SUMMARY
Three (BLM, WRN, RECQ4) of the five human RecQ helicases are linked to genetic disorders characterized by genomic instability, cancer, and accelerated aging [1]. RECQ1, the first human RecQ helicase discovered [2–4] and most abundant [5], was recently implicated in breast cancer [6,7]. RECQ1 is an ATP-dependent DNA unwinding enzyme (helicase) [8,9] with roles in replication [10–12] and DNA repair [13–16]. RECQ1 is highly expressed in various tumors and cancer cell lines (for review, see [17]) and its suppression reduces cancer cell proliferation [14], suggesting a target for anti-cancer drugs. RECQ1’s assembly state plays a critical role in modulating its helicase, branch-migration (BM), or strand annealing [18,19]. The crystal structure of truncated RECQ1 [20,21] resembles that of E. coli RecQ [22] with two RecA-like domains, a RecQ-specific zinc-binding and winged-helix domains, the latter implicated in DNA strand separation and oligomer formation. In addition, a conserved aromatic loop (AL) is important for DNA unwinding by bacterial RecQ [23,24] and truncated RECQ1 helicases [21]. To better understand the roles of RECQ1, two AL mutants (W227A, F231A) in full-length RECQ1 were characterized biochemically and genetically. The RECQ1 mutants were defective in helicase or BM, but retained DNA binding, oligomerization, ATPase, and strand annealing. RECQ1-depleted HeLa cells expressing either AL mutant displayed reduced replication tract length, elevated dormant origin firing, and increased double-strand breaks that could be suppressed by exogenously expressed Replication Protein A (RPA). Thus, RECQ1 governs RPA’s availability in order to maintain normal replication dynamics, suppress DNA damage, and preserve genome homeostasis.
Graphical abstract

RESULTS
Biochemical characterization of RECQ1 aromatic loop mutants
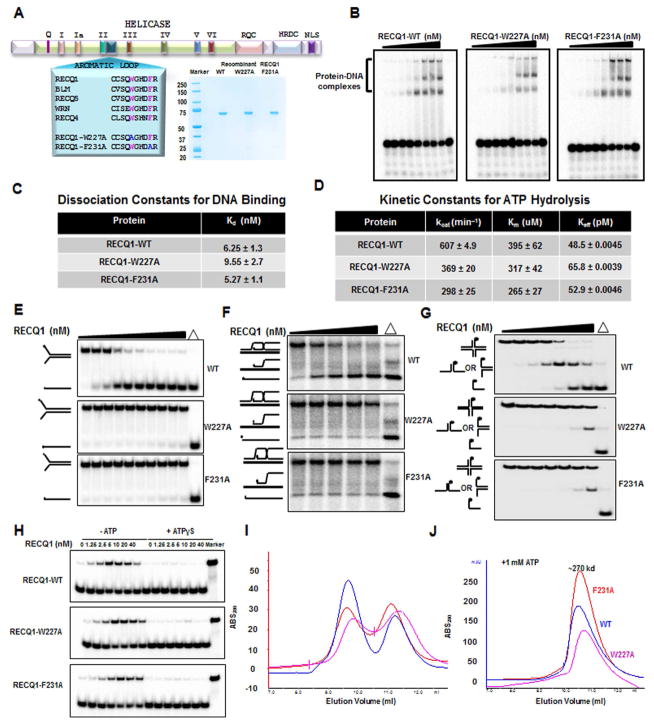
Two invariant aromatic loop (AL) residues, Tryptophan (W) 227 and Phenylalanine (F) 231, located within the helicase core domain of full-length RECQ1 were replaced with Alanine (A) (Figure 1A, S1A). Electrophoretic mobility shift assays (EMSA) demonstrated the purified recombinant mutant RECQ1 proteins were proficient in DNA binding (Figure 1B, 1C, S1B). Km values for ATP hydrolysis by RECQ1-W227A and RECQ1-F231A were comparable to RECQ1-WT, suggesting normal ATP binding (Figure 1D). Turnover rate constants for ATP hydrolysis (kcat) were reduced 1.6-fold for RECQ1-W2227A and 2.0-fold for RECQ1-F231A (Figure 1D). Keff values for ATP hydrolysis as a function of DNA effector concentration for the RECQ1 proteins were comparable (Figure 1D), consistent with their similar Kd values. The relatively modest effects of either AL substitution on RECQ1 DNA binding and ATPase led us to ask if RECQ1 unwinding (helicase) or BM were affected. Both W227A and F231A showed minimal unwinding activity on a forked duplex (19 bp) DNA substrate efficiently unwound by RECQ1-WT (Figure 1E, S1C).
Figure 1. Biochemical analysis of purified recombinant human RECQ1 proteins.
(A) Schematic of human RECQ1 showing conserved domains and helicase core signature motifs. The conserved aromatic-rich sequence between motifs II and III is shown with a sequence alignment of the five human RecQ helicases. The two invariant aromatic amino acids and corresponding amino acid substitutions in RECQ1 are indicated in pink and blue, respectively (left). Coomassie-stained SDS polyacrylamide gel showing recombinant human RECQ1 proteins purified from insect cells (right). (B) Autoradiogram showing representative gel from EMSA experiments with RECQ1 proteins (0.15, 0.31, 0.62, 1.25, 2.5, 5, 10, and 20 nM monomer) incubated with a forked 19 bp duplex DNA substrate. (C) Apparent dissociation constant values (Kd) estimating binding affinities of RECQ1 proteins for the forked duplex substrate. (D) Kinetic constants for ATP hydrolysis by RECQ1 proteins. (E) Autoradiogram showing representative native polyacrylamide gel from helicase assays with RECQ1 proteins (0.15, 0.31, 0.62, 1.25, 2.5, 5, 10, 20 and 40 nM monomer) and the 19 bp forked duplex DNA substrate. (F) Autoradiogram showing representative native polyacrylamide gel from BM assays with RECQ1 proteins (25, 50,100, and 200 nM monomer) and mobile three-stranded D-loop DNA substrate. (G) Autoradiogram showing representative native polyacrylamide gel from BM assays with RECQ1 proteins (0.62, 1.25, 2.5, 5, 10, and 20 nM monomer) and the four-stranded HJ DNA substrate. (H) Autoradiogram showing representative native polyacrylamide gel from strand annealing assays with RECQ1 proteins (1.25, 2.5, 5, 10, 20, and 40 nM monomer) and complementary single-stranded oligonucleotides in the absence or presence of 1 mM ATPγS as indicated. (I, J) Size exclusion chromatogram showing distribution of purified RECQ1 proteins incubated in the absence (I) or presence (J) of 1 mM ATP.
RECQ1 plays an important role in homologous recombination (HR) repair [8,14,25]; therefore, we characterized the effect of RECQ1 AL mutations on BM of HR DNA intermediates. Both AL mutations almost completely eliminated RECQ1’s ability to resolve a closed mobile D-loop (Figure 1F, S1D) or open three-stranded BM structure (Figure S1F, S1G). The RECQ1 mutants displayed reduced but detectable BM activity on a four-stranded HJ substrate (Figure 1G, S1E). Thus, despite fairly modest effects of AL mutations on DNA binding and ATPase, AL integrity is critical for RECQ1-catalyzed BM and helicase activity.
RECQ1 aromatic loop mutants retain strand annealing and oligomerization
RECQ1, like other RecQ helicases, anneals complementary single-stranded DNA molecules in an ATP-regulated manner [8]. RECQ1-W227A annealing was very similar to RECQ1-WT (Figure 1H, S1H, S1I). RECQ1-F231A also performed strand annealing; however, maximal product formation was reduced. Kinetic analysis of RECQ1 strand annealing kinetics correlated with protein titrations (Figure S1J). Strand annealing by RECQ1-W227A or RECQ1-F231A was inhibited by ATPγS in a manner comparable to RECQ1-WT (Figure 1H), suggesting ATP binding induced a conformational change in the AL mutants as observed for RECQ1-WT [8,18], consistent with their similar Km values for ATP hydrolysis (Figure 1D).
ATP binding affects not only RECQ1 conformational state [8], but also oligomerization [18]. In the presence of ATP (1 mM), RECQ1 was observed to shift from a tetramer capable of strand annealing to a dimer with optimal helicase activity [19]. These observations led us to ask if the RECQ1 AL mutant proteins behaved in a similar ATP-regulated manner. Size exclusion chromatography with 1 mM ATP present demonstrated comparable shift of wild-type and mutant RECQ1 proteins from two species of ~400 kD and ~200 kD to a single species of intermediate size (~270 kD) (Figure 1I, 1J). We conclude that the AL mutations in full-length RECQ1 do not perturb ATP-regulated oligomerization.
RECQ1 aromatic loop mutations interfere with normal cellular replication
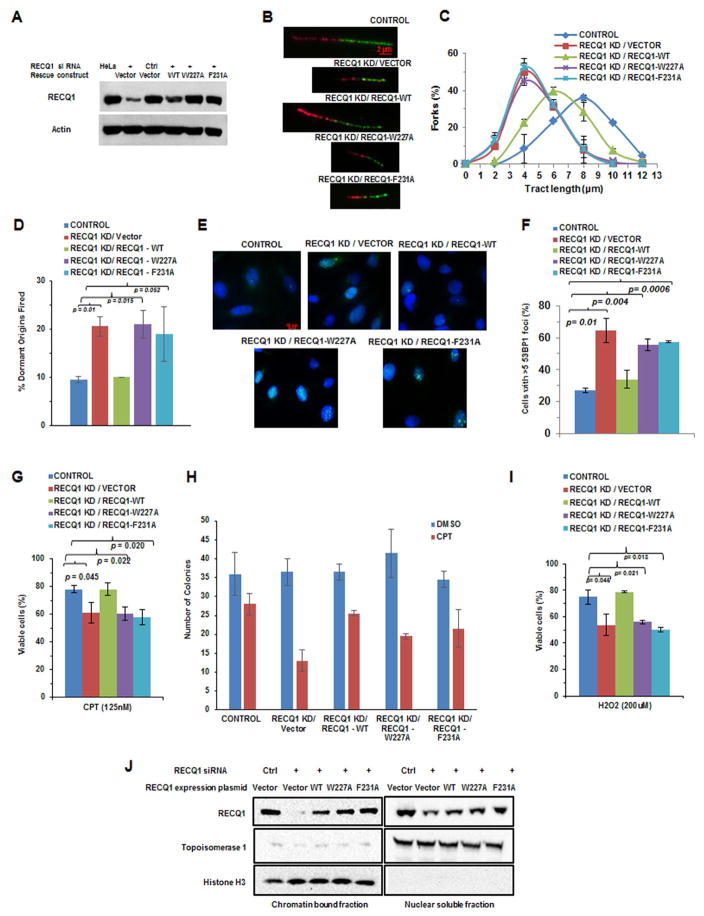
Approximately 75% of endogenous RECQ1 was depleted by RECQ1-siRNA, and plasmid-based expression for either RECQ1-W227A or RECQ1-F231A in RECQ1-depleted cells was comparable to endogenous RECQ1, whereas exogenous expression of RECQ1-WT was slightly less than endogenous RECQ1 (Figure 2A). HeLa cells co-transfected with a control siRNA and blank vector displayed a greater percentage of replication tracts in the range of 8–10 μm (Figure 2B, 2C). RECQ1-depleted cells co-transfected with empty vector displayed much reduced tract length with the peak shifted to 2–6 μm, indicating slower replication. RECQ1-depleted cells exogenously expressing RECQ1-WT showed a greater number of forks with replication tract lengths in the 4–10 μm range (Figure 2B, 2C). In contrast, RECQ1-depleted cells expressing RECQ1-W227A or RECQ1-F231A displayed replication tract lengths similar to RECQ1-depleted cells co-transfected with empty vector, indicating the RECQ1 AL mutants were defective in restoring normal replication.
Figure 2. Perturbed replication and genomic instability of human cells expressing RECQ1-W227A or RECQ1-F231A aromatic loop mutant proteins.
(A) Representative Western blot from lysates of HeLa cells transfected with control siRNA or siRNA against 3′-UTR of RECQ1 and an expression construct encoding the indicated RECQ1 protein. Actin serves as a loading control. (B) RECQ1-depleted cells (RECQ1 KD) exogenously expressing RECQ1-WT, RECQ1-W227A, or RECQ1-F231A protein were labeled with CldU or IdU indicated by red and green tracts, respectively. Representative DNA fiber tracts from microfluidic-assisted tract analysis are shown. (C) Statistical analysis of percentage replication forks with the indicated tract lengths (μm). At least 100 tracts were analyzed for each cell line. (D) Percentage of dormant origins fired for indicated cell lines. (E) Representative 53BP1 fluorescence staining images of indicated cell lines. (F) Quantitative analysis of immunofluorescence data as shown in (E). Percentage of cells with >5 53BP1 foci is shown. (G) Western blot showing the relative binding of RECQ1 proteins to chromatin isolated from CPT-treated cells. (H) The indicated cell line was exposed to CPT (125 nM, 16 hr) and percent viable cells determined by WST-1 assay. (I) Indicated cell lines were exposed to 100 nM CPT for 1 hr and monitored after 13 days for colony formation. (J) Cell lines were exposed to H2O2 (200 μM, 30 min) and percent viable cells was determined by WST-1 assay.
Slower replication causes dormant replication origin firing to compensate for the delay [26,27], leading us to ask if RECQ1 deficiency resulted in elevated dormant origin firing. RECQ1-depleted cells co-transfected with empty vector displayed double the number of dormant origins fired compared to control cells or RECQ1-depleted cells exogenously expressing RECQ1-WT (Figure 2D). RECQ1-depleted cells expressing RECQ1-W227A or RECQ1-F231A showed a two-fold increase in dormant origins fired. Thus, expression of either RECQ1 AL mutant failed to rescue the reduced replication fork rate and elevated dormant origin firing characteristic of RECQ1-deficient cells.
Based on the abnormal replication dynamics of RECQ1-deficient cells ([10,11,13], current study), we asked if RECQ1 deficiency resulted in increased double-strand breaks (DSB), and if expression of the RECQ1 AL mutants could restore this defect. RECQ1-depleted cells co-transfected with empty vector or plasmids encoding RECQ1-W227A or RECQ1-F231A displayed an increased percentage of cells with >5 53BP1 foci compared to control cells or RECQ1-depleted cells exogenously expressing RECQ1-WT (Figure 2E, 2F), indicative of DSB accumulation.
Compromised DNA damage resistance of RECQ1-depleted cells expressing RECQ1 aromatic loop mutants
RECQ1’s role in replication restart after exposure to the topoisomerase inhibitor camptothecin (CPT) [11] led us to ask if the RECQ1 AL mutants could restore CPT resistance. Chromatin-associated wild-type or AL mutant RECQ1 in the CPT-treated RECQ1 transfected cell lines was comparable (Figure 2G); however, neither RECQ1 mutant restored cellular resistance to CPT (Figure 2H, 2I). Because CPT-induced blocked replication forks are known to incur single-stranded and double-stranded DNA breaks [28] and RECQ1-deficient cells are sensitive to hydrogen peroxide (H2O2) that induces strand breaks and oxidative damage [15], we tested for H2O2 sensitivity and found that neither RECQ1 AL mutant could restore viability to the level of control cells or RECQ1-depleted cells exogenously expressing RECQ1-WT (Figure 2J).
RECQ1 suppresses dormant origin firing and maintains free RPA pool
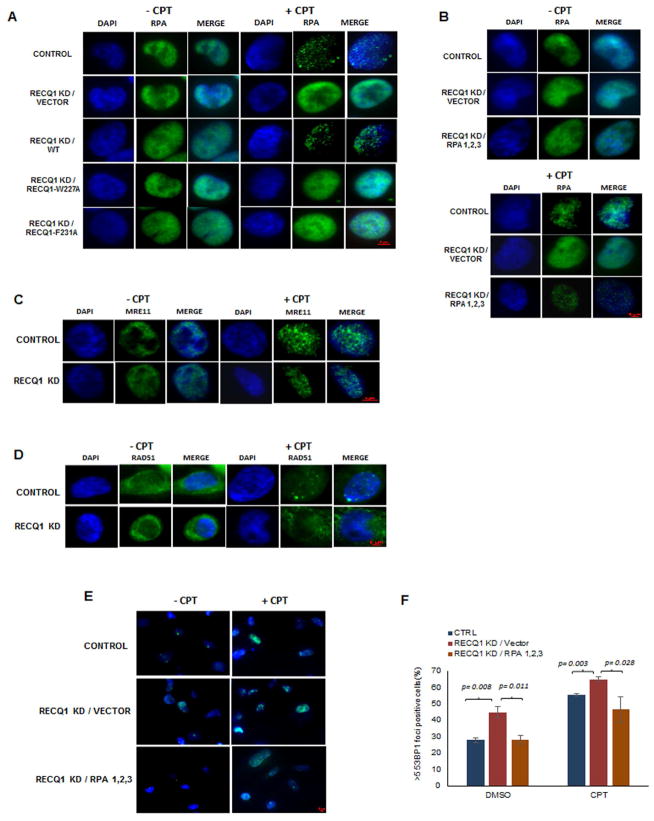
Dormant origin firing was shown to cause RPA recruitment to sites of nascent DNA synthesis, thereby exhausting the global RPA pool, resulting in single-stranded DNA exposure vulnerable to strand breaks [29]. This led us to investigate if RECQ1 played a role in determining RPA availability during the DNA damage response. RPA foci were induced upon CPT challenge in RECQ1-proficient HeLa cells (Figure 3A, S2B). However, cells depleted of endogenous RECQ1 and transfected with either blank vector or RECQ1 AL mutant poorly formed RPA foci upon CPT exposure (Figure 3A, S2B). Exogenous RPA expression (Figure S2A) restored formation of CPT-induced RPA foci in RECQ1-depleted cells (Figure 3B, S2B). MRE11 foci were able to form in CPT-treated RECQ1-depleted cells, suggesting that there was not a defect upstream of RPA (Figure 3C). In contrast, the downstream player RAD51 poorly formed foci in CPT-treated RECQ1-deficient cells (Figure 3D, S2C). Spontaneous 53BP1 foci were reduced in cells overexpressing RPA in the RECQ1-depleted condition (Figure 3E, 3F), suggesting that DNA damage accumulation in RECQ1-deficient cells is due to RPA sequestration by nascent DNA synthesis sites at activated dormant origins.
Figure 3. DNA damage in HeLa cells expressing catalytically defective RECQ1 is a function of RPA exhaustion.
(A) Representative immunofluorescence images of RPA staining in indicated HeLa cell lines treated with either DMSO or 200 nM CPT for 30 min. Representative immunofluorescence images of RPA (B), MRE11 (C), RAD51 (D), and 53BP1 (E) staining in indicated HeLa cell lines treated with either DMSO or 200 nM CPT for 30 min. (F) Quantitative analysis of 53B1 staining in cells represented in E.
Dominant negative phenotypes exerted by RECQ1 aromatic loop mutants
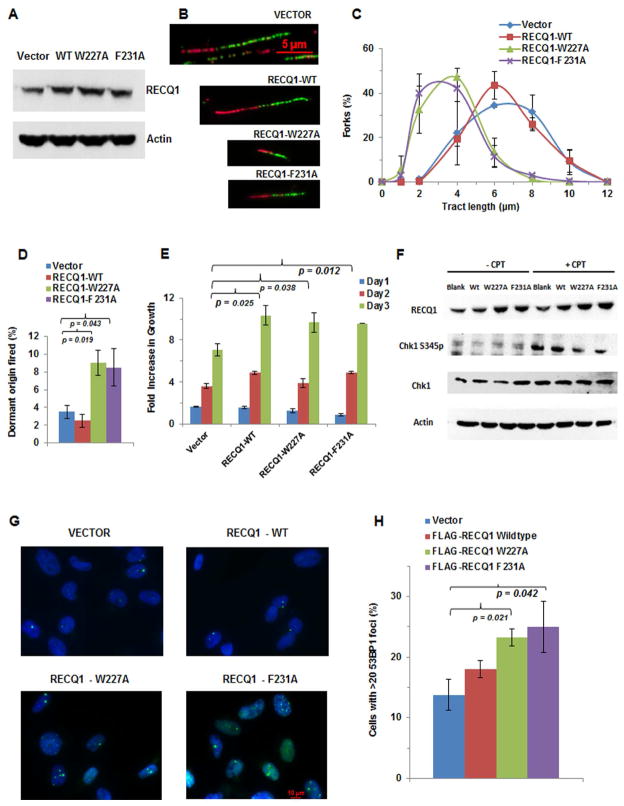
Given the importance of tumor suppressor heterozygosity in cancer [30–32], we assessed if the RECQ1 AL mutants exerted dominant negative effects. HeLa cells transfected with wild-type or mutant RECQ1 expression constructs showed a 2-fold increase of RECQ1 protein compared to cells transfected with empty vector (Figure 4A). DNA fiber analysis demonstrated that expression of RECQ1-W227A or RECQ1-F231A in the wild-type background resulted in shorter replication tract lengths (Figure 4B, 4C). Consistent with slower replication, dormant origin firing in cells expressing either RECQ1 mutant was increased 2-fold (Figure 4D).
Figure 4. Dominant negative effects exerted by expression of RECQ1 aromatic loop mutant proteins.
(A) Representative Western blot from lysates of HeLa cells transfected with an expression construct encoding the indicated RECQ1 protein. Actin serves as a loading control. (B) HeLa cells exogenously expressing RECQ1-WT, RECQ1-W227A, or RECQ1-F231A protein were labeled with CldU or IdU indicated by red and green tracts, respectively. Representative DNA fiber tracts from microfluidic-assisted tract analysis are shown for each of the cell lines. (C) Statistical analysis of percentage replication forks with the indicated tract lengths (μm). At least 100 tracts were analyzed for each cell line. (D) Percentage of dormant origins fired for the indicated cell lines. (E) Graph comparing the growth pattern of HeLa cells over-expressing exogenous wild-type or either of the two RECQ1 AL mutant proteins over a period of 3 days. (F) Western blot showing Chk1 S345 phosphorylation in untreated and CPT-treated (2 μM, 1 hr) HeLa cells transfected with blank vector, wild-type RECQ1 or either AL mutant RECQ1 expressing construct. CPT-treated RECQ1-W227A and RECQ1-F231A transfected cells displayed 47% and 79% less phosphorylated Chk1S345, respectively, compared to CPT-treated cells that were transfected with blank vector, as normalized against total Chk1. (G) Representative fluorescence images from 53BP1 staining of indicated cell lines. (H) Quantitative analysis of immunofluorescence data as shown in (G). Percentage of cells with >20 53BP1 foci is shown.
The abnormal replication dynamics of HeLa cells expressing RECQ1-W227A or RECQ1-F231A led us to examine their growth. Cells expressing RECQ1-W227A or RECQ1-F231A displayed greater proliferation compared to cells transfected with empty vector and at a level comparable to cells exogenously expressing RECQ1-WT (Figure 4E). It was reported that RECQ1 depletion resulted in slow growth of cancer cells accompanied by reduced DNA synthesis [14], consistent with slower fork progression; therefore, it was surprising to find that RECQ1 AL mutant expression increased cell proliferation given their negative effect on replication tract length (Figure 4B, 4C), suggesting a possible defect in checkpoint activation. CPT-treated HeLa cells expressing either RECQ1 AL mutant displayed decreased phosphorylated Chk1S345 (Figure 4F), and Chk1S317p or Chk2T68p (Figure S3A, S3B). Consistent with a checkpoint defect, CPT-treated cells expressing the RECQ1 AL mutant did not show an increased S-phase population (Figure S3C).
HeLa cells expressing either RECQ1 AL mutant showed significantly greater spontaneous DNA damage compared to cells transfected with empty vector or RECQ1-WT plasmid (Figure 4G, 4H). Moreover, RECQ1-W227A or RECQ1-F231A expressed in a wild-type RECQ1 background rendered cells more sensitive to either CPT (Figure S3D, S3E) or H2O2 (Figure S3F), also observed for DSB induction (Figure S3G–J). CPT-treated HeLa cells expressing RECQ1 AL mutants failed to form discreet RPA foci; however, exogenous RPA expression alleviated this effect (Figure S3K). Formation of RAD51 foci (Figure S3K), but not MRE11 foci (data not shown), was impaired by expression of RECQ1 AL mutants, consistent with a direct effect on free RPA pool.
Biochemical mixing experiments with mutant and wild-type RECQ1 proteins
In vitro mixing experiments with RECQ1-WT and either RECQ1-W227A or RECQ1-F231A were performed to assay effects on HJ resolution or forked duplex DNA unwinding. The presence of either RECQ1 AL mutant in reaction mixtures with RECQ1-WT inhibited unwinding of forked duplex DNA; however, inhibition on the HJ substrate was only observed in conversion of splayed arm product to single-stranded DNA, not HJ BM (Figure S3L–SA–F). The RECQ1 AL mutants retained HJ substrate binding (Figure S3T), consistent with their residual HJ BM activity (Figure 1G, S1E, S3L–O). Taking together these and published results [19], we conclude that either RECQ1 AL mutant protein poisons forked duplex unwinding by RECQ1 dimer (and possibly monomer), but not HJ BM catalysed by RECQ1 tetramer.
DISCUSSION
Biochemical characterization of RECQ1-W227A or RECQ1-F231A revealed a critical role of the conserved AL to couple ATP hydrolysis to DNA BM or helicase activity. The severe reduction of RECQ1 BM despite a modest reduction in ATPase activity suggests that the surface exposed AL of RECQ1 may very well interact with single-stranded DNA during concomitant ATPase-dependent coupled strand separation and annealing, consistent with predictions from analysis of truncated RECQ1-DNA crystal structures [19,20] (Figure S1J).
The reduced replication kinetics of RECQ1 AL mutant expressing cells is accompanied by increased dormant origin firing, presumably to help them complete DNA synthesis in a timely manner during S phase. Reduced replication rate is also likely to contribute to elevated DSBs. Nascent DNA synthesis at dormant origins activated as a result of slow replication fork progression in cells expressing either RECQ1 AL mutant sequester free RPA, thereby exposing single-stranded DNA and making it more vulnerable to damage. RECQ1-depleted cells expressing RECQ1 AL mutant protein show increased sensitivity to DNA damage induced by H2O2 or collision of replication forks with CPT drug-topoisomerase complexes [33,34], suggesting for the first time that catalytic strand separation activities of RECQ1 are required for a robust DNA damage response. Based on the elevated sister chromatid exchange and IR sensitivity of RECQ1-deficient mouse embryonic fibroblasts [13] or human cells [14], we propose that RECQ1 supports or facilitates HR repair of frank or replication-associated DSBs, consistent with its ability to BM HJs [8] and D-loop intermediates [25]. Furthermore, RECQ1 governs the RPA-mediated response to replication stress to maintain chromosomal stability.
Expression of either RECQ1 AL mutant in a genetic background of wild-type RECQ1 resulted in reduced replication rate accompanied by increased dormant origin firing, elevated DSBs, and compromised H2O2 or CPT resistance, similar to that observed for RECQ1-depleted cells expressing either AL mutant. Biochemical mixing experiments demonstrated that either RECQ1 AL mutant poisoned DNA unwinding by RECQ1-WT, presumably by hetero-oligomerization or static mutant RECQ1-DNA complexes. Further studies are required to ascertain mechanism of helicase inhibition and dominant negative effects in vivo.
Our finding that cancer cells expressing helicase-defective RECQ1 accumulate DNA damage and bypass replication checkpoint suggests that RECQ1 variants may contribute to tumorigenesis. RECQ1 overexpression is observed in glioblastomas and ovarian cancers and suggested to play a role in tumor cell proliferation [35–37]. Several RECQ1 polymorphisms are associated with lower pancreatic cancer survival [38], and RECQ1 is a proposed therapeutic target and proliferative marker for head and neck squamous cell carcinoma [39] and hepatocellular carcinoma [39,40]. The growth advantage of cancer cells expressing catalytically defective mutant RECQ1 is most likely through a mechanism involving checkpoint bypass distinct from that involving overexpression of wild-type RECQ1 which probably helps cancer cells cope with replicative lesions.
Clinically important missense mutations have been identified in helicase genes [41]. RECQ1 missense variants reside in the core helicase domain (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=5965) (Figure S3U), and adjacent to the AL (http://exac.broadinstitute.org/gene/ENSG00000004700). RECQ1 SNPs may contribute to tumorigenesis [42] or act as genetic modifiers to influence pathogenesis, as proposed for other DNA damage response proteins [43].
Given that CPT-induced replication blocks closely mimic the spectra of normal spontaneously collapsed replication forks [44], our studies are informative for understanding how RECQ1 helps cancer cells deal with endogenous replication stress. RECQ1 directs a productive RPA-dependent response to endogenous replication stress by insuring a normal replication rate and suppressing dormant origin firing. By maintaining the global RPA pool, RECQ1 provides cancer cells an opportunity to protect exposed single-stranded DNA in periods of heightened replication stress to enable fork progression. Further work may help to elucidate mechanisms of RECQ1 that could be targeted for inhibition in anti-cancer therapies.
EXPERIMENTAL PROCEDURES
Plasmids and cell lines
RECQ1 site-directed mutagenesis was performed using pFastBac-RECQ1 as template by Loftstrand Lab Ltd (MD, USA). RECQ1 cDNAs were cloned into pFLAG-CMV-2 expression vector from Sigma (MO, USA). HeLa cells were from ATCC. pGFP-RPA expression plasmids were kindly provided by Dr. M. Wold (Univ. Iowa).
Recombinant proteins
Full-length His-tagged RECQ1 recombinant proteins were purified as described in Supplementary Experimental Procedures.
Size Exclusion Chromatography
Size exclusion chromatography was performed with purified RECQ1 proteins using a 24 ml Superdex 200 size exclusion column (GE Healthcare) by AKTA FPLC (GE Healthcare) (Supplementary Experimental Procedures).
DNA substrates
The 19 base-pair (bp) forked duplex and HJ(X12) DNA substrates were prepared as described [8]. Mobile D-loops and open three-stranded DNA substrates were prepared by sequential annealing [25] (Supplementary Experimental Procedures).
DNA helicase and branch migration assays
DNA helicase [8] and BM assays for HJ [8], 3-stranded open [25], and D-loop [25] DNA substrates were as described.
DNA binding and strand annealing assays
DNA binding and strand annealing assays were as previously described [8].
ATP hydrolysis assays
ATP hydrolysis was measured using [γ-32P] ATP (PerkinElmer Life Sciences) and analyzed by thin layer chromatography on polyethyleneimine-cellulose plates (Mallinckrodt Baker) (Supplementary Experimental Procedures).
DNA spreading
Replication track analyses were performed as described in Supplementary Experimental Procedures.
Immunostaining
Immunostaining of transfected cells treated was performed as described in Supplementary Experimental Procedures.
Survival assays
Cell viability assays were performed using WST-1 reagent (Roche) according to manufacturer’s protocol.
Chromatin fractionation
Chromatin fractionation was performed using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Scientific) according to manufacturer’s protocol. Western blots were done using antibodies against RECQ1 (Santa Cruz Biotechnology), Topoisomerase I (BD Biosciences), and Histone H3 (Abcam).
Supplementary Material
Highlights.
RECQ1 aromatic loop mutants disable DNA branch-migration and helicase activities
DNA repair and replication is governed by RECQ1-dependent RPA dynamics
RECQ1 missense mutations exert dominant negative effects on genomic stability
Mutant RECQ1-expressing cancer cells accrue DNA damage and bypass S phase checkpoint
Acknowledgments
We wish to thank Dr. M. Wold (University of Iowa) for RPA expression constructs, and Drs. M. Scheibye-Knudsen and K. Aly (NIA-NIH) for critical comments on the manuscript.
FUNDING
This work was supported by National Institutes of Health, NIA, Intramural Research Program.
Footnotes
AUTHOR CONTRIBUTIONS
TB, RMB and MMS devised experiments. TB and JAS performed experiments. TB, JAS and RMB analysed data. JH helped with DNA fibre assay. TB and RMB wrote the manuscript. All authors reviewed and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brosh RM., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13:542–548. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seki M, Yanagisawa J, Kohda T, Sonoyama T, Ui M, Enomoto T. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J Biochem. 1994;115:523–531. doi: 10.1093/oxfordjournals.jbchem.a124369. [DOI] [PubMed] [Google Scholar]
- 3.Puranam KL, Kennington E, Sait SN, Shows TB, Rochelle JM, Seldin MF, Blackshear PJ. Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics. 1995;26:595–598. doi: 10.1016/0888-7543(95)80181-k. [DOI] [PubMed] [Google Scholar]
- 4.Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 5.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 6.Cybulski C, Carrot-Zhang J, Kluzniak W, Rivera B, Kashyap A, Wokolorczyk D, Giroux S, Nadaf J, Hamel N, Zhang S, et al. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet. 2015;47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, et al. Mutations in RECQL gene Are associated with predisposition to breast cancer. PLoS Genet. 2015;11:e1005228. doi: 10.1371/journal.pgen.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 9.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human Replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. The human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst) 2012;11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Parvathaneni S, Hara T, Lal A, Sharma S. Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol Cancer. 2013;12:29. doi: 10.1186/1476-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S. An appraisal of RECQ1 expression in cancer progression. Front Genet. 2014;5:426. doi: 10.3389/fgene.2014.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pike AC, Gomathinayagam S, Swuec P, Berti M, Zhang Y, Schnecke C, Marino F, von DF, Renault L, Costa A, et al. Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures. Proc Natl Acad Sci U S A. 2015;112:4286–4291. doi: 10.1073/pnas.1417594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci U S A. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucic B, Zhang Y, King O, Mendoza-Maldonado R, Berti M, Niesen FH, Burgess-Brown NA, Pike AC, Cooper CD, Gileadi O, et al. A prominent {beta}-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 2010;39:1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33:6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manthei KA, Hill MC, Burke JE, Butcher SE, Keck JL. Structural mechanisms of DNA binding and unwinding in bacterial RecQ helicases. Proc Natl Acad Sci U S A. 2015;112:4292–4297. doi: 10.1073/pnas.1416746112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugreev DV, Brosh RM, Jr, Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem. 2008;283:20231–20242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst) 2010;9:1249–1255. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yekezare M, Gomez-Gonzalez B, Diffley JF. Controlling DNA replication origins in response to DNA damage - inhibit globally, activate locally. J Cell Sci. 2013;126:1297–1306. doi: 10.1242/jcs.096701. [DOI] [PubMed] [Google Scholar]
- 28.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 30.Kechagioglou P, Papi RM, Provatopoulou X, Kalogera E, Papadimitriou E, Grigoropoulos P, Nonni A, Zografos G, Kyriakidis DA, Gounaris A. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer Res. 2014;34:1387–1400. [PubMed] [Google Scholar]
- 31.Fam HK. Delineating the effects BRCA1 and BRCA2 loss of heterozygosity in pancreatic cancer progression. Clin Genet. 2014;85:18–20. doi: 10.1111/cge.12306. [DOI] [PubMed] [Google Scholar]
- 32.Mizoguchi M, Kuga D, Guan Y, Hata N, Nakamizo A, Yoshimoto K, Sasaki T. Loss of heterozygosity analysis in malignant gliomas. Brain Tumor Pathol. 2011;28:191–196. doi: 10.1007/s10014-011-0038-0. [DOI] [PubMed] [Google Scholar]
- 33.Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 34.Driessens N, Versteyhe S, Ghaddhab C, Burniat A, De DX, Van SJ, Dumont JE, Miot F, Corvilain B. Hydrogen peroxide induces DNA single- and double-strand breaks in thyroid cells and is therefore a potential mutagen for this organ. Endocr Relat Cancer. 2009;16:845–856. doi: 10.1677/ERC-09-0020. [DOI] [PubMed] [Google Scholar]
- 35.Mendoza-Maldonado R, Faoro V, Bajpai S, Berti M, Odreman F, Vindigni M, Ius T, Ghasemian A, Bonin S, Skrap M, et al. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol Cancer. 2011;10:83. doi: 10.1186/1476-4598-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanada S, Futami K, Terada A, Yonemoto K, Ogasawara S, Akiba J, Yasumoto M, Sumi A, Ushijima K, Kamura T, et al. RECQL1 DNA repair helicase: a potential therapeutic target and a proliferative marker against ovarian cancer. PLoS ONE. 2013;8:e72820. doi: 10.1371/journal.pone.0072820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao J, Tao S, Han J, Zhou Z, Zhang X, Wang H, Chen R, Ji F, Zhu Y. RECQL1 plays an important role in the development of tongue squamous cell carcinoma. Cell Physiol Biochem. 2014;33:1579–1590. doi: 10.1159/000358721. [DOI] [PubMed] [Google Scholar]
- 38.Li D, Liu H, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 2006;66:3323–3330. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arai A, Chano T, Futami K, Furuichi Y, Ikebuchi K, Inui T, Tameno H, Ochi Y, Shimada T, Hisa Y, et al. RECQL1 and WRN proteins are potential therapeutic targets in head and neck squamous cell carcinoma. Cancer Res. 2011;71:4598–4607. doi: 10.1158/0008-5472.CAN-11-0320. [DOI] [PubMed] [Google Scholar]
- 40.Futami K, Ogasawara S, Goto H, Yano H, Furuichi Y. RecQL1 DNA repair helicase: A potential tumor marker and therapeutic target against hepatocellular carcinoma. Int J Mol Med. 2010;25:537–545. doi: 10.3892/ijmm_00000375. [DOI] [PubMed] [Google Scholar]
- 41.Suhasini AN, Brosh RM., Jr Disease-causing missense mutations in human DNA helicase disorders. Mutat Res. 2012;752:138–152. doi: 10.1016/j.mrrev.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li XL, Lu X, Parvathaneni S, Bilke S, Zhang H, Thangavel S, Vindigni A, Hara T, Zhu Y, Meltzer PS, et al. Identification of RECQ1-regulated transcriptome uncovers a role of RECQ1 in regulation of cancer cell migration and invasion. Cell Cycle. 2014;13:2431–2445. doi: 10.4161/cc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS Genet. 2012;8:e1002644. doi: 10.1371/journal.pgen.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.