Abstract
Approximately 50% of epithelial ovarian cancers (EOCs) exhibit defective DNA repair via homologous recombination (HR) due to genetic and epigenetic alterations of HR pathway genes. Defective HR is an important therapeutic target in EOC as exemplified by the efficacy of platinum analogues in this disease, as well as the advent of poly-ADP ribose polymerase inhibitors which exhibit synthetic lethality when applied to HR deficient cells. Here, we describe the genotypic and phenotypic characteristics of HR deficient EOCs, discuss current and emerging approaches for targeting these tumors, and present challenges associated with these approaches focusing on development and overcoming resistance.
INTRODUCTION
Epithelial ovarian cancer (EOC) remains the most lethal gynecologic malignancy and the fifth most frequent cause of cancer-related mortality in women in United States (1). Approximately 75% of EOC patients are diagnosed with advanced disease which is curable only in a minority of the cases resulting in a modest 5-year overall survival rate of 20–30% (2, 3). The standard of care management of EOC consists of primary surgical cytoreduction followed by platinum-based chemotherapy (3, 4). Platinum analogues have been used to treat ovarian cancer since the late 1970s when clinical trials demonstrated that cisplatin was capable of achieving almost double the overall response rates and the number of complete responses compared with non-platinum agents (5, 6). Since then, platinum agents (initially cisplatin, then carboplatin which is better tolerated but equally effective (7)) have constituted the backbone of chemotherapy used in EOC and have defined the comparison arms for the majority of the clinical trials conducted in this disease. However, despite important advancements in the efficacy of platinum chemotherapy achieved by incorporation of taxanes (8) in the 1990s and by administration of chemotherapy via the intraperitoneal (IP) route (9) in early 2000, the plateau of the survival curve has not changed appreciably (3, 8, 10–12), suggesting that alternative approaches are urgently needed.
Platinum analogs induce intrastrand and interstrand cross-links (ICLs) between purine bases of the DNA. ICLs are extremely deleterious lesions that covalently tether both duplex DNA strands and pose formidable blocks to DNA repair (13). Repair of ICLs is dependent on both Fanconi Anemia (FA) and BRCA proteins, which act in a common DNA repair pathway (also referred to as the Fanconi Anemia/BRCA pathway) that involves homologous recombination (HR) (14, 15) (Figure 1). The striking platinum sensitivity of EOC tumors is thought to be related to an underlying defect in HR-mediated DNA repair, particularly in those with high grade serous histology (approximately 70% of all EOCs). In this regard, a plethora of genetic studies, and most recently The Cancer Genome Atlas (TCGA) project, have consistently shown that high grade serous ovarian cancers (HGSOCs) are characterized by frequent genetic and epigenetic alterations of HR pathway genes, most commonly BRCA1 and BRCA2 genes (16, 17). Defective HR is an important therapeutic target in EOC, as exemplified by the central role of platinum agents in the management of this disease as well as the advent of poly-ADP ribose polymerase inhibitors (PARPis), a novel class of anticancer agents which exhibit synthetic lethal effects when applied to cells with defective HR (18–21). In this review, we discuss the molecular alterations and clinical phenotype of HR deficient EOCs, describe current and emerging approaches for targeting HR deficient ovarian cancers, and present the challenges associated with these approaches focusing on development and overcoming drug resistance.
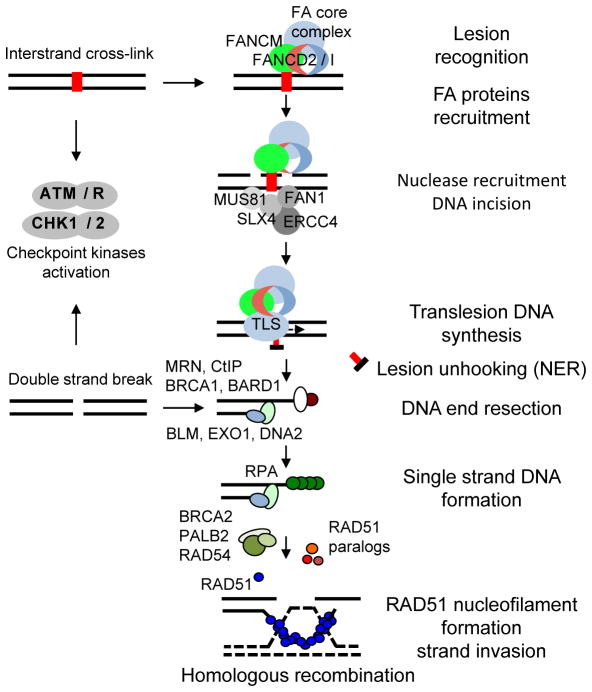
Figure 1. Cooperation of the Fanconi Anemia (FA) and BRCA1/2 proteins in a common ICL repair pathway.
Stalling of replication forks on DNA ICLs induces lesion recognition by the FANCM–FAAP24–MHF1/2 complex and subsequent recruitment of the FA core complex, which in turn recruits the mono-ubiquitinated FANCD2-FANCI to the ICL region. FANCM also initiates checkpoint response, which phosphorylates multiple FA proteins. Ubiquitinated FANCD2 acts as a landing pad for recruiting several nucleases to coordinate nucleolytic incisions. Unhooking the DNA leaves the cross-linked nucleotides tethered to the complementary strand, which are bypassed by TLS polymerases. DNA incisions create a DSB, which is then repaired by HR. Downstream FA proteins such as BRCA1, BRCA2, and PALB2 promote RAD51-dependent strand invasion and resolution of recombinant intermediates.
HR PATHWAY ALTERATIONS IN EOC
Approximately 50% of HGSOCs exhibit genetic or epigenetic alterations in the FA/BRCA pathway (Figure 2) (16). Although these alterations are most commonly encountered in high grade serous histology, nonserous histologies including clear cell, endometrioid and carcinosarcomas have also been shown to harbor such alterations (22). Germline BRCA1 and BRCA2 mutations are the most common alterations, and are present in 14–15% of all EOCs (23, 24) and as high as 22.6% of HGSOCs (16, 23, 24) while somatic BRCA1 and BRCA2 mutations have been identified in 6–7% of high grade serous EOCs (16, 25). Although in the TCGA dataset there was a similar incidence of germline and somatic BRCA1 and BRCA2 mutations, BRCA1 mutations are more commonly observed (60% of all BRCA mutations) in other datasets (23, 24). Importantly, 81% of BRCA1 and 72% of BRCA2 mutations are accompanied by heterozygous loss (26) indicating that both alleles are inactivated, as predicted by Knudson’s two-hit hypothesis. The majority of germline and somatic BRCA1/2 mutations are frameshift insertions or deletions, while missense mutations are rare; mutations have been identified in all functional domains of BRCA1 (RING, coiled coil and BRCT domains) and BRCA2 (BRC, DNA binding, oligonucleotide-binding folds, and tower domains) genes (27).
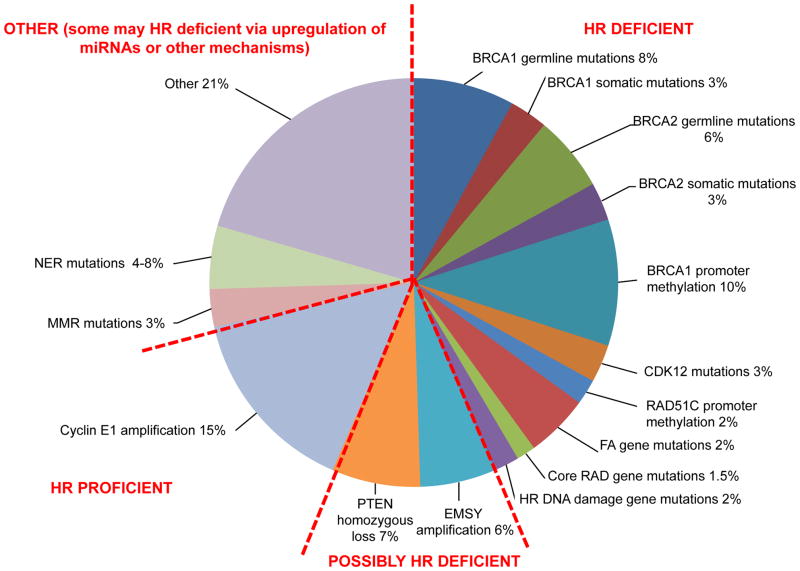
Figure 2. Approximately 50% of high grade serous EOC have alterations in HR repair genes.
Frequency of genetic and epigenetic changes involving HR pathway genes or non-HR pathway genes that modulate HR pathway. FA/BRCA pathway alterations have been experimentally found to be associated with HR deficiency (HR deficient tumors on the right). PTEN deletion and EMSY amplification have been reported to confer HR deficiency but data are evolving (Possibly HR deficient tumors on the bottom). Tumors with cyclin E1 (CCNE1) amplification are enriched for HR proficiency (HR proficient tumors on the left) and are associated with inferior outcome and response to platinum based chemotherapy. Among the remaining tumors, some may be HR deficient via miRNA upregulation or other unknown mechanisms.
Epigenetic silencing via promoter hypermethylation occurs for BRCA1, but not BRCA2, in EOC. BRCA1 promoter hypermethylation has been reported in approximately 10–20% of HGSOCs and is mutually exclusive of BRCA1/2 mutations suggesting that there is strong selective pressure to inactivate BRCA via either mutation or epigenetic silencing in this disease (26, 28, 29). Other HR pathway alterations include mutations in several FA genes (mainly PALB2, FANCA, FANCI, FANCL and FANCC), in core HR RAD genes such as RAD50, RAD51, RAD51C and RAD54L, and in DNA damage response genes involved in HR such as ATM, ATR, CHEK1 and CHEK2 (Figure 2). Interestingly, pathogenic germline RAD51C and RAD51D mutations have been identified in families with both breast and ovarian tumors but not in families with breast cancer (30, 31). RAD51C was also epigenetically silenced via promoter hypermethylation in about 2% of the cases in the TCGA dataset.
Defective HR in EOC may also occur via alterations in non-bona fide HR genes which are known to modulate the HR pathway and indirectly cause HR deficiency. PTEN deficiency has been reported to be synthetically lethal with PARP inhibition and one of the proposed mechanisms is transcriptional downregulation of RAD51(32, 33). A focal deletion region at 10q23.31 that includes only PTEN has been found in approximately 7% of high grade serous EOCs; these tumors exhibit homozygous PTEN deletion which is also associated with downregulation of PTEN at the mRNA level (26). Furthermore, several studies have reported both overexpression and amplification of EMSY as another mechanism of HR deficiency in as high as 17% of high grade sporadic EOC (34). EMSY was identified in a yeast two-hybrid screen to interact with the transactivation domain of BRCA2 leading to inhibition of its transcriptional activity (34). EMSY also colocalizes with BRCA2 at DNA damage sites and interacts with several chromatin remodeling proteins. However, EMSY is located at 11q13, a region known to be amplified in multiple cancers which contains a multiple different oncogenes including LRRC32 (GARP), and PAK112(35). Additionally, EMSY amplification has been associated with worse outcome (35); a finding that would be inconsistent if it caused HR deficiency. For these reasons, although EMSY alterations are commonly cited as a mechanism underlying deficient HR, its role remains controversial. Unlike PTEN and EMSY, the association between inactivating mutations of CDK12 and HR deficiency is clearly established (36, 37). CDK12 is one of the only 9 significantly mutated genes in ovarian cancer (3% of cases in the TCGA dataset) and is known to promote the transcription of several HR pathway genes including BRCA1. Inactivation of CDK12 leads to suppression of HR via reduced expression of BRCA1 and other HR genes, and confers PARPi sensitivity.
It is important to underscore that there may be additional mechanisms underlying defective HR in EOC. Overexpression of specific miRNAs which induce HR deficiency have been identified in breast cancer (such as mir-182 which targets BRCA1)(38) and analogous miRNAs have also been identified in ovarian cancer (such as miR-1255b, miR-148b* and miR-193b* which target BRCA1, BRCA2 and RAD51)(39). Finally, alterations in other DNA repair pathways such as nucleotide excision repair (NER) and mismatch repair (MMR) have been reported in up to 8% and 3% of high grade serous EOCs respectively (Figure 2) (40).
CLINICAL PHENOTYPE OF HR DEFICIENT EOC
Several studies have highlighted a distinct clinical phenotype associated with HR-deficient cancers, especially those with BRCA1/2 mutations. Patients with germline BRCA1/2 mutations are associated with the hereditary breast/ovarian cancer syndrome, which is characterized by familial clustering of breast and ovarian tumors (41). This syndrome has also been linked to germline mutations in other HR genes such as BARD1, BRIP1, MRE11A, NBN, RAD50, CHEK2, ATM, PALB2, RAD51C and RAD51D although the exact penetrance of these genes in terms of breast and/or ovarian cancer remains unknown (22). Large studies have also consistently demonstrated that patients with BRCA1/2-mutated ovarian cancers exhibit significantly improved overall survival compared to patients with non-BRCA mutated tumors; this effect is more pronounced for BRCA2 mutation carriers who exhibit even longer survival compared to BRCA1 carriers (26, 42, 43). Interestingly, the survival advantage of BRCA1 carriers was shown to be dependent on the location of the mutation; worse survival was observed as the mutation site moved from 5′ to 3′ end of the BRCA1 gene (42). Unlike BRCA1/2 mutations, EOCs with epigenetic silencing of BRCA1 through promoter hypermethylation appear not to respond as favorably to platinum and not to exhibit improved survival suggesting that different mechanisms of HR deficiency may confer distinct clinical phenotypes (26, 44). The survival advantage of BRCA-mutated tumors is at least partly related to their enhanced responsiveness to platinum based chemotherapy although a more indolent natural history due to intrinsic biologic differences compared to non BRCA mutated tumors may also play a role. In this regard, although available data suggest that HR deficiency may be both a predictive factor of response to first line platinum chemotherapy and a prognostic factor in EOC, it is unclear whether the prognostic significance of HR deficiency in EOC is solely due to its association with increased sensitivity to chemotherapy or due to other independent factors. For example, several lines of evidence indicate that BRCA1/2-mutated tumors may harbor more tumor-infiltrating lymphocytes and thus be more immunogenic compared to HR proficient EOCs, which may relate to a greater number of mutations observed in these tumors (45, 46). In this regard, the increased immunogenicity of HR deficient tumors may explain their prognostic significance independent of their predictive association with response to 1st line chemotherapy. Whether additional mechanisms may explain the improved survival of HR deficient tumors independently of their enhanced sensitivity to chemotherapy remains to be determined.
BRCA1/2 mutated tumors are also associated with higher grade (grade 2 or grade 3), poorly differentiated or undifferentiated tumors, higher stage (stage III or IV) at presentation, and serous histology (as opposed to endometrioid, clear cell or mucinous histologies)(23, 24, 42, 47). Finally, in terms of pattern of recurrence, BRCA1/2-mutated tumors are more likely to develop visceral metastases (parenchymal liver, lung, adrenal, spleen and brain metastases) and this effect appears more prominent for BRCA1-mutated tumors (23, 48). These clinical features may be at least partly related to the high degree of genomic instability that is characteristic of BRCA1/2-mutated tumors.
BIOMARKERS OF HR DEFICIENCY
Development of a robust biomarker which adequately captures the diverse genetic and epigenetic mechanisms of HR deficiency and is compatible with formalin fixed and paraffin embedded (FFPE) specimens remains elusive. Several approaches have been proposed, including application of gene expression profiles of BRCAness (49) or DNA repair (50), evaluating BRCA1 protein expression by immunohistochemistry (51), and assessing the wider tumor genome nucleotide sequences and mutational spectrums, or ‘sequence scars’, that may be characteristic of defective DNA repair via HR (52). Targeted mutational profiling of HR genes using next-generation sequencing has also been evaluated. BROCA is a targeted capture and massively parallel sequencing assay which accurately identifies all types of mutations of key HR genes including single-base substitutions, small insertions and deletions, and large gene rearrangements (22, 53). Identification of HR gene mutations by BROCA is highly predictive of improved primary response to platinum chemotherapy and longer overall survival in EOC (22). Alternative multigene, next generation sequencing assays are also offered in several cancer centers in the US and routinely include assessment of core HR genes (54).
HR deficient tumors exhibit large (>15Mb) sub-chromosomal deletions and harbor allelic imbalance extending to the telomeric end of the chromosomes with or without changes in overall DNA copy number. Recently, three quantitative metrics of these structural chromosomal aberrations have been developed using single nucleotide polymorphism (SNP) array data. These include: i) the whole genome tumor loss of heterozygosity (LOH) score (55), ii) the telomeric allelic imbalance (TAI) score (56) and iii) the large-scale state transitions (LST) score which quantifies chromosomal breaks between adjacent regions of at least 10 Mb (57). All three scores are highly correlated with alterations in BRCA1/2 and other HR pathway genes in ovarian cancer, and with sensitivity to platinum and PARP-inhibitors (58). These scores have been implemented either alone or in combination with targeted sequencing approaches such as the BROCA assay to achieve better sensitivity in capturing HR deficiency (58). Of note, genomic LOH was recently shown to correlate well with response to the PARPi rucaparib in a phase 2 clinical trial in EOC (ARIEL2). Specifically, among the women without the BRCA 1/2 mutations, those with high genomic LOH had an overall response rate of between 32–40%, while those without LOH had an overall response rate of just 8% (59).
A major limitation of these assays is that they are largely insensitive to reversion of HR deficiency which may occur upon development of resistance to platinum and PARP-inhibitors. When reversion of HR deficiency to HR proficiency occurs, the cumulative defects that had occurred in the cancer genome as the result of the original HR deficiency do not reverse; therefore these assays still interpret these HR proficient tumors as HR deficient. This phenomenon has been observed in BRCA1/2 mutated cell lines with BRCA1/2 reversion mutations which restore BRCA1/2 and HR function; these lines are still interpreted as HR deficient by the aforementioned assays (55). One way to overcome this problem is by development of dynamic, functional biomarkers of HR deficiency, whereby the HR pathway is mechanistically evaluated by directly assessing RAD51 foci formation via immunofluorescence or by assessing other DNA repair complexes via immunohistochemistry (60–62). The challenge of functional biomarkers of HR deficiency is that they require the cancer specimen to be exposed to some form of DNA damage (i.e., radiation or chemotherapy) ex vivo before the RAD51 foci or other DNA repair complexes can be evaluated. This requirement precludes use of FFPE specimens, increases the technical complexity, and limits the reproducibility of these assays. Overall, there is currently no prospectively validated biomarker of HR deficiency that has been incorporated in clinical practice, and this remains an active area of investigation.
TARGETING HR DEFICIENT TUMORS
a. Conventional chemotherapy
A number of conventional chemotherapy agents that are used routinely in the management of EOC exhibit significant cytotoxicity against HR deficient tumors. Platinum analogues, which have formed the backbone of first line chemotherapy of EOC for more than 30 years, induce ICLs which are highly lethal against tumors with defective HR (Figure 1). The integral role of platinum based chemotherapy in the clinical management of EOC is further evident by the fact that management of relapsed disease is stratified based on the platinum free interval (i.e. the time between completion of platinum-based treatment and the detection of relapse; PFI>=6 months is assigned as platinum sensitive and PFI<6 months as platinum resistant disease). Clinically, patients with BRCA1/2-mutated tumors are associated with significantly higher response rates and prolonged progression-free survival after platinum based chemotherapy (26, 42, 63). These patients commonly exhibit good responses after retreatment with platinum upon development of recurrence and many of them end up receiving multiple lines of platinum chemotherapy. Of note, the enhanced sensitivity of BRCA-mutated tumors to platinum agents challenges the traditional clinical definition of platinum resistance because many of these patients respond well to platinum rechallenge even within 6 months from the end of first line platinum therapy (23).
The high correlation between HR deficiency and response to platinum chemotherapy is also highlighted by the fact that development of platinum resistance is commonly related to restoration of proficient HR via various mechanisms (discussed in more detail below). Platinum sensitivity has been used as a clinical surrogate of HR deficiency and clinical trials of PARPis have used platinum sensitivity as an eligibility criterion for selecting patients that are enriched for HR deficient tumors that would respond to PARPis (64, 65). However, it is important to underscore that platinum sensitivity may also result from defective nucleotide excision repair, and in that case it does not necessarily translate into PARPi sensitivity (40).
Non-platinum cytotoxic agents that induce double strand breaks have also been shown to be active against HR deficient tumors. Topotecan, a semisynthetic water-soluble camptothecin (CPT) analogue is FDA approved for recurrent ovarian cancer and has demonstrated response rates ranging from 13%–33% in phase II trials depending on platinum sensitivity (66, 67). In a phase III trial, topotecan demonstrated an overall objective response rate (CR+PR) of 17.0% (28.8% in platinum sensitive and 6.5% in the platinum resistant/refractory disease)(66). Topotecan inhibits the religation step of the breakage/reunion reaction of topoisomerase I (TopI) resulting in accumulation of topotecan-TopI-DNA covalent complexes which are converted to DNA double-strand breaks (DSBs) when replication forks encounter the single-strand breaks (SSBs). Studies in yeast have demonstrated that the DSBs induced by TopI inhibitors are repaired by HR during S phase (68); the nonhomologous end joining (NHEJ) pathway is significantly less involved in repair of TopI inhibitor induced DSBs (69).
Similar to TopI inhibitors, topoisomerase II (TopII) inhibitors such as doxorubicin and etoposide are also more active in HR deficient cells and are routinely used in the management of relapsed EOC (pegylated liposomal doxorubicin (PLD) is also FDA-approved for this indication)(66, 67, 70). TopII (Top2a and b) cleave both strands of one DNA duplex simultaneously and form transient tyrosyl-DNA cleavage complex intermediates to allow another duplex to pass through the TopII-linked DSB; TopII generates DSBs in cycling cells especially during mitosis phase, in which both HR and NHEJ are available for repair (69). Etoposide and doxorubicin are TopII poisons which inhibit the religation step of the breakage/reunion reaction of TopII and trap the TopII cleavage complex intermediates. Etoposide is a non-intercalating drug which acts mainly as a TopII trap, while doxorubicin is an intercalator which not only traps TopII but also kill cells by intercalation and generation of oxygen radicals (71). Although etoposide and doxorubicin are more active in HR deficient cells, one striking difference from TopI inhibitors, is that the NHEJ pathway is significantly more involved in repair of DSBs induced by TopII than TopI inhibitors. Consistent with their enhanced activity in HR deficient cells, the activity of both etoposide and doxorubicin is much higher in the platinum sensitive compared to the platinum resistant setting (66, 70). Furthermore, treatment of BRCA-associated EOC patients with PLD has been shown to result in higher response rates, longer time to treatment failure and improved overall survival compared with non-BRCA mutated patients (72).
Finally, HR deficient cells are also sensitive to antimetabolites which induce base lesions and/or replication fork stalling such as gemcitabine. Gemcitabine is a nucleoside analog, whose metabolites (diphosphate and triphosphate nucleosides) facilitate incorporation of gemcitabine nucleotide into DNA which blocks further extension of the nascent strand and causes stalling of replication forks. Furthermore, gemcitabine irreversibly inhibits the ribonucleotide reductase enzyme leading to cell’s inability to produce the deoxyribonucleotides required for DNA replication and repair, and thus inducing apoptosis. Gemcitabine is currently FDA approved in combination with carboplatin, for the treatment of EOC that has relapsed at least 6 months after completion of platinum-based therapy (i.e. in platinum sensitive disease)(73). However, gemcitabine has also been studied and is one of the standard treatment options as a single agent in platinum resistant disease, although the response is less in that setting (74).
b. PARP-inhibition as a synthetic lethal strategy against HR-deficient cancers
i. Mechanism of action of PARPis
HR deficient cells have been shown to be extremely sensitive to poly(ADP ribose) polymerase (PARP) inhibitors (PARPis)(18, 75, 76). Different aspects of PARP1 biology have been proposed to explain the synthetic lethal interaction between PARPi and HR deficiency, but the mechanistic basis is incompletely understood (77).
Since PARP1 was originally shown to be essential for base excision repair (BER) (78, 79), preventing the repair of DNA single strand breaks by PARPis (SSBs, normally repaired by BER) would convert them into the more cytotoxic double strand breaks (DSBs) that are normally repaired by HR (80). In that scenario, an HR-proficient cell will repair these DSBs by HR, whereas these lesions will remain unrepaired and cause cytotoxicity in HR deficient cells (Figure 3(I)) (81, 82). However, some findings do not fit with this model; for instance, knockdown of XRCC1, a downstream effector of PARP1 in BER, does not affect the survival of HR-deficient cells suggesting that BER activity is not critical for HR-deficient cell survival (83).
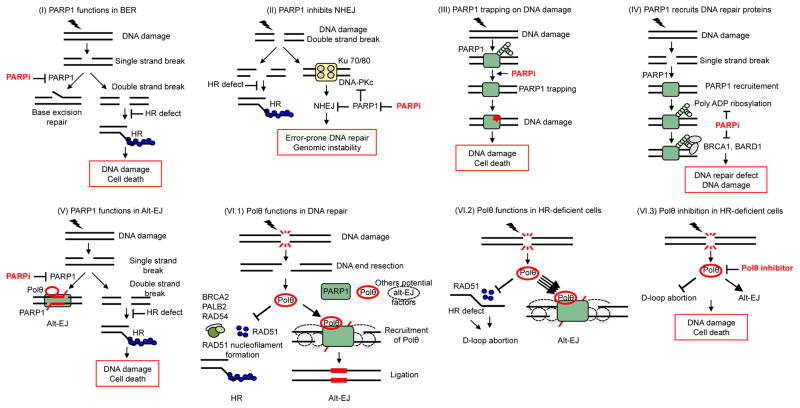
Figure 3. Mechanisms of synthetic lethality between PARP1 or POLQ inhibition and HR deficiency.
Inhibition of PARP1 activity in BER (I) and C-NHEJ (II) is toxic in HR-deficient cells and explains the observed PARPi-HR synthetic lethality. (III) PARPi’s induce PARP1 trapping on DNA lesions which is highly toxic in HR-deficient cells. (IV) PAR-mediated recruitment of the BARD1-BRCA1 complex is impaired by PARPi, resulting in the persistence of DNA lesions that are toxic to HR-deficient cells. (V) Inhibition of PARP1/Polθ-mediated Alt-EJ is toxic in HR-deficient cells. (VI.1) Under physiological conditions, Polθ expression is low and its impact on the repair of DNA double-strand breaks (DSB) is limited. Polθ limits RAD51-ssDNA filament assembly and subsequent HR activity; at the same time, it promotes alt-EJ through its polymerase domain.. (VI.2) Upon an HR defect, Polθ expression increases substantially and channels DSB repair into alt-EJ. (VI.3) In the case of an HR defect, inhibition of Polθ causes cell death through the persistence of toxic RAD51 intermediates and inhibition of alt-EJ.
Another proposed explanation for the PARPi-HR synthetic lethality pertains to the role of PARP1 in limiting classical-NHEJ (C-NHEJ) repair activity (83, 84). C-NHEJ is error-prone and induces genomic instability, which is believed to be particularly deleterious for HR-deficient cells (Figure 3(II)). Thus, PARPi-mediated inhibition of PARP1 would promote C-NHEJ and genome instability. Experimental evidence supporting this model are found in studies showing that the genomic instability induced by PARPi treatment in HR-deficient cells is reduced by concomitant inhibition of DNA-PK, a critical factor of C-NHEJ (18, 83). Several C-NHEJ proteins such as Ku70, Ku80, and DNA-PKcs, bind to poly(ADP ribose) polymers that are generated by PARP enzymes, and these interactions are critical for suppressing C-NHEJ (85–89); furthermore, PARP1 and Ku80 compete for DNA ends in vitro (83). Even though the role of PARP1 in limiting C-NHEJ is now well established, the link between PARPi-mediated C-NHEJ activation and the HR-deficient/PARPi synthetic lethal interaction remains to be fully elucidated.
It has also been suggested that the extreme sensitivity of HR-deficient cells to PARPi might result from the trapping of PARP1 at sites of endogenous damage (Figure 3(III))(90). When DNA damage activates PARP1(91), the PAR-dependent recruitment of additional repair proteins (92–94) simultaneously reduces PARP1 affinity for DNA (95), thereby ensuring tight control of the repair process. Since mutant PARP1 that is unable to synthesize poly(ADP ribose) polymers has been shown to be trapped on DNA and to inhibit DNA repair (95), PARPi-mediated inactivation of PARP1 may likewise induce PARP1 trapping and inhibition of DNA repair. Accordingly, recent studies have demonstrated that PARPi-mediated trapping of PARP1-DNA complexes showed higher cytotoxicity than unrepaired SSBs caused by knockdown of PARP1 (90). This mechanism may explain the cytotoxic effect of certain drug combinations such as PARPi and temozolomide or topotecan (96–98), and is believed to account for the observation that PARP1 knockdown selectively kills HR-deficient cells (18, 76, 83), as these PARP-trapping-induced DNA lesions are thought to be mostly toxic in an HR-deficient setting.
An alternative model might provide an explanation for the cytotoxicity of PARPis in certain BRCA1-deficient cells. DNA damage activates PARP1 (91), which in turn poly(ADP-ribosyl)ates or PARylates many proteins at DNA break sites to orchestrate repair (93). During the HR process, BRCA1 is recruited to damage sites by the PAR- and BRCA1-binding protein BARD1 (99). Indeed, the BARD1 PAR-binding domain is critical for BRCA1 localization to DNA damage sites, particularly when the additional mode of BRCA1 recruitment through γH2AX binding is impaired (for instance in certain BRCA1 mutated tumors). While this model (Figure 3(IV)) may explain the PARPi hypersensitivity of certain BRCA1-mutated tumors, it cannot be expanded to other HR-deficient contexts to serve as a global mechanism for PARPi sensitivity.
A clue in this regard, is the recent finding that HR-deficient cells are dependent on the alternative end joining (alt-EJ) DSB repair pathway for survival. Inhibition of proteins functioning in alt-EJ, such as PARP1 or the polymerase Polθ, is synthetically lethal with defective HR. Thus, the HR-deficiency/PARPi synthetic lethality likely stems from the simultaneous loss of HR and alt-EJ (Figure 3 (V), also discussed below)(100, 101). Finally, some PARPis likely inhibit all PARP family members and the observed synthetic lethality could arise from a compound effect, not solely that of PARP1 inhibition.
ii. PARP inhibitors in the clinical management of HR deficient ovarian cancers
PARPis, including olaparib (AZD2281), rucaparib (CO338, AG014699 and PF01367338), veliparib (ABT888), niraparib (MK4827) have been extensively studied in EOC (102). Iniparib was also initially evaluated but it is now clear that it exhibits very low PARP inhibition in vitro, and its mechanism of action in vivo remains to be elucidated. All aforementioned PARPis inhibit PARP-1 and PARP-2 in vitro at nanomolar concentrations but differ in their ability to trap PARP1 and PARP2 on the DNA SSB sites; niraparib and the newer PARPi BMN673 exhibit higher potency in trapping PARPs than olaparib and rucaparib, while ABT-888 is the least potent of all PARPis in terms of its PARP trapping ability (90). In December 2014, olaparib was granted accelerated approval by the U.S. FDA for use in EOC patients with germline BRCA1/2 mutations who have received three or more chemotherapy regimens based on the results of an international single-arm trial which demonstrated an objective response rate (ORR) of 34% and a median duration of response of 7.9 months (19, 103–105). Olaparib was previously approved in Europe (in October 2014) by the European Medicines Agency (EMA) for a different indication, i.e. for use in the maintenance treatment of patients with platinum-sensitive relapsed BRCA-mutated (germline and/or somatic) high-grade serous EOC who had a complete or partial response to platinum-based chemotherapy. This approval by the EMA was based on a randomized, double blind, phase 2 clinical trial which showed that olaparib maintenance therapy significantly prolonged progression-free survival, compared to placebo, in patients with BRCA-mutated (germline or somatic) ovarian cancer with a hazard ratio of 0.18(65, 106). Strikingly, 40% of patients with BRCA-mutated tumors that were treated with olaparib derived long term benefit, without developing progressive disease for at least 3 years after randomization. Furthermore, exposure to olaparib did not decrease subsequent sensitivity to platinum or other chemotherapies in BRCA1/2-mutated tumors; upon development of PARPi resistance, subsequent response to platinum based chemotherapy has been reported to be as high as 40% by RECIST (107). Randomized phase III trials of maintenance niraparib, rucaparib and olaparib are currently ongoing in patients with high grade serous ovarian cancer who had demonstrated a response and platinum sensitivity for both the ultimate and the penultimate platinum regimens (Table 1).
Table 1.
Important previous and ongoing Phase II/III studies of PARPis in EOC
| Setting | Study Identifier | Agents | Design | Patients/Accrual | Primary Endpoint | Results/Status |
|---|---|---|---|---|---|---|
| First Line/Maintenance | NCT01844986 | Olaparib Vs Placebo | Phase III, Randomized, Double Blind, Placebo Controlled | BRCA mutated high grade serous or high grade endometrioid ovarian cancer N = 344 |
PFS | Ongoing |
| Recurrent/Maintenance | NCT00753545 | Olaparib Vs Placebo | Phase II, Randomized, Double Blind, Placebo Controlled | Recurrent platinum sensitive high grade serous N=265 |
PFS | BRCAm PFS: 11.2 vs 4.3 months, HR=0.18 NonBRCAm PFS: 7.4 vs 5.5 months, HR=0.54 |
| Recurrent/Maintenance | NCT01847274 | Niraparib Vs Placebo | Phase III, Randomized, Double Blind, Placebo Controlled | Platinum sensitive recurrent high grade serous endometrioid/BRCA stratified N=360 |
PFS | Ongoing |
| Recurrent/Maintenance | NCT01874353 | Olaparib Vs Placebo | Phase III, Randomized, Double Blind, Placebo Controlled | Recurrent platinum sensitive BRCAm high grade serous or endometrioid EOC N=264 |
PFS | Ongoing |
| Recurrent/Maintenance | NCT01968213 | Rucaparib Vs Placebo | Phase III, Randomized, Double Blind, Placebo Controlled | Platinum sensitive recurrent high grade serous endometrioid/BRCA stratified N=540 |
PFS | Ongoing |
| Recurrent/Maintenance | NCT01081951 | Olaparib/Carbo/Taxol Vs Carbo/Taxol | Randomized Phase II, Open Label | Platinum sensitive recurrent HGSC (both BRCAm and nonBRCAm) N=90 |
PFS | ALL: 12.2 vs 9·6 months, HR=0·51 BRCAm: HR=0.21 |
| Recurrent | NCT01116648 | Olaparib/Cediraniib Vs Olaparib | Randomized Phase II, Open Label | Platinum sensitive recurrent HGSC (both BRCAm and nonBRCAm) N=90 |
PFS | ALL: 17·7 vs 9·0 months, HR=0·42 BRCAm: 19.4 vs 16.5 months, HR=0.55 nonBRCAm: 16.5 vs 5.7 months, HR=0.32 |
PARPis have also demonstrated activity in non-BRCA mutated EOC patients although they are not approved for these patients in any setting, either in the US or Europe. This is consistent with the fact that HR deficiency may occur in EOC via multiple mechanisms in the absence of BRCA1/2-mutations (Figure 2). In the aforementioned phase II study of olaparib maintenance, patients whose tumors lacked a BRCA1/2 mutation also derived a benefit from olaparib with hazard ratio for PFS of 0.53(65, 106). Furthermore, in a Phase II study of high-grade serous EOC with unknown/non-mutated BRCA status, olaparib was associated with a 24% ORR and a 30% combined RECIST or CA125 response rate (105). Olaparib sensitivity was higher in platinum-sensitive compared with platinum-resistant non-BRCA-mutated tumors; 50% of non-BRCA-mutated platinum sensitive tumors responded to olaparib as compared to only 4% of non-BRCA-mutated platinum resistant tumors suggesting that platinum sensitivity may be a good surrogate of HR deficiency and PARPi response among non-BRCA-mutated EOCs. A similar correlation between olaparib and platinum sensitivity has also been found for BRCA-mutated tumors but the difference is less pronounced (105) (60% among platinum sensitive vs 33% among platinum resistant BRCA-mutated tumors) suggesting that platinum resistance cannot be used as an exclusion criterion for PARPi therapy in BRCA-mutated cancers because these tumors may be PARPi sensitive even if they are platinum resistant. PARPi studies investigating candidate biomarkers of PARPi response are currently being performed in non-BRCA-mutated EOCs.
Finally, combinations of PARPis with conventional chemotherapy such as platinum compounds and topoisomerase inhibitors have been explored in BRCA-mutated EOCs (108–110). Given that PARPis inhibit base excision repair which is partly responsible for repair of the damage caused by these chemotherapy agents, addition of PARPis may potentiate the action of these agents. However, when PARPis are combined with chemotherapy, achievement of full dose chemotherapy has been challenging because of the overlapping myelosuppression of PARP inhibitors and chemotherapy (111). In a recently reported randomized, open-label, phase 2 study, in patients with platinum-sensitive, recurrent, high-grade serous ovarian cancer who had received up to three previous courses of platinum-based chemotherapy, olaparib plus paclitaxel and carboplatin (at lower than standard doses) followed by maintenance olaparib monotherapy significantly improved progression-free survival versus paclitaxel plus carboplatin alone (given at their standard doses) with the greatest clinical benefit in BRCA-mutated tumors (PFS hazard ratio 0.22), and had an acceptable and manageable tolerability profile (109). PARPis are currently not part of the initial standard of care chemotherapy regimen for BRCA-mutated EOC (which still remains a platinum and taxane doublet), although clinical trials are exploring their incorporation into first line chemotherapy (Table 1).
c. Inhibition of the Polθ-dependent alternative end-joining (Alt-EJ) pathway as a synthetic lethal strategy against HR-deficient cancers
Recent observations indicate that PARP1 functions in a pathway required for the repair of DNA DSBs, referred to as error-prone alternative end joining (alt-EJ) or microhomology-mediated end-joining (MMEJ) (112). Furthermore, recent studies have shown that HR-deficient ovarian and breast tumors have a compensatory increase in the Polθ/PARP1-mediated alt-EJ pathway that appears to occupy a key role for their survival and proliferation (100, 101). The importance of this pathway in addition to classical NHEJ (C-NHEJ) is now increasingly appreciated (113).
Early evidence for alt-EJ came from studies in yeast and mammalian cells deficient in C-NHEJ that were still able to repair DSBs via end-joining (114, 115) and by the observation that mice deficient in C-NHEJ still exhibited chromosomal translocations and V(D)J recombination (116, 117). Molecular characterization of alt-EJ revealed that the XRCC1/DNA ligase III complex and PARP1 were involved (85, 118, 119). Initially, alt-EJ was considered merely a backup repair pathway for C-NHEJ for end-joining of chromosomal DSBs (119–121), but subsequent studies have demonstrated that alt-EJ might have a role in repairing chromosomal DSBs, depending on the biological context, such as HR deficiency (100, 101). However the use of alt-EJ for repairing DSB poses a particular threat to genomic stability because of its predilection for joining DNA breaks on different chromosomes, generating chromosomal translocations (122–124). Indeed, fill-in synthesis in alt-EJ is likely mediated by the Polθ polymerase, which is error-prone and likely produces point mutations, as well as random insertions and deletions (indels) (125)(126). Indeed, up-regulation of budding yeast Polθ appears to generate random deletions or insertions of 20–200 base pairs (127, 128). Thus, the use of alt-EJ, which could be indicative of an HR-defect, is likely to leave a mutational signature comprising indels at sites of microhomology. Characterization of such a mutational signature may ultimately define a biomarker of HR deficiency (127).
The alt-EJ genetic signature likely hinges upon Polθ, which has two distinct functions in DNA repair. First, Polθ prevents RAD51 assembly on ssDNA, and thus toxicity in HR-deficient cells. This function is mediated by the RAD51-binding domain and is distinct from the polymerase domain. Second, Polθ mediates PARP1-dependent alt-EJ replication rescue though its polymerase domain. Cells expressing a mutant Polθ polymerase exhibit reduced survival when BRCA1 is knocked down. Given the synthetic lethal interaction between HR deficiency and inhibition of Polθ (40, 101), it is important to determine which Polθ function(s) (RAD51 binding vs polymerase) should be targeted to efficiently impair the survival of HR-deficient cells (Figure 3(VI)). Although both the RAD51-binding motifs and the polymerase domain of Polθ contribute to the survival of HR-deficient cells, the exact relative contribution of each domain remains to be elucidated in order to induce selective killing of HR deficient tumors.
d. Cell cycle and DNA damage checkpoint inhibitors against HR deficient tumors
Checkpoint signaling facilitates the coordination between DNA damage response and cell cycle control to allow ample time for repair and prevent permanent DNA damage produced by replication and mitosis. Two of the PI3K-related protein kinases (PIKKs), Ataxia-telangiectasia mutated (ATM) and Ataxia-telangiectasia and Rad3-related (ATR), occupy a central role in signaling DNA damage to cell cycle checkpoints and DNA repair pathways (129). The ATM–CHK2 pathway primarily responds to double strand breaks (DSBs) to induce G1 arrest via phosphorylating and activating CHK2 and p53, while the ATR–CHK1 pathway triggers S and G2 phase arrest. ATM promotes HR by recruiting BRCA1 to DSBs but can also antagonize BRCA1 and promote NHEJ by recruiting p53 binding protein 1 (53BP1), and these antagonistic functions are cell cycle regulated. ATR is activated by DNA single-strand–double-strand junctions that arise as intermediates in nucleotide excision repair (NER), by replication stress which is defined as the slowing or stalling of replication fork progression, and at resected DSBs. ATR triggers the intra-S phase and the G2 checkpoints via phosphorylation of CHK1 at Ser345 and Ser317 leading to its activation (130). Activated Chk1 in turns phosphorylates WEE1 (which activates this kinase) and cell division cycle 25 (CDC25A and CDC25C) phosphatases (which inhibits them) to inhibit cell cycle progression through the coordinated suppression of cyclin-dependent kinase activity (131). ATR and CHK1 also phosphorylate a number of proteins involved in HR and ICL repair, including BRCA2, RAD51, FANCD2 and FANCE. Importantly, there is significant crosstalk between the ATM/Chk2 and ATR/Chk1 pathways, and they share many substrates (131).
Abrogation of cell-cycle checkpoints leads to accumulation of DNA damage and cellular death, and this approach has shown significant promise as anticancer strategy. HR deficient EOCs are p53 mutated (which is also case for almost all high grade serous cancers) and have lost G1 checkpoint control which makes them hyper-dependent on the S and G2 checkpoints to prevent DNA damage triggering cell death (26, 132). In this regard, targeting the S and G2 checkpoints by inactivation of the ATR/CHK1/WEE1 pathway will inhibit the DNA damage-induced G2 checkpoint arrest leading to mitotic catastrophe and cell death (132). HR deficient tumors are even more sensitive to combinations of checkpoint inhibitors with DNA damaging chemotherapy drugs because they are both deficient in repairing the DNA damage caused by chemotherapy as well as susceptible to abrogation of S and G2 checkpoints.
Importantly, even in the absence of cytotoxic chemotherapy, unrepaired endogenous DNA damage in HR deficient EOC cells may sensitize them to checkpoint inhibition (132). In this regard, it has been shown that FA deficient tumor cells are hypersensitive to inhibition of CHK1, which is more pronounced when combined with platinum therapy (133). DNA repair through the FA pathway occurs primarily during S phase of the cell cycle and FA tumor cells acquire extensive DNA damage in S phase. These lesions persist throughout the remainder of the S and G2 phase, ultimately activating the G2/M checkpoint; increased accumulation of cells in the G2 phase of the cell cycle is a useful diagnostic feature of FA cells and correlates with the hyperactivation of the G2/M checkpoint (134). FA pathway deficient cancer cells have a greater requirement for CHK1 function than DNA repair proficient cells thereby supporting the presence of a therapeutic window that could be exploited in treating DNA repair deficient cancers with CHK1 inhibitors, while sparing toxicity in normal, DNA repair proficient cells (133). Although FA deficient cells are hypersensitive to cisplatin, addition of a CHK1 inhibitor further increases cytotoxicity to a significant degree. Besides abrogation of G2 and S checkpoints, it has been shown that FA deficient cells are hypersensitive to ATM inhibition suggesting that ATM and FA pathways also function in a compensatory manner to maintain genome integrity (135). As with CHK1 inhibition, the selectivity of ATM inhibition alone for FA deficient cells is modest, but the effect of combining ATM inhibition with platinum is significantly augmented in FA deficient cells (135). Importantly, a synthetic lethal interaction also exists between the ATM and ATR signaling pathways, i.e. ATR inhibitors exhibit significant antitumor activity in ATM-deficient but not ATM-proficient backgrounds (136). Taken together, between the 3 pathways, i.e. ATM, ATR and FA pathways, synthetic lethal interactions exist between all individual pairs i.e. all FA/ATM, FA/ATR and ATR/ATM combinations are synthetically lethal.
Several approaches to inhibit the ATR/CHK1/WEE1 pathway including ATR inhibitors (such as VX-970 and AZD6738), WEE1 inhibitors (such as AZD1775) and CHK1 inhibitors (GDC-0425 and LY2606368) are currently in early clinical trial evaluation in EOC. Of note, AZD1775 has already shown clinical activity as monotherapy in BRCA-mutated tumors (137). In these trials, these agents are combined with chemotherapy, primarily drugs that cause replication stress, such as antimetabolites (particularly nucleoside analogues that cause replication arrest, such as gemcitabine), topoisomerase I poisons and DNA crosslinking agents, such as platinum agents. However, although cell cycle checkpoint inhibition offers the advantage of selective cytotoxicity by exploiting molecular alterations (p53 mutations, HR defects) that are present only in tumors cells, there is always concern for toxicity especially when they are combined with chemotherapy. In this regard, phase I trials of combinations of these agents with chemotherapeutic agents have started at lower doses of chemotherapy which are being escalated to assess for safety. Overall, abrogation of the S and G2 checkpoint via inhibitors of the ATR/CHK1/WEE1 pathway in combination with chemotherapy appears to exert a synthetic lethal interaction with HR deficient EOCs and may thus be an attractive therapeutic strategy against these tumors.
MECHANISMS OF RESISTANCE IN HR DEFICIENT EOCs
In BRCA1/2-mutated tumors, the most common acquired mechanism of resistance to cisplatin or PARPis is secondary intragenic mutations restoring the BRCA1 or BRCA2 protein functionality (Figure 4) (138, 139). Restoration of BRCA1/2 functionality occurs either by genetic events that cancel the frameshift caused by the original mutation and restore the open reading frame (ORF) leading to expression of a functional nearly-full-length protein, or by genetic reversion of the inherited mutation which also restores full-length wild-type protein. These genetic events were originally observed in BRCA2- and BRCA1-mutated cancer cells under selective pressure due to exposure to cisplatin or PARPis, and were associated with secondary genetic changes on the mutated allele that restored a functional protein and conferred platinum and PARPi resistance (Figure 4) (140–143). This mechanism of resistance is highly clinically relevant for patients with BRCA-mutated EOC who are treated with platinum-based therapy; 46% of platinum resistant BRCA-mutated EOCs exhibit tumor-specific secondary mutations that restore the ORF of either BRCA1 or BRCA2(144). Similar observations have been made in biopsies from olaparib-resistant tumors in which acquisition of secondary BRCA2 mutations restored a functional BRCA2 protein (145). Of note, multiple reversion events in BRCA1/2 genes have also been reported as a mechanism of platinum resistance in a recent study of whole-genome characterization of chemoresistant ovarian cancer (146). Strikingly, in one patient with BRCA2-mutated EOC, 12 independent BRCA2 reversion events were identified with multiple reversion events occurring even in individual tumor deposits. Furthermore, in the same study (146), reversal of BRCA1 promoter methylation has also been reported in one patient as a mechanism of platinum resistance. In that case, the primary sensitive sample showed extensive promoter methylation and low BRCA1 expression, while the sample from the relapsed disease had lost BRCA1 methylation and BRCA1 gene was expressed at comparable levels to homologous recombination proficient tumors. Of note, a specific rather than generalized patter of altered methylation was noted at relapse in this patient. Even though BRCA mutations remain the strongest predictor for sensitivity to PARPis, not every mutation will result in the same functional defect and response to these agents. Analysis of BRCA1 missense mutations suggests that the conserved N- and C-terminal domains are most important for the response to HR-deficiency targeted therapies (147). Specifically, tumors carrying the BRCA1-C61G mutation which disrupts the N-terminal RING domain responded poorly to platinum drugs and PARPis, and rapidly developed resistance (148). Similarly, Brca1Δ11/Δ11; p53−/− mouse mammary tumors, which only express the BRCA1-Δ11 isoform (generated by the exon 11 splicing) can acquire resistance to cisplatin (149, 150) showing that some hypomorphic BRCA alleles, although unable to prevent tumor development, can affect response to therapy. Interestingly, mutations in the BRCA C-terminal (BRCT) domain of BRCA1 commonly create protein products that are subject to protease-mediated degradation as they are unable to fold. HSP90 may stabilize the BRCT domain of these mutant BRCA1 proteins under PARP inhibitor selection pressure (151); the HSP90-stabilized mutant BRCA1 proteins can efficiently interact with PALB2-BRCA2-RAD51, form RAD51 foci and confer PARP inhibitor and cisplatin resistance. Treatment of resistant cells with the HSP90 inhibitor 17-dimethylaminoethylamino-17-demethoxygeldanamycin reduced mutant BRCA1 protein levels and restored their sensitivity to PARP inhibition (151).
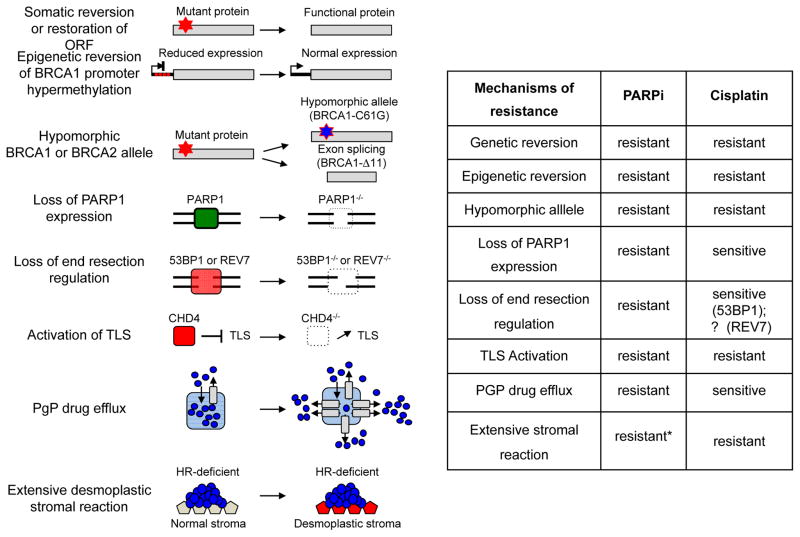
Figure 4. Mechanisms of PARPi resistance in HR deficient cells.
Known mechanisms conferring PARPi resistance in tumors cells and cross-resistance to cisplatin are indicated. An acquired genetic reversion of the original truncating mutations restores functional protein expression inducing PARPi resistance. Alternatively, acquired epigenetic reversion of BRCA1 promoter hypermethylation can restore normal BRCA1 protein expression levels conferring PARPi resistance. Hypomorphic alleles, such as BRCA1-C61G or BRCA1-Δ11, are unable to prevent tumor development but confer resistance to PARPi. Tumor cells may also become PARPi resistant through loss of PARP1 expression. Rescue of DNA end-resection in BRCA1-deficient tumors through loss of 53BP1 or REV7, increases HR capacity and confers resistance to PARPi. Loss of CHD4, a negative regulator of translesion synthesis (TLS), enhances DNA damage tolerance and induces PARPi resistance. Increased in P-glycoprotein (PGP)–mediated efflux, notably through ABCB1 upregulation (via fusion with SLC25A40) reduces intracellular PARPi concentrations inducing resistance. Desmoplastic stromal reaction is associated with reduced drug uptake conferring chemoresistance. (*: Although this mechanism has been described for cisplatin it might also apply for PARPi).
Since PARPis function by blocking the enzymatic action of PARP enzymes, another possible mechanism of PARPi resistance may be decreased expression of PARP enzymes (Figure 4). This mechanism of resistance may be particularly relevant to the PARP-trapping mechanism of action of PARPis. A mutagenesis screen designed to identify mechanisms of resistance to PARPis revealed PARP1 loss-of-function as a potent mechanism of resistant to olaparib in mouse embryonic stem cells and in human tumors cells (152). Accordingly, PARP1 levels have also been shown to be low in human cancer cell lines that have acquired resistance to the PARPi veliparib (96). Taken together, these observations suggest the possibility that tumor-specific mutation or inhibition of PARP1 (for instance, by epigenetic silencing or increased turnover of the protein) would result in resistance and disease progression, a hypothesis that has not yet been validated in patients.
Several mechanisms of resistance involving reacquisition of DNA end resection capacities have also been described. Discovery of these mechanisms came from the observation that the requirement of BRCA1 for HR can be alleviated by concomitant loss of 53BP1. 53BP1 blocks CtIP-mediated DNA end resection via downstream effectors like Rif1 and PTIP (153–158) and thus commits DNA repair to C-NHEJ (159, 160). Loss of 53BP1 partially restores the HR defect of Brca1-deleted mouse embryonic stem cells and reverts their hypersensitivity to DNA-damaging agents (159, 160). However, a deficiency in Ligase IV, another component of the C-NHEJ pathway, does not rescue cell proliferation or HR defect in Brca1-deficient cells. Authors showed that loss of 53BP1 but not Ligase IV was able to promote ssDNA formation competent for RPA phosphorylation. These data suggest that loss of 53BP1 but not Ligase IV promotes activation of DNA end resection. This discordance might explain why combined deficiencies in Brca1/53BP1 but not Brca1/Ligase IV reverse the HR defect in Brca1-deficient cells (159). Recently, an shRNA screen for hairpins promoting survival of BRCA1-deficient mouse mammary tumors to PARPi identified REV7 and 53BP1 as the top hits (161). REV7 was shown to promote C-NHEJ by inhibiting DNA end resection downstream of Rif1. Loss of REV7 in BRCA1-deficient cells induces CtIP-dependent end resection, leading to HR restoration and PARPi resistance (161, 162). Even though there is little evidence of such resistance mechanisms in human EOCs, a mouse model of BRCA1-associated breast cancer demonstrated low 53BP1 expression in a few olaparib-resistant BRCA1-deficient mouse tumors, suggesting that an acquired change in 53BP1 expression could occur in vivo as a resistance mechanism (163). In BRCA1-mutant cells, loss of 53BP1 confers resistance to PARPi. However, whether loss of 53BP1 confers cross-resistance to cisplatin is still elusive to date. In Brca1-deficient cell lines, shRNA-mediated loss of 53BP1 fully abolished the cisplatin sensitivity (160). However, in the olaparib-resistant BRCA1-deficient mouse tumors, the HR restoration conferred by 53BP1 loss is only partial (measured by RAD51 foci formation), and may explain the lack of cross-resistance to cisplatin (163).
Further studies are necessary to fully address whether 53BP1 loss in vivo can also confer resistance to cisplatin, to assess whether loss of REV7 also confers resistance to platinum therapy and whether these resection-dependent resistance mechanisms (described only in BRCA1-deficient cells so far) may also be relevant to other HR-deficient settings such as BRCA2 mutated cells.
Apart from the mechanisms of resistance intrinsic to the DNA damage response, pharmacological effects that alter the cellular response to PARPis may also be relevant. Several studies have shown that PARPi responses may be modified by ATP-binding cassette (ABC) transporters (164). Increased expression in tumor cells of ABC transporters, such as the P-glycoprotein (PgP) efflux pump (also known as multidrug resistance protein 1 (MDR1) or ATP-binding cassette sub-family B member 1 (ABCB1)) have been implicated in reducing the efficacy of many compounds by enhancing their extracellular translocation (Figure 4). In a genetically-engineered mouse model for BRCA1-mutated breast cancer, PARPi resistance was mediated via upregulation of the Abcb1a and Abcb1b genes encoding PgP pumps. Of note, up-regulation of ABCB1 gene through promoter fusion and translocation involving the 5′ region of the gene (most frequently with SLC25A40 gene) was found in approximately 8% of HGSOC recurrence samples in another study (146). Although not relevant to platinum analogues, this mechanism of resistance may be relevant to PARPi resistance and other drugs such as etoposide, paclitaxel and doxorubicin. Furthermore, resistance to PARPi could be reversed by coadministration of PgP inhibitors arguing that upregulation of PgP may be a clinically relevant and druggable acquired mechanism of PARPi resistance (165, 166). Of note, PgP can be inhibited in clinic (such as with tariquidar) (165); furthermore novel PARPis (such as AZD2461) have been developed that have lower affinity to PgP thereby circumventing this mechanism of PARPi resistance (163, 167).
Extensive tumor desmoplasia has also been suggested as a mechanism of resistance in a BRCA1 mutated tumor without BRCA1 reversion (146). In that case, an extensive desmoplastic stromal reaction was observed at autopsy; tumor desmoplasia has been associated with chemoresistance and suboptimal drug uptake in pancreatic cancer and may have accounted for resistance despite persistence of HR deficiency due to the BRCA1 mutation. In another study, loss of the nucleosome remodeling factor CHD4 was found to be associated with cisplatin resistance in BRCA2 mutated tumors (168). Restoration of cisplatin resistance was independent of HR but correlated with restored cell cycle progression, reduced chromosomal aberrations, and enhanced DNA damage tolerance. Of note, BRCA2 mutant ovarian cancers with reduced CHD4 expression significantly correlated with shorter progression-free survival and shorter overall survival (168).
Interestingly, in a genetically engineered mouse model for BRCA1-deficient tumors, in which genetic reversion was made impossible by the large intragenic BRCA1 deletion, no acquired platinum drug resistance was observed (169). This raises the question whether mechanisms other than genetic BRCA1/2 reversions can result in resistance to platinum-based chemotherapy in patients (170).
Finally, besides the aforementioned resistance mechanisms, an important question relates to the nature of resistance of residual tumor cells which may respond again to platinum drugs (i.e. recurrence of disease that responds again to platinum chemotherapy). Several approaches have been attempted to target these residual cells that are not killed by first line chemotherapy. First, maintenance chemotherapy, i.e. continuing chemotherapy after achievement of complete clinical remission to first line chemotherapy has been widely studied but no chemotherapy regimen has been associated with an improved overall survival or cure rate in that setting (171). Second, increasing dose intensity approaches have also been attempted, including increasing the dose of platinum, combining platinum agents, increasing the number of cycles of chemotherapy, or using high dose chemotherapy also incorporating alternative DNA cross-linking agents such as melphalan and cyclophosphamide in combination with bone marrow transplantation or with peripheral blood stem cell support; all these approaches failed to improve outcome compared to standard chemotherapy (172). The only dose intensity approach that has been associated with improved survival was administration of chemotherapy via the intraperitoneal route which is capable of achieving high local concentrations of chemotherapy with acceptable systemic side effects (10). However, definitive data regarding comparison of IP chemotherapy versus IV dose dense chemotherapy (GOG252 study) are still pending.
In conclusion, understanding the mechanisms of resistance to PARPis in HR deficient EOCs is critical in order to identify approaches that may overcome resistance and/or minimize the emergence of secondary resistant clones.
OVERCOMING DENOVO AND ACQUIRED HR PROFICIENCY
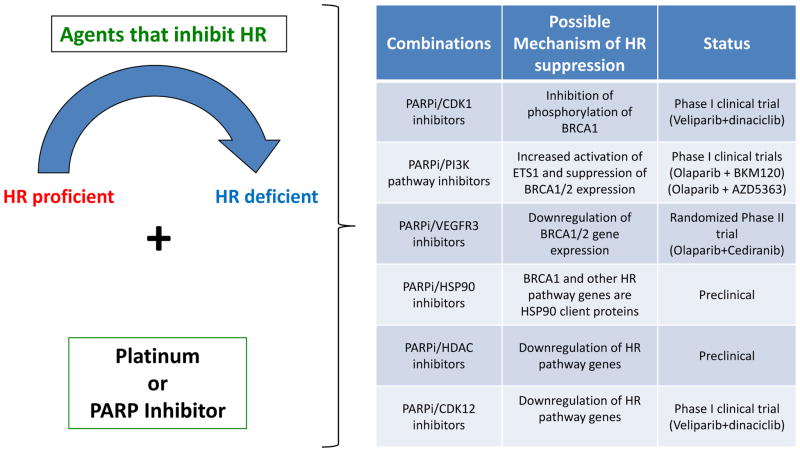
The promise of platinum agents and PARPis in the management of ovarian cancers is tempered by the fact that HR-proficient tumors do not respond to these agents, suggesting that as many as 50% of ovarian patients (i.e. those with tumors which are de novo HR proficient) do not benefit from these drugs. Furthermore, even the 50% of EOCs, which are initially HR-deficient, eventually become HR-proficient as a result of development of resistance to platinum or PARPis. Combination of platinum or PARPis with agents that inhibit HR may therefore represent an effective strategy to sensitize HR proficient tumors to platinum and PARPis, and thus potentially expand use of these agents into EOCs with denovo or acquired HR proficiency. Multiple strategies designed to selectively disrupt HR in cancer cells and sensitize them to PARPis or platinum have been evaluated both preclinically as well as in early clinical trials in EOC (Figure 5). Such strategies include combinations of platinum and PARPis with i) CDK1 inhibitors (inhibition of CDK1 induces HR deficiency via inhibition of phosphorylation of BRCA1 by CDK1 (173, 174)), ii) with PI3K or AKT inhibitors (inhibition of PI3K pathway leads to ERK activation/phosphorylation, increased activation of ETS1 and suppression of BRCA1/2 expression and of HR (175, 176)), iii) CDK12 inhibitors (abrogation of CDK12 leads to downregulation of HR genes as discussed above), iv) HDAC inhibitors (which induce coordinated down-regulation of HR pathway genes (177)) and v) HSP90 inhibitors (which induce HR deficiency because multiple HR proteins including BRCA1 are HSP90 clients (178) and they may also overcome HSP90-mediated stabilization of BRCA1 mutant proteins as a mechanism of PARPi resistance (151)). Preclinical evaluation has demonstrated that CDK1-, CDK12-, PI3K-, AKT-, HDAC- and HSP90-inhibitors are able to inhibit HR and sensitize HR proficient cells to PARPis and/or platinum. Of note, phase I clinical evaluation of olaparib with PI3K inhibitors (BYL719 or BKM120) or the AKT inhibitor (AZD5363) in ovarian and breast cancers provided evidence of response in patients who were expected to have HR proficient tumors thereby providing proof of principle for this approach.
Figure 5. PARPi combinations against HR proficient tumors.
Rationale behind use of specific PARPi combinations as a strategy against HR proficient tumors. Specifically, use of agents that inhibit HR such as CDK1- or HSP90 inhibitors may render HR proficient tumors into HR deficient and thus sensitize them to platinum or PARPis. The proposed mechanism of HR suppression and the clinical status of these PARPi combinations are presented in the right panel.
Interestingly, in a randomized, open-label, phase 2 study, the combination of olaparib plus cediranib (which is a VEGFR1, VEGFR2 and VEGFR3 inhibitor) significantly improved PFS in recurrent platinum sensitive EOC compared to olaparib alone, and the greatest benefit was observed among patients without BRCA1/2 mutations (64). Although a number of mechanisms may explain this result, this finding may also indicate that there is greater synergism between these two drugs in the setting of HR proficient tumors. In this regard, VEGFR3 inhibition has been shown to downregulate BRCA gene expression, reverse chemotherapy resistance and restore chemosensitivity in resistant cell lines in which a BRCA2 mutation had reverted to wild type (179). It is therefore possible that cediranib may be enhancing the response to olaparib in HR proficient tumors via inhibition of HR (due to VEGFR3 inhibition).
An important challenge for the clinical development of these combinations of HR inhibitors with PARPis is the potentially low therapeutic window between normal and cancer cells and thus the risk of enhanced toxicity. Therefore, careful Phase I evaluation of these combinations will be required, with increased focus on proof of mechanism pharmacodynamic studies. Nonetheless, thus far, the clinical trials of olaparib combinations with PI3K pathway inhibitors have not shown any alarming signals besides the expected non-overlapping toxicities of these agents.
CONCLUSIONS
Approximately 50% of EOCs exhibit defective DNA repair via HR and represent a distinct EOC subtype with unique clinical characteristics that have important implications for management. Germline and somatic BRCA1/2 mutations are the most common mechanisms of HR deficiency but multiple alternative mechanisms also contribute to this phenomenon in EOC. HR deficiency explains the enhanced sensitivity of EOC to platinum based chemotherapy and is an important therapeutic strategy in this disease. The striking activity of PARPis in HR deficient EOCs highlights the potential of synthetic lethality as anticancer strategy and is first molecular targeted therapy approved in this disease. Additional, potentially non-cross resistant, synthetic lethal approaches such as inhibition of Alt-EJ pathway and cell cycle checkpoint inhibition are exciting novel approaches against HR deficient cancers.
Although PARPis are now FDA approved in patients with BRCA1/2-mutated EOCs, patients with HR deficient/non-BRCA-mutated tumors do not have access to these agents outside a clinical trial. This highlights the importance of development of a robust and prospectively validated biomarker of HR deficiency that is capable to identify non-BRCA-mutated patients who may benefit from these agents. Another challenge is denovo and acquired resistance which are often encountered in clinic and have tempered the enthusiasm for the potential of PARPis in HR deficient EOCs. Understanding the mechanisms of PARPi resistance and their relation to platinum resistance may aid the development of novel non-cross resistant therapies and may help optimize the sequence of how these agents are incorporated in the clinical management of HR deficient EOC. Finally, combinations of PARPis with agents that inhibit HR are exciting strategies to sensitize HR proficient tumors to platinum and PARPis, and thus potentially expand use of these agents into EOCs with denovo or acquired HR proficiency. Initial reports from the clinical evaluation of these combinations provide clinical proof of principle for this approach without prohibitive toxicities.
STATEMENT OF SIGNIFICANCE.
Defective DNA repair via homologous recombination is a pivotal vulnerability of epithelial ovarian cancer, particularly of the high grade serous histologic subtype. Targeting defective HR offers the unique opportunity of exploiting molecular differences between tumor and normal cells, thereby inducing cancer-specific synthetic lethality; the promise and challenges of these approaches in ovarian cancer are discussed in this review.
Acknowledgments
This research was supported by a Stand Up To Cancer – Ovarian Cancer Research Fund-Ovarian Cancer National Alliance-National Ovarian Cancer Coalition Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT16-15). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. This work was also supported by grants from the U.S. National Institutes of Health (P50CA168504), and the Breast Cancer Research Foundation (to A.D.D.). As well as support from the Susan Smith Center for Women’s Cancers, and the Department of Defense Ovarian Cancer Academy Award (W81XWH-10-1-0585) (to P.A.K.).
Footnotes
CONFLICTS OF INTEREST
Dr. Shapiro and Dr. Konstantinopoulos have served as consultants/advisory board members for Vertex. Dr. Shapiro and Dr. D’Andrea serve as consultants and receive research funding from Eli Lilly and Company.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Herzog TJ. The current treatment of recurrent ovarian cancer. Curr Oncol Rep. 2006;8:448–54. doi: 10.1007/s11912-006-0074-9. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinopoulos PA, Awtrey CS. Management of ovarian cancer: a 75-year-old woman who has completed treatment. JAMA. 2012;307:1420–9. doi: 10.1001/jama.2012.269. [DOI] [PubMed] [Google Scholar]
- 4.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–29. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 5.Rossof AH, Talley RW, Stephens R, Thigpen T, Samson MK, Groppe C, Jr, et al. Phase II evaluation of cis-dichlorodiammineplatinum(II) in advanced malignancies of the genitourinary and gynecologic organs: a Southwest Oncology Group Study. Cancer Treat Rep. 1979;63:1557–64. [PubMed] [Google Scholar]
- 6.Thigpen T, Shingleton H, Homesley H, LaGasse L, Blessing J. cis-Dichlorodiammineplatinum(II) in the treatment of gynecologic malignancies: phase II trials by the Gynecologic Oncology Group. Cancer Treat Rep. 1979;63:1549–55. [PubMed] [Google Scholar]
- 7.Ozols RF, Bundy BN, Greer BE, Fowler JM, Clarke-Pearson D, Burger RA, et al. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: a Gynecologic Oncology Group study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:3194–200. doi: 10.1200/JCO.2003.02.153. [DOI] [PubMed] [Google Scholar]
- 8.McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, et al. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334:1–6. doi: 10.1056/NEJM199601043340101. [DOI] [PubMed] [Google Scholar]
- 9.Cannistra SA. Intraperitoneal chemotherapy comes of age. N Engl J Med. 2006;354:77–9. doi: 10.1056/NEJMe058308. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 11.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 12.Winter WE, 3rd, Maxwell GL, Tian C, Carlson JW, Ozols RF, Rose PG, et al. Prognostic factors for stage III epithelial ovarian cancer: a Gynecologic Oncology Group Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:3621–7. doi: 10.1200/JCO.2006.10.2517. [DOI] [PubMed] [Google Scholar]
- 13.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 14.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010;362:1909–19. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:3799–808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 16.Integrated genomic analyses of ovarian carcinoma. Nature. 2012;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 19.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 20.Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–40. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 21.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:1383–8. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 22.Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:764–75. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:2654–63. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal T, Permuth-Wey J, Betts JA, Krischer JP, Fiorica J, Arango H, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–16. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 25.Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD, 2nd, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3570–6. doi: 10.1200/JCO.2009.27.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313:1347–61. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baldwin RL, Nemeth E, Tran H, Shvartsman H, Cass I, Narod S, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer research. 2000;60:5329–33. [PubMed] [Google Scholar]
- 29.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 30.Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–82. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–4. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 32.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–22. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–70. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 34.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–35. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 35.Brown LA, Kalloger SE, Miller MA, Shih Ie M, McKinney SE, Santos JL, et al. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–9. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 36.Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer research. 2013;74:287–97. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. The Journal of biological chemistry. 2014;289:9247–53. doi: 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Molecular cell. 2011;41:210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi YE, Pan Y, Park E, Konstantinopoulos P, De S, D’Andrea A, et al. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. Elife (Cambridge) 2014;3:e02445. doi: 10.7554/eLife.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceccaldi R, O’Connor KW, Mouw KW, Li AY, Matulonis UA, D’Andrea AD, et al. A Unique Subset of Epithelial Ovarian Cancers with Platinum Sensitivity and PARP Inhibitor Resistance. Cancer research. 2015;75:628–34. doi: 10.1158/0008-5472.CAN-14-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–6. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 42.Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–90. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, et al. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA. 2000;283:2260–5. doi: 10.1001/jama.283.17.2260. [DOI] [PubMed] [Google Scholar]
- 44.Ruscito I, Dimitrova D, Vasconcelos I, Gellhaus K, Schwachula T, Bellati F, et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients--a study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD) Eur J Cancer. 2014;50:2090–8. doi: 10.1016/j.ejca.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Soslow RA, Han G, Park KJ, Garg K, Olvera N, Spriggs DR, et al. Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod Pathol. 2011;25:625–36. doi: 10.1038/modpathol.2011.183. [DOI] [PubMed] [Google Scholar]
- 46.Strickland K, Howitt BE, Rodig SJ, Ritterhouse L, D’Andrea AD, Matulonis UA, et al. Tumor infiltrating and peritumoral T cells and expression of PD-L1 in BRCA1/2-mutated high grade serous ovarian cancers. J Clin Oncol. 2015;33(suppl):abstr 5512. [Google Scholar]
- 47.Soegaard M, Kjaer SK, Cox M, Wozniak E, Hogdall E, Hogdall C, et al. BRCA1 and BRCA2 mutation prevalence and clinical characteristics of a population-based series of ovarian cancer cases from Denmark. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:3761–7. doi: 10.1158/1078-0432.CCR-07-4806. [DOI] [PubMed] [Google Scholar]
- 48.Gourley C, Michie CO, Roxburgh P, Yap TA, Harden S, Paul J, et al. Increased incidence of visceral metastases in scottish patients with BRCA1/2-defective ovarian cancer: an extension of the ovarian BRCAness phenotype. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2505–11. doi: 10.1200/JCO.2009.25.1082. [DOI] [PubMed] [Google Scholar]
- 49.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3555–61. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang J, D’Andrea AD, Kozono D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J Natl Cancer Inst. 2012;104:670–81. doi: 10.1093/jnci/djs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weberpals JI, Tu D, Squire JA, Amin MS, Islam S, Pelletier LB, et al. Breast cancer 1 (BRCA1) protein expression as a prognostic marker in sporadic epithelial ovarian carcinoma: an NCIC CTG OV.16 correlative study. Ann Oncol. 2011;22:2403–10. doi: 10.1093/annonc/mdq770. [DOI] [PubMed] [Google Scholar]
- 52.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 53.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–7. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–82. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birkbak NJ, Wang ZC, Kim JY, Eklund AC, Li Q, Tian R, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer discovery. 2012;2:366–75. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popova T, Manie E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer research. 2012;72:5454–62. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 58.Timms KM, Abkevich V, Hughes E, Neff C, Reid J, Morris B, et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014;16:475. doi: 10.1186/s13058-014-0475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swisher E, Brenton J, Kaufmann S, Oza A, Coleman RL, O’Malley D, et al. 215 Updated clinical and preliminary correlative results of ARIEL2, a Phase 2 study to identify ovarian cancer patients likely to respond to rucaparib. European Journal of Cancer. 50:73. [Google Scholar]
- 60.Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:6159–68. doi: 10.1158/1078-0432.CCR-10-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukhopadhyay A, Elattar A, Cerbinskaite A, Wilkinson SJ, Drew Y, Kyle S, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:2344–51. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
- 62.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–65. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu JF, Barry WT, Birrer M, Lee JM, Buckanovich RJ, Fleming GF, et al. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol. 2014;15:1207–14. doi: 10.1016/S1470-2045(14)70391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–92. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 66.Gordon AN, Fleagle JT, Guthrie D, Parkin DE, Gore ME, Lacave AJ. Recurrent epithelial ovarian carcinoma: a randomized phase III study of pegylated liposomal doxorubicin versus topotecan. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19:3312–22. doi: 10.1200/JCO.2001.19.14.3312. [DOI] [PubMed] [Google Scholar]
- 67.Naumann RW, Coleman RL. Management strategies for recurrent platinum-resistant ovarian cancer. Drugs. 2011;71:1397–412. doi: 10.2165/11591720-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 68.Nitiss J, Wang JC. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci U S A. 1988;85:7501–5. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maede Y, Shimizu H, Fukushima T, Kogame T, Nakamura T, Miki T, et al. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol Cancer Ther. 2014;13:214–20. doi: 10.1158/1535-7163.MCT-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ozols RF. Oral etoposide for the treatment of recurrent ovarian cancer. Drugs. 1999;58(Suppl 3):43–9. doi: 10.2165/00003495-199958003-00007. [DOI] [PubMed] [Google Scholar]
- 71.Doroshow JH. Role of hydrogen peroxide and hydroxyl radical formation in the killing of Ehrlich tumor cells by anticancer quinones. Proc Natl Acad Sci U S A. 1986;83:4514–8. doi: 10.1073/pnas.83.12.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Safra T, Borgato L, Nicoletto MO, Rolnitzky L, Pelles-Avraham S, Geva R, et al. BRCA Mutation Status and Determinant of Outcome in Women with Recurrent Epithelial Ovarian Cancer Treated with Pegylated Liposomal Doxorubicin. Mol Cancer Ther. 2011;10:2000–7. doi: 10.1158/1535-7163.MCT-11-0272. [DOI] [PubMed] [Google Scholar]
- 73.Pfisterer J, Plante M, Vergote I, du Bois A, Hirte H, Lacave AJ, et al. Gemcitabine plus carboplatin compared with carboplatin in patients with platinum-sensitive recurrent ovarian cancer: an intergroup trial of the AGO-OVAR, the NCIC CTG, and the EORTC GCG. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:4699–707. doi: 10.1200/JCO.2006.06.0913. [DOI] [PubMed] [Google Scholar]
- 74.Lorusso D, Di Stefano A, Fanfani F, Scambia G. Role of gemcitabine in ovarian cancer treatment. Ann Oncol. 2006;17(Suppl 5):v188–94. doi: 10.1093/annonc/mdj979. [DOI] [PubMed] [Google Scholar]
- 75.Brody LC. Treating cancer by targeting a weakness. The New England journal of medicine. 2005;353:949–50. doi: 10.1056/NEJMcibr052331. [DOI] [PubMed] [Google Scholar]
- 76.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 77.Scott CL, Swisher EM, Kaufmann SH. Poly (adp-ribose) polymerase inhibitors: recent advances and future development. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:1397–406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochemical pharmacology. 2012;84:137–46. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 79.Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, De La Rubia G, et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 80.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer research. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 81.Iglehart JD, Silver DP. Synthetic lethality--a new direction in cancer-drug development. The New England journal of medicine. 2009;361:189–91. doi: 10.1056/NEJMe0903044. [DOI] [PubMed] [Google Scholar]
- 82.Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA: a cancer journal for clinicians. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 83.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3406–11. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Lorenzo SB, Patel AG, Hurley RM, Kaufmann SH. The Elephant and the Blind Men: Making Sense of PARP Inhibitors in Homologous Recombination Deficient Tumor Cells. Frontiers in oncology. 2013;3:228. doi: 10.3389/fonc.2013.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic acids research. 2006;34:6170–82. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic acids research. 2008;36:6959–76. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS genetics. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. The EMBO journal. 2006;25:1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paddock MN, Bauman AT, Higdon R, Kolker E, Takeda S, Scharenberg AM. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA repair. 2011;10:338–43. doi: 10.1016/j.dnarep.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer research. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langelier MF, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Current opinion in structural biology. 2013;23:134–43. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–5. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 93.Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic acids research. 2012;40:7788–805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Molecular and cellular biology. 1998;18:3563–71. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–8. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 96.Liu X, Han EK, Anderson M, Shi Y, Semizarov D, Wang G, et al. Acquired resistance to combination treatment with temozolomide and ABT-888 is mediated by both base excision repair and homologous recombination DNA repair pathways. Molecular cancer research : MCR. 2009;7:1686–92. doi: 10.1158/1541-7786.MCR-09-0299. [DOI] [PubMed] [Google Scholar]
- 97.Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. The Journal of pharmacology and experimental therapeutics. 2014;349:408–16. doi: 10.1124/jpet.113.210146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. The Journal of biological chemistry. 2012;287:4198–210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–62. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–7. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu JF, Konstantinopoulos PA, Matulonis UA. PARP inhibitors in ovarian cancer: Current status and future promise. Gynecol Oncol. 2014;133:362–9. doi: 10.1016/j.ygyno.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 103.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 104.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2512–9. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 105.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–61. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 106.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–61. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 107.Ang JE, Gourley C, Powell CB, High H, Shapira-Frommer R, Castonguay V, et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: a multi-institutional study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:5485–93. doi: 10.1158/1078-0432.CCR-13-1262. [DOI] [PubMed] [Google Scholar]
- 108.Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer research. 2011;71:5626–34. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oza AM, Cibula D, Benzaquen AO, Poole C, Mathijssen RH, Sonke GS, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 2014;16:87–97. doi: 10.1016/S1470-2045(14)71135-0. [DOI] [PubMed] [Google Scholar]
- 110.Lee JM, Hays JL, Annunziata CM, Noonan AM, Minasian L, Zujewski JA, et al. Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2 mutation-associated breast or ovarian cancer with biomarker analyses. J Natl Cancer Inst. 2014;106:dju089. doi: 10.1093/jnci/dju089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rajan A, Carter CA, Kelly RJ, Gutierrez M, Kummar S, Szabo E, et al. A phase I combination study of olaparib with cisplatin and gemcitabine in adults with solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2344–51. doi: 10.1158/1078-0432.CCR-11-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 113.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annual review of genetics. 2013;47:433–55. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 114.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. The EMBO journal. 1996;15:5093–103. [PMC free article] [PubMed] [Google Scholar]
- 115.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic acids research. 1998;26:5333–42. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–21. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 117.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–6. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 118.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 119.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer research. 2005;65:4020–30. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 120.Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. European journal of immunology. 2002;32:701–9. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 121.Wang H, Perrault AR, Takeda Y, Qin W, Wang H, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic acids research. 2003;31:5377–88. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology. 2010;17:410–6. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nature structural & molecular biology. 2011;18:80–4. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Advances in immunology. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA repair. 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nature structural & molecular biology. 2015;22:230–7. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–38. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 130.Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–40. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen T, Stephens PA, Middleton FK, Curtin NJ. Targeting the S and G2 checkpoint to treat cancer. Drug Discov Today. 2012;17:194–202. doi: 10.1016/j.drudis.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 132.Evers B, Helleday T, Jonkers J. Targeting homologous recombination repair defects in cancer. Trends Pharmacol Sci. 2010;31:372–80. doi: 10.1016/j.tips.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 133.Chen CC, Kennedy RD, Sidi S, Look AT, D’Andrea A. CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pulsipher M, Kupfer GM, Naf D, Suliman A, Lee JS, Jakobs P, et al. Subtyping analysis of Fanconi anemia by immunoblotting and retroviral gene transfer. Mol Med. 1998;4:468–79. [PMC free article] [PubMed] [Google Scholar]
- 135.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, et al. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–9. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015;149:124–38. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 137.Do KT, Wilsker D, Balasubramanian P, Zlott J, Jeong W, Lawrence SM, et al. Phase I trial of AZD1775 (MK1775), a wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol. 2014;60:4009. [Google Scholar]
- 138.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nature medicine. 2013;19:1381–8. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 139.Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:540–7. doi: 10.1158/1078-0432.CCR-13-0225. [DOI] [PubMed] [Google Scholar]
- 140.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 142.Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer research. 2008;68:2581–6. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer research. 2009;69:6381–6. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3008–15. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barber LJ, Sandhu S, Chen L, Campbell J, Kozarewa I, Fenwick K, et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. The Journal of pathology. 2013;229:422–9. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- 146.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 147.Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, Prasetyanti P, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer discovery. 2013;3:1142–55. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- 148.Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 149.Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, et al. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer research. 2008;68:3243–50. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tammaro C, Raponi M, Wilson DI, Baralle D. BRCA1 exon 11 alternative splicing, multiple functions and the association with cancer. Biochemical Society transactions. 2012;40:768–72. doi: 10.1042/BST20120140. [DOI] [PubMed] [Google Scholar]
- 151.Johnson N, Johnson SF, Yao W, Li YC, Choi YE, Bernhardy AJ, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc Natl Acad Sci U S A. 2013;110:17041–6. doi: 10.1073/pnas.1305170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pettitt SJ, Rehman FL, Bajrami I, Brough R, Wallberg F, Kozarewa I, et al. A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PloS one. 2013;8:e61520. doi: 10.1371/journal.pone.0061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Molecular cell. 2013;49:858–71. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–5. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. The Journal of biological chemistry. 2013;288:11135–43. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Molecular cell. 2013;49:872–83. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 157.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–4. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–80. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature structural & molecular biology. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, Bouwman P, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–4. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, Wevers BA, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature. 2015;521:537–40. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer discovery. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Choi YH, Yu AM. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Current pharmaceutical design. 2014;20:793–807. doi: 10.2174/138161282005140214165212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17079–84. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J, et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell cycle. 2012;11:3837–50. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li X, Delzer J, Voorman R, de Morais SM, Lao Y. Disposition and drug-drug interaction potential of veliparib (ABT-888), a novel and potent inhibitor of poly(ADP-ribose) polymerase. Drug metabolism and disposition: the biological fate of chemicals. 2011;39:1161–9. doi: 10.1124/dmd.110.037820. [DOI] [PubMed] [Google Scholar]
- 168.Guillemette S, Serra RW, Peng M, Hayes JA, Konstantinopoulos PA, Green MR, et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015;29:489–94. doi: 10.1101/gad.256214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rottenberg S, Nygren AO, Pajic M, van Leeuwen FW, van der Heijden I, van de Wetering K, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci U S A. 2007;104:12117–22. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Borst P, Rottenberg S, Jonkers J. How do real tumors become resistant to cisplatin? Cell cycle. 2008;7:1353–9. doi: 10.4161/cc.7.10.5930. [DOI] [PubMed] [Google Scholar]
- 171.Markman M, Liu PY, Wilczynski S, Monk B, Copeland LJ, Alvarez RD, et al. Phase III randomized trial of 12 versus 3 months of maintenance paclitaxel in patients with advanced ovarian cancer after complete response to platinum and paclitaxel-based chemotherapy: a Southwest Oncology Group and Gynecologic Oncology Group trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:2460–5. doi: 10.1200/JCO.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 172.Ozols RF. Ovarian cancer: is dose intensity dead? J Clin Oncol. 2007;25:4157–8. doi: 10.1200/JCO.2007.12.1723. [DOI] [PubMed] [Google Scholar]
- 173.Johnson N, Li YC, Walton ZE, Cheng KA, Li D, Rodig SJ, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nature medicine. 2011;17:875–82. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Johnson SF, Johnson N, Chi D, Primack B, D’Andrea AD, Lim E, et al. Abstract 1788: The CDK inhibitor dinaciclib sensitizes triple-negative breast cancer cells to PARP inhibition. Cancer Res. 2013;73:1788. [Google Scholar]
- 175.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer discovery. 2012;2:1036–47. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer discovery. 2012;2:1048–63. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Konstantinopoulos PA, Wilson AJ, Saskowski J, Wass E, Khabele D. Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer. Gynecol Oncol. 2014;133:599–606. doi: 10.1016/j.ygyno.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Choi YE, Battelli C, Watson J, Liu J, Curtis J, Morse AN, et al. Sublethal concentrations of 17-AAG suppress homologous recombination DNA repair and enhance sensitivity to carboplatin and olaparib in HR proficient ovarian cancer cells. Oncotarget. 2014;5:2678–87. doi: 10.18632/oncotarget.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lim JJ, Yang K, Taylor-Harding B, Wiedemeyer WR, Buckanovich RJ. VEGFR3 inhibition chemosensitizes ovarian cancer stemlike cells through down-regulation of BRCA1 and BRCA2. Neoplasia. 2014;16:343–53. e1–2. doi: 10.1016/j.neo.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]