Abstract
Some familial platelet disorders are associated with predisposition to leukemia, myelodysplastic syndrome (MDS) or dyserythropoietic anemia.1,2 We identified a family with autosomal dominant thrombocytopenia, high erythrocyte mean corpuscular volume (MCV) and two occurrences of B-cell precursor acute lymphoblastic leukemia (ALL). Whole exome sequencing identified a heterozygous single nucleotide change in ETV6 (Ets Variant Gene 6), c.641C>T, encoding a p.Pro214Leu substitution in the central domain, segregating with thrombocytopenia and elevated MCV. A screen of 23 families with similar phenotype found two with ETV6 mutations. One family had the p.Pro214Leu mutation and one individual with ALL. The other family had a c.1252A>G transition producing a p.Arg418Gly substitution in the DNA binding domain, with alternative splicing and exon-skipping. Functional characterization of these mutations showed aberrant cellular localization of mutant and endogenous ETV6, decreased transcriptional repression and altered megakaryocyte maturation. Our findings underscore a key role for ETV6 in platelet formation and leukemia predisposition.
ETV6 (also known as TEL) encodes a transcriptional repressor in the ETS family.3 ETV6 was initially identified as a tumor suppressor by involvement in somatic translocations in childhood leukemia, including ETV6-RUNX1. These often co-occur with a somatic mutation in the intact ETV6 allele, suggesting that loss of ETV6 function contributes to the development of leukemia.4–6 Somatic ETV6 mutations have also been described in patients with myelodysplastic syndrome (MDS) and T-cell leukemias,7,8 but until recently germline mutations have not been described.9 ETV6 also plays a critical role in hematopoiesis, as demonstrated in animal models.10,11 Erythroid/megakaryocytic conditional Etv6 knockout mice are thrombocytopenic, and megakaryocyte colony formation is absent in homozygous knockout hematopoietic cells and decreased in heterozygotes, indicating Etv6 involvement in thrombopoiesis.12,13 Etv6 is also one of four transcription factors shown to be sufficient to differentiate mouse fibroblasts into hematopoietic lineage cells.14
We present here three families with germline mutations in ETV6 and defects in hematopoiesis. Affected members (n=5) in the original family from the United States (Family 1 Fig 1a) have variable thrombocytopenia (67,000–132,000 platelets/uL) and elevated red cell MCV (92.5–101.5 fL), suggesting a defect affecting megakaryocytic-erythroid precursors. Hematocrit and other hematologic indexes are within normal ranges (Supplementary Table 1). Platelets have normal mean volume and ultrastructure, with some elongated alpha granules (Supplementary Fig 1). Patients exhibit mild to moderate bleeding and two developed precursor B cell ALL at ages 3 (III-1) and 37 (II-7). Histopathological analysis of bone marrow from affected individuals without leukemia revealed small, hypolobulated megakaryocytes and abnormal red blood cell precursors (Fig 1b).
Figure 1.
Mutation analysis of ETV6. (a) Pedigrees for three affected families. Solid symbols represent affected individuals with thrombocytopenia and high red cell mean corpuscular volume. Asterisks represent individuals that developed B cell leukemia. (See Supplementary Table 1 for CBC values). (b) Representative images of a bone marrow aspirate for one of the affected individuals without leukemia that shows mild dyserythropoiesis in the left panel (yellow arrows) and an immature hypolobulated megakaryocyte (yellow arrow) on the right panel. (c) Schematic of ETV6, which is composed of 8 exons encoding untranslated regions (yellow) and protein coding sequences (blue). Two mutations, c.641C>T in exon 5 and c.1252A>G in exon 7 are depicted. In the lower section of the panel a schematic of ETV6 is represented with its different domains, pointed (PNT), central and ETS (which is the DNA-binding domain) and the location of the amino acid changes. (d) Effects of ETV6 mutant alleles (p.Pro214Leu, p.Arg418Gly, and p.385_418del) in the activity of an ETV6-responsive reporter plasmid (pGL2-754TR) when expressed alone or with the wild type allele. Luciferase activity, represented here by fold repression, is shown relative to empty vector and normalized using an internal control plasmid expressing Renilla luciferase. Wild type ETV6 (WT) repressed luciferase expression by approximately 7.5 fold compared to empty vector, whereas p.P214L ETV6, p.R418G, and p.385_418del repressed transcription 4.5, 1.5, and 1.5 fold, respectively (p<0.0001, one way ANOVA) (Figure 1d). Co-expression of WT and mutant proteins caused intermediate repression of the stromelysin-1 promoter. Experiments were done in triplicates and repeated 4 times (error bars = standard deviation).
Whole exome sequencing of Family 1 revealed a heterozygous single nucleotide change in ETV6, which segregated completely with thrombocytopenia and high red blood cell MCV. This missense variant, c.641C>T, encodes a proline to leucine amino acid change (p.Pro214Leu) in the central domain of ETV6 (Fig 1c) that was not found in 1000 Genomes or dbSNP databases and was predicted to be possibly damaging by PolyPhen2. Twenty-three additional European families with autosomal dominant thrombocytopenia, high red blood cell MCV and in some increased incidence of leukemia were screened via Sanger sequencing of exons and exon-intron boundaries of ETV6. This approach found two families with germline ETV6 mutations. One (Family 2, Fig 1a) had affected members with platelet counts of 44,000–115,000 platelets/uL, MCV of 88–97 fL and ALL in individual I-2 at age 14, all exhibiting the identical c.641C>T mutation. In the other (Family 3, Fig 1a), having individuals with platelet counts of 99,000–101,000 platelets/uL and MCV of 93–98 fL but no malignancies, a mutation in the DNA-binding domain (c.1252A>G, p.Arg418Gly) was detected that was not observed in 1000 Genomes and was predicted to be highly damaging by PolyPhen2. Sequence alignment demonstrated that both mutations affect amino acids that are highly conserved across multiple species (Supplementary Note). Both mutations were present in the COSMIC database, indicating that somatic acquisition of these mutations may be oncogenic.
The c.1252A>G mutation is located in the last codon of exon 7, which is split with exon 8, raising the possibility of this variant disrupting a splice site. To test this possibility RNA was isolated from peripheral blood cells from two individuals with the c.1252A>G mutation. RT-PCR detected two different transcripts, one of expected size (386 bp) and another of 285 bp, indicating an alternatively-spliced product (Supplementary Fig 2). Sequencing of the 285bp product revealed skipping of exon 7. This c.1153_1253del mutation is predicted to cause a partial deletion of the putative DNA binding domain (aa 385–418, p.385_418del) and a subsequent p.Asn385Valfs*7 frameshift alteration resulting in a premature stop codon, raising the possibility of a truncated protein. Although this truncated protein was expressed in transfection assays in HEK293T cells (Supplementary Fig 3) it was not detected in patients' platelets (Supplementary Fig 4), suggesting it is not functional in megakaryocytes. The alternative splicing did not affect all mutant RNA, since sequencing of the 386 bp RT-PCR product, as well as plasmids where this product was cloned, showed the presence of the G nucleotide in addition to the wild type A, indicating that the p.Arg418Gly form of ETV6 is likely to be expressed in patients from this family. This type of genetic aberration, in which an exonic mutation creates an alternative splice site as well as an amino acid change, has been observed in other hematological disorders such as Hemoglobin E disease.15
ETV6 is a 57kD protein with 452 amino acids and three functional domains: N-terminal pointed (PNT), central regulatory and C-terminal DNA-binding (ETS; Fig 1c). Nuclear localization and transcriptional repression activity of ETV6 require homodimerization via the pointed domain.16 ETV6 modulates the activity of other ETS transcription factors such as FLI1 - where hemizygous deletions cause Paris-Trousseau related thrombocytopenia -17,18 and several other known ETV6 interaction partners that are present in platelets,19,20 and presumably megakaryocytes. The function of the central domain is not well understood, but it has been shown to undergo posttranslational modifications and to be essential for the repressive function of ETV6 in in vitro reporter assays.21
To investigate the effect of the p.Pro214Leu, p.Arg418Gly, and p.385_418del mutations on ETV6 transcriptional repression we used an in vitro reporter assay to measure luciferase expression induced by the promoter of the known ETV6 target stromelysin-1.8,22 As expected, wild type ETV6 (WT) repressed luciferase expression compared to empty vector. Expression of each of the three mutant forms of ETV6 resulted in less transcriptional repression when expressed alone or WT ETV6 (Figure 1d). Protein expression of WT and all ETV6 mutants in transfected cells were determined to be equivalent by immunoblot, suggesting that these mutations do not influence protein stability (Supplementary Fig 3).
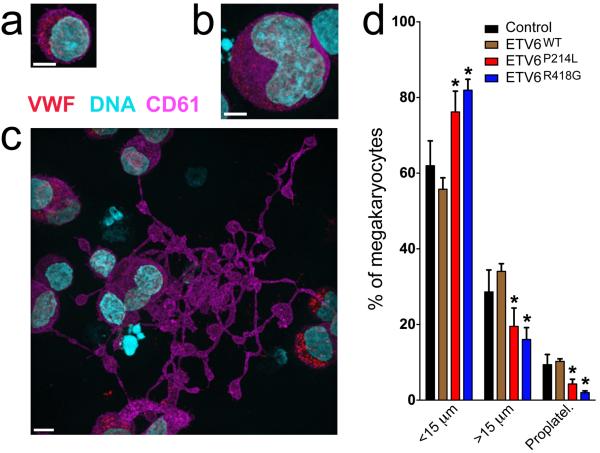
Thrombopoiesis involves a complex sequence of cellular events in bone marrow megakaryocytes culminating with the elaboration of proplatelet extensions that release platelets into the circulation. Approximately 1011 platelets must be produced daily to maintain normal concentrations of 150–400 × 109 platelets per liter of human blood.23 To determine the effect of P214L and R418G ETV6 on megakaryocyte differentiation, human CD34+ cells were transduced with lentivirus containing WT, P214L or R418G ETV6 alleles and cultured with thrombopoietin to support megakaryocyte development. P214L and R418G ETV6-transduced cells showed delayed/decreased maturation when compared to those transduced with WT ETV6, indicated by increased numbers of small, immature megakaryocytes (similar to the abnormalities observed in the bone marrow of affected individuals, Fig 1b) and decreased generation of proplatelets (Fig 2 a–d). These findings indicate that these mutations affect megakaryocyte development and likely platelet production.
Figure 2.
Abnormal development of day 12 cultured megakaryocytes expressing mutant ETV6. Control cells and those transduced with lentivirus to express myc-tagged forms of ETV6 (ETV6WT, ETV6P214L and ETV6R418G) were assessed via immunofluorescence microscopy imaging; scale bars = 5 μm. (a–c) Megakaryocytes (control cells shown) were identified via expression of CD61 (violet) and VWF (red) and staged by diameter: <15 μm (a), >15μm (b), or the presence of proplatelets (c). (d) Population distributions showed no significant differences between control and ETV6WT-transduced cells, while ETV6P214L-transduced megakaryocytes showed a significantly higher proportion (*P<0.05, 2-tailed t-test) of cells in the earlier developmental stage (<15 μm) and fewer in the mature proplatelet-forming stage (control: n=7, WT: n=3, P214L: n=3, R418G: n=4 cultures with >300 cells for each; error bars = standard deviation). Images for control cells and those transduced with WT, P214L and R418G forms of ETV6 are shown in Supplementary Fig 6.
Since ETV6 requires dimerization to exert transcriptional repression, it is possible that the P214L, R418G and p.385-418del mutants may affect this interaction in a dominant negative manner. All mutants were shown to dimerize with wild type ETV6, demonstrated by concomitant “pull down” of both differentially tagged his-ETV6 WT and myc-ETV6 mutants (Supplementary Fig 5). The intracellular distribution of protein produced by transduced P214L or R418G and WT forms of ETV6 in megakaryocytes was examined by immunofluorescence staining for their myc tag, which showed expected nuclear localization for WT ETV6, while P214L and R418G ETV6 were concentrated in the cytoplasm (Supplementary Figure 6). Staining for ETV6 (Figure 3) showed this protein concentrated in the nuclei of an untransduced control and WT-transduced cells, and largely in the cytoplasm of cells expressing mutant ETV6 (see also Videos 1–4). These results indicate that the mutant forms of ETV6 mislocalize inside cells, and by dimerizing with endogenous WT protein, may also prevent nuclear localization, further compromising ETV6 repression activity.
Figure 3.
Aberrant cytoplasmic localization of ETV6 in cultured megakaryocytes transduced with ETV6 mutants. Comparison via immunofluorescence microscopy of ETV6 localization in Day 12 cultured control megakaryocytes and those transduced with lentiviral wild-type (ETV6WT) or mutant (ETV6P214L, ETV6R418G) myc-tagged forms of ETV6. Confocal Z-sections of representative mature cells (>15μm) are shown stained for DNA (blue), ETV6 (both endogenous and myc-tagged; green) and myc (red). In control cells endogenous ETV6 is concentrated in the nucleus, as is endogenous and myc-tagged WT ETV6 in cells transduced with ETV6WT. In contrast, cells transduced with mutant myc-tagged ETV6 (ETV6P214L, ETV6R418G) show this protein concentrated in the cytoplasm with little visible in the nucleus. Scale bars = 5 μm. See also Videos 1–4.
Several cases of leukemia involving somatic ETV6 alterations have been associated with loss of ETV6 function.4–6 In order to identify additional genetic aberrations contributing to leukemogenesis, whole exome and RNA sequencing were performed on leukemia blast cells from time of diagnosis, and from bone marrow cells after remission in the same patient (III-1, Family 1). RNA sequencing revealed that the wild type ETV6 allele was structurally normal and that wild type and p.P214L transcripts were expressed at equal amounts in the bone marrow samples. Several genetic variants and a novel gene fusion between PAX5 and SHB were found in the leukemia sample and were not present in the remission sample (Supplementary Table 2). These variants may represent candidate cooperative mutations with the germline ETV6 mutation in the development of leukemia. 24,25
In summary, we present novel germline mutations in ETV6, a gene known to be involved in human leukemogenesis and murine and zebra fish hematopoiesis, in three families with thrombocytopenia and high red cell MCV, and leukemia predisposition in two of the three families (with p.Pro214Leu mutation). These mutations partially disrupt ETV6 transcriptional repression in vitro and cause aberrant cytoplasmic localization of both mutant and endogenous ETV6, suggesting a dominant negative effect. Recently, while this manuscript was under review, a manuscript by Zhang et al. showed similar findings.9 These mutations also impair megakaryocyte development and proplatelet formation in culture. Furthermore, platelet RNA seq of patients with the p.Pro214Leu mutation revealed decreased expression of platelet-specific transcripts associated with the mutation, including significant reduction in several cytoskeletal transcripts (Supplementary Note, Supplementary Figs 7 and 8). The mutations in these families reveal the implications of viable germline mutations in ETV6, and provide insights into the importance of ETV6 for platelet biology and tumor suppressor activity.
Online Methods
Patient tissue acquisition
All patients in the original cohort (family 1) were recruited at the Hematology Clinic at Children's Hospital of Michigan. The study received Institutional Review Board approval from the University of Colorado Anschutz Medical Campus, and informed consent was obtained for all participants. Patients in families 2 and 3 were recruited in clinical centers in the Czech Republic and Italy as part of a European Consortium focused on inherited platelet disorders. The Institutional Review Board of the IRCCS Policlinico San Matteo Foundation of Pavia, Italy, approved the study protocol. Informed consent was obtained from all patients in the study. Studies were performed in accordance with the Declaration of Helsinki. Genomic DNA was extracted from whole blood using the Gentra Puregene DNA Extraction Kit (Qiagen). Platelets were purified from whole blood and negatively selected using CD45+ MACS microbeads (Miltenyi Biotec). Bone marrow aspirates were obtained for diagnosis and permission was obtained from families to use images.
Exome Sequencing
Genomic DNA (3–5 micrograms) was sheared, size selected (~400–600 bp), ligated to sequencing adapters, and PCR amplified following standard library preparation. The post-PCR library was then used for exome capture using the Agilent SureSelect Human All Exon v4 Capture kit (Santa Clara, CA, USA). Eight exome enriched samples were sequenced using 2 (2×100bp) lanes on an Illumina HiSeq 2000 (Illumina, Inc, San Diego, CA). Exome sequencing was performed in 9 individuals from Family 1: II1, II2, II3, II5, II7, III1, III2, III9 and III10. We screened for ETV6 mutations in the European families by Sanger sequencing of promoter regions, exons and exon flanking regions of ETV6.
Bioinformatics of Exome Sequencing
Reads passing Illumina chastity filter were subjected to a quality filter step that removed low quality bases from the 3′ end and retained pairs of reads if the trimmed reads for both members of the pair were 50 bp or longer. Paired reads that passed the quality filter were mapped to the reference human genome sequence (hg19) with GSNAP (Genomic Short-read Nucleotide Alignment Program, version 2012-07-20).26 Sequence calls for single-nucleotide polymorphisms (SNPs) and insertions and deletions (indels) were performed using the GATK (Broad's Genome Analysis Toolkit, v2.1-8-g5efb575). 27
The program ANNOVAR (Annotate Variation, version 2012-03-08) was used to classify variants and to cross-reference all variants across various genetic variation databases. Included in ANNOVAR are databases to determine nonsynonymous and splice site variants (refGene.txt), variants in conserved genomic regions (phastConsElements46way.txt), variants in segmental duplications (genomicSuperDups.txt), and variant frequencies from 1000 genomes database (hg19_ALL.sites.2012_02.txt). Variants located outside of conserved regions, or with frequencies >1% were excluded from further analysis._ENREF_228 Only non-synonymous changes (SNPs and indels), splice site variants, and/or an aberrant stop codon changes were considered for further analysis. All insertion and deletion variants were considered damaging, whereas SNP variants were cross-referenced to the dbNSFP (database for nonsynonymous SNPs' functional predictions, version 2.0) to determine whether the changes would be considered tolerable or damaging using four algorithms (Sorting Intolerant From Tolerant (SIFT), PolyPhen-2 (Polymorphism Phenotyping v2), likelihood ratio test (LRT), MutationTaster).29
The final filtered list of variants for each affected family member was then intersected to find putative causal variants.
Platelet RNA Sequencing
RNA was isolated from leukoreduced platelet preparations stored in Trizol as previously described.30 RNA-seq libraries were prepared and bar-coded using Illumina TruSeq V2 with oligo dT selection. 50 cycle single end reads were generated on a single lane of the HiSeq 2000, and aligned using Novoalign (Novocraft Technologies, Malaysia) to UCSC genome version hg19 with known and shuffled splice junctions included. Normalization of read counts and differential expression analysis was performed with DESeq2 (http://biorxiv.org/). Sample to sample variability in the level of leukocyte transcripts, which can significantly alter read counts (JWR, unpublished observations), was corrected for by including the ratio of PTPRC (the leukocyte marker CD45)/ITGA2B (platelet marker) as a factor in the model for significance testing. Relationship to affected was also included in the model. Euclidean distance computation, clustering (complete linkage), and heatmap analysis of the regularized log transformed read counts was performed in R as described in the DESeq2 vignette. A list of 351 expressed transcripts involved in platelet biogenesis or function was curated from Reactome and from transcripts enriched in platelets compared to all other tissues in Illumina's human body map 2.0. Gene Set Enrichment Analysis (GSEA)31 of the 177 differentially expressed targets included testing against GO BP, Biocarata, Kegg, and Reactome gene sets.
RNA Sequencing (bone marrow)
Total RNA was isolated from samples taken from bone marrow at diagnosis of leukemia, and again during remission. cDNA libraries were constructed for each sample using the TruSeq mRNA sample preparation kit (Illumina, San Diego CA). Poly-A containing mRNA from the total RNA is isolated using poly-T oligo-attached magnetic bead selection. RNA was chemically fragmented and random primed prior to reverse transcription and cDNA generation. cDNA fragments then undergo end repair, the addition of a single ‘A’ base to the 3' end, and ligation of adapters. Finally, the products are purified and PCR-enriched to create the double-stranded cDNA library. The uniquely indexed libraries were pooled and sequenced in a single lane of an Illumina HiSeq 2000 flow cell, yielding 2×100bp reads.
Bioinformatics of RNA Sequencing
Reads passing Illumina chastity filter were subjected to a quality filter step as described above. Paired reads that passed the quality filter were mapped to the reference human genome sequence (hg19) with GSNAP (Genomic Short-read Nucleotide Alignment Program, version 2012-07-20).26 The aligned reads were then searched for gene fusions using 2 separate algorithms: TopHat-Fusion, v2.0.9 32 and FusionMap, version 7.0.1.25.33 Intersection of the resulting gene fusion predictions from the 2 programs resulted in a single high-confidence candidate.
Plasmids and mutagenesis
A pCMV-Entry vector containing a C-terminal Myc/DDK tagged Human ets variant 6 (ETV6) cDNA was obtained from OriGene. Site-directed mutagenesis was done using the QuikChange Site-Directed Mutagnesis Kit (Agilent Technologies) and primers designed to contain the mutations of interest (c.641C>T, c.1252A>G) with flanking complementary sequence. WT ETV6 was also cloned into the pCMV-Entry vector containing a C-terminal His tag (Origene). A construct designed to encode the alternatively spliced/truncated (p.385_418del, p.Asn385Valfs*7) ETV6 was made by GenScript and cloned into the pCMV-Entry C-terminal Myc/DDK tagged vector. The luciferase reporter construct (pGL2-754TR) was originally published by S. Hiebert and was obtained from A. Ferrando.8 The pCMV-Renilla was also obtained from A. Ferrando. All constructs were transformed into Mix & Go Competent E.Coli cells (Zymogen Research) and grown up with 100ug/mL Kanamycin or Ampicillin.
Luciferase reporter assays
HEK293T cells (a gift from David Ginsburg, University of Michigan) were cultured in Minimum Essential Medium (MEM, Invitrogen) supplemented with 10% Fetal Bovine Serum (FBS). HEK293T cells were seeded at 1.5×106 cells/60mm dish and transfected using Trans-IT 293 (Mirus) 24 hours after seeding with either 2ug empty pCMV-Entry vector, WT ETV6, p.P214L ETV6, p.R418G ETV6, p.385_418del ETV6 or 1ug of WT ETV6 and 1ug of mutant ETV6 cDNA. 2ug pGL2-754TR luciferase reporter was co-transfected into cells, along with 15ng pCMV-Renilla as a transfection control. Cells were harvested 48 hours after transfection, lysed with Passive Lysis Buffer (Promega), and analyzed using the Dual Luciferase Reporter assay kit (Promega). Before experiments cells were tested for mycoplasma contamination, and contamination was ruled out.
Pull down assay
HEK293T cells were transfected and harvested as previously described with WT ETV6-myc/DDK, WT ETV6-His, or co-transfected with WT ETV6-His and myc/DDK tagged WT ETV6, p.P214L ETV6, p.R418G ETV6, or p.385_418del ETV6. Protein complexes containing the myc/DDK tag were purified from cell lysate using the c-Myc tagged Protein Mild Purification Kit (Medical and Biological Laboratories). Protein was eluted by the c-myc peptide, and eluate was run on SDS-PAGE and probed using a DDK or His primary antibody.
Lentiviral CD34+ cell transduction
Lentiviral generation and CD34+ cell transduction were done following methods previously described.34 Specifically, C-myc tagged ETV6WT, ETV6P214L and ETV6R418G were cloned into the lentiviral vector PLJM1 and co-transfected into HEK293T cells with PMD2.G, PMDLg/pRRE and pRSV-Rev. Supernatants were collected every 24 hours for up to 72 hours and virus concentrated via ultracentrifugation. CD34+ cells were affinity purified from G-CSF-mobilized blood progenitor cells from bone marrow transplant donors using the CD34 Microbead Kit (Miltenyi Biotec) and cultured with IMDM (Gibco) supplemented with 10% BIT 9500 (Stemcell Technologies), 2 mM L-Glutamine (Wisent), 1% Penicillin/Streptomycin (Wisent) and 50ng/mL thrombopoietin (gift from Kirin Brewery Company). On day 2 cells were pelleted and re-suspended in culture media containing either ETV6WT, ETV6P214L or ETV6R418G viruses. Cell-virus mixtures were placed onto Retronectin (Clontech) coated plates for transduction, which was repeated 12 hours later. Cells were transferred to a new culture dish 12 hours later. On culture day 8 cells were seeded onto coverslips coated with a 1:6 dilution of Matrigel (Corning) in DMEM as previously described.35
High resolution laser immunofluorescence confocal microscopy
On culture day 12, cells growing in Matrigel on coverslips were fixed with paraformaldehyde, permeabilized and immunostained with human specific antibodies: megakaryocyte-specific proteins were detected with rabbit anti-CD61 (Abcam) and sheep anti-VWF (AbD Serotec), myc-tagged ETV6 was detected using mouse anti-c-myc (Covance) and endogenous/tagged ETV6 via rabbit anti-ETV6 (Prestige Antibodies, Sigma-Aldrich). Antibodies were visualized using donkey secondary antibodies: anti-rabbit Alexa Fluor 647 or 488, anti-sheep Alexa Fluor 555 and anti-mouse Alexa Fluor 488 or 546 (Life Technologies). DNA was stained with DAPI and samples were prepared with fluorescent mounting medium (Dako).
Images were obtained with an oil immersion objective (60×/1.35) using a Quorum Olympus spinning disc confocal inverted epifluorescence microscope equipped with 4 solid-state lasers (Spectral Applied Research): 405 nm, 491 nm, 561 nm, 642 nm, an Improvision Piezo Focus Drive, 1.5× magnification lens (Spectral Applied Research), a Hamamatsu C9100-13 back-thinned EM-CCD camera and Yokogawa CSU X1 spinning disk confocal scan head with Spectral Aurora Borealis upgrade. Acquisition, image processing and cell counting were done using Perkin Elmer Volocity software (versions 5.5 – 6.1). Confocal images were deconvolved and exported as z-sections or extended focus images to Adobe Photoshop and/or Illustrator for final presentation. Images were rendered as 3D volumes (maximum intensity) and animated using Imaris 7.6 to create videos.
Transmission electron microscopy and immunoblots
Transmission electron microscopy of platelets and preparation of platelet lysate immunoblots were done as previously described.36 Antibodies used in immunoblots presented throughout were polyclonal rabbit anti-ETV6 (Thermo Scientific), mouse monoclonal anti-GAPDH (Millipore), mouse monoclonal 4C5 anti-DDK antibody (OriGene) and rabbit polyclonal anti-His antibody (Cell Signaling).
Supplementary Material
Acknowledgments
We are grateful to the families studied for their contribution to this project. We are also grateful to Tamim Shaikh, Rich Spritz and Jeff Murray for their insightful comments. This work was supported by the Postle Family Chair in Pediatric Cancer and Blood Disorders (JDP), NIH HL112311 (ASW), GM103806 (JWR). WHAK was supported by operating grants from the Canadian Institutes of Health Research (CIHR; MOP-81208 and MOP-259952). PN and AS were supported by grant GGP13082 from Telethon Foundation.
Footnotes
Author Contributions L.N., R.L., A.B.L-S., A.S.W., W.H.K., C.C.P., J.D.P. conceived and designed the experiments. L.N., R.W.L., A.B.L-S., R.L., F.G.P., L.L., L.L, A.S., C.G. and D.D.R. performed experiments and provided critical data. M.C., M.R., P.N., C.B., A.P., M.D., A.G-H., L.X. and C.L-M provided patients samples, study materials and collected and assembled data. S.H., P.H. and A.G-H. analyzed data. K.J., K.G. and J.W.R analyzed genomic and transcriptome data. L.N., R.L., A.B.L-S., F.G.P., A.S.W., W.H.K., C.C.P. and J.D.P. wrote the manuscript. All authors reviewed and contributed to the final version of the manuscript. A.S.W., W.H.K., C.C.P. and J.D.P. jointly supervised the research.
The authors declare no competing financial interests.
Accession codes BioProject ID: PRJNA275981 Sequence Read Archive (SRA)
References
- 1.Song WJ, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–75. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 2.Nichols KE, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–70. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kar A, Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit Rev Biochem Mol Biol. 2013;48:522–43. doi: 10.3109/10409238.2013.838202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romana SP, et al. Deletion of the short arm of chromosome 12 is a secondary event in acute lymphoblastic leukemia with t(12;21) Leukemia. 1996;10:167–70. [PubMed] [Google Scholar]
- 5.Patel N, et al. Expression profile of wild-type ETV6 in childhood acute leukaemia. Br J Haematol. 2003;122:94–8. doi: 10.1046/j.1365-2141.2003.04399.x. [DOI] [PubMed] [Google Scholar]
- 6.Barjesteh van Waalwijk van Doorn-Khosrovani S, et al. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene. 2005;24:4129–37. doi: 10.1038/sj.onc.1208588. [DOI] [PubMed] [Google Scholar]
- 7.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Vlierberghe P, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208:2571–9. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang MY, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47:180–5. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasighaemi P, Onnebo SM, Liongue C, Ward AC. ETV6 (TEL1) regulates embryonic hematopoiesis in zebrafish. Haematologica. 2014 doi: 10.3324/haematol.2014.104091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LC, et al. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–83. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang LC, et al. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hock H, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–41. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira CF, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–18. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orkin SH, et al. Abnormal RNA processing due to the exon mutation of beta E-globin gene. Nature. 1982;300:768–9. doi: 10.1038/300768a0. [DOI] [PubMed] [Google Scholar]
- 16.Green SM, Coyne HJ, 3rd, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J Biol Chem. 2010;285:18496–504. doi: 10.1074/jbc.M109.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwiatkowski BA, et al. The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J Biol Chem. 1998;273:17525–30. doi: 10.1074/jbc.273.28.17525. [DOI] [PubMed] [Google Scholar]
- 18.Raslova H, et al. FLI1 monoallelic expression combined with its hemizygous loss underlies Paris-Trousseau/Jacobsen thrombopenia. J Clin Invest. 2004;114:77–84. doi: 10.1172/JCI21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Million RP, Harakawa N, Roumiantsev S, Varticovski L, Van Etten RA. A direct binding site for Grb2 contributes to transformation and leukemogenesis by the Tel-Abl (ETV6-Abl) tyrosine kinase. Mol Cell Biol. 2004;24:4685–95. doi: 10.1128/MCB.24.11.4685-4695.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roukens MG, Alloul-Ramdhani M, Moghadasi S, Op den Brouw M, Baker DA. Downregulation of vertebrate Tel (ETV6) and Drosophila Yan is facilitated by an evolutionarily conserved mechanism of F-box-mediated ubiquitination. Mol Cell Biol. 2008;28:4394–406. doi: 10.1128/MCB.01914-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez RG, et al. TEL is a sequence-specific transcriptional repressor. J Biol Chem. 1999;274:30132–8. doi: 10.1074/jbc.274.42.30132. [DOI] [PubMed] [Google Scholar]
- 22.Fenrick R, et al. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stromelysin-1. Mol Cell Biol. 2000;20:5828–39. doi: 10.1128/mcb.20.16.5828-5839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machlus KR, Italiano JE., Jr The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785–96. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 25.Strehl S, Konig M, Dworzak MN, Kalwak K, Haas OA. PAX5/ETV6 fusion defines cytogenetic entity dic(9;12)(p13;p13) Leukemia. 2003;17:1121–3. doi: 10.1038/sj.leu.2402923. [DOI] [PubMed] [Google Scholar]
- 26.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Human Mutation. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowley JW, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–11. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Salzberg S. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge H, et al. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- 34.Wilcox DA, et al. Induction of megakaryocytes to synthesize and store a releasable pool of human factor VIII. J Thromb Haemost. 2003;1:2477–89. doi: 10.1111/j.1538-7836.2003.00534.x. [DOI] [PubMed] [Google Scholar]
- 35.Kahr WH, et al. Abnormal megakaryocyte development and platelet function in Nbeal2(−/−) mice. Blood. 2013;122:3349–58. doi: 10.1182/blood-2013-04-499491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urban D, et al. The VPS33B-binding protein VPS16B is required in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2012;120:5032–40. doi: 10.1182/blood-2012-05-431205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.