Lytic polysaccharide monooxygenase from J. denitrificans was purified and crystallized. Room-temperature X-ray and neutron diffraction data were collected to 1.1 and 2.1 Å resolution, respectively.
Keywords: lytic polysaccharide monooxygenase, chitin, Jonesia denitrificans, biofuel, neutron crystallography
Abstract
Bacteria and fungi express lytic polysaccharide monooxgyenase (LPMO) enzymes that act in conjunction with canonical hydrolytic sugar-processing enzymes to rapidly convert polysaccharides such as chitin, cellulose and starch to single monosaccharide products. In order to gain a better understanding of the structure and oxidative mechanism of these enzymes, large crystals (1–3 mm3) of a chitin-processing LPMO from the Gram-positive soil bacterium Jonesia denitrificans were grown and screened for their ability to diffract neutrons. In addition to the collection of neutron diffraction data, which were processed to 2.1 Å resolution, a high-resolution room-temperature X-ray diffraction data set was collected and processed to 1.1 Å resolution in space group P212121. To our knowledge, this work marks the first successful neutron crystallographic experiment on an LPMO. Joint X-ray/neutron refinement of the resulting data will reveal new details of the structure and mechanism of this recently discovered class of enzymes.
1. Introduction
Lytic polysaccharide monooxygenases (LPMOs) are recently discovered enzymes that use an unusual oxidative mechanism to cleave polysaccharides which differs from the canonical mode of action used by glycoside hydrolases (Vaaje-Kolstad et al., 2010 ▸; Horn et al., 2012 ▸). In addition to being of interest from the standpoint of basic biochemical research, these enzymes have potentially wide-ranging applications in biofuel production strategies owing to their ability to efficiently break down cellulosic materials. However, despite the availability of several X-ray crystal and NMR structures of LPMOs, in addition to extensive biochemical analyses, the detailed catalytic mechanism of these enzymes has remained elusive. LPMOs are metalloenzymes that have an active site near the surface of the protein where copper is coordinated (Quinlan et al., 2011 ▸; Aachmann et al., 2012 ▸). Two histidine residues bind the metal in a conserved structural arrangement known as the histidine brace (Quinlan et al., 2011 ▸). Water molecules and an aromatic residue also play key roles in coordinating the metal (Forsberg et al., 2014 ▸). Although the mechanism of LPMOs and the intermediates formed during the reaction remain speculative, it is clear that the activity of LPMO involves the reduction of dioxygen by copper(I) and the subsequent attack of an activated oxygen species on either the C1 or C4 position of the scissile bond in the polysaccharide substrate, ultimately leading to its cleavage (Vaaje-Kolstad et al., 2010 ▸; Beeson et al., 2012; Kim et al., 2014 ▸; Kjaergaard et al., 2014 ▸). In the case of C1 oxidation, the product δ-1,5 lactones are thought to be subsequently hydrated to form aldonic acids (Vaaje-Kolstad et al., 2010 ▸; Forsberg et al., 2011 ▸), while for C4 oxidation the product 4-ketoaldoses are subsequently hydrated to form gem-diols (Forsberg et al., 2014 ▸; Isaksen et al., 2014 ▸). A putative electron-transfer pathway has also been proposed to reduce copper(II) back to copper(I) in order to complete the catalytic cycle (Beeson et al., 2012 ▸; Hemsworth et al., 2013 ▸; Lo Leggio et al., 2015 ▸).
In order to gain a better understanding of the structure and mechanism of LPMOs, we targeted several cellulose- and chitin-processing LPMOs for neutron crystallographic studies. Following extensive crystallization and screening efforts using neutron sources at both Los Alamos National Laboratory and Oak Ridge National Laboratory, the chitin-processing N-terminal LPMO domain of a three-domain chitinase from Jonesia denitrificans, referred to here as JdLPMO10A, was found to produce crystals of a suitable size and quality for neutron diffraction studies. Neutron structural studies will allow an intricately detailed analysis of LPMOs in terms of hydrogen-bonding networks, water structure and the protonation states of the residues involved in catalysis and the putative electron-transfer pathway.
2. Materials and methods
2.1. Macromolecule production
A codon-optimized gene fragment encoding the N-terminal LPMO domain of J. denitrificans Jd_1381 (UniProt ID C7R4I0, 651 amino acids in total) was synthesized (GenScript). The gene fragment encoding the LPMO domain with its native signal sequence (residues 1–143 followed by a stop codon) was amplified for cloning into the pUCBB-eGFP expression vector (Vick et al., 2011 ▸). The pUCBB-eGFP vector was pre-cut with restriction endonucleases (BamHI and NotI) to remove nucleotides encoding the eGFP protein prior to ligation of the lpmo gene using the T4 DNA ligase kit (Invitrogen). The resulting expression vector was transformed by heat shock into chemically competent One Shot BL21 Star (DE3) Escherichia coli cells (Invitrogen). Fresh colonies were inoculated in LB-Amp (50 µg ml−1 ampicillin) medium and grown at 30°C for 16 h. Cells were harvested by centrifugation (9800g for 10 min) and the periplasmic fraction was extracted using the cold osmotic shock method (Manoil & Beckwith, 1986 ▸). The periplasmic extract was sterilized by filtration (0.22 µm) and adjusted to 50 mM Tris–HCl pH 8.0 before loading it onto a 5 ml anion-exchange HiTrap DEAE FF column connected to an ÄKTApurifier FPLC system (GE Healthcare). Protein in Tris buffer was eluted by applying a 60-column-volume linear salt gradient of 0–500 mM NaCl. LPMO-containing fractions were pooled and concentrated to approximately 1 ml using Amicon Ultra centrifugal filters with a molecular-weight cutoff of 10 kDa (Millipore) and were then loaded onto a HiLoad 16/60 Superdex 75 size-exclusion column (GE Healthcare) with a running buffer consisting of 20 mM Tris–HCl pH 8.0, 250 mM NaCl. Protein-containing fractions were analyzed by SDS–PAGE and subsequently concentrated using an Amicon Ultra centrifugal filter (Millipore). Details of the cloning and protein-production procedures are summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | J. denitrificans strain ATCC 14870 |
| DNA source | Synthetic DNA |
| Forward primer | 5-CCGTAGCAATGGATCCATGAAGAAGAGAAAGTTGAGAGCGTCAGC-3 |
| Reverse primer | 5-TCGTAATGCCGCGGCCGCTCATGAGACCACAACATCCATACAGTTG-3 |
| Expression vector | pUCBB-eGFP |
| Expression host | E. coli BL21 Star (DE3) |
| Complete amino-acid sequence of the construct produced | HGWVTDPPSRQALCASGETSFDCGQISYEPQSVEAPKGATTCSGGNEAFAILDDNSKPWPTTEIASTVDLTWKLTAPHNTSTWEYFVDGQLHQTFDQKGQQPPTSLTHTLTDLPTGEHTILARWNVSNTNNAFYNCMDVVVS |
2.2. Crystallization
Prior to crystallization, the purified enzyme was incubated with a threefold molar excess of CuSO4 for 30 min at room temperature as described previously (Forsberg et al., 2014 ▸). To remove excess copper, the protein was loaded onto a PD MidiTrap G-25 desalting column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 8.0. Crystals were grown by sitting-drop vapor diffusion in grease-sealed nine-well sandwich boxes with 25 ml well solution. For the JdLPMO10A crystal grown for neutron data collection, 14 µl protein solution at 48 mg ml−1 was mixed with 14 µl reservoir crystallization buffer consisting of 1.9 M dl-malic acid pH 7.0 (adjusted with NaOH), resulting in a crystal with dimensions of 3 × 1.8 × 0.5 mm. The JdLPMO10A crystal grown for X-ray data collection was obtained under the same experimental conditions but with a total drop size of 60 µl, resulting in a crystal with dimensions of 2 × 0.6 × 0.6 mm. Prior to data collection, the crystals were mounted in quartz capillaries, the surrounding mother liquor was thoroughly wicked away and a plug of precipitant solution prepared in D2O was inserted adjacent to the crystal. The ends of the capillary were sealed with wax and the crystal was allowed to exchange with the deuterated buffer at room temperature for two weeks. Details of protein crystallization are summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Sitting-drop vapor diffusion |
| Plate type | 9-well |
| Temperature (K) | 293 |
| Protein concentration (mgml1) | 48 |
| Buffer composition of protein solution | 20mM TrisHCl pH 8.0 |
| Composition of reservoir solution | 1.9M DL-malic acid pH 7.0 |
| Volume and ratio of drop | 14:14l (neutron); 30:30l (X-ray) |
| Volume of reservoir (ml) | 25 |
2.3. Data collection and processing
2.3.1. Neutron data
Initial screening of crystals was performed at the Protein Crystallography Station (PCS) located at the Los Alamos Neutron Science Center (Los Alamos National Laboratory). A room-temperature neutron diffraction data set was collected from a single crystal using the MaNDi instrument at the Spallation Neutron Source (Oak Ridge National Laboratory; Coates et al., 2010 ▸, 2015 ▸). A single ω angle was chosen for data collection and each fixed image was separated by a 10° φ rotation, resulting in 12 fixed images spanning 120°. These images were processed and integrated with MANTID and wavelength-normalized and merged with LAUENORM (Campbell et al., 1998 ▸).
2.3.2. X-ray data
A room-temperature X-ray data set was collected on beamline 4.2.2 at the Advanced Light Source, Berkeley using a RDI 8M CMOS detector in shutterless mode. 100 images were collected in nine segments along the length of the crystal with a 0.10 s exposure time and a 0.20° oscillation angle, with a total of 20° per segment. These images were integrated using XDS (Kabsch, 2010a ▸), merged using XSCALE (Kabsch, 2010b ▸) and converted to MTZ format using SCALA (Evans, 2006 ▸). Data-collection and processing statistics are presented in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| X-rays | Neutrons | |
|---|---|---|
| Diffraction source | Beamline 4.2.2, ALS, Berkeley | MaNDi, ORNL |
| Wavelength () | 1.000 | 24 |
| Temperature (K) | 295 | 295 |
| Detector | RDI 8M CMOS | 27 SNS Anger cameras |
| Crystal-to-detector distance (mm) | 100 | 450 |
| Rotation range per image () | 0.2 | Fixed |
| Total rotation range () | 180 | 120 |
| Exposure time per image (s) | 0.1s | 20h |
| Space group | P212121 | P212121 |
| Unit-cell parameters (, ) | a = 32.0, b = 75.6, c = 120.4, = = = 90.0 | a = 32.5, b = 76.4, c = 122.1, = = = 90.0 |
| Mosaicity () | 0.07 | NA |
| Resolution range () | 32.041.10 (1.161.10) | 11.972.10 (2.172.10) |
| Total No. of reflections | 1248110 (170145) | 62823 (2757) |
| No. of unique reflections | 116274 (16352) | 13989 (1091) |
| Completeness (%) | 97.3 (95.0) | 76.02 (59.4)† |
| Multiplicity | 10.7 (10.4) | 4.5 (2.5) |
| I/(I) | 13.7 (1.5) | 9.4 (2.7) |
| R p.i.m. | 0.031 (0.436) | 0.089 (0.196) |
| Overall B factor from Wilson plot (2) | 9.2 | 24.7 |
Owing to the limited availability of neutron instrumentation and beam time, it is often not possible to collect data sets that have the data-completeness levels of X-ray data.
3. Results and discussion
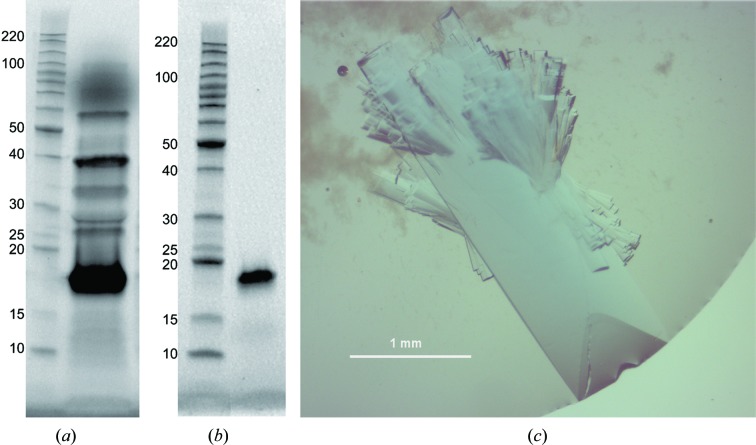
Recombinant JdLPMO10A was expressed in E. coli and highly pure enzyme was obtained (Figs. 1 ▸ a and 1 ▸ b). JdLPMO10A crystals that were large enough for neutron diffraction studies (Fig. 1 ▸ c) were grown using a protein concentration of 48 mg ml−1 in sitting-drop vapor-diffusion experiments, in which the drop sizes were as large as 60 µl. Crystals grew in 1–2 weeks under these conditions, and the resulting crystals had a noticeable blue color which was attributable to copper(II) bound in the enzyme active site (Fig. 1 ▸ c). Representative diffraction data images for both the X-ray and neutron diffraction experiments are shown in Fig. 2 ▸. In order to assess the potential X-ray radiation damage during data collection, 180° of data were initially collected at the tip of the large crystal and processed in XDS. Analysis using XDSSTAT showed an increase in the slope of the plot of R d versus image number after ∼30°, indicating radiation damage. Data were thus collected in 20° wedges, translating 1.5 beam widths across the sample after each wedge. After processing, each wedge was individually analyzed in XDSSTAT and the R d versus image number plot was flat in each case. The space group was assigned as P212121, with unit-cell parameters a = 32.0, b = 75.6, c = 120.4 Å for the X-ray data and a = 32.5, b = 76.4, c = 122.1 Å for the neutron data. The minor variation in unit-cell parameters can be attributed to the fact that a different crystal was used for each data-set collection and cell refinement was performed using different software. It should be noted that the B-factor values from the Wilson plot from the X-ray and neutron data are not comparable, as the data are from vastly different resolution ranges. Furthermore, the neutron experiment used a Laue-based technique, whereas the X-ray data were collected using a monochromatic technique. Laue experiments in general are not able to produce R merge values similar to those produced by monochromatic experiments. The predicted theoretical molecular weight based on the amino-acid sequence of JdLPMO10A is 15 531.9 Da. Considering the molecular weight of two protein molecules in the asymmetric unit, this gives a Matthews coefficient of 2.4 Å3 Da−1 (Matthews, 1968 ▸; Kantardjieff & Rupp, 2003 ▸) and an estimated solvent content of 47.6% (from the X-ray data) or 49.6% (from the neutron data). The lower solvent content of the JdLPMO10A crystals compared with other LPMO crystals tested (>60%), in addition to the relatively large size of the crystals, is likely to account for the better diffraction of these crystals at neutron sources. Growing crystals that are large enough for neutron diffraction studies is one of the main difficulties that impede structure determination using this method (Ng et al., 2015 ▸). The amount of available beamtime for neutron diffraction data collection from this sample was limited by operational availability. In the future more data-collection time can be used to collect a more complete data set. Additionally, more detectors have recently been added to MaNDi to increase the detector solid-angle coverage, thereby increasing the number of data collected per setting and increasing the data completeness.
Figure 1.
SDS–PAGE analysis of JdLPMO10A production and the resulting crystals. In (a) lane 1 contains Benchmarker protein ladder (Life Technologies) with molecular weights labelled in kDa. Lane 2 contains the E. coli periplasmic extract containing overexpressed JdLPMO10A (15.5 kDa). In (b), lane 1 contains protein ladder and lane 2 contains JdLPMO10A purified by anion-exchange and size-exclusion chromatography used for protein crystallization. (c) A typical crystal grown for neutron diffraction studies of JdLPMO10A. Smaller crystals attached to the large crystal were either removed with a loop or were displaced during the process of mounting. The remaining satellite crystals did not affect the diffraction of the major crystal.
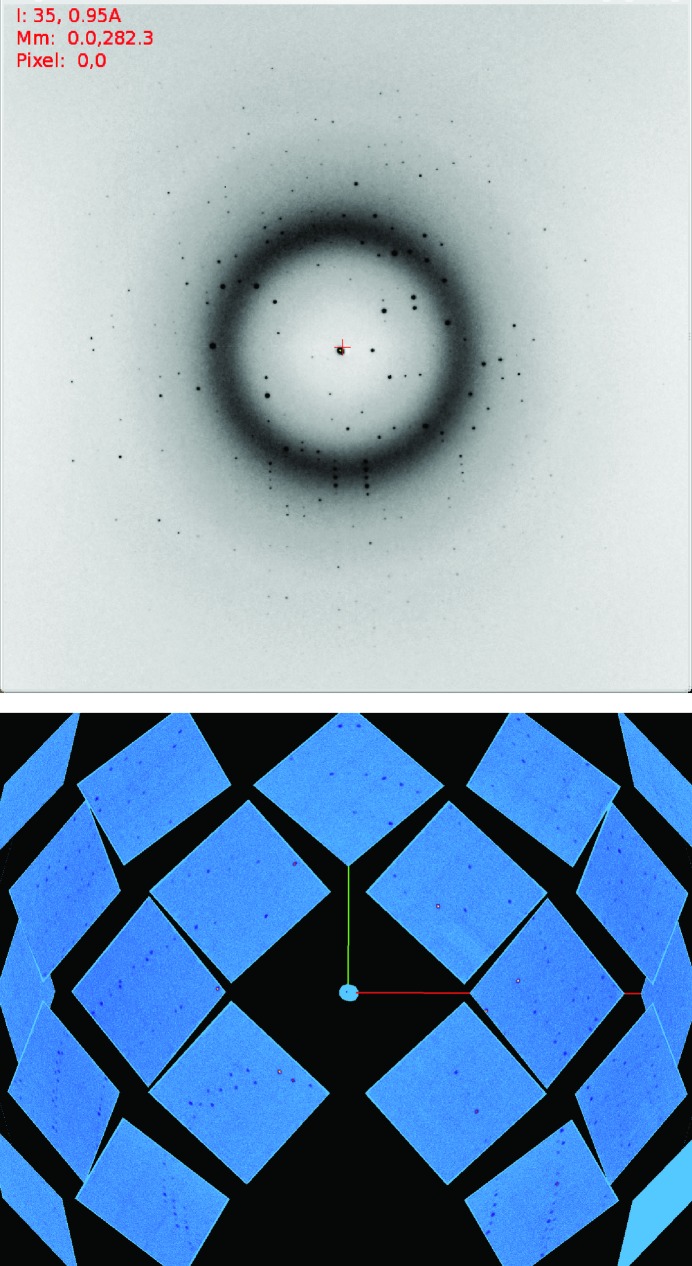
Figure 2.
X-ray (top) and neutron diffraction images (bottom) of a JdLPMO10A crystal.
Structure determination was successfully performed using the recently deposited X-ray crystal structure of JdLPMO10A (PDB entry 5aa7; S. Mekasha, Z. Forsberg, B. Dalhus, S. Choudhary, C. Schmidt-Dannert, G. Vaaje-Kolstad & V. Eijsink, unpublished work) as the starting structure for initial rigid-body refinement. Refinement of the protein model using both the neutron and high-resolution X-ray data is ongoing. A general problem related to X-ray diffraction crystallography of redox-active proteins is photoreduction of catalytic transition metals by the high-energy X-ray beam (Gudmundsson et al., 2014 ▸). Conveniently, the neutron beam does not influence the redox state of transition metals. Thus, fully refined structures of JdLPMO10A can yield detailed insights into the coordination of the copper ion in its oxidized or its reduced state, depending on the experimental conditions. Furthermore, the neutron structure will help in elucidating the protonation states of titratable residues and may allow the discovery of unusual bonding interactions and hydration patterns, as have been reported in other neutron structures (Chen et al., 2012 ▸; Michalczyk et al., 2015 ▸) and which may also be influenced by the active-site redox status. Future planned experiments will involve the determination of neutron structures of LPMOs in the apo form (without a metal bound) and with other metals such as zinc bound, and of structures of LPMOs bound to dioxygen mimics such as cyanide, which has been shown to inhibit the function of LPMO (Vaaje-Kolstad et al., 2010 ▸). These studies will yield new insights into the structure and mechanism of LPMOs and may aid in future engineering efforts towards optimizing these enzymes in biofuel production strategies.
Acknowledgments
JPB, JCHC and CJU were partially funded through the Protein Crystallography Station from the Department of Energy Office of Biological and Environmental Research. The research at the Spallation Neutron Source at Oak Ridge National Laboratory was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy. The Office of Biological and Environmental Research supported the research at Oak Ridge National Laboratory’s Center for Structural Molecular Biology (CSMB) using facilities supported by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy. We thank Claudia Schmidt-Dannert and colleagues at the University of Minnesota for help in enzyme cloning. This work was supported by grants 214138 and 221576 from the Research Council of Norway and by grant 6510 from the VISTA program of the Norwegian Academy of Science and Letters.
References
- Aachmann, F. L., Sørlie, M., Skjak-Braek, G., Eijsink, V. G. H. & Vaaje-Kolstad, G. (2012). Proc. Natl Acad. Sci. USA, 109, 18779–18784. [DOI] [PMC free article] [PubMed]
- Beeson, W. T., Phillips, C. M., Cate, J. H. D. & Marletta, M. A. (2012). J. Am. Chem. Soc. 134, 890–892. [DOI] [PubMed]
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst. 31, 496–502.
- Chen, J. C.-H., Hanson, B. L., Fisher, S. Z., Langan, P. & Kovalevsky, A. Y. (2012). Proc. Natl Acad. Sci. USA, 109, 15301–15306. [DOI] [PMC free article] [PubMed]
- Coates, L., Cuneo, M. J., Frost, M. J., He, J., Weiss, K. L., Tomanicek, S. J., McFeeters, H., Vandavasi, V. G., Langan, P. & Iverson, E. B. (2015). J. Appl. Cryst. 48, 1302–1306.
- Coates, L., Stoica, A. D., Hoffmann, C., Richards, J. & Cooper, R. (2010). J. Appl. Cryst. 43, 570–577.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Forsberg, Z., Mackenzie, A. K., Sørlie, M., Røhr, A. K., Helland, R., Arvai, A. S., Vaaje-Kolstad, G. & Eijsink, V. G. H. (2014). Proc. Natl Acad. Sci. USA, 111, 8446–8451. [DOI] [PMC free article] [PubMed]
- Forsberg, Z., Vaaje-Kolstad, G., Westereng, B., Bunaes, A. C., Stenstrøm, Y., MacKenzie, A., Sørlie, M., Horn, S. J. & Eijsink, V. G. H. (2011). Protein Sci. 20, 1479–1483. [DOI] [PMC free article] [PubMed]
- Gudmundsson, M., Kim, S., Wu, M., Ishida, T., Momeni, M. H., Vaaje-Kolstad, G., Lundberg, D., Royant, A., Ståhlberg, J., Eijsink, V. G. H., Beckham, G. T. & Sandgren, M. (2014). J. Biol. Chem. 289, 18782–18792. [DOI] [PMC free article] [PubMed]
- Hemsworth, G. R., Taylor, E. J., Kim, R. Q., Gregory, R. C., Lewis, S. J., Turkenburg, J. P., Parkin, A., Davies, G. J. & Walton, P. H. (2013). J. Am. Chem. Soc. 135, 6069–6077. [DOI] [PMC free article] [PubMed]
- Horn, S. J., Vaaje-Kolstad, G., Westereng, B. & Eijsink, V. G. H. (2012). Biotechnol. Biofuels, 5, 45. [DOI] [PMC free article] [PubMed]
- Isaksen, T., Westereng, B., Aachmann, F. L., Agger, J. W., Kracher, D., Kittl, R., Ludwig, R., Haltrich, D., Eijsink, V. G. H. & Horn, S. J. (2014). J. Biol. Chem. 289, 2632–2642. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Kim, S., Ståhlberg, J., Sandgren, M., Paton, R. S. & Beckham, G. T. (2014). Proc. Natl Acad. Sci. USA, 111, 149–154. [DOI] [PMC free article] [PubMed]
- Kjaergaard, C. H., Qayyum, M. F., Wong, S. D., Xu, F., Hemsworth, G. R., Walton, D. J., Young, N. A., Davies, G. J., Walton, P. H., Johansen, K. S., Hodgson, K. O., Hedman, B. & Solomon, E. I. (2014). Proc. Natl Acad. Sci. USA, 111, 8797–8802. [DOI] [PMC free article] [PubMed]
- Lo Leggio, L. et al. (2015). Nature Commun. 6, 5961. [DOI] [PMC free article] [PubMed]
- Manoil, C. & Beckwith, J. (1986). Science, 233, 1403–1408. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Michalczyk, R., Unkefer, C. J., Bacik, J.-P., Schrader, T. E., Ostermann, A., Kovalevsky, A. Y., McKenna, R. & Fisher, S. Z. (2015). Proc. Natl Acad. Sci. USA, 112, 5673–5678. [DOI] [PMC free article] [PubMed]
- Ng, J. D., Baird, J. K., Coates, L., Garcia-Ruiz, J. M., Hodge, T. A. & Huang, S. (2015). Acta Cryst. F71, 358–370. [DOI] [PMC free article] [PubMed]
- Quinlan, R. J. et al. (2011). Proc. Natl Acad. Sci. USA, 108, 15079–15084.
- Vaaje-Kolstad, G., Westereng, B., Horn, S. J., Liu, Z., Zhai, H., Sørlie, M. & Eijsink, V. G. H. (2010). Science, 330, 219–222. [DOI] [PubMed]
- Vick, J. E., Johnson, E. T., Choudhary, S., Bloch, S. E., Lopez-Gallego, F., Srivastava, P., Tikh, I. B., Wawrzyn, G. T. & Schmidt-Dannert, C. (2011). Appl. Microbiol. Biotechnol. 92, 1275–1286. [DOI] [PubMed]