Abstract
Tetracyclines, a class of antibiotics that target bacterial translation, are commonly used in research for inducible gene expression using Tet-ON/Tet-OFF systems. However, such tetracycline-inducible systems carry a risk. Given that mitochondria have a “bacterial” ancestry, these antibiotics also target mitochondrial translation and impair mitochondrial function. Indeed, treatment with doxycycline—a tetracycline derivative—disturbs mitochondrial proteostasis and metabolic activity, and induces widespread gene expression changes. Together, this affects physiology in well-established model systems ranging from cultured cells to simple organisms and to mice and plants. These changes are observed with doxycycline doses that are widely used to regulate gene expression. In light of these findings, and bearing in mind the conserved role of mitochondria in metabolism and whole organism homeostasis, we caution against the use of tetracyclines in experimental approaches. The use of newly developed tetracycline-based systems that are more sensitive could be an alternative; however, even if no overt mitochondrial toxicity is detected, widespread changes in gene expression may sensitize cells to the intended tetracycline-controlled loss- or gain-of-function, thereby introducing a “two-hit model”. This is highly relevant for cancer research, as mitochondrial metabolism holds a central position in the reallocation of nutrients for biomass production known as the Warburg effect.
Keywords: antibiotics, doxycycline, mitochondrial dysfunction, Tet-on/Tet-off, cancer metabolism
Introduction
Mitochondria are at the core of cellular metabolism, as they participate in the final breakdown of nutrients and produce ATP after a series of oxidation reactions that take place in the mitochondrial sub-compartments. Next to their vital metabolic role, they are also involved in many other important processes of the cell, such as calcium homeostasis, apoptosis and intra-cellular signaling (1). Considering the versatile functions of mitochondria, it is no surprise that dysfunction of these organelles can lead to manifestation of different diseases affecting various tissues and with symptoms of ranging severity that can even be life-threatening (2). In recent years, mitochondrial dysfunction has also been implicated in multifactorial age-related diseases such as diabetes mellitus, Parkinson’s disease and cancer. Consequently, there has been an ever-growing interest in mitochondria as targets for pharmacological interventions (3).
Mitochondria as bacteria in our cells
An important feature of mitochondria, which is reflected in both its unique ultrastructure and physiology, is its eubacterial ancestry. Mitochondria are derived from the endosymbiosis of an α-proteobacterium with a host amitochondriate cell, an event that is considered to have given rise to the modern eukaryotic cell (4). Throughout evolution, most of the endosymbiont’s genome was transferred to the nucleus of the host cell, but mitochondria still harbor remnants of the ancestral bacterial genome and have retained their own transcriptional and translational machinery. Mitochondria contain several copies of this circular double stranded molecule (mitochondrial DNA or mtDNA) that encodes for 13 core subunits of the oxidative phosphorylation (OXPHOS) complexes and for specific mitochondrial tRNAs and rRNAs (3). Because all other mitochondrial proteins—including the remainder of the OXPHOS subunits—are encoded by nuclear DNA (nDNA), the stoichiometric balance between both genomes is tightly regulated (5). Disruption of this balance, a state we previously termed mitonuclear protein imbalance, occurs when either the nuclear or the mitochondrial protein synthesis is perturbed. Such imbalance leads to the activation of the mitochondrial unfolded protein response (UPRmt), an adaptive stress-response pathway working to restore protein homeostasis within the mitochondrial matrix (6, 7). Moderate mitochondrial dysfunction and UPRmt activation is beneficial as it allows mitochondria to recuperate, while prolonged mitochondrial stress can ignite more terminal responses including apoptosis. For example, RNAi-mediated knockdown of nDNA-encoded OXPHOS protein subunits led to UPRmt activation and lifespan extension in both fruit flies and the nematode C. elegans (8, 9). Similar effects were observed after inhibiting mtDNA replication and transcription with ethidium bromide treatment or after attenuation of mitochondrial translation by knockdown of components of the mitoribosome in C. elegans (5, 10). On the other hand, pathogenic mutations in these pathways are linked to severe disorders that often present in infancy (2), emphasizing the delicate intergenomic balance.
Given the “bacterial” origin of mitochondria, it is not surprising that antibiotics targeting bacterial translation—such as tetracyclines and chloramphenicol—can also inhibit mitochondrial translation (11). Indeed, similar to RNAi-mediated down-regulation of the mitochondrial translational machinery, treatment with doxycycline—an antibiotic belonging to the family of tetracyclines—induced mitonuclear protein imbalance, impaired mitochondrial respiration, activated UPRmt and dose-dependently extended the nematode lifespan (5). Similar effects on mitochondrial function and proteostasis were observed in tissues of mice fed with doxycycline, as well as in human and murine cell-based models, where doxycycline reduced mitochondrial respiration and induced UPRmt (5, 12).
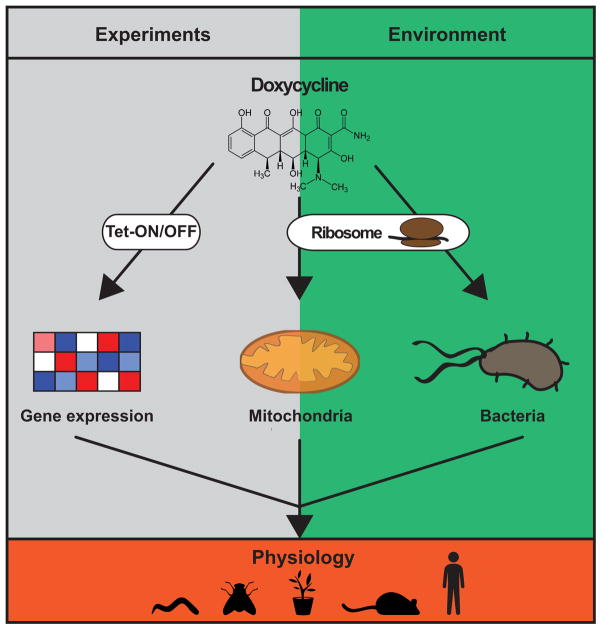
On an organismal level, doxycycline proved beneficial on some physiological aspects, as it increased motility in worms and flies, and extended the lifespan of worms (5, 12). At the same time, however, both doxycycline-treated models presented with developmental delay and physiological impairment related to body size and fecundity (12). Even plants (Arabidopsis thaliana) treated with doxycycline displayed marked growth retardation, while on a molecular level they presented with mitochondrial dysfunction and activated UPRmt (12). These findings underscore the immense effects of doxycycline on mitochondrial function and whole-body physiology through a mechanism that is conserved from plants and invertebrates to mice and humans (Fig. 1).
Figure 1. Experimental and therapeutic use of doxycycline carries the risk of altering mitochondrial function and organismal physiology.
Doxycycline is a tetracycline antibiotic that blocks the bacterial ribosomes and is commonly used to treat infections. At the same time it is also utilized in research to control gene expression in Tet-ON/Tet-OFF inducible systems. What is often overlooked however, is the fact that doxycycline inhibits not only bacterial but also mitochondrial translation, altering mitochondrial metabolism. The combined effects of tetracycline antibiotics have a major impact on organismal physiology.
Mitochondria in cancer
Accumulating evidence suggests that functional mitochondria are essential for cancer cell survival and proliferation. The emerging role of mitochondrial bioenergetics, biogenesis and signaling pathways on tumorigenesis has drawn the attention to the organelle’s function as a potentially fruitful field for cancer therapy development. Tumor cells have in essence an opposite metabolic profile from that of normal cells in differentiated tissues. In the mid-1920s, Otto Warburg observed that cancer cells produce large quantities of lactate even in the presence of ample oxygen, a state he termed “aerobic glycolysis” (13). We now know that increased glycolytic rates ensure faster supply of metabolic intermediates essential for fatty acid, amino acid and nucleotide synthesis, facilitating rapid growth and division of tumor cells (14). At the same time, the mitochondrial tricarboxylic acid (TCA) cycle is highly active to provide precursors for biomass production and bioenergetics, a situation that can only be maintained through efficient replenishment of TCA cycle intermediates, for instance through glutamine addiction, a phenomenon called anaplerosis (14). Thus, mitochondria in cancer cells remain metabolically active, meeting their bioenergetic needs by utilizing alternative substrates while also supplying metabolic intermediates to sustain their proliferative capacity.
The relevance of mitochondrial metabolism in cancer is emphasized by the identification of tumors with mtDNA mutations (15); these mtDNA mutations seem to alter mitochondrial metabolism, enhance tumorigenesis and allow cancer cell adaptation to changing environment. In addition, mutations in nDNA-encoded mitochondrial genes, including TCA cycle enzymes succinate dehydrogenase subunits (SDH), fumarate hydratase (FH) and isocitrate dehydrogenase 2 (IDH2), contribute to tumorigenesis as well as tumor progression (16). Another example of the vital role of mitochondria for tumor progression, is the finding that reduction of systemic copper has anti-proliferative effects, by reducing activity of the mitochondrial enzyme cytochrome c oxidase which leads to a decrease in ATP levels despite increased glycolysis rates (17). In addition to these metabolic switches, mitochondrial metabolism can also promote the production of mitochondrial reactive oxygen species (ROS) and change the cellular redox status. Mitochondrial ROS are important signaling molecules in the context of tumorigenesis by modifying the transcriptional activities of factors such as HIF-1, FOS and JUN to change gene expression and stimulate cancer cell proliferation (15). These findings strongly support the fact that mitochondrial function is fundamental for cancer cells survival and proliferation, and in this light it is interesting that specific antibiotics—those that target mitochondrial function—were identified as anti-proliferative treatment (18). As such, the reemergence of mitochondria as a central metabolic hub required for tumorigenesis highlights its potential as a target for cancer therapy.
Tetracyclines in cancer research
The development of anti-cancer regimens is promising but at the same time it is important to consider that tetracyclines not only serve as potential cancer therapy, but are also widely used in experimental systems, amongst others in the cancer field. The rise of the genomic era has allowed researchers to investigate the molecular function of many genes in great detail, but the drastic adverse effects of complete gain- or loss-of-function called for a more subtle approach. To serve this purpose, tetracycline inducible gene expression systems have been widely adopted to regulate the spatiotemporal expression of oncogenes and tumor suppressor genes in cancer development (19). These Tet-ON/Tet-OFF gene regulation systems are based on Escherichia coli tetracycline resistance operon. E. coli that express this operon acquire a membrane efflux pump that pumps out metal-bound tetracycline in exchange for a proton. Overexpression of the efflux pump would protect the bacterium against tetracycline, but eventually cause a deleterious loss of the membrane potential and hence requires strict regulation (20). It is the operon’s structural organization and mode of regulation that made it ideal for applications in eukaryotic cells. The operon contains an operator sequence (TetO) and two genes encoding the efflux pump and tetracycline repressor protein (TetR). When the operon is expressed under normal conditions, the TetR protein binds to the TetO sequence and thereby inhibits expression of the operon. The presence of tetracyclines however, interrupts TetR binding to TetO and allows expression of the operon (19). TetR and TetO were the foundation of the commonly used Tet-ON/Tet-OFF system (21, 22). The Tet-OFF system was established by fusing TetR to a VP16 transcription activation domain that binds to multiple TetO sequences to drive transcription of a transgene with a minimal promoter (e.g. TATA box) (21). In this case, the tetracycline-controlled transactivator (tTA) loses its binding potential to TetO upon tetracycline supplementation, and thereby prevents transgene expression. In the Tet-ON set-up or reversed transactivation (rtTA), the transgene transcription is only induced by rtTA binding to TetO upon addition of tetracycline or its more stable derivative doxycycline (22). Because gene expression can be regulated in a spatiotemporal manner, both the Tet-ON and Tet-OFF have been popular tools in cell culture models as well as model organisms including mice (19).
Despite all the enthusiasm, the use of tetracyclines in such experimental setup comes at a cost. The inhibition of mitochondrial translation through tetracyclines, for instance doxycycline, instigates adverse effects on mitochondria at the concentration generally used in Tet-ON/Tet-OFF systems (Fig. 1). Doxycycline exposure, even at low concentrations, increased glycolytic metabolism and markedly reduced the oxygen consumption of several human cell lines, including lung, prostate and cervical cancer cell lines (23). We recently demonstrated that exposure to low concentration doxycycline elicits similar adverse effects, i.e. mitonuclear protein imbalance and decreased oxygen consumption (12). This mitochondrial dysfunction was found in several of the commonly used cell lines, such as HeLa, HEK293T, and Hepa 1–6. Additionally, this metabolic stress was conserved in other model organisms, including worms, flies, mice and even plants. Strikingly, a reanalysis of microarray transcriptome profiles of human bladder cancer cell line RT112 exposed to doxycycline showed a 10% change in global gene expression (12), confirming the molecular basis for the widespread adverse effects of this treatment (Fig. 1).
Perspectives
The Tet-ON/Tet-OFF system has allowed a more intricate study of gene function without the dramatic adverse effects associated with gain- or loss-of-function mutants. Nevertheless, it has become apparent that gene expression regulation using doxycycline or similar antibiotics also induces molecular changes that can seriously confound the experimental outcome. Therefore, we advocate against the use of doxycycline in such systems. This not only applies for researchers investigating mitochondrial function specifically, but given the central role of mitochondria in cellular physiology, this is also relevant for other fields, including cancer. So what is the alternative? New tetracycline-responsive systems have been developed that are more sensitive and hence require lower doxycycline doses that do not cause overt adverse effects on mitochondrial function or global gene expression (24). Nevertheless, we propose that mild, even non-measureable, changes may sensitize a cell or organism to the intended alteration in gene expression, and thereby introduce a “two-hit model” (12). Other inducible systems, such as the tamoxifen-responsive estrogen receptor, also suffer from adverse effects, particularly in the liver of mice that have been engineered with this type of regulation (25). The only setup that so far appears to be safe is the light-switchable that is responsive to blue light (26), although further research is necessary to establish whether absorption of this light by specific proteins indeed does not interfere with cellular function.
Finally, the extensive use of tetracycline in the clinic and in livestock should be carefully evaluated and monitored. In the United States and countries in the European Union, the supply of tetracycline destined for animal use exceeds 8.5 million kilograms a year (27, 28). Similar to the exogenously added antibiotics, e.g. in the Tet-ON/Tet-OFF, these antibiotics may end up in the serum used in cell culture media and thereby confound experimental outcomes in research. More importantly, at least from a global health perspective, high concentrations of antibiotics are ending up in the environment. Little is known about the exact antibiotic concentrations in soils, but tetracycline concentrations up to 300 mg/kg have been reported in specific contexts, such as swine manure (29) or agricultural fields in Beijing suburbs (30). The magnitude of the effects on plants, animals, and humans requires more investigation, but it is clear that the excessive use of antibiotics can have a major impact on public health (Fig. 1). This involves the potential hazards of antibiotic resistance, disturbed gut microbiota, but also mitochondrial toxicity.
Acknowledgments
Financial support: Work in the Houtkooper group is financially supported by a grant from the Rembrandt Institute for Cardiovascular Science, an ERC Starting grant (no. 638290), and a VENI grant from ZonMw (no. 91613050). Work in the J.A. laboratory is supported by the Ecole Polytechnique Federale de Lausanne, the NIH (R01AG043930), the Swiss National Science Foundation (31003A-124713), and Systems X (51RTP0-151019). L.M. is supported by an FRM fellowship, and I.A.C. by an AMC PhD Scholarship.
Footnotes
Conflict of interest: The authors disclose no conflict of interest related to this work.
References
- 1.Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–22. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 2.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12:465–83. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–81. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 5.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–7. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–55. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 7.Held NM, Houtkooper RH. Mitochondrial quality control pathways as determinants of metabolic health. BioEssays. 2015 doi: 10.1002/bies.201500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–66. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 11.Clark-Walker GD, Linnane AW. In vivo differentiation of yeast cytoplasmic and mitochondrial protein synthesis with antibiotics. Biochem Biophys Res Commun. 1966;25:8–13. doi: 10.1016/0006-291x(66)90631-0. [DOI] [PubMed] [Google Scholar]
- 12.Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, et al. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 2015;10:1681–91. doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 14.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2013;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frezza C, Pollard PJ, Gottlieb E. Inborn and acquired metabolic defects in cancer. J Mol Med (Berl) 2011;89:213–20. doi: 10.1007/s00109-011-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishida S, Andreux P, Poitry-Yamate C, Auwerx J, Hanahan D. Bioavailable copper modulates oxidative phosphorylation and growth of tumors. Proc Natl Acad Sci USA. 2013;110:19507–12. doi: 10.1073/pnas.1318431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, et al. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget. 2015;6:4569–84. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–55. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 20.Berens C, Hillen W. Gene regulation by tetracyclines: Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur J Biochem. 2003;270:3109–21. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 21.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 23.Ahler E, Sullivan WJ, Cass A, Braas D, York AG, Bensinger SJ, et al. Doxycycline alters metabolism and proliferation of human cell lines. PLoS One. 2013;8:e64561. doi: 10.1371/journal.pone.0064561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao JS, Chao CC, Chang CL, Chiu YR, Yuan CJ. Development of single-vector Tet-on inducible systems with high sensitivity to doxycycline. Mol Biotechnol. 2012;51:240–6. doi: 10.1007/s12033-011-9461-z. [DOI] [PubMed] [Google Scholar]
- 25.Lelliott CJ, López M, Curtis RK, Parker N, Laudes M, Yeo G, et al. Transcript and metabolite analysis of the effects of tamoxifen in rat liver reveals inhibition of fatty acid synthesis in the presence of hepatic steatosis. FASEB J. 2005;19:1108–19. doi: 10.1096/fj.04-3196com. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–9. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 27.FDA. 2011 summary report on antimicrobials sold or distributed for use in food-producing animals. 2014 http://www.fda.gov/downloads/ForIndustry/UserFees/
- 28.ESVAC. Sales of veterinary antimicrobial agents in 25 EU/EEA countries in 2011. 2013 http://www.ema.europa.eu/docs/en_GB/document_libra.
- 29.Hamscher G, Sczesny S, Höper H, Nau H. Determination of Persistent Tetracycline Residues in Soil Fertilized with Liquid Manure by High-Performance Liquid Chromatography with Electrospray Ionization Tandem Mass Spectrometry. Anal Chem. 2002;74:1509–18. doi: 10.1021/ac015588m. [DOI] [PubMed] [Google Scholar]
- 30.Xie X, Zhou Q, Lin D, Guo J, Bao Y. Toxic effect of tetracycline exposure on growth, antioxidative and genetic indices of wheat (Triticum aestivum L.) Environ Sci Pollut Res Int. 2011;18:566–75. doi: 10.1007/s11356-010-0398-8. [DOI] [PubMed] [Google Scholar]