The ligand-binding region of human uPARAP, an endocytic collagen receptor, was overexpressed in Drosophila S2 cells and crystallized at pH 5.3 and pH 7.4. Structural characterization of this receptor at two pH values will reveal the structural basis of a pH-induced conformational change in the mannose receptor family.
Keywords: uPARAP, collagen endocytic receptor, ligand-binding region, crystallization
Abstract
Urokinase plasminogen activator receptor-associated protein (uPARAP) is an endocytic receptor that internalizes collagen for lysosomal degradation and plays an important role in matrix remodelling. Previous recombinant protein production of uPARAP in Pichia pastoris generated protein with highly heterogeneous glycans that was prone to proteolytic degradation, resulting in highly twinned crystals. In this study, the uPARAP ligand-binding region was expressed in stably transfected Drosophila S2 insect cells. The recombinant protein was homogeneous after purification by metal-affinity and anion-exchange chromatography. Crystals were obtained at two different pH values (5.3 and 7.4) and diffracted to 2.44 and 3.13 Å resolution, respectively. A model of the ligand-binding region of uPARAP was obtained by molecular replacement combined with autobuilding. As the first multidomain crystal structure of the mannose receptor family, structural characterization of the uPARAP ligand-binding region will provide insight into the pH-induced conformational rearrangements of the mannose receptor family.
1. Introduction
Urokinase plasminogen activator receptor (uPAR)-associated protein (uPARAP/Endo180) is a member of the macrophage mannose receptor family that is expressed on fibroblasts, macrophages and a subset of endothelial cells (Engelholm et al., 2003 ▸). This endocytic receptor has been found to be important for numerous processes involving degradation of the major types of collagen, notably type I, type II, type IV and type V. In vivo, uPARAP-dependent collagen turnover has been shown to be central to the development of bone in cattle (Fasquelle et al., 2009 ▸) and mice (Wagenaar-Miller et al., 2007 ▸; Engelholm et al., 2001 ▸). uPARAP deficiency leads to excess collagen deposition as a matrix in murine lung (Bundesmann et al., 2012 ▸), kidney (Eddy, 2009 ▸) and liver fibrosis (Madsen et al., 2012 ▸) models as well as the accumulation of collagen I and collagen IV inside tumours in breast carcinoma (Curino et al., 2005 ▸). Increased expression of uPARAP is associated with tumour progression in the stroma of several types of cancer (Nielsen et al., 2002 ▸; Kogianni et al., 2009 ▸; Takahashi et al., 2011 ▸; Sulek et al., 2006 ▸) and in wound healing (Honardoust et al., 2006 ▸; Rohani et al., 2014 ▸) in humans.
uPARAP consists of a large ectodomain, a transmembrane segment and a short cytoplasmic region. The ectodomain of uPARAP includes, from its amino-terminus, a single cysteine-rich (CysR) domain, a single fibronectin type II (FnII) domain and eight C-type lectin-like domains (CTLDs) (Behrendt et al., 2000 ▸; East & Isacke, 2002 ▸). The collagen-binding activity of uPARAP has been ascribed to the FnII domain, but the adjacent domains have been shown to stabilize the interaction with collagen (Jürgensen et al., 2014 ▸). CTLD2 is the only CTLD with lectin activity among the eight CTLDs of this receptor. It has recently been found to interact directly with glycosylated collagens and modulates the endocytosis of glycosylated collagens such as basement membrane collagen IV (Jürgensen et al., 2011 ▸). The first four domains of uPARAP are important for the binding of collagen and glycans, and we refer to these domains as the ligand-binding region (LBR) in this study. Conformational rearrangements of the LBR of uPARAP (uPARAP-LBR) as well as the other member of the mannose family dependent on the pH have previously been indicated by electron-microscopy studies (Rivera-Calzada et al., 2003 ▸; Boskovic et al., 2006 ▸; He & Bjorkman, 2011 ▸; Cao et al., 2015 ▸). However, detailed structural information on uPARAP and its domain orientation is currently not available, but will be critical for the further understanding of collagen binding and the pH-induced conformational changes.
A previous structural study using uPARAP-LBR expressed in Pichia pastoris was hindered by the highly heterogeneous glycans in the recombinant protein and proteolytic degradation by endogenous proteases from P. pastoris (unpublished results). To overcome these problems, a Drosophila S2 insect expression system was used for uPARAP-LBR expression owing to its capability to generate homogeneously glycosylated recombinant proteins. Up to 100 mg of uPARAP-LBR per litre of culture was obtained after induction. uPARAP-LBR was purified to homogeneity and crystals were obtained that were appropriate for further structural studies.
2. Materials and methods
2.1. Macromolecule production
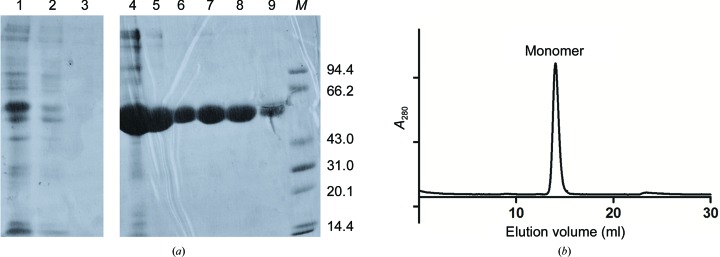
The open reading frame encoding the first four domains of human uPARAP was PCR-amplified using specific primers introducing BglI and MluI restriction sites at the 5′ and 3′ ends, respectively, and was subcloned into pMT/Bip/V5-His vector (Invitrogen) (Table 1 ▸). The recombinant plasmid encoding uPARAP-LBR was then co-transfected with the pCoBlast vector into Drosophila S2 cells (Invitrogen) according to the manufacturer’s instructions. To establish stable clones, blasticidin (Invitrogen) was added to the medium to 25 µg ml−1 2 d after transfection. The selective medium was replaced every 3 d and stably transformed polyclonal cell populations were isolated after 14 d. For large-scale production, recombinant proteins were expressed using EX-CELL 420 medium (Sigma) at a constant temperature of 298 K and 115 rev min−1 in a shaker incubator. Expression of the protein was induced by the addition of 0.75 mM CuSO4, and the culture supernatant was harvested after one week. 700 ml medium was mixed with 10 ml Ni Sepharose Excel resin (GE Healthcare Life Sciences) and incubated at room temperature for 30–60 min with rocking to keep the beads in suspension. After washing with 50 ml buffer containing 5 mM imidazole, the target protein was eluted with 30 ml buffer containing 200 mM imidazole and was dialyzed with standard dialysis membranes (molecular-weight cutoff 10–14 kDa) against 4 l buffer solution consisting of 20 mM Tris–HCl pH 7.4, 50 mM NaCl for 4 h twice at 277 K. The eluted protein was further purified on a Resource Q column (GE Healthcare Life Sciences) with a linear gradient of 50–300 mM NaCl. All fractions were analysed on 12% polyacrylamide gels (Fig. 1 ▸ a). Fractions containing pure uPARAP-LBR were pooled and concentrated to 6 mg ml−1. For deglycosylation of the protein, recombinant MBP-PNGase was expressed and purified using an Ni–NTA column and then dialysed against 20 mM Tris–HCl pH 7.4, 50 mM NaCl. 10 mg pure uPARAP-LBR was subjected to deglycosylation using 100 µg recombinant MBP-PNGase for 4 h at 277 K before anion-exchange chromatography. Protein for crystallization was analysed by size-exclusion chromatography (SEC) on a Superdex 200 10/300 GL column (GE Healthcare Life Sciences) using an ÄKTA FPLC system (Fig. 1 ▸ b).
Table 1. Macromolecule-production information.
| Source organism | Homo sapiens |
| DNA source | cDNA |
| Forward primer† | GGAAGATCTGGCGCCCCTGGGGAC |
| Reverse primer‡ | CGACGCGTCAGCTGGCCTGCCTTCTTG |
| Cloning vector | pMT/Bip/BSD |
| Expression vector | pMT/Bip/BSD |
| Expression host | Drosophila S2 cells |
| Complete amino-acid sequence of the construct produced§ | RSGAPGDAALPEPNIFLIFSHGLQGCLEAQGGQVRVTPACNTSLPAQRWKWVSRNRLFNLGTMQCLGTGWPGTNTTASLGMYECDREALNLRWHCRTLGDQLSLLLGARTSNISKPGTLERGDQTRSGQWRIYGSEEDLCALPYHEVYTIQGNSHGKPCTIPFKYDNQWFHGCTSTGREDGHLWCATTQDYGKDERWGFCPIKSNDCETFWDKDQLTDSCYQFNFQSTLSWREAWASCEQQGADLLSITEIHEQTYINGLLTGYSSTLWIGLNDLDTSGGWQWSDNSPLKYLNWESDQPDNPSEENCGVIRTESSGGWQNRDCSIALPYVCKKKPNATAEPTPPDRWANVKVECEPSWQPFQGHCYRLQAEKRSWQESKKACLRGGGDLVSIHSMAELEFITKQIKQEVEELWIGLNDLKLQMNFEWSDGSLVSFTHWHPFEPNNFRDSLEDCVTIWGPEGRWNDSPCNQSLPSICKKAGQLTRTGHHHHHH |
The BglII restriction site is underlined.
The MluI restriction site is underlined.
His6-tag and vector residues are underlined.
Figure 1.
Purification of recombinant uPARAP-LBR. (a) SDS–PAGE analysis of samples taken at different steps during purification of uPARAP-LBR. Lane 1, supernatant from medium; lane 2, flowthrough from Ni Sepharose Excel; lanes 3, wash from Ni Sepharose Excel; lanes 4–5, elution fractions from Ni Sepharose Excel; lanes 6–9, elution fractions from Resource Q; lane M, molecular-weight marker (labelled in kDa). (b) Size-exclusion chromatography of uPARAP-LBR on a Superdex 200 10/300 GL column. The protein eluted with a retention volume of 14.4 ml, which corresponds to a molecular weight of approximately 57 kDa as a monomer.
2.2. Crystallization
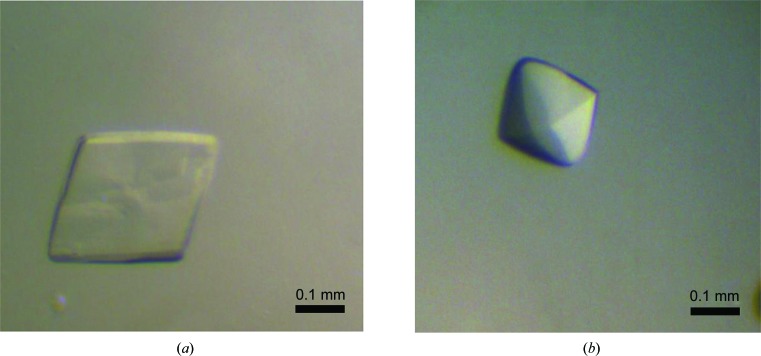
There are three potential N-linked glycosylation sites in uPARAP-LBR and two predicted calcium-binding sites in CTLD2. The glycosylated and deglycosylated proteins from the Drosophila S2 expression system were thus screened for crystallization in the presence or absence of 20 µM calcium using a Phoenix crystallization robot (Art Robbins Instruments) with commercial screening kits from XtalQuest (BioXtal) and Hampton Research (Index). The preliminary crystallization screening was performed using the sitting-drop vapour-diffusion method in 96-well plates, with each well containing 60 µl reservoir solution and with drops consisting of 0.4 µl protein solution mixed with 0.4 µl reservoir solution. After optimization, well formed crystals of uPARAP-LBR (Fig. 2 ▸) were obtained as detailed in Table 2 ▸. Various cryoprotectants (glycerol, 2-methyl-2,4-pentanediol, ethylene glycol and sucrose) were screened at various concentrations (10–30%). Crystals were found to be stable in reservoir solution containing 25%(v/v) ethylene glycol. Crystallization information is summarized in Table 2 ▸.
Figure 2.
Crystals of recombinant uPARAP-LBR. (a) Crystal of human uPARAP-LBR obtained at pH 5.3. The approximate dimensions of the crystal used for data collection were 0.25 × 0.25 × 0.02 mm. (b) Crystal of human uPARAP-LBR obtained at pH 7.4. The approximate dimensions of the crystal used for data collection were 0.2 × 0.22 × 0.15 mm. The scale bar represents 0.1 mm.
Table 2. Crystallization of uPARAP-LBR at two different pH values.
| Protein | Deglycosylated uPARAP-LBR | Glycosylated uPARAP-LBR |
|---|---|---|
| Crystallization pH | 5.3 | 7.4 |
| Method | Sitting-drop vapour diffusion | Sitting-drop vapour diffusion |
| Plate type | 48-well plate | 48-well plate |
| Temperature (K) | 293 | 293 |
| Protein concentration (mgml1) | 6 | 6 |
| Buffer composition of protein solution | 5mM TrisHCl pH 7.4, 200mM NaCl | 5mM TrisHCl pH 7.4, 100mM NaCl |
| Composition of reservoir solution | 812%(w/v) PEG 3350, 200mM NaCl, 35mM CaCl2 in 50mM sodium acetate pH 5.3 | 57% PEG 3350, 1mM CaCl2 in 100mM HEPES pH 7.4 |
| Volume and ratio of drop | 2l (1:1 ratio of protein solution and reservoir solution) | 2l (1:1 ratio of protein solution and reservoir solution) |
| Volume of reservoir (l) | 100 | 100 |
2.3. Data collection and processing
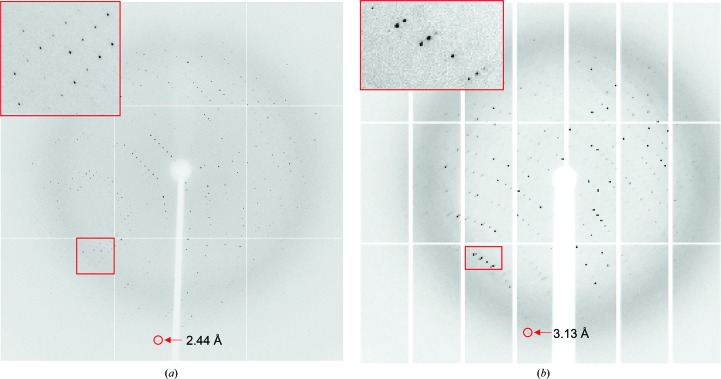
Prior to data collection, the crystals were picked up in a cryoloop (0.3–0.4 mm; Hampton Research), immediately soaked in cryoprotectant solution for 10–20 s and then flash-cooled in liquid nitrogen. For the crystals obtained at pH 5.3, diffraction experiments were conducted at 100 K using an ADSC Q315 detector on beamline BL17U at Shanghai Synchrotron Radiation Facility (SSRF). A total of 360 images were collected at a crystal-to-detector distance of 350 mm with 1 s exposure for every 0.5° oscillation frame (Fig. 3 ▸ a). Diffraction data sets were processed using the xia2 automated data-processing pipeline (Winter, 2010 ▸) running XDS (Kabsch, 2010 ▸). For the crystals obatined at pH 7.4, diffraction experiments were conducted at 100 K using a PILATUS3 6M detector on beamline BL19U at SSRF. A total of 720 images were collected at a crystal-to-detector distance of 350 mm with 0.2 s exposure for every 0.5° oscillation frame (Fig. 3 ▸ b). Intensity data were integrated and scaled using the HKL-3000 package (Minor et al., 2006 ▸). Additional data-collection and processing statistics are summarized in Table 3 ▸.
Figure 3.
Representative diffraction images obtained from crystals grown at pH 5.3 (a) and pH 7.4 (b). The inset is a magnification of the region of the diffraction pattern in the red box.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Crystallization pH | pH 5.3 | pH 7.4 |
|---|---|---|
| Diffraction source | BL17U, SSRF | BL19U, SSRF |
| Wavelength () | 0.979 | 0.979 |
| Temperature (K) | 100 | 100 |
| Detector | ADSC Q315 | PILATUS3 6M |
| Crystal-to-detector distance (mm) | 350 | 350 |
| Rotation range per image () | 0.5 | 0.5 |
| Total rotation range () | 180 | 360 |
| Exposure time per image (s) | 1 | 0.2 |
| Space group | P21 | P41212 |
| a, b, c () | 74.27, 102.7, 87.76 | 74.46, 74.46, 222.36 |
| , , () | 90.00, 94.54, 90.00 | 90.00, 90.00, 90.00 |
| Mosaicity () | 0.35 | 0.42 |
| Resolution range () | 43.742.44 (2.5272.44) | 503.13 (3.183.13) |
| Total No. of reflections | 373017 | 2973493 |
| No. of unique reflections | 48904 (4828) | 42565 (2070) |
| Completeness (%) | 99.9 (99.9) | 100.0 (100.0) |
| Multiplicity | 7.6 (7.7) | 13.8 (13.1) |
| I/(I) | 16.2 (2.9) | 22.3 (2.1) |
| R r.i.m. | 0.085 (0.711) | 0.102 (0.970) |
| Overall B factor from Wilson plot (2) | 39.7 | 73.9 |
3. Results and discussion
Recombinant uPARAP-LBR with a C-terminal His6 tag was overexpressed in Drosophila S2 cells and purified to homogeneity by two chromatographic steps (Fig. 1 ▸ a), yielding approximately 70 mg of target protein per litre of expression medium. SDS–PAGE analysis revealed that uPARAP-LBR was homogenous, with little glycosylation and with a molecular weight near to 57 kDa (the calculated molecular weight of uPARAP-LBR; Fig. 1 ▸ a). The protein was further confirmed to exist as a monomer based on its elution volume (14.4 ml) from a gel-filtration column (Fig. 1 ▸ b). Using this protein, we obtained crystals with a good morphology (Fig. 2 ▸) compared with the highly twinned crystals obtained using deglycosylated protein expressed in P. pastoris (unpublished results). Crystals obtained at two pH values (5.3 and 7.4) diffracted to 2.44 Å (Fig. 3 ▸ a) and 3.13 Å (Fig. 3 ▸ b) resolution, respectively. Two data sets were collected and the statistics of data collection are listed in Table 3 ▸. The crystal obtained at pH 5.3 belonged to space group P21, with unit-cell parameters a = 74.27, b = 102.7, c = 87.76 Å. The most probable Matthews coefficient was 2.96 Å3 Da−1 (Matthews, 1968 ▸) and corresponded to two protein molecules per asymmetric unit, with a solvent content of 58.43%. The crystal obtained at pH 7.4 belonged to space group P41212, with unit-cell parameters a = b = 74.46, c = 222.36 Å. The most probable Matthews coefficient was 2.72 Å3 Da−1 and corresponded to one protein molecule per asymmetric unit, with a solvent content of 54.78%.
No multidomain structure of any member of the mannose receptor family is currently available as a search template for molecular replacement (MR). Therefore, we used the structures of similar individual domains as molecular-replacement models. A BLAST search gave the following best hits for each domain: the cysteine-rich domain of a mannose receptor (Liu et al., 2001 ▸; PDB entry 1fwu; 29% identity over 130 residues to the CysR domain), the gelatin-binding domain of fibronectin (Bocquier et al., 1999 ▸; PDB entry 1qo6; 52% identity over 51 residues in the FnII domain) and the c-type lectin domain from rat aggrecan (Lundell et al., 2004 ▸; PDB entry 1tdq; 42% identity over 127 residues to CTLD2 and 42% identity over 127 residues to CTLD1). MR search models for each domain were constructed with CHAINSAW from the CCP4 suite (Winn et al., 2011 ▸) using the above structures as templates, and MR trials for the pH 5.3 data set were performed using Phaser (McCoy et al., 2007 ▸). Only the CTLD1 and FnII models yielded reasonable MR solutions, which were improved by morphing (Terwilliger et al., 2012 ▸) within the PHENIX suite of programs (Adams et al., 2010 ▸). The AutoBuild (Terwilliger et al., 2008 ▸) routine in PHENIX was used to extend the model, yielding the majority of the residues in the first three domains (CysR, FnII and CTLD1). The RESOLVE (Terwilliger, 2000 ▸) and Buccaneer (Cowtan, 2006 ▸) options for density modification and model building were used during AutoBuild. The CTLD2 model then yielded a good MR solution with the first three domains fixed during molecular replacement. The pH 7.4 structure was solved using the low-pH model. Manual model building and structure refinement of both structures are currently in progress.
Acknowledgments
We thank Shanghai Synchrotron Radiation Facility beamlines BL17U and BL19U for X-ray data collection. We also wish to thank Dr Niels Behrendt from the Biotech Research and Innovation Centre for the gift of the cDNA clone for human uPARAP. This work was supported by grants from NSFC (31570745, 31170707 and 31370737) and SRF for ROCS, SEM.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Behrendt, N., Jensen, O. N., Engelholm, L. H., Mørtz, E., Mann, M. & Danø, K. (2000). J. Biol. Chem. 275, 1993–2002. [DOI] [PubMed]
- Bocquier, A. A., Potts, J. R., Pickford, A. R. & Campbell, I. D. (1999). Structure, 7, 1451–1460. [DOI] [PubMed]
- Boskovic, J., Arnold, J. N., Stilion, R., Gordon, S., Sim, R. B., Rivera-Calzada, A., Wienke, D., Isacke, C. M., Martinez-Pomares, L. & Llorca, O. (2006). J. Biol. Chem. 281, 8780–8787. [DOI] [PubMed]
- Bundesmann, M. M., Wagner, T. E., Chow, Y.-H., Altemeier, W. A., Steinbach, T. & Schnapp, L. M. (2012). Am. J. Respir. Cell Mol. Biol. 46, 233–239. [DOI] [PMC free article] [PubMed]
- Cao, L., Shi, X., Chang, H., Zhang, Q. & He, Y. (2015). Proc. Natl Acad. Sci. USA, 112, 7237–7242. [DOI] [PMC free article] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Curino, A., Engelholm, L., Yamada, S., Holmbeck, K., Lund, L., Molinolo, A., Behrendt, N., Nielsen, B. S. & Bugge, T. H. (2005). J. Cell Biol. 169, 977–985. [DOI] [PMC free article] [PubMed]
- East, L. & Isacke, C. M. (2002). Biochim. Biophys. Acta, 1572, 364–386. [DOI] [PubMed]
- Eddy, A. A. (2009). Thromb. Haemost. 101, 656–664. [PMC free article] [PubMed]
- Engelholm, L. H., List, K., Netzel-Arnett, S., Cukierman, E., Mitola, D. J., Aaronson, H., Kjøller, L., Larsen, J. K., Yamada, K. M., Strickland, D. K., Holmbeck, K., Danø, K., Birkedal-Hansen, H., Behrendt, N. & Bugge, T. H. (2003). J. Cell Biol. 160, 1009–1015. [DOI] [PMC free article] [PubMed]
- Engelholm, L. H., Nielsen, B. S., Netzel-Arnett, S., Solberg, H., Chen, X.-D., Lopez Garcia, J. M., Lopez-Otin, C., Young, M. F., Birkedal-Hansen, H., Danø, K., Lund, L. R., Behrendt, N. & Bugge, T. H. (2001). Lab. Invest. 81, 1403–1414. [DOI] [PubMed]
- Fasquelle, C., Sartelet, A., Li, W., Dive, M., Tamma, N., Michaux, C., Druet, T., Huijbers, I. J., Isacke, C. M., Coppieters, W., Georges, M. & Charlier, C. (2009). PLoS Genet. 5, e1000666. [DOI] [PMC free article] [PubMed]
- He, Y. & Bjorkman, P. J. (2011). Proc. Natl Acad. Sci. USA, 108, 12431–12436. [DOI] [PMC free article] [PubMed]
- Honardoust, H. A., Jiang, G., Koivisto, L., Wienke, D., Isacke, C. M., Larjava, H. & Häkkinen, L. (2006). Histopathology, 49, 634–648. [DOI] [PubMed]
- Jürgensen, H. J., Johansson, K., Madsen, D. H., Porse, A., Melander, M. C., Sørensen, K. R., Nielsen, C., Bugge, T. H., Behrendt, N. & Engelholm, L. H. (2014). J. Biol. Chem. 289, 7935–7947. [DOI] [PMC free article] [PubMed]
- Jürgensen, H. J., Madsen, D. H., Ingvarsen, S., Melander, M. C., Gårdsvoll, H., Patthy, L., Engelholm, L. H. & Behrendt, N. (2011). J. Biol. Chem. 286, 32736–32748. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kogianni, G., Walker, M. M., Waxman, J. & Sturge, J. (2009). Eur. J. Cancer, 45, 685–693. [DOI] [PubMed]
- Liu, Y., Misulovin, Z. & Bjorkman, P. J. (2001). J. Mol. Biol. 305, 481–490. [DOI] [PubMed]
- Lundell, A., Olin, A. I., Mörgelin, M., al-Karadaghi, S., Aspberg, A. & Logan, D. T. (2004). Structure, 12, 1495–1506. [DOI] [PubMed]
- Madsen, D. H., Jürgensen, H. J., Ingvarsen, S., Melander, M. C., Vainer, B., Egerod, K. L., Hald, A., Rønø, B., Madsen, C. A., Bugge, T. H., Engelholm, L. H. & Behrendt, N. (2012). J. Pathol. 227, 94–105. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Nielsen, B. S., Rank, F., Engelholm, L. H., Holm, A., Danø, K. & Behrendt, N. (2002). Int. J. Cancer, 98, 656–664. [DOI] [PubMed]
- Rivera-Calzada, A., Robertson, D., MacFadyen, J. R., Boskovic, J., Isacke, C. M. & Llorca, O. (2003). EMBO Rep. 4, 807–812. [DOI] [PMC free article] [PubMed]
- Rohani, M. G., Chow, Y.-H., Razumova, M. V., Ash, S., Hung, C. F. & Schnapp, L. M. (2014). PLoS One, 9, e92660. [DOI] [PMC free article] [PubMed]
- Sulek, J., Wagenaar-Miller, R. A., Shireman, J., Molinolo, A., Madsen, D. H., Engelholm, L. H., Behrendt, N. & Bugge, T. H. (2006). J. Histochem. Cytochem. 55, 347–353. [DOI] [PubMed]
- Takahashi, S., Yamada-Okabe, H., Hamada, K., Ohta, S., Kawase, T., Yoshida, K. & Toda, M. (2011). J. Neurooncol. 103, 267–276. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Read, R. J., Adams, P. D., Brunger, A. T., Afonine, P. V., Grosse-Kunstleve, R. W. & Hung, L.-W. (2012). Acta Cryst. D68, 861–870. [DOI] [PMC free article] [PubMed]
- Wagenaar-Miller, R. A., Engelholm, L. H., Gavard, J., Yamada, S. S., Gutkind, J. S., Behrendt, N., Bugge, T. H. & Holmbeck, K. (2007). Mol. Cell. Biol. 27, 6309–6322. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.