUC57 vector with ampicillin resistance (pUC57-cgl). Following the standard protocol of Gateway cloning (Hartley et al., 2000 ▸), assisted by an intermediary sequence confirmation, the CGL coding sequence was subcloned into pDest-527 vector (http://www.addgene.org/11518/; Addgene, Cambridge, Massahusetts, USA). The resulting expression plasmid encodes an N-terminal His6 tag followed by a stretch of nucleotides specific to Gateway cloning, as well as a TEV protease cleavage site (all elements are contributed by the pDest-527 vector). For expression, this plasmid was transformed into E. coli BL21(DE3) competent cells (Novagen, USA) and plated onto a Luria–Bertani (LB) agar plate supplied with 100 µg ml−1 ampicillin. A single colony was cultured overnight in LB medium supplied with 100 µg ml−1 ampicillin at 37°C with shaking (250 rev min−1). The following morning, the overnight culture was diluted 50-fold with fresh pre-warmed (30°C) LB medium supplied with 100 µg ml−1 ampicillin (a production culture) and culture growth continued until the optical density at 600 nm (OD600) reached 0.8. At this point, the culture was cooled on ice (with stirring) to 18°C and transferred to a shaker equilibrated at the same temperature. After 30 min, expression of CGL was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM. After induction, the culture was maintained under the abovementioned conditions for a further 16 h. All subsequent procedures were conducted at 5°C. The cells were harvested following standard procedures and were resuspended in 50 mM Na2HPO4/NaH2PO4 buffer, 500 mM NaCl pH 7.4. After three passes through a pressure homogenizer, the lysed suspension was cleared by centrifugation (30 min, 25 000g) followed by filtration through a 0.45 µm filtering membrane and the filtrate was applied onto a 20 ml column packed with TALON metal-affinity resin (Clontech Laboratories Inc.). The protein was eluted with a gradient of imidazole. Combined fractions containing CGL were concentrated and dialyzed against 50 mM Na2HPO4/NaH2PO4 buffer pH 7.4 with 150 mM NaCl. Dialyzed protein solution was applied onto a 5 ml HiTrap Heparin HP column (GE Healthcare Life Sciences) and the CGL-containing fractions were eluted with a gradient of NaCl using an ÄKTA FPLC system (GE Healthcare Life Sciences), with the elution profile monitored by UV absorption at 280 nm. The final purification step utilized size exclusion on a Superdex 75 16/60 PG column (GE Healthcare Life Sci.), with a solution consisting of 50 mM Na2HPO4/NaH2PO4 pH 7.4, 150 mM NaCl acting as an elution buffer. After analysis by gel electrophoresis, fractions containing pure CGL were combined and concentrated to 20 mg ml−1. The purity of the protein solution is illustrated in Fig. 1 ▸(a) and the production information for CGL is summarized in Table 1 ▸.
Figure 1.
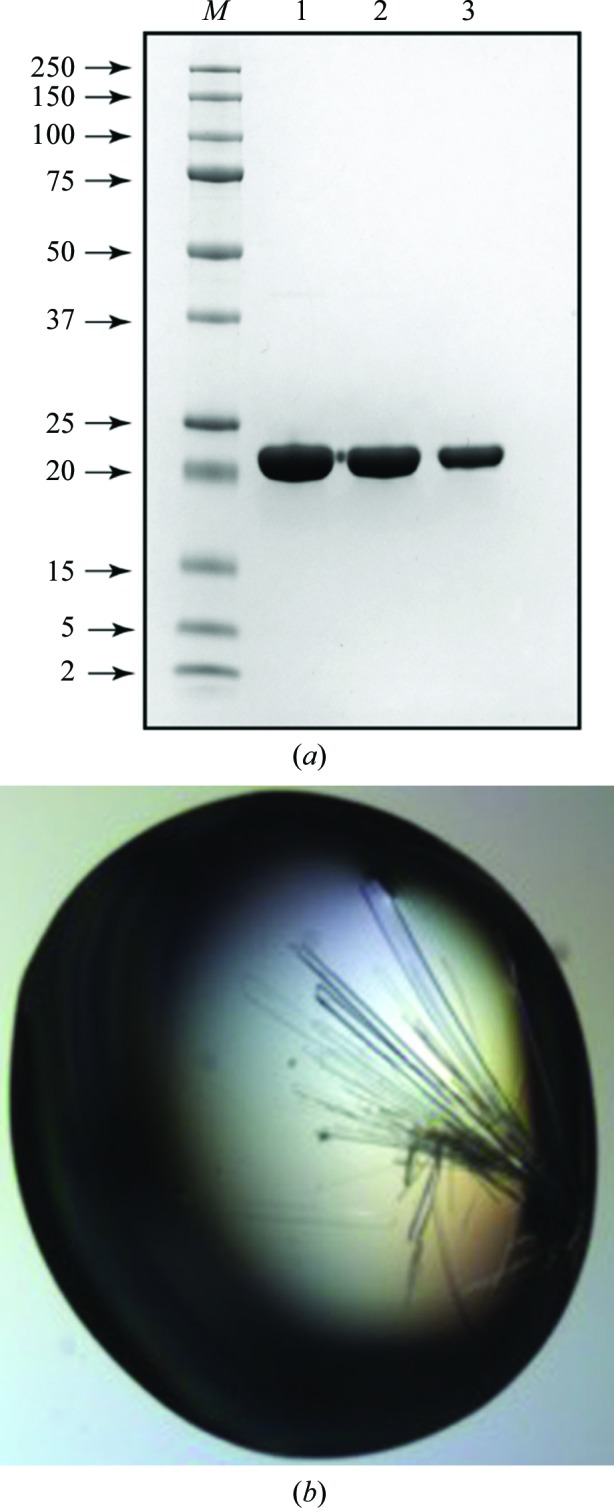
(a) SDS–PAGE analysis of the final, pure preparation of the recombinant CGL. Lane M contains molecular-mass marker (labeled in kDa). Lanes 1, 2 and 3 contain 10, 5 and 2.5 µg 6×His-CGL, respectively. (b) Crystals of CGL.
Table 1. Details of the production of recombinant CGL.
| Source organism/DNA source | C. grayanus (mollusk) |
| Expression vector | Synthetic pDest-527 |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MRSGSHHHHHHRSDITSLYKKAGSENLYFQSMTTFLIKHKASGKFLHPYGGSSNPANNTKLVLHSDIHERMYFQFDVVDERWGYIKHVASGKIVHPYGGQANPPNETNMVLHQDRDRALFAMDFFNDNIMHKGGKYIHPKGGSPNPPNNTETVIHGDKHAAMEFIFVSPKNKDKRVLVYA |
2.2. Crystallization
The initial crystallization conditions were found in trials conducted at 20°C using four MCSG crystallization screens (Microlytic, USA) representing 384 individual conditions. The sitting droplets, composed of equal volumes (200 nl) of protein solution (20 mg ml−1 in 50 mM Na2HPO4/NaH2PO4 pH 7.4, 150 mM NaCl) and of a respective screen solution, were set up using a Phoenix crystallization robot (Art Robbins Instruments, California, USA) and equilibrated against 80 µl reservoir solution. Subsequent optimization of the crystallization conditions was performed in 15-well plates using a hanging-drop setup, and needle-like crystals that reached dimensions of 0.5 × 0.08 × 0.04 mm were obtained after ∼10 d (see Fig. 1 ▸ b). A summary of the crystallization conditions is shown in Table 2 ▸.
Table 2. Crystallization of CGL.
| Method | Hanging-drop vapor diffusion |
| Plate type | EasyXtal 15-well |
| Temperature (K) | 293 |
| Protein concentration (mgml1) | 20 |
| Buffer composition of protein solution | 50mM Na2HPO4/NaH2PO4 pH 7.4, 150mM NaCl |
| Composition of reservoir solution | 0.1M HEPES pH 7.0, 18%(w/v) PEG 4000, 0.1M sodium acetate, 0.1M lithium sulfate |
| Composition of droplet | 2l (protein) + 3l (reservoir) |
| Volume of reservoir (ml) | 1.0 |
2.3. Data collection, processing, structure solution and refinement
Crystals of CGL were washed and cryoprotected in the mother liquor containing 20%(v/v) glycerol. After flash-cooling a single crystal in liquid nitrogen, diffraction data were collected on beamline 22-ID (SER-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory, USA. Data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▸). Data-collection and processing statistics are shown in Table 3 ▸.
Table 3. Diffraction data collection and processing.
Values in parentheses are for the highest resolution shell.
| Diffraction source | Beamline 22-ID, SER-CAT, APS |
| Wavelength () | 1.000 |
| Temperature (K) | 100 |
| Detector | Rayonix 300HS high-speed CCD detector |
| Crystal-to-detector distance (mm) | 300 |
| Rotation range per image () | 0.5 |
| Total rotation range () | 250 |
| Exposure time per image (s) | 0.25 |
| Space group | C2 |
| a, b, c () | 137.24, 68.33, 131.49 |
| , , () | 90, 110.5, 90 |
| Mosaicity () | 0.6 |
| Resolution range () | 50.02.12 (2.162.12) |
| Total No. of reflections | 335480 |
| No. of unique reflections | 63465 |
| Completeness (%) | 97.7 (80.2) |
| Multiplicity | 5.3 (4.7) |
| I/(I) | 21.2 (3.9) |
| R merge † | 0.070 (0.421) |
| R r.i.m ‡ | 0.033 (0.210) |
| Overall B factor from Wilson plot (2) | 23.2 |
| CC1/2 | 0.980 (0.898) |
R
merge = 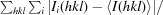
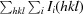 .
.
Estimated R r.i.m. = R merge[N/(N 1)]1/2, where N is the data multiplicity.
The values of F obs were obtained using SCALEPACK2MTZ (Winn et al., 2011 ▸). The structure of CGL was solved by the molecular-replacement method using Phaser (McCoy et al., 2007 ▸), followed by model extension and rebuilding with phenix.mr_rosetta (Terwilliger et al., 2012 ▸). The molecular-replacement solution was refined with REFMAC5 (Murshudov et al., 2011 ▸) and manually improved with Coot (Emsley et al., 2010 ▸). MolProbity (Chen et al., 2010 ▸) was used for Ramachandran analysis. The refinement statistics are shown in Table 4 ▸.
Table 4. CGL structure refinement.
Values in parentheses are for the highest resolution shell.
| Resolution range () | 38.002.12 (2.1752.12) |
| Completeness (%) | 97.6 (81.4) |
| Cutoff | None |
| No. of reflections, working set | 61539 (3736) |
| No. of reflections, test set | 1922 (133) |
| Final R cryst | 0.140 (0.168) |
| Final R free | 0.185 (0.196) |
| Cruickshank DPI | 0.1758 |
| No. of non-H atoms | |
| Protein | 7276 |
| Ion | 0 |
| Ligand | 138 |
| Water | 743 |
| Total | 8157 |
| R.m.s. deviations | |
| Bonds () | 0.019 |
| Angles () | 1.640 |
| Average B factors (2) | |
| Protein | 17.7 |
| Ion | 0.0 |
| Ligand | 32.8 |
| Water | 27.2 |
| Ramachandran plot | |
| Favored regions (%) | 96.6 |
| Additionally allowed (%) | 3.4 |
| PDB code | 5duy |
3. Results and discussion
A diffraction data set extending to a resolution of 2.12 Å was measured using only one crystal of CGL. The contents of the asymmetric unit could not be unambiguously determined based only on the size of the protein and of the asymmetric unit of the crystal, although it was clear that multiple copies of the CGL molecule must be present. Based on the most likely values of the Matthews coefficient (V M), the asymmetric unit could contain as few as four monomers of CGL (V M = 4.43 Å3 Da−1, solvent content 72.3%) or as many as eight (V M = 2.22 Å3 Da−1, solvent content 51.5%). Initial attempts to solve the structure by molecular replacement (MR) were conducted with three different programs, MOLREP (Vagin & Teplyakov, 2010 ▸), Phaser and EPMR (Kissinger et al., 1999 ▸), and utilized several different search models. The first group of search models was based on the structure of an artificial lectin with an idealized β-trefoil topology, ThreeFoil (PDB entry 3pg0; Broom et al., 2012 ▸). The second group of models was generated with the aid of the web-based servers Phyre2 (Kelley et al., 2015 ▸) and I-TASSER (Yang et al., 2015 ▸). None of these searches identified a correct solution, even for a substructure of CGL.
To prepare a more extensive list of models, we performed homology searches with the web-based server SALAMI (Margraf et al., 2009 ▸), in which we used the ThreeFoil structure as a query against the entries from the PDB present in the server’s database. We selected the top 39 nonredundant PDB entries, all representing proteins with the trefoil topology, as models for further MR searches. A table listing the selected PDB entries is provided as Supporting Information. The monomers representative of each entry were structurally superimposed in the common unit cell. Using the SFALL and MAPMASK utilities from the CCP4 suite (Winn et al., 2011 ▸), we calculated the molecular electron density averaged over the assembly of all monomers. This average molecular electron density was used as a template in the MR searches performed with Phaser. The coordinates of ThreeFoil aligned with the electron-density template were supplied for concurrent packing analysis and refinement. Searches were conducted in an automatic mode and aimed at the identification of a partial solution for the first two monomers of CGL. The rationale behind this strategy was that a signal originating from just a single monomer could be difficult to identify if it represented as little as just one eighth of the asymmetric unit content. On the other hand, attempting to identify a more complete MR solution in automatic mode could be unnecessarily time-consuming. Searches were conducted within the resolution range 35–2.6 Å and the putative structural similarity between the model and a solution, expressed as the r.m.s. deviation between equivalent Cα atoms, was assumed to be 1.3 Å.
Using this approach, we identified a strong signal, described by log-likelihood (LLG) and Z-score values of 91 and 8.4, respectively, which was interpreted as representing a correct partial solution for the two CGL molecules. It is worth noting that for unsuccessful MR searches conducted under the same conditions using Phaser the LLG and Z-score values usually did not exceed 35 and 4.8, respectively. The MR searches were continued to identify solutions for additional CGL monomers, one per search. Using this approach, we could successfully place four monomers in the asymmetric unit (LLG = 257, Z-score = 12.9). However, our attempts to further extend the CGL model did not succeed, although analysis of the crystal packing indicated that the four monomers of CGL did not represent a complete solution. Owing to the presence of extensive regions that lacked any molecular components, the unit cell was filled by non-interacting layers of CGL molecules. However, it was also evident that the correct content of the asymmetric unit corresponds to six or five CGL molecules rather than eight. Furthermore, when subjected to structural refinement with REFMAC5, the four-molecule model of CGL could be refined only partially (R cryst of ∼0.38 and R free of ∼0.49). While the 2F o − F c difference electron density correlated well with a four-molecule solution, significant bias and insufficient phasing power prevented further manual extension of the model. A representative example of the 2F o − F c electron density for the MR solution, together with an equivalent density for the complete, refined model of CGL is shown in Fig. 2 ▸. Subsequently, the model was subjected to rebuilding and extension with phenix.mr_rosetta, which identified the coordinates for two additional complete monomers of CGL and several water molecules. The improved solution was characterized by R cryst and R free values of 0.244 and 0.282, respectively. This solution was subjected to extensive rebuilding and refinement using Coot and REFMAC5 (R cryst = 0.153 and R free = 0.203), respectively, resulting in the final model describing six monomers of CGL in the asymmetric unit.
Figure 2.
A stereoimage showing six monomers of CGL (A–F, the content of the asymmetric unit) in ribbon representation, together with the 2F o − F c electron-density maps covering residues 50–70 in each of the monomers. The electron-density maps are contoured at the 1.3σ level. (a) Maps were calculated with the best phases obtained from the molecular replacement. Notice a lack of interpretable electron-density peaks in the regions of monomers E and F. (b) Maps were calculated using the phases corresponding to the final refined model of CGL.
It is interesting to understand the reasons behind the failure to identify a complete solution for all six monomers using MR. Fig. 3 ▸ illustrates the assembly of 39 superimposed monomers used to calculate the averaged molecular electron density used in MR searches. Even though the β-trefoil topology and the associated intramolecular threefold symmetry of these molecules are clearly visible, structural differences between the conformations of subdomains in specific molecules are quite striking, both between the subdomains of different molecules as well as within the same molecule. As a result, the molecular electron density for this assembly lacked information describing structural details. Thus, an extension of the MR solution by subsequent monomers led to an increase in the noise introduced by regions of the model electron density inaccurately representing structural details of CGL, especially at the medium-to-higher resolution range of the diffraction data. The phasing power of the partial solution was further weakened by the fact that the sequence of the CGL polypeptide chain was not correctly ‘locked’ in the electron density, since the coordinates assisting the model electron density represented a different lectin, ThreeFoil. As we found in post-analysis, by placing correctly oriented molecules of refined CGL in the positions determined for partial solution, it was possible to recover a sufficiently strong signal (electron-density peaks) for further model extension and refinement. While such a task would be quite daunting by manual rebuilding, the computing power provided by phenix.mr_rosetta passed this barrier quite swiftly.
Figure 3.
A stereoimage illustrating the assembly of 39 superimposed monomers with a β-trefoil topology oriented along a pseudo-threefold. The component monomers in the assembly were selected from the PDB based on structural similarity to the idealized lectin ThreeFoil. This set of molecules was included in calculations of the average molecular electron density used as a template in the MR searches. Monomers shown in this figure are represented by Cα coils; however, for the electron-density calculations all atoms were included. A complete list of the PDB entries contributing to this assembly is given in Supplementary Table S1.
In the final model, the nearly complete molecules of six CGL monomers were successfully modeled. The electron density for the N-terminal Met residue was defined in only one monomer (C), while monomer D was missing the first two N-terminal amino acids, Met1 and Thr2. The r.m.s. deviations between the positions of equivalent Cα atoms in different monomers of CGL vary between 0.106 Å (between monomers C and E) and 0.131 Å (between monomers B and C), with an average value of 0.119 Å. The N-terminal addition, composed of a His6 affinity tag, an artifact of the Gateway cloning and a TEV protease cleavage site, together consisting of 31 amino acids, was not modeled.
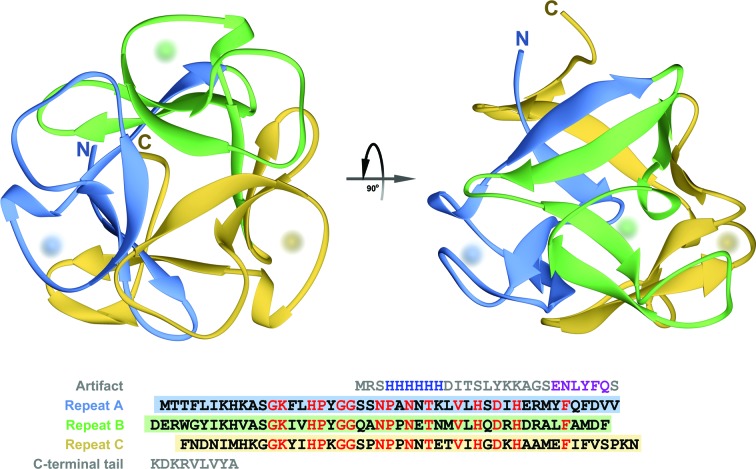
Analysis of the crystal packing of CGL as well as the profile of elution through the size-exclusion column (not shown) indicate that the biological form of this lectin is a monomer. The overall structure of a CGL monomer is shown in Fig. 4 ▸. It is very typical of all β-trefoil domains formed by three structurally conserved subdomains. Each of these subdomains is composed of four β-strands, with two strands from each module collectively forming a six-stranded β-barrel and the remaining two from each module together forming a β-hairpin triplet that caps one end of the barrel. In CGL, sections consisting of three residues each (Glu37–Met39, Asp85–Ala87 and Ala99–Met101) of the coils connecting the third and fourth β-strand within each subdomain form a single 310-helical turn. The putative glycan-binding site, one in each subdomain, is formed by the residues located in a region spanning over about 25 amino acids from the second β-strand to the 310-helical turn. In all six molecules of CGL present in the asymmetric unit, each of three glycan-binding sites is occupied by a molecule of glycerol.
Figure 4.
A ribbon representing the monomer of CGL is shown in two orthogonal orientations. Sections of the ribbon shown in three different colors correspond to subdomains of CGL. The amino-acid sequence of the lectin is shown beneath the ribbons. The N- and C-termini are labeled and locations of putative carbohydrate-binding sites are indicated by diffuse spheres. In the sequence representation, three subdomains are aligned and highlighted using the same color scheme as in the cartoon. Residues colored red, representing 37% of the aligned sections on average, are common to all three repeats.
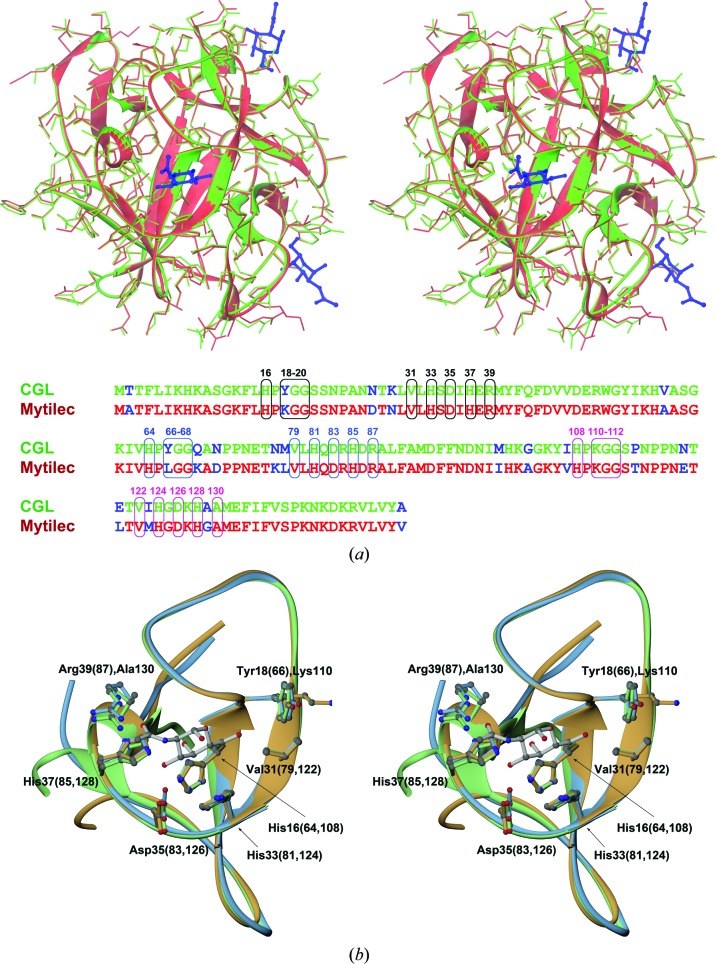
While this work was being finalized, the coordinates of two high-resolution structures (PDB entry 3wmu at 1.1 Å and PDB entry 3wmv at 1.05 Å; D. Terada, F. Kawai, H. Noguchi, S. Unzai, S.-Y. Park, Y. Ozeki & J. R. H. Tame, unpublished work) of a very closely related lectin, MytiLec, were deposited in the Protein Data Bank. However, we found no associated publication describing these structures. MytiLec is another mollusk lectin, isolated from the mussel M. galloprovincialis, that shares 83% amino-acid sequence identity with CGL. The proteins differ at only 19 positions (Fig. 5 ▸ a). When the monomers of CGL and MytiLec are superimposed, the similarity of both structures, including not only the main chain but also the side chains, is quite striking. This observation is particularly notable as both proteins were produced and purified following different protocols, crystallized under different conditions and refined completely independently. Even more significant is the finding that the glycan-binding sites of both lectins are nearly identical. Therefore, these lectins are very likely to share the same pattern of specificity for carbohydrate moieties. As in MytiLec, a glycan-binding pocket in CGL is fully defined by the residues from a single subdomain. A more detailed representation of the binding pockets is shown in Fig. 5 ▸(b), where three structurally aligned subdomains of CGL are displayed together with the molecule of α-GalNAc placed identically as in the structure of the MytiLec complex.
Figure 5.
(a) A stereoimage showing the superposition of monomers of CGL (shown in green) and MytiLec (shown in red; PDB entry 3wmv) in an orientation displaying one of the carbohydrate-binding sites of MytiLec at the front. In addition to the main chains, shown as ribbons, all side chains are depicted in stick representation. Three molecules of α-GalNAc identified in the structure of MytiLec are also shown and are colored dark blue. At the bottom of this figure the amino-acid sequences of both lectins are aligned, which differ at 19 positions (shown in blue). Residues forming the α-GalNAc-binding sites in MytiLec and their equivalents in CGL are shown in boxes and labeled using different colors for each site. (b) A stereoimage of the three aligned CGL subdomains, colored according to the scheme introduced in Fig. 4 ▸. Fragments are shown from the view of a putative carbohydrate-binding site together with a model of the α-GalNAc molecule, a likely ligand of CGL (see text). Side chains of amino acids interacting with α-GalNAc are also shown for all three repeats and are labeled according to their positions in the CGL sequence.
The putative glycan-binding pocket in the first CGL subdomain is formed by the side chains of His16, Tyr18, Val31, His33, Asp35, His37 and Arg39 and the backbone of Gly19 and Gly20. Thus, if the definition of the first binding pocket of CGL is abbreviated as CGLI(HYGGVHDHR), the remaining two pockets can be represented as CGLII(HYGGVHDHR) and CGLIII(HKGGVHDHA). As not all of the residues lining the glycan-binding site are conserved across the three subdomains of the CGL polypeptide chain, the pocket signature can be represented as CGL(H,Y/K,GGVHDH,R/A). Similarly, the glycan-binding pocket in MytiLec can be defined as MytiLec(H,K/L,GGVHDH,R/A). In both proteins, Arg is one of two residues that are not fully conserved. In the complex of MytiLec with α-GalNAc, the side chain of this residue forms a hydrogen bond to the carbonyl O atom of the acetyl group of the ligand, either directly (first subdomain) or via a water molecule (second subdomain). A similar arrangement is found in the modeled complex for CGL. In both domains the Arg side chain is stabilized by an adjacent acidic residue: either Glu (first subdomain) or Asp (second subdomain). The Asp–Arg interaction is also mediated through a water molecule, suggesting that the overall stabilization and interaction of Asp–Arg–(α-GalNAc) may be weaker than in the case of Glu–Arg–(α-GalNAc). Furthermore, in the third subdomain of both lectins, where Arg is replaced by Ala, an adjacent acidic residue is also not present and its place is taken by either Ala (CGL) or Gly (MytiLec). This coordinated change is a good example of evolution-based modifications to the sequences of both lectins. Also, one could conclude that any contribution of the Arg residue to the ligand binding is only secondary or, perhaps, that the glycan affinities (specificities) of different subdomains are not completely identical. A similar analysis for the second nonconserved residue in the binding pocket (Tyr, Lys or Leu) suggests that the most common denominator appears to be a contribution of hydrophobic properties, as the amino group of Lys, if present, points away from the carbohydrate ligand. Again, a possibility of some differences in the affinity or specificity for glycan moieties between subdomains should not be excluded.
In this report, we present a detailed structural description of a novel mollusk lectin CGL from the mussel C. grayanus, which has a very unique amino-acid sequence. Although CGL shares glycan preferences with other galectins, its structure is different. Both CGL and its homologous counterpart MytiLec have been shown to be capable of either killing or inhibiting growth of certain cancer cells. Therefore, the structural results presented in this report may help in understanding and rationally enhancing the anticancer properties of these lectins.
Supplementary Material
PDB reference: CGL, 5duy
Models used in the molecular-replacement searches as identified by the SALAMI server.. DOI: 10.1107/S2053230X15019858/sw5091sup1.pdf
Acknowledgments
We would like to thank Mr Mi Li for assistance with X-ray data collection. We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) located at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and in part by funds from the NIH Intramural AIDS Targeted Antiviral Program (to AW, BRO and MJ).
References
- Belogortseva, N. I., Molchanova, V. I., Kurika, A. V., Skobun, A. S. & Glazkova, V. E. (1998). Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 119, 45–50. [DOI] [PubMed]
- Broom, A., Doxey, A. C., Lobsanov, Y. D., Berthin, L. G., Rose, D. R., Howell, P. L., McConkey, B. J. & Meiering, E. M. (2012). Structure, 20, 161–171. [DOI] [PubMed]
- Chikalovets, I. V., Chernikov, O. V., Pivkin, M. V., Molchanova, V. I., Litovchenko, A. P., Li, W. & Lukyanov, P. A. (2015). Fish Shellfish Immunol. 42, 503–507. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fujii, Y. et al. (2012). J. Biol. Chem. 287, 44772–44783. [DOI] [PMC free article] [PubMed]
- Furtak, V. A., Kurika, A. V., Belogortseva, N. I., Chikalovets, I. V. & Kleshch, Y. (1999). Bull. Exp. Biol. Med. 128, 1039–1041. [PubMed]
- Hartley, J. L., Temple, G. F. & Brasch, M. A. (2000). Genome Res. 10, 1788–1795. [DOI] [PMC free article] [PubMed]
- Iordache, F., Ionita, M., Mitrea, L. I., Fafaneata, C. & Pop, A. (2015). Curr. Pharm. Biotechnol. 16, 152–161. [DOI] [PubMed]
- Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. (2015). Nature Protoc. 10, 845–858. [DOI] [PMC free article] [PubMed]
- Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999). Acta Cryst. D55, 484–491. [DOI] [PubMed]
- Koharudin, L. M. & Gronenborn, A. M. (2014). Curr. Opin. Virol. 7, 95–100. [DOI] [PMC free article] [PubMed]
- Kovalchuk, S. N., Chikalovets, I. V., Chernikov, O. V., Molchanova, V. I., Li, W., Rasskazov, V. A. & Lukyanov, P. A. (2013). Fish Shellfish Immunol. 35, 1320–1324. [DOI] [PubMed]
- Luk’yanov, P. A., Chernikov, O. V., Kobelev, S. S., Chikalovets, I. V., Molchanova, V. I. & Li, W. (2007). Russ. J. Bioorg. Chem. 33, 161–169. [PubMed]
- Margraf, T., Schenk, G. & Torda, A. E. (2009). Nucleic Acids Res. 37, W480–W484. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Oliver, H., Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951). J. Biol. Chem. 193, 265–275. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ozeki, Y., Matsui, T., Suzuki, M. & Titani, K. (1991). Biochemistry, 30, 2391–2394. [DOI] [PubMed]
- Renko, M., Sabotič, J. & Turk, D. (2012). Biol. Chem. 393, 1043–1054. [DOI] [PubMed]
- Sharon, N. & Lis, H. (2004). Glycobiology, 14, 53R–62R. [DOI] [PubMed]
- Terwilliger, T. C., DiMaio, F., Read, R. J., Baker, D., Bunkóczi, G., Adams, P. D., Grosse-Kunstleve, R. W., Afonine, P. V. & Echols, N. (2012). J. Struct. Funct. Genomics, 13, 81–90. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Varrot, A., Basheer, S. M. & Imberty, A. (2013). Curr. Opin. Struct. Biol. 23, 678–685. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J. & Zhang, Y. (2014). Nature Methods, 12, 7–8. [DOI] [PMC free article] [PubMed]
- Yau, T., Dan, X., Ng, C. C. W. & Ng, T. B. (2015). Molecules, 20, 3791–3810. [DOI] [PMC free article] [PubMed]
- Zarogoulidis, P. et al. (2015). J. Cancer, 6, 9–18.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: CGL, 5duy
Models used in the molecular-replacement searches as identified by the SALAMI server.. DOI: 10.1107/S2053230X15019858/sw5091sup1.pdf