Abstract
Quantification of miRNAs in blood can be potentially used for early disease detection, surveillance monitoring and drug response evaluation. However, quantitative and robust measurement of miRNAs in blood is still a major challenge in large part due to their low concentration and complicated sample preparation processes typically required in conventional assays. Here, we present the ‘Integrated Comprehensive Droplet Digital Detection’ (IC 3D) system where the plasma sample containing target miRNAs is encapsulated into microdroplets, enzymatically amplified and digitally counted using a novel, high-throughput 3D particle counter. Using Let-7a as a target, we demonstrate that IC 3D can specifically quantify target miRNA directly from blood plasma at extremely low concentrations ranging from 10s to 10,000 copies/mL in ≤ 3 hours without the need for sample processing such as RNA extraction. Using this new tool, we demonstrate that target miRNA content in colon cancer patient blood is significantly higher than that in healthy donor samples. Our IC 3D system has the potential to introduce a new paradigm for rapid, sensitive and specific detection of low-abundance biomarkers in biological samples with minimal sample processing.
Introduction
MicroRNAs (miRNAs) are a class of short (approximately 18–23 nucleotides (nt)), single-stranded, non-coding endogenous RNAs which play important roles in regulating gene expression via target mRNA degradation or translational repression.1 When cells undergo apoptosis or necrosis in the body, significant amounts of miRNA can be shed into the blood stream.2 The released miRNAs can circulate in blood in a remarkably stable form, mainly due to the protection from RNase degradation by Ago2 and exosomes.3 It has been demonstrated that circulating miRNA expression levels are correlated to the disease states of various diseases including cancer, neurodegenerative disorders and cardiovascular diseases.2, 4, 5 Therefore, miRNAs can potentially be used as convenient blood test markers for early disease detection, surveillance monitoring and drug response evaluation.2, 4, 5
However, accurate quantification of circulating miRNA biomarkers in blood that often exist at low concentrations for early-stage, metastatic or recurrent diseases is still a major challenge. Due to the low abundance of target miRNAs, large input clinical sample volumes (100s µLs to mLs) are typically required, which few conventional detection systems can directly handle without sample preparation and volume reduction. Moreover, analytical sensitivity and specificity are further hindered by the high background signal because target miRNAs are surrounded by other circulating nucleic acids that are typically >1,000 times more abundant in blood. Indeed, conventional miRNA detection methods, such as reverse transcription real-time polymerase chain reaction (RT-qPCR) and microarray, suffer from non-specific amplification and lack of valid internal controls.6 More recently, a number of new methods including droplet digital PCR (ddPCR), nanopore-based detection, sequencing and electrochemical sensing methods have also been developed for miRNA measurement.7–13 In particular, ddPCR is an absolute quantification method where extracted nucleic acid samples in a small volume are partitioned to picoliter or nanoliter droplets, resulting in one or no molecule in each droplet reaction.7 Compared to RT-qPCR, ddPCR has greater precision and improved day-to-day reproducibility because it uses digital counting (1 or 0) without the need of normalization.7 However, despite of recent advances14, 15, fluorescent droplet quantification using flow cytometry, 1D (or 2D) on-chip counting in the current commercialized ddPCR systems (e.g., Bio-Rad, Life technologies (QuantStudio) and Raindance) is limited by its low throughput in droplet count (~1000 droplets/s) and is only able to analyze small sample volumes (10s microliters (µLs) or ~104–106 droplets).16 Importantly, with few exceptions12, 17, the current miRNA detection methods including ddPCR typically require complex sample processing steps including miRNA isolation, purification, and reverse transcription, etc.7, 18 These pre-analysis manipulations significantly affect assay robustness and data reproducibility due to loss of targets (especially when dealing with low-abundance markers), and variations in reagents and kits and technical expertise.7 Therefore, there is a great need for a new technology that is more sensitive, robust and can directly detect miRNAs in a large volume of patient blood samples without the need for complex sample preparation.19
Recently, we have developed a platform technology termed ‘Integrated Comprehensive Droplet Digital Detection (IC 3D)’ that integrates real-time fluorescent sensors, droplet microencapsulation and a high-throughput 3D particle counter for rapid digital analysis.20 In our previous work, we demonstrated that the IC 3D can selectively detect bacteria directly from diluted blood at single-cell sensitivity using microencapsulated DNAzyme sensors.20 Here, we demonstrate that the IC 3D can be used to accurately quantify target miRNAs directly from blood plasma within 3 hours. Specifically, the IC 3D system combines microencapsulation for enabling digital quantification; isothermal exponential amplification (EXPAR) for specific miRNA detection; and a 3D particle counter for rapid, high-throughput droplet counting (Scheme 1). Briefly, the plasma sample and sensing elements are injected and mixed in a microfluidic device and then immediately encapsulated into hundreds of millions of picoliter-sized droplets. The EXPAR reaction, which eliminates the need for thermocycler and reverse transcription, would produce a real-time rapid fluorescence signal only in the droplets that contain target miRNA. Our rationale is that the confinement of samples in picoliter droplets can significantly increase the effective miRNA concentration and minimize the number of background molecules (interference) from plasma.21 Therefore, microencapsulation permits high sensitivity and digital detection (i.e., each droplet has 0 or 1 target miRNA). This is supported by extensive previous droplet-based detection methods including ddPCR and digital RT-LAMP.22–25 In our IC 3D, the generated droplets (‘microreactors’) are allowed to incubate at an isothermal temperature for a certain period of time to generate fluorescence signal. As we mentioned earlier, we want to analyze unprocessed patient plasma with a clinical sample volume of typically 100s µLs to mLs, which translates to hundreds of millions to billions of droplets. To effectively detect single fluorescent, target miRNA-containing droplets among hundreds of millions of blank ones, in the IC 3D system, we integrate a novel 3D particle counter system that can detect fluorescent particles from mL volumes at single-particle sensitivity within only several minutes.26, 27
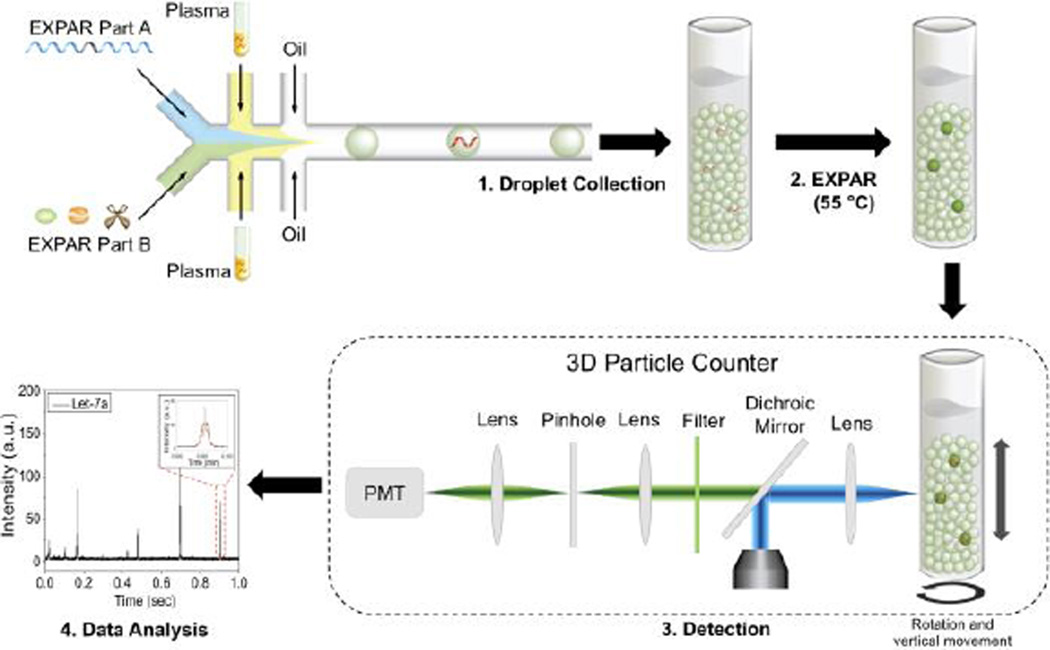
Scheme 1.
Schematic diagram of IC 3D technology for miRNA detection. Plasma samples and sensing elements (EXPAR part A and EXPAR part B, see details in the experimental section) are mixed and encapsulated in hundreds of millions of picoliter-sized droplets using a flow-focusing microfluidic device (step 1). Droplets are then collected in a tube and incubated to conduct isothermal exponential amplification (step 2). After incubation, droplets are detected (step 3) and analysed (step 4) using a high-throughput 3D particle counter that can robustly and accurately detect single fluorescent droplets from milliliter (mL) volumes of sample within several minutes.
Materials and Methods
Apparatus
ViiA™ 7 Real-Time PCR system (Life technologies, Carlsbad, CA, USA) was used to control reaction temperature and measure real-time fluorescent signal from exponential amplification reaction (EXPAR) for target miRNA detection. RT-qPCR was performed with the same instrument. Inverted fluorescence microscope (Eclipse Ti, Nikon, Tokyo, Japan) was used to image the fluorescent droplets when studying EXPAR reaction kinetics in droplet. 3D particle counting prototype system (described in details below) 26, 27 was used to count fluorescent droplets in a high throughput.
Reagents and Materials
All synthetic, HPLC-purified DNA and miRNA (Let-7a, Let-7b, Let-7c, miR-39) (sequences in Table S1) were purchased from IDT (Integrated DNA Technologies, Coralville, IA, USA). Vent (exo-) polymerase, nicking endonuclease Nt.BstNBI, NEbuffer 3.1 and ThermoPol buffer were purchased from NEB (New England Biolabs, Ipswich, MA, USA). RNase inhibitor, Taqman® miRNA reverse transcription kit, Taqman® probes for miRNA including Let-7a and miR-39 (an external control used in RT-qPCR), ROX reference dye and Countess® cell counting chamber slides (for microscopic imaging of the droplets) were purchased from Life Technologies (Carlsbad, CA, USA). EvaGreen® DNA staining dye was purchased from Biotium (Hayward, CA, USA). PCR supermix was purchased from Bio-Rad (Hercules, CA, USA). miRNeasy Serum/Plasma Kit was purchased from QIAGEN (Netherlands). Diethylpyrocarbonate (DEPC) was purchased from Sigma-Aldrich (St. Louis, MO, USA). All solutions for EXPAR were prepared in DEPC-treated deionized water. ExoQuick exosome isolation kit was purchased from System Biosciences (SBI, Mountain View, CA, USA).
Cancer patient and healthy donor plasma samples
Healthy human blood samples were obtained from University of California, Irvine Institute for Clinical and Translational Science (ICTS) through the blood donor program (IRB # 2012-9023). Human blood samples from patients with metastatic colon cancer were collected with written informed consent at University of California, Irvine Medical Center (IRB # 2013-9627). All blood samples were collected with Light Blue-Top Tube (3.2% Sodium Citrate, Becton Dickinson, Franklin Lakes, NJ, USA) and plasma was isolated by centrifugation at 2000 rcf (relative centrifugal force) for 20 min. Isolated plasma samples were then stored at −80°C. For the experiments where we use synthetic miRNA spiked healthy donor plasma to evaluate IC 3D performance (Figure 3, a–d), the endogenous miRNAs were first depleted prior to spiking with synthetic miRNA in healthy donor plasma (see below for details). When we analyzed miRNA to compare colon cancer plasma and healthy donor plasma (Figure 3f), we analyzed the raw plasma sample without miRNA depletion from either samples.
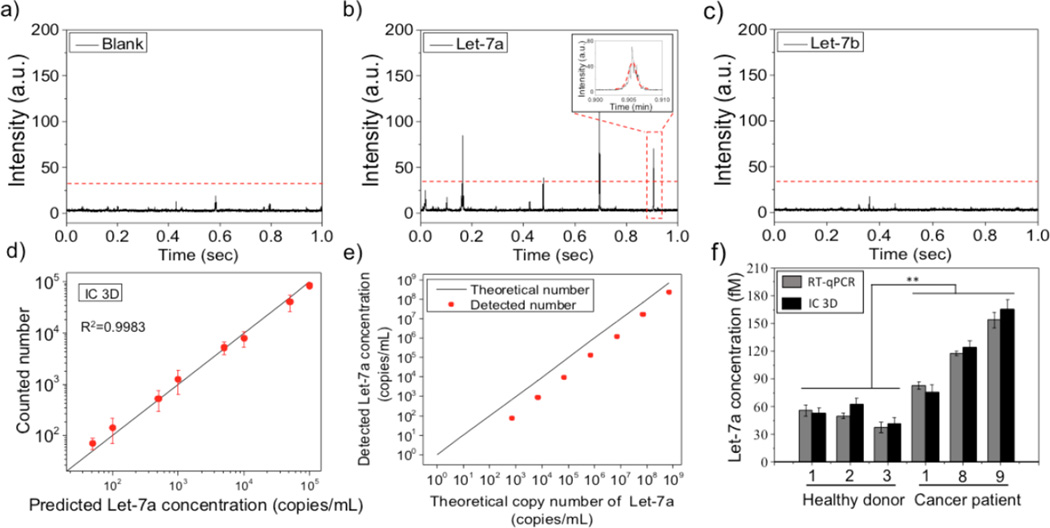
Figure 3.
Let-7a quantification in plasma using the IC 3D. a–c) Representative time trace with fluorescence intensity profiles of droplets obtained from blank (a), Let-7a (b) and Let-7b (c). Only the target Let-7a group generates fluorescence intensity spikes, which demonstrates the specificity of the IC 3D assay. In b) and c), the miRNA concentration is 10fM in plasma before encapsulation. Threshold was obtained by ‘Three sigma rule’ (three times s.d. of the baseline) d) Actually counted Let-7a number using IC 3D (y axis) vs. spiked Let-7a concentration (x axis). 103 copy/mL = 1.66aM. Error bar is based on triplicate experiments. Mean ±s.e.m. n=3. e) RT-qPCR for Let-7a detection in plasma (after miRNA purification and reverse transcription).Note: 10 and 100 copies/mL are undetectable. Error bars are based on triplicate experiments and are too small to see. Mean ±s.e.m. f) Let-7a concentration quantification in 3 healthy donor plasma samples and 3 colon cancer patient plasma samples detected by RT-qPCR and IC 3D. Error bar is based on triplicate experiments. Mean ± s.e.m. n=3. P value<0.05 (Student T test).
Preparation of endogenous miRNA depleted health donor plasma
In order to test EXPAR using synthetic miRNA in 100% plasma, we eliminated endogenous miRNAs from plasma samples to avoid interference. First, we removed exosomal miRNAs using Exoquick kit (SBI (System Biosciences), CA, USA) according to the manufacturer’s protocol. Briefly, the plasma was centrifuged at 3000 rcf for 15 min to remove cell debris. Then the supernatant was transferred to a sterile vessel. ExoQuick exosome precipitation solution (120 µL) was added to the plasma (500 µL) and mixed thoroughly, which was then refrigerated overnight. The ExoQuick/plasma mixture was centrifuged at 1500 rcf for 30 min. Following centrifugation, exosome-depleted plasma was separated for subsequent experiments. Non-exosomal miRNAs in plasma and residue exosomal miRNAs were then removed by running the exosome-depleted plasma through a miRNA isolation column. Specifically, exosome-depleted plasma was heated at 60°C for 10 min to release the miRNAs from residual exosomes and exosome-protein complexes.28 Plasma samples were then loaded on a miRNA isolation column (miRNeasy Mini prep, QIAGEN, Netherlands) and centrifuged at 2000 rcf for 1 min to remove miRNA. The collected solution was then filtered two more times using new miRNA isolation columns to remove the remaining miRNA. The resultant, miRNA depleted plasma was subsequently used to prepare synthetic miRNA spiked plasma samples (below).
miRNA detection in plasma by bulk EXPAR
EXPAR for miRNA detection in bulk was performed based on modifications of a previously reported protocol.29 Different amounts of Let-7a miRNA were spiked in miRNA-depleted plasma (final concentration: 1 fM, 10 fM, 100 fM, 1 pM, 10 pM, 100 pM or 1 nM). To suppress non-specific background amplification, the reaction mixture for EXPAR was prepared separately on ice as part A and part B. Part A consisted of 20% miRNA-spiked plasma, 1× NEBuffer 3.1, 0.2µM amplification template, 500µM dNTPs and 1.6U/µL RNase inhibitor. Part B consisted of 2× ThermoPol buffer, 0.8U/µL Nt.BstNBI nicking endonuclease, 0.1U/µL Vent (exo-) DNA polymerase, 2× EvaGreen®, 2× ROX reference dye. Part A and Part B were pre-heated at 55°C before mixing. EXPAR was performed at 55°C and fluorescence intensity was monitored immediately after mixing Part A and Part B solutions at intervals of 1min for 70 min using a ViiA™ 7 Real-Time PCR system. All experiments were performed in triplicate. In another set of experiments, endogenous Let-7a miRNA in healthy donor or colon cancer plasma samples were analyzed using bulk EXPAR as described above.
Fabrication and manipulation of microfluidic device
Microfluidic chip design and operating mechanism are shown in Figure 2a and Figure s3. Microchannel architectures were designed using AutoCAD (Autodesk, San Rafael, CA, USA) and transferred to high-resolution photomasks (CAD/Art Services, Bandon, OR, USA). The microfluidic device with one oil inlet and three aqueous inlets (one for the plasma sample and the others for the sensing elements, i.e., EXPAR part A and B) has a flow-focusing structure (width: 30 µm, depth: 40 µm) to form droplets through a flowing oil phase (See the microscope image in Figure 2a for structures and channel size). All microfluidic channels are 40 µm in depth but it has different width (15 µm for the injection inlet channel and 30 µm for the droplet generation structure). The microfluidic channels were fabricated with polydimethylsiloxane (PDMS) using standard soft lithographic techniques30. Briefly, silicon elastomer base and curing agent (Sylgard 184; Dow Corning, Wiesbaden, Germany) were mixed in a ratio of 10:1 w/w, degassed, decanted onto SU8-on-Si wafer master (IDB Technologies Ltd, Wiltshire, UK), and fully cured overnight on a heating plate at 65 °C. After thermal curing, the PDMS layer was peeled off, followed by the punching of inlet and outlet holes with a 1 mm sized punch (BIOPSY punch, Kay Industries co, Gifu, Japan). The PDMS layer and a glass slide were bonded immediately after plasma exposure. Another glass slide, containing inlets and outlet, was bounded on top of the PDMS layer to make three layers of glass-PDMS-glass, which was tightened with two binder clips to prevent detachment of the PDMS and glass layers due to high pressures within the microfluidic channel. To make the inlets and outlet on the glass slide (1mm thickness), 1mm halls were formed using a press drill with a 1 mm diamond-coated drill bit. HFE-7500 fluorocarbon oil (Dow Corning, Midland, MI, USA) containing 1.8% (w/w) perfluorinated polyetherspolyethyleneglycol (PFPE-PEG) surfactant was used as a continuous phase. Aqueous and oil phases were injected into the microfluidic device via pressure equalization (PE) tubes (Smith medical, Kent, UK). In all microfluidic experiments, PHD 2000 syringe pumps (Harvard Apparatus, Holliston, MA, USA) were used to inject liquids.
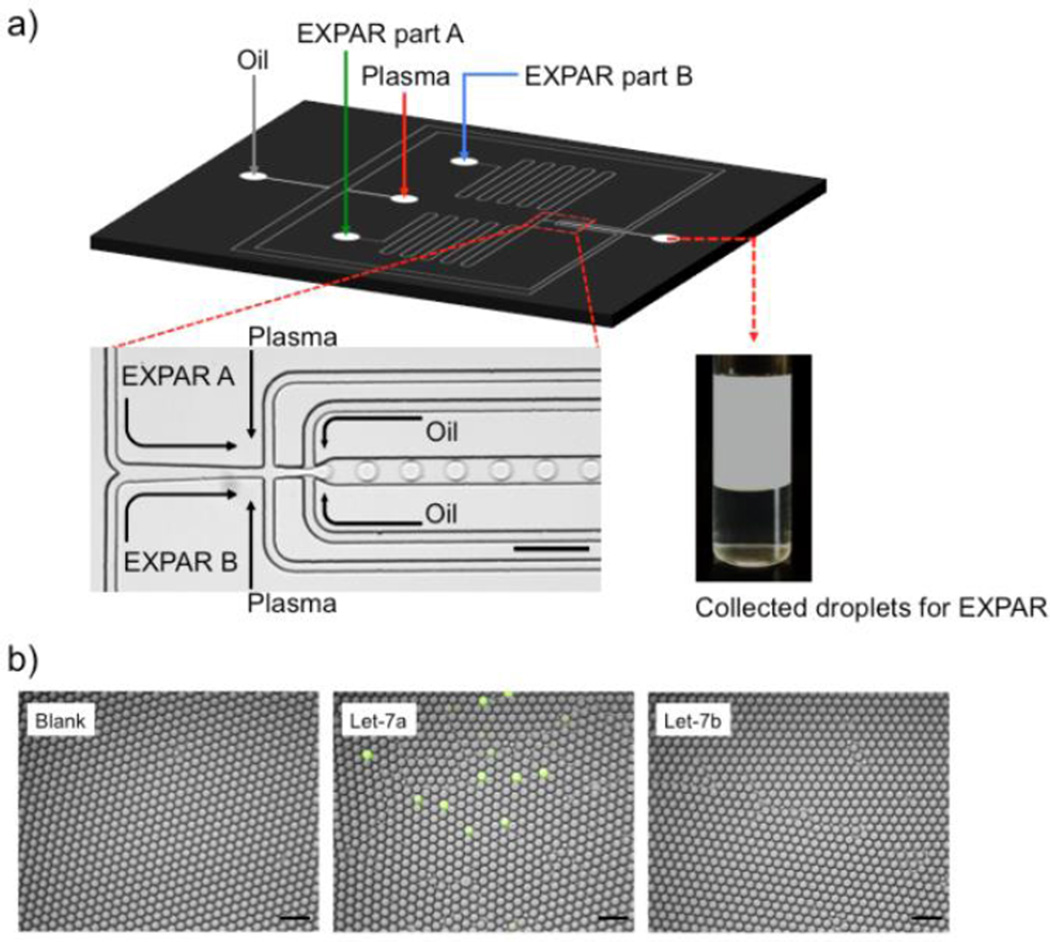
Figure 2.
miRNA detection by droplet EXPAR. a) CAD design of the droplet-based microfluidic device (Top) and microscopic image of plasma microencapsulation (Bottom). Scale bar: 100 µm. b) Representative fluorescence microscope images of miRNA-depleted plasma (100%) alone, spiked with Let-7a, or spiked with Let-7b that were encapsulated with EXPAR reagent. The collected droplets were incubated at 55°C for 50 min before imaging. Scale bar: 100 µm.
Droplet generation of synthetic miRNA Let-7a spiked plasma
Different copy numbers (10, 50, 100, 500, 10000, 50000 or 100000 copies) of synthetic miRNA were spiked in miRNA-depleted plasma samples (100%) which were encapsulated with EXPAR reagents to form 30 µm diameter droplets. As such, the plasma sample was diluted 10× on-chip with the EXPAR reagents. miRNA containing Plasma, EXPAR part A (1× NEBuffer 3.1, 0.2µM amplification template, 500µM dNTPs and 1.6U/µL RNase inhibitor) and EXPAR part B (2× ThermoPol buffer, 0.8U/µL Nt.BstNBI nicking endonuclease, 0.1U/µL Vent (exo-) DNA polymerase, 2× EvaGreen®, 2× ROX reference dye) were injected independently into the microfluidic device (Figure 2a) via respective inlets at flow rates of 0.3 µL/min for plasma and 1.35 µL/min for part A and part B respectively, while the oil phase was injected at a flow rate of 4 µL/min. Uniform 30 µm diameter droplets were generated by flow focusing the continuous flow, and the generated droplets were collected in 1 mL syringes while pulling with the syringe pump to decrease the pressure inside the microfluidic channel. To increase the throughput of the droplet generation, four of the microfluidic devices were used at the same time and droplets were collected for 2 hours to generate 1.4 mL droplets in total from the four devices. Droplet generation and droplet collection were all performed in a cold room at 4°C to avoid non-specific EXPAR.
Droplet detection using fluorescent microscope to monitor EXPAR kinetics
Collected droplets were then transferred into 1.5 mL Eppendorf tubes for EXPAR at 55°C and 3 µL of droplets were taken out from the tube every 10 min for imaging with a fluorescent microscope (Eclipse Ti, Nikon, Japan). Microscopic images were analyzed by manually for the quantification of fluorescent droplets in the image.
3D particle counting system
A 3D particle counting prototype instrument built to our specifications 26, 27 (ISS Inc, Champaign, IL, USA) was used for the rapid quantification of fluorescent droplets in mLs of sample (Scheme 1).20 The apparatus consists of a small microscope and two motors providing rotational and vertical motion of the cuvette. The excitation light generated by lasers is focused at the volume positioned inside the cuvette, at a distance of about 1 mm from the cuvette wall. This distance can be adjusted so that detection of particles and analysis could be done even in highly scattering media. The excitation sources are two diode lasers emitting at 469 nm (ISS Inc) or at 532 nm (Aquaplan, Busto Arsizio, Italy). A confocal microscope was used in combination with simple mechanical motions of the sample container in front of the objective which provides the way to move and analysis a sample containing particles through an observation region without requiring a complex optical system with moveable optical components. The excitation lights from the laser are combined in one path through a set of dichroic filters Z470rdc (Chroma Technology Corporation, Rockingham, VT, USA) and directed through a 20× 0.4 NA air objective (Newport, Newport Beach, CA, USA) to the same volume in cuvette. Fluorescence emitted from fluorescent droplets is collected by the same objective, transmitted through the set of dichroic filters, focused by a lens into a large pinhole (diameter = 2 mm), and then collimated by a second lens to the detectors. Emission filters (FF01-HQ 500/24- 25, Semrock, Rochester, NY, USA) are located in front of each PMT. The signal from the PMT is sent to the analog-to-digital converter (ADC) and acquisition card (IOTech, Cleveland, OH, USA). The sampling frequency is set to 100,000 Hz, corresponding to a time resolution of 10 µs.
Droplet detection using 3D particle counter
After 50 min incubation at 55°C, droplets were transferred to a glass cuvette and the number of fluorescent droplets was quantified by the 3D particle counter. We typically analyze 1 mL of droplet solution for 10 minutes. In practice, we calibrate the system with a known concentration of particles (for example 100/mL) and we fix the time for the measurement (can be any time, for example 2 minutes or 10 minutes) and record the number of hits. Collected intensity profiles from the 3D particle counter were analysed with a pattern recognition filter by SimFCS software (Laboratory for Fluorescence Dynamics, Irvine, CA, USA: www.lfd.uci.edu). Patterns from fluorescent droplets were identified and quantified by matching with predetermined patterns using fluorescent droplets that contain reacted EXPAR products, which include distributions (e.g. Gaussian) of intensities as a function of time, the speed of the particles, and the rate of signal sampling.26, 27 The number of detected fluorescent droplets containing miRNA in actual samples was converted into a value of concentration using the calibration factor obtained previously with known numbers of fluorescent miRNA-containing droplets.
RT-qPCR for miRNA detection
Taqman probe based RT-qPCR was performed for miRNA detection following a reported protocol.31 Briefly, 14 – 140,000,000 copies of Let-7a miRNA were spiked into 200 µL of miRNA-depleted plasma. Total miRNA was extracted and isolated from plasma samples using miRNeasy Serum/Plasma Kit (Qiagen, USA) according to the manufacturer’s protocol. Reverse transcription was performed using the Taqman miRNA reverse transcription kit with miRNA-specific stem loop primers. For the real time PCR reaction, a 30 µL reaction mixture consisted of 15 µL 2× PCR supermix, 1.5 µL 20× miRNA-specific probe, 0.6 µL 50× ROX reference dye, 3µL reverse transcription product and 10µL H2O, which was prepared for triplicate experiments. Standard curve, obtained with spiked synthetic miR-39 at known concentrations, was used to estimate of target miRNA concentration in samples.
Results and discussion
Bulk EXPAR for miRNA detection in plasma
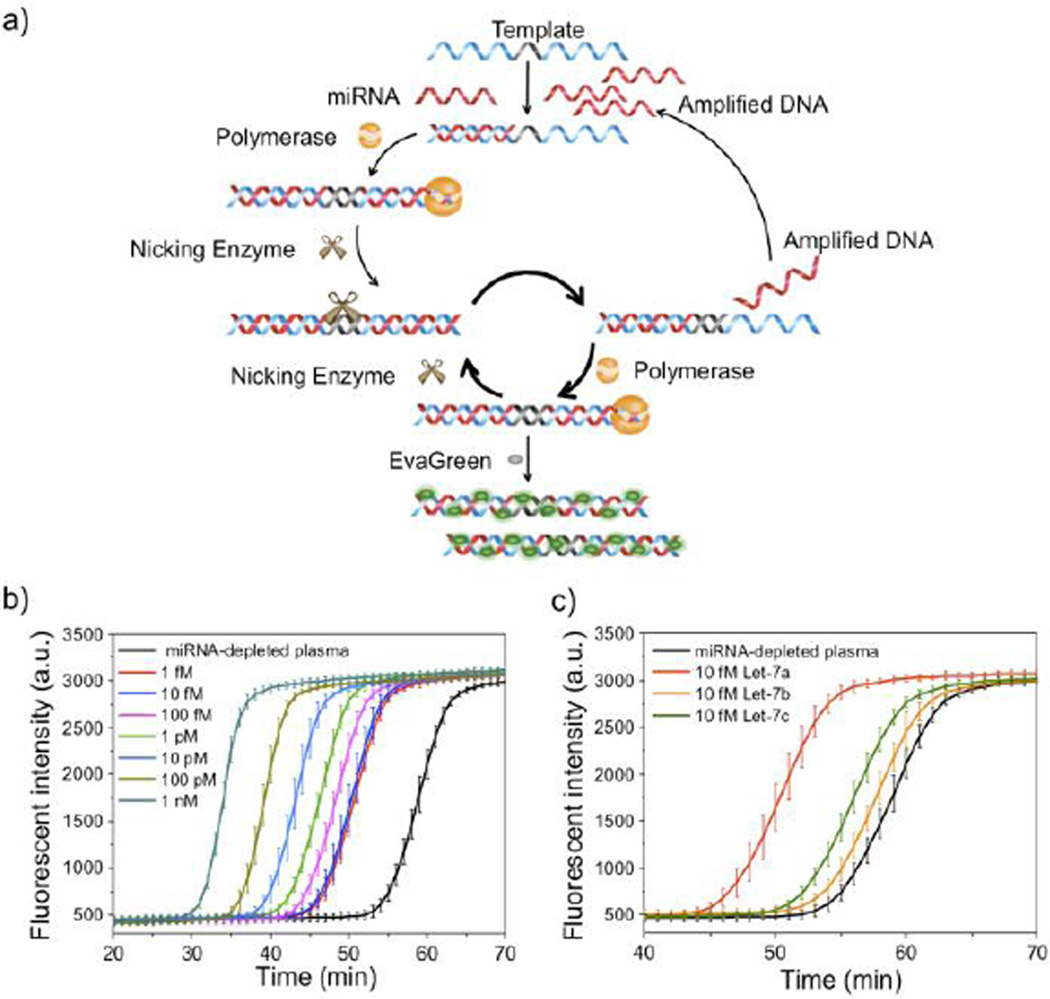
We first validated EXPAR for rapid and real-time miRNA detection in bulk. EXPAR is a homogenous and ultrasensitive isothermal amplification method which has been used for DNA and miRNA detection (Figure 1a).29 EXPAR can provide high output signal which is optimal for digital detection. Since there is significant amount of miRNA in exosomes in plasma, the choice of EXPAR was additionally motivated by the fact that its operating temperature (55 °C) can release target miRNA from exosomes. 28 In our study, Let-7a was chosen as a model target because of its critical roles in regulating cellular processes as a tumor suppressor by inhibiting actively translating polyribosomes and interfering with the accumulation of growing polypeptides.32 We first demonstrate that, in reaction buffer (1X ThermoPol buffer, 0.5X NEBuffer 3.1), Let-7a can be specifically detected at the fM level by real-time fluorescent measurement using EXPAR (Figure s1). We then examined EXPAR performance in plasma: various amounts of synthetic Let-7a were spiked into RNase inhibitor-treated, endogenous miRNA-depleted healthy donor plasma (see Methods).We confirmed that our miRNA depletion protocol effectively removed endogenous Let-7a content in plasma (Figure 1b). As shown in Figure 1b, EXPAR in bulk can detect 10 fM or higher concentrations of Let-7a in plasma but cannot robustly distinguish concentrations below 10 fM (compared to 1fM in reaction buffer) in bulk reactions because of the interference from plasma. To evaluate the specificity of EXPAR in plasma, Let-7b and Let-7c were selected as controls because of their sequence homology with Let-7a (sequences are shown in Table s1). Real-time fluorescence measurement shows that EXPAR can specifically distinguish Let-7a from Let-7b and Let-7c in plasma, which only differ by one or two nucleotides (Figure 1c).
Figure 1.
Exponential amplification reaction (EXPAR) for miRNA detection in plasma. a) Mechanism of how EXPAR generates fluorescent signal in the presence of target miRNA. Designed DNA templates recognize the target miRNA which triggers downstream amplification in the presence of Vent (exo−) polymerase and nicking enzyme. The template strand is designed with two identical sequences at each end of the template that are complementary to the target miRNA and are separated by a central domain composed of a nicking recognition site. Binding of the target to the 3’ end of the template triggers extension and nicking by Vent (exo-) polymerase and nicking enzyme, respectively. Exponential amplification is obtained by that 1) the nicked strand is continuously extended by strand displacement and polymerization, 2) and the displaced strand can also trigger further reactions with more template molecules. Together, these cyclical processes yield exponential amplification. A dsDNA-binding dye such as EvaGreen® in the reaction mixture binds to the amplified sequences and generates a fluorescent signal that can be monitored in a real-time fashion. b) Real-time fluorescence measurement of EXPAR triggered by spiked 1fM to 1nM synthetic Let-7a miRNA in plasma. Error bar is based on triplicate experiments. Mean ±s.e.m. c) Real-time fluorescence measurement of EXPAR triggered by spiked 10fM synthetic Let-7a, Let-7b and Let-7c in miRNA-depleted plasma. Error bar is based on triplicate experiments. Mean ± s.e.m.
Droplet digital (dd) EXPAR for miRNA detection
EXPAR in droplet for miRNA detection was subsequently investigated. The droplet microfluidic device was designed and fabricated using standard soft lithography and operated following previously established procedures (see Methods).30 As shown in Figure 2a, 100% raw plasma with various amount of spiked miRNA was introduced through one of three aqueous inlets of the fabricated microfluidic device (also see Figure s3 for the CAD design). EXPAR reagents were introduced separately as part A (NEBuffer 3.1, DNA template, deoxyribonucleotides (dNTPs) and RNase inhibitor) and part B (ThermoPol buffer, DNA polymerase (Vent (exo-)), nicking endonuclease (Nt.BstNBI), EvaGreen and ROX reference dye) to avoid non-specific amplification before introduction of the target miRNA. The reagents and plasma are merged within the microfluidic channel and encapsulated into uniform droplets (30 µm in diameter in this work, supplementary movie) using a flow-focusing structure (Bottom right panel in Figure 2a). The plasma sample was diluted 10 times on-chip with the EXPAR reagents after mixing. Since the EXPAR reaction will yield nonspecific background amplification if given sufficient time (Figure 1b)33, kinetics studies for single miRNA detection in droplet were first performed using fluorescence microscopy to identify the optimal detection time that generates maximum target-specific fluorescence signal with minimum background (Figure s4). In this set of experiments, the bulk concentration of spiked miRNA before encapsulation was 10 fM, which translates to 8.85% of droplets with 1 miRNA per droplet on average after encapsulation. To avoid non-specific EXPAR reaction, droplet generation and collection were all performed in a cold room at 4°C prior to the reaction. We observed that after a 50 min EXPAR reaction the number of fluorescent droplets that contain single Let-7a molecules increases closely to the predicted number while Let-7b and blank samples have no fluorescent droplets, although non-specific signals begin to arise after 60 min (Figure s4a, b). The background signal is generated from non-specific amplification likely due to the interaction between DNA polymerase and the single-stranded EXPAR template sequence, which can spontaneously form trigger sequences.33 This set of data allow us to 1) demonstrate the feasibility of single miRNA detection using ddEXPAR, and 2) identify 50 min as the optimal ddEXPAR reaction time, which was used in subsequent experiments when we detect miRNA in blood using IC 3D (Figure 2b).
Digital quantification of Let-7a in plasma by the IC 3D
To robustly and accurately count fluorescent droplets that contain target miRNA among hundreds of millions of blank ones in a short period of time (which cannot be achieved by existing flow cytometry or 1D on-chip droplet detection), in our IC 3D system, we incorporated a 3D particle counter which is able to detect fluorescent particles from mL volumes at single-particle sensitivity within minutes.26, 27 The innovative design of this instrument allows for rapid scanning of mLs of collected droplets in a cuvette (Figure 2a) in a spiral motion in front of an objective of the confocal microscope (Scheme 1). The optics of the particle counter is designed to measure a relatively large volume (100pL) in about 0.01 ms (or 100 seconds for about 1mL). As we described earlier, droplets were formed by encapsulating miRNA-spiked raw plasma with different miRNA concentration (10, 50, 100, 500, 10000, 50000 or 100000 copies/mL) and EXPAR reagents in the microfluidic device. In addition, as we used a single-channel droplet generation device in this paper, in order to rapidly analyse a large sample volume, we used four parallel devices that allowed us to produce 1 mL droplets within 2 hours. This also helps us to minimize the interference resulted from potential nonspecific EXPAR reaction during droplet production. The droplets were then collected, incubated at 55 °C for 50 min off-chip to allow the EXPAR reaction to take place, and then analyzed by the 3D particle counter (Scheme 1).
Using this system, we have demonstrated that fluorescent droplets that contain single target miRNA, can be detected at single-droplet sensitivity. Figure 3b shows a typical time trace with fluorescence intensity spikes obtained from Let-7a-containing droplets but there were no signal peaks above the threshold (three times s.d. of the baseline) from control droplets that contained only plasma (Figure 3a) or Let-7b spiked-plasma (Figure 3c). To determine the concentration of the droplets in the sample, the temporal profile generated by the photodetector is analyzed with a pattern recognition algorithm (inserted box in Figure 3b) implemented in the SimFCS software. The pattern recognition algorithm matches amplitude and shape features in the temporal profile to a predetermined pattern that is characteristic of the time-dependent fluorescence intensity of droplets passing through the observation volume. Such pattern recognition allows us to achieve exceptionally reliable and accurate detection of a low concentration of target fluorescent particles or droplets in large sample volumes that are often surrounded with nonspecific background signals.26, 27 We next demonstrate that IC 3D can provide digital quantification of target Let-7a at a broad range of extremely low concentrations from ~10s to 100,000 copies/mL (Figure 3d, raw data are presented in Table s2). There is an excellent linear correlation between actually detected number of droplets and the predicted concentration of Let-7a spiked in plasma with accuracy and relative standard deviation (i.e., precision) typically of ~60–95% and ~10–50%, respectively (Table s2). Because our current particle counter prototype is configured to analyze 1 mL sample and the raw plasma samples are subject to a 10 times on-chip dilution, we practically only analyze 100 μL of each raw sample in this set of experiments. Under this condition, our limit of detection (LOD) is 50 copies/mL for miRNA spiked in 100% raw plasma. We had inconsistent result when we tried to detect 10 copies/mL, i.e., 1 copy in 100 μL that was actually analyzed, which is likely due to the inherent difficulty in single molecule sample preparation, handling and measurement among a large volume. However, we emphasize that our particle counter can robustly detect as low as 1–10 fluorescent particles or droplets in an mL solution.20, 26 We believe the LOD can be further improved in the future by minimizing the plasma dilution factor or miRNA enrichment, which allows us to analyze a larger volume of raw plasma sample per measurement than the current 100 µL used in this paper.
We next directly compared our IC 3D with the current gold standard, RT-qPCR for Let-7a measurement in plasma. The RT-qPCR experiments were performed following standard protocols where total miRNA were first isolated using a conventional miRNA isolation kit and then analyzed by TaqMan probe-based RT-qPCR (Figure 3e). The LOD of the IC 3D assay is two to three orders of magnitude lower than that of RT-qPCR, which is ~103 copies/mL according to our experimental data (Figure 3e).We observed that the RT-qPCR consistently detected smaller amounts of let-7a than the spiked amounts, while the IC 3D in Fig. 3d did not show such bias. We contribute the artifact in RT-qPCR to the loss of target miRNA during sample processing, which is one of the challenges our new IC 3D_EXPAR aims to address as it can directly detect miRNA from unprocessed plasma. In addition, the artifact resulted from sample processing in RT-qPCR is particularly profound when dealing with target miRNA at low concentrations compared to high concentrations (Figure 3e and 3f below). In addition, we found that RT-qPCR typically only detected approximately 11–32% of the actual targets spiked in the plasma sample. The loss in target detection is likely due to the inefficiency of the miRNA isolation step. It should also be noted that RT-qPCR cannot operate directly using plasma samples and typically requires miRNA extraction and purification (see Methods). Furthermore, RT-qPCR takes approximately 8 hours to conduct the manual experimental procedures including miRNA isolation, reverse transcription, and quantitative PCR, while our IC 3D approach allows us to get a result within 3 hours. In summary, despite of recent advances in qPCR-based RNA analysis that could achieve LOD of 50 copies/mL using expensive and sophisticated sample processing and analysis equipment (e.g., Life technologies and Qiagen)34, our simple IC 3D technology exhibits superior performance than routine RT-qPCR methods with respect to LOD, robustness, assay time, and simplified work-flow.
Clinical validation of IC 3D using clinical colon cancer samples
To demonstrate the potential clinical applicability of the IC 3D system, plasma samples from colon cancer patients and healthy donors were analyzed (these clinical samples were utilized after review and approval from the IRB (see Methods)). The plasma samples were first tested in bulk using EXPAR and demonstrated that EXPAR can be used for direct Let-7a detection in plasma, although the fluorescent amplification curves between healthy donor and colon cancer patient samples cannot be well distinguished (Figure s2). IC 3D was then used to quantify Let-7a concentration in 3 representative colon cancer patient samples and also three healthy donor controls. Our hypothesis is that that the confinement of samples containing low-abundance miRNAs in picoliter droplets can significantly increase the effective miRNA concentration, provide a means for digital quantification (i.e., each droplet has 0 or 1 target miRNA) and minimizes the number of background molecules (interference) from plasma.25 Indeed, we demonstrated that the IC 3D can robustly quantify target miRNA directly from unprocessed plasma as validated by RT-qPCR for the same samples (Figure 3f). RNase-treated plasma was also included as a negative control to confirm that the fluorescent droplets are due to the target Let-7a (Figure s5). Interestingly, we found that the Let-7a content in colon cancer samples is statistically significantly higher than that in healthy donor samples as digitally quantified by IC 3D (Figure 3f) (although it was not distinguishable by bulk EXPAR (Figure s2)). This is further confirmed by RT-qPCR using more patient samples (Figure s6). The higher level of Let-7a (although known as a tumor suppressor) 35 in cancer samples could be due to the higher content of exosomes and miRNAs that shed from tumors into the blood stream, 36 which presents a topic of future inquiry. Although Let-7a is associated within exosome37, some other miRNAs are known to be resistant to degradation by complexing with proteins such as Ago2.28 To demonstrate the sequence universality of IC 3D_EXPAR for miRNA detection directly from plasma, we also performed IC 3D_EXPAR that targets miRNA-92a (Figure s7), which is known to form a complex with the Ago2 protein. Moreover, miRNA-92a is also considered as one of the most important biomarkers for colorectal cancer.38, 39 For this experiment, IC 3D_EXPAR was directly compared with RT-qPCR as we described previously for Let-7a. We found that miRNA-92a can be quantified by IC 3D_EXPAR but the quantified miRNA-92 concentration was about 17.2–22.9% of that of RT-qPCR (Figure s7b). We suspect this is due to that miRNA-92 are stably associated with Ago2 protein and therefore not accessible for EXPAR reaction. Arroyo et al., described that exosomal miRNA Let-7a can be released by heating at 55 °C but Ago2-associated miRNA-92a required protease K treatment for dissociation. We tried to release miRNA-92a from Ago2 by protease K treatment followed by denaturing of protease K at 75°C for 10 minutes prior to encapsulation with the EXPAR components. However, protease K could not be fully inactivated by heating and thus inhibited the IC 3D_EXPAR (Data is not shown). In an alternative approach, plasma-containing droplets were heated at 70 °C for 5 min to release miRNA-92a from Ago2 and IC 3D_EXPAR was performed immediately after. Our attempt to dissociate miRNA from Ago2 by pre-heating within the droplets has resulted in significantly increased counted numbers although still lower than that obtained from RT-qPCR. At this stage, it is unclear whether the lower miRNA count in IC 3D_EXPAR even after pre-heating is due to incomplete disassociation of miRNA from Ago2 or actually degradation of miRNA and/or inactivation of polymerase/nicking enzyme by heating.
Conclusions
In summary, we have developed a new technology termed ‘Integrated Comprehensive Droplet Digital Detection (IC 3D)’ that can provide digital quantification of target miRNA in unprocessed plasma at a broad range of low concentrations from 10s to 100,000 copies/mL within 3 hours and a LOD of ~ 50 copies/mL. The innovative integration of real-time isothermal sensing, microencapsulation and the 3D particle counter in the IC 3D system surpasses existing methods for miRNA detection with respect to sensitivity, rapidness, and, importantly, minimal sample processing (i.e., without the need for RNA isolation and purification, as typically required by other systems).7, 8, 10, 11, 40, 41 Indeed, the conventional methods including RT-PCR typically require sample preparation or conducted in buffer with typical LOD of 103 copies/mL.18, 34 In addition, compared to other 1D on-chip7, 42, 43 or 2D droplet detection systems14, 15 such as wide-field fluorescence imaging, the major advantages of our simple 3D particle counter include 1) high throughput: 100,000s droplets/s or an effective volume of observation of 0.1 ml/min, which allows us to rapidly interrogate a large volume (mL) of droplets, and 2) robustness: the detection of a ‘hit’ is defined by a pattern recognition algorithm20 besides threshold intensity that alone often suffers from higher false-positive/negative rates.23, 24 Additionally, our IC 3D can distinguish single molecules because IC 3D operates using “digital” measurements, which droplets are counted as either ‘0 (negative)’ or ‘1 (positive)’. Because of the “digital” nature and pattern recognition algorithm for a “hit”, our assay is consistently able to distinguish a droplet that contains zero or a single miRNA. The other advantage of our IC 3D is that we can avoid loss of miRNA during sample preparation, which further improves sensitivity. The different blood miRNA levels between cancer and healthy individuals observed in this study suggest that using miRNA as a biomarker, while using the IC 3D as a sensitive and robust detection system, can potentially be used for cancer monitoring and therapeutic response evaluation. In addition, advantages of IC 3D overcurrent commercially available ddPCR systems such as Bio-Rad, Life technologies (Quanta Studio) and Raindance technologies, are 1) miRNA can be quantified directly in plasma, 2) IC 3D can accommodate large sample volume (1 mL or hundreds of millions of droplets), compared to typical sample volumes of 14~50µL in the commercial system.7, 44, 45 This not only allows us to improve detection throughout but further increase detection sensitivity of target nucleic acids from vast background, 3) Our system operates under isothermal condition without the need of reverse-transcription and thermocycler, and 4) IC 3D is less expensive because the particle counter is basically miniaturized confocal microscope system that consists of inexpensive diode lasers and PMT detector. The cost of particle counter can be as low as several thousands dollars as we described in our previous report.20 Finally, our ongoing effort aims to develop a fully automated system that is able to perform multiplex assays using multiple sensors for different targets. In particular, we will integrate a high-throughput droplet generation system (256 droplet-generating channels) which is able to convert 1mL plasma to 30 µm droplets at a generation rate of 1000Hz per channel within 10 min. Finally, we are performing head-to-head comparison between our IC 3D and other digital detection systems (e.g., ddPCR) for miRNA detection with respect to sensitivity, sample preparation, robustness and throughput.
Supplementary Material
Acknowledgements
This work is supported by the start-up fund from the Department of Pharmaceutical Sciences, Sue and Bill Gross Stem Cell Research Center and the Chao Family Comprehensive Cancer Center at UC Irvine provided to W.Z. The project was additionally supported by NCI Cancer Center Support Grant P30CA062203. The project described was in part supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR000153. EG and MAD acknowledge NIH funding from grants P41 GM103540, NIH P50-GM076516 and the National Natural Science Foundation of China (No. 21235004, No. 21327806). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. K. Z is supported by a scholarship from the China Scholarship Council (CSC 201206210111).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Contributor Information
Jinghong Li, Email: jhli@mail.tsinghua.edu.cn.
Weian Zhao, Email: weianz@uci.edu.
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. P Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasso M, Piscopo P, Confaloni A, Denti MA. Molecules. 2014;19:6891–6910. doi: 10.3390/molecules19056891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayed AS, Xia K, Salma U, Yang T, Peng J. Heart, lung & circulation. 2014 doi: 10.1016/j.hlc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. Chemical reviews. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 7.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campuzano S, Torrente-Rodriguez RM, Lopez-Hernandez E, Conzuelo F, Granados R, Sanchez-Puelles JM, Pingarron JM. Angewandte Chemie. 2014;53:6168–6171. doi: 10.1002/anie.201403270. [DOI] [PubMed] [Google Scholar]
- 9.Duan RX, Zuo XL, Wang ST, Quan XY, Chen DL, Chen ZF, Jiang L, Fan CH, Xia F. J Am Chem Soc. 2013;135:4604–4607. doi: 10.1021/ja311313b. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Zheng DL, Tan QL, Wang MX, Gu LQ. Nature Nanotechnology. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labib M, Khan N, Ghobadloo SM, Cheng J, Pezacki JP, Berezovski MV. J Am Chem Soc. 2013;135:3027–3038. doi: 10.1021/ja308216z. [DOI] [PubMed] [Google Scholar]
- 12.Das J, Ivanov I, Montermini L, Rak J, Sargent EH, Kelley SO. Nature Chemistry. 2015;7:569–575. doi: 10.1038/nchem.2270. [DOI] [PubMed] [Google Scholar]
- 13.Murtaza M, Dawson SJ, Tsui DWY, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong ASC, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 14.Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, Yang DL, Lee AP. Lab on a chip. 2011;11:3838–3845. doi: 10.1039/c1lc20561g. [DOI] [PubMed] [Google Scholar]
- 15.Kim M, Pan M, Gai Y, Pang S, Han C, Yang C, Tang SKY. Lab on a Chip. 2015;15:1417–1423. doi: 10.1039/c4lc01465k. [DOI] [PubMed] [Google Scholar]
- 16.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang SL, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. Analytical chemistry. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Kermekchiev MB, Barnes WM. J Mol Diagn. 2010;12:152–161. doi: 10.2353/jmoldx.2010.090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Nucleic acids research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Hoon DSB, Pantel K. Nature Reviews Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 20.Kang DK, Ali MM, Zhang KX, Huang SS, Peterson E, Digman MA, Gratton E, Zhao WA. Nature communications. 2014;5:5427. doi: 10.1038/ncomms6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang DK, Ali MM, Zhang KX, Pone EJ, Zhao WA. Trac Trend Anal Chem. 2014;58:145–153. [Google Scholar]
- 22.Selck DA, Karymov MA, Sun B, Ismagilov RF. Analytical chemistry. 2013;85:11129–11136. doi: 10.1021/ac4030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nixon G, Garson JA, Grant P, Nastouli E, Foy CA, Huggett JF. Analytical chemistry. 2014;86:4387–4394. doi: 10.1021/ac500208w. [DOI] [PubMed] [Google Scholar]
- 24.Sun B, Shen F, McCalla SE, Kreutz JE, Karymov MA, Ismagilov RF. Analytical chemistry. 2013;85:1540–1546. doi: 10.1021/ac3037206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witters D, Sun B, Begolo S, Rodriguez-Manzano J, Robles W, Ismagilov RF. Lab on a chip. 2014;14:3225–3232. doi: 10.1039/c4lc00248b. [DOI] [PubMed] [Google Scholar]
- 26.Altamore I, Lanzano L, Gratton E. Meas Sci Technol. 2013;24:65702. doi: 10.1088/0957-0233/24/6/065702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skinner JP, Swift KM, Ruan QQ, Perfetto S, Gratton E, Tetin SY. Review of Scientific Instruments. 2013;84:074301. doi: 10.1063/1.4812782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia HX, Li ZP, Liu CH, Cheng YQ. Angewandte Chemie. 2010;49:5498–5501. doi: 10.1002/anie.201001375. [DOI] [PubMed] [Google Scholar]
- 30.McDonald JC, Whitesides GM. Acc Chem Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 31.Schmittgen TD, Lee EJ, Jiang JM, Sarkar A, Yang LQ, Elton TS, Chen CF. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nottrott S, Simard MJ, Richter JD. Nature structural & molecular biology. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 33.Tan E, Erwin B, Dames S, Ferguson T, Buechel M, Irvine B, Voelkerding K, Niemz A. Biochemistry. 2008;47:9987–9999. doi: 10.1021/bi800746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D’Andrade P, DeMayo M, Dennis L, Derveaux S, Feng Y, Fulmer-Smentek S, Gerstmayer B, Gouffon J, Grimley C, Lader E, Lee KY, Luo S, Mouritzen P, Narayanan A, Patel S, Peiffer S, Ruberg S, Schroth G, Schuster D, Shaffer JM, Shelton EJ, Silveria S, Ulmanella U, Veeramachaneni V, Staedtler F, Peters T, Guettouche T, Wong L, Vandesompele J. Nature methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 35.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Brit J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. PloS one. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creemers EE, Tijsen AJ, Pinto YM. Circulation Research. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 38.Ng EKO, Chong WWS, Jin H, Lam EKY, Shin VY, Yu J, Poon TCW, Ng SSM, Sung JJY. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 39.Huang ZH, Huang D, Ni SJA, Peng ZL, Sheng WQ, Du X. International Journal of Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 40.Lee JM, Cho H, Jung Y. Angewandte Chemie. 2010;49:8662–8665. doi: 10.1002/anie.201004000. [DOI] [PubMed] [Google Scholar]
- 41.Degliangeli F, Kshirsagar P, Brunetti V, Pompa PP, Fiammengo R. J Am Chem Soc. 2014;136:2264–2267. doi: 10.1021/ja412152x. [DOI] [PubMed] [Google Scholar]
- 42.Srisa-Art M, deMello AJ, Edel JB. Chem Commun. 2009:6548–6550. doi: 10.1039/b917721c. [DOI] [PubMed] [Google Scholar]
- 43.Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Nat Protoc. 2013;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurent-Puig P, Pekin D, Normand C, Kotsopoulos SK, Nizard P, Perez-Toralla K, Rowell R, Olson J, Srinivasan P, Le Corre D, Hor T, El Harrak Z, Li XY, Link DR, Bouche O, Emile JF, Landi B, Boige V, Hutchison JB, Taly V. Clin Cancer Res. 2015;21:1087–1097. doi: 10.1158/1078-0432.CCR-14-0983. [DOI] [PubMed] [Google Scholar]
- 45.Chen R, Mias GI, Li-Pook-Than J, Jiang LH, Lam HYK, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.