Abstract
The fruit, which develops from the fertilised gynoecium formed in the innermost whorl of the flower, is the reproductive organ and one of the most complex structures of an angiosperm plant. Phytohormones play important roles during flower and fruit patterning, morphogenesis and growth, and there is emerging evidence for a cross-talk between different classes of plant hormones throughout these processes. Here, we show that the bHLH transcription factors HECATE 1 (HEC1), HEC2 and HEC3, which have previously been identified as essential components of transmitting tract formation, affect both auxin and cytokinin responses during reproductive tissue development. We find that HEC1 interacts with SPATULA (SPT) to control carpel fusion and that both transcription factors restrict sensitivity to cytokinin in the gynoecium. In addition, HEC1 is tightly integrated into the auxin-signalling network at the levels of biosynthesis, transport and transcriptional response. Based on this data, we propose that HEC1 acts as a local modulator of auxin and cytokinin responses to control gynoecium development in Arabidopsis.
KEY WORDS: Gynoecium development, HECATE, SPATULA, Phytohormones, Auxin, Cytokinin, Arabidopsis
Highlighted article: In the developing reproductive tissue of plants, HECATE 1 and SPATULA coordinate auxin and cytokinin signalling to orchestrate the development of the gynoecium.
INTRODUCTION
The flower is the defining structure of all angiosperms and has been studied in great detail in Arabidopsis. It consists of four types of organs: The outer, leaf-like sepals and petals, and the gametophyte-producing stamens and carpels. Flower development begins with the specification of floral meristem identity in a subgroup of cells at the flank of the shoot apical meristem (SAM). Later, flower primordia emerge, become separated from the main stem cell system of the shoot apical meristem and floral organ primordia arise. Fruits originate from the female reproductive organ, the gynoecium, which consists of two fused carpels. The gynoecium is capped by specialised epidermal cells called stigmata that function in pollen reception and neighbour the cylindrical style, which connects the apical stigma with the large central ovary. The outer layers of the ovary form the valves (carpel walls) and the replum (Fig. 1A,B). At pollination (flower stage 13), pollen grains germinate on the stigma and growing pollen tubes are guided through the transmitting tract to the ovules where fertilisation takes place (Edlund et al., 2004; Ferrándiz et al., 1999; Østergaard, 2009; Smyth et al., 1990).
Fig. 1.
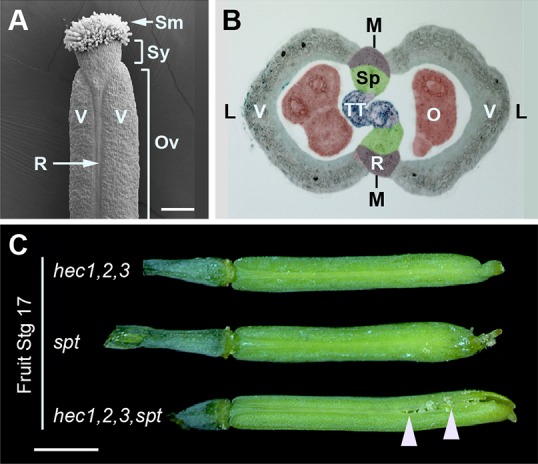
HEC1 and SPT genetically interact to control gynoecium development. (A,B) Longitudinal (A) and cross section view (B) of a wild-type Arabidopsis gynoecium. Stigma (Sm), style (Sy), ovary (Ov), valve (V), replum (R), septum (Sp), transmitting tract (TT), ovules (O), lateral (L) and medial (M) regions are shown. (C) Stage 17 fruits of hec1,2,3, spt and hec1,2,3 spt mutant plants. The apical part of spt fruits is unfused, and this phenotype is dramatically enhanced in hec1,2,3 spt quadruple mutant (arrowheads). Scale bars: 200 µm in A; 1 mm in C. See also supplementary material Fig. S1.
Phytohormones play important roles in flower and fruit development. Auxin has been shown to act as a morphogen during gynoecium formation (Nemhauser et al., 2000). Several components of auxin biosynthesis, homeostasis and signalling are active in apical-basal fruit patterning, including the efflux facilitator PIN-FORMED 1 (PIN1), the protein kinase PINOID (PID), the auxin response factor ETTIN (ETT), and the RING-finger-like proteins STYLISH 1 (STY1) and STY2 (Bennett et al., 1995; Nemhauser et al., 2000; Okada et al., 1991; Sohlberg et al., 2006). STY1 promotes the production of auxin by inducing YUCCA (YUC) gene expression, and sty1,2 double mutants can be partially rescued by exogenous auxin application (Cheng et al., 2006; Eklund et al., 2010; Ståldal et al., 2008). In addition, the specification of a local auxin minimum is crucial for the formation of the valve margin separation layer where fruit dehiscence takes place (Sorefan et al., 2009). HECATE 1 (HEC1), HEC2 and HEC3 genes are involved in transmitting tract and stigma development and code for closely related basic helix-loop-helix (bHLH) transcription factors. Overexpression of any of these genes leads to pin-like phenotypes, and consequently it has been thought that they might coordinate auxin signalling in the gynoecium, but so far no direct evidence has been reported (Gremski et al., 2007). SPATULA (SPT), another bHLH transcription factor that controls carpel margin tissue development by promoting growth of the style, stigma and septum, interacts with INDEHISCENT (IND) to control PID expression (Alvarez and Smyth, 1999; Girin et al., 2011; Heisler et al., 2001). Interestingly, both SPT and HEC genes are under negative control of ETT, which prevents expression of these genes in abaxial regions during gynoecium development (Gremski et al., 2007; Heisler et al., 2001).
In addition to the established roles for auxin, it has been suggested that also cytokinin is important for gynoecium and fruit patterning, on the one hand by promoting proliferation at earlier stages of reproductive tract development, and on the other hand during valve margin morphogenesis at later stages (Marsch-Martínez et al., 2012a). Mutations in the CYTOKININ OXIDASE/ DEHYDROGENASE 3 (CKX3) and CKX5 genes, which catalyse the degradation of cytokinin, lead to increased gynoecium size and seed yields (Bartrina et al., 2011). Most interesting, there is emerging evidence that the balance between auxin and cytokinin, which has been shown to be essential in maintaining root and shoot stem cell systems, might also play a role during the development of the female reproductive tract (Jones et al., 2010; Marsch-Martínez et al., 2012b; Müller and Sheen, 2008; Zhao et al., 2010).
Recently, we showed that HEC1 coordinates the balance between proliferation and differentiation in the shoot apical meristem by promoting stem cell proliferation, while antagonising niche cell activity through physical interaction with SPT. In the SAM, HEC1 activates the expression of several type-A ARABIDOPSIS RESPONSE REGULATOR (ARR) genes and we proposed that these negative regulators of cytokinin signalling act as mobile signals to non-cell-autonomously interfere with the expression of the stem cell regulator WUSCHEL (Schuster et al., 2014).
Here, we investigate the function of HEC1 during reproductive tissue development. Our data reveals that HEC1, in the same way as in the SAM, interacts with SPT, and that both transcriptional regulators buffer cytokinin signals in the gynoecium. We also show that HEC1 controls auxin distribution during gynoecium development, via the direct regulation of PIN1 and PIN3 auxin transporters. This mechanism does not appear to be relevant for HEC activity in shoot stem cells, illustrating an exquisite spatial specificity. Together, our data highlight the conserved function of the interaction between HEC1 and SPT in modulating cytokinin signalling in diverse plant tissues, and suggests that both transcription factors might orchestrate the cross-talk between the two essential phytohormones auxin and cytokinin during reproductive development.
RESULTS
HEC1 and SPT act together during gynoecium development
We have recently shown that HEC1 physically interacts with SPT in vivo to regulate stem cell proliferation in the SAM (Schuster et al., 2014). Because both bHLH transcription factors additionally play important roles during female reproductive development (Gremski et al., 2007; Heisler et al., 2001), we hypothesised that this interaction might also be relevant in the developing gynoecium. During Arabidopsis development, HEC1 and SPT are co-expressed in the SAM, early flower primordia and in the carpel (supplementary material Fig. S1) (Gremski et al., 2007; Heisler et al., 2001; Schuster et al., 2014). To further characterise the expression of both transcription factors during carpel and fruit development at high spatio-temporal resolution, we performed β-glucuronidase assays on reporter lines and found that the promoters of HEC1 and SPT exhibit very similar activity patterns (supplementary material Fig. S1). This result, together with the mutant phenotypes reported previously, supported the idea that HEC1 and SPT might functionally interact throughout female reproductive development. To test this interaction genetically, we went on to create the hec1,2,3 spt quadruple mutant. Both parents displayed short fruits, with hec1,2,3 being completely infertile, whereas spt sustained moderate fertility but showed unfused carpels at the stylar region (Fig. 1C) (Alvarez and Smyth, 1999; Gremski et al., 2007). The hec1,2,3 spt quadruple mutant exhibited a dramatically enhanced phenotype compared with both parents, with carpels being completely unfused at the apical part, up to one third of the entire fruit, illustrating the synergistic activities of these transcription factors (Fig. 1C). To trace the defects of triple and quadruple mutants during early morphogenesis of the gynoecium, we used scanning electron microscopy (SEM) and found that at stage 9-10, hec1,2,3 triple mutants showed a retarded growth of the gynoecial tube in the medial region compared with wild type, similar to what has been reported for spt (Fig. 2A-C) (Alvarez and Smyth, 2002). This phenotype was severely enhanced in the hec1,2,3 spt quadruple mutant (Fig. 2D). At later developmental stages, carpel fusion defects of the hec1,2,3 spt quadruple mutant became even more prominent (Fig. 2E-P) and remarkably, hec1,2,3 mutants failed to form any stigmatic papillae at the apex at the onset of stage 11 (Fig. 2E,F). Together, these results confirm that HEC and SPT genetically interact during reproductive tissue formation and provide direct evidence that HEC genes are required for carpel fusion in the developing gynoecium.
Fig. 2.
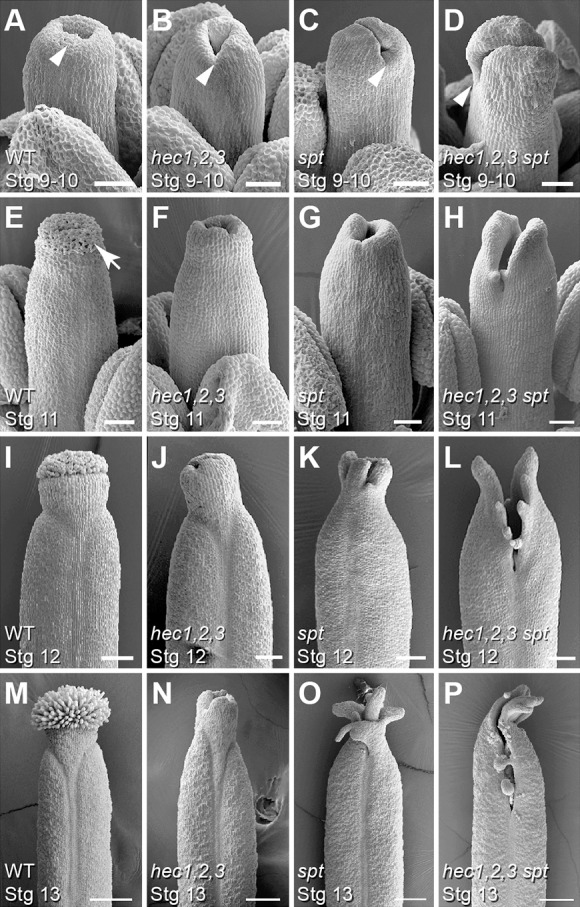
Development of the hec1,2,3, spt and hec1,2,3 spt mutant gynoecia. (A-D) Stage 9-10 gynoecia of wild type (A), hec1,2,3 (B), spt (C) and hec1,2,3 spt (D) mutant plants. Whereas the gynoecial tube in the medial regions of wild-type plants (A) is extending, hec1,2,3 (B) and spt (C) mutant gynoecia show a retarded growth in this region (arrowheads). This phenotype is enhanced in hec1,2,3 spt mutants (D). (E-H) Stage 11 gynoecia of wild type (E), hec1,2,3 (F), spt (G) and hec1,2,3 spt (H) mutant plants. Stigmatic papillae (arrow) appear at the top of the wild-type gynoecium (E), but are absent in hec1,2,3 (F). Carpel fusion defects of the hec1,2,3 spt quadruple mutant become more prominent (H). (I-P) Stage 12 (I-L) and stage 13 (M-P) gynoecia of wild type (I,M), hec1,2,3 (J,N), spt (K,O) and hec1,2,3 spt (L,P). Note that developing ovules are visible externally in the quadruple mutant (L,P). Scale bars: 50 µm in A-H; 100 µm in I-L; 200 µm in M-P.
HEC1 regulates auxin biosynthesis and transport in the gynoecium and fruit
In a recent study, it was found that SPT and IND cooperate to locally modulate auxin signalling output by directly regulating the expression of PID (Girin et al., 2011). A number of observations suggested that HEC genes could also be involved in mediating auxin signalling during gynoecium development: First, hec1,2,3 mutants showed enlarged petals and complete infertility (Fig. 3A,B; supplementary material Fig. S2) (Gremski et al., 2007), traits which have been linked to defects in the regulation of auxin transport, signalling, or synthesis in numerous studies (Cheng et al., 2006; Okada et al., 1991; Varaud et al., 2011). Second, Gremski and colleagues showed that overexpressing any of the HEC genes using the p35S promoter resulted in plants with pin-like inflorescences, very similar to pin1 and pid auxin transport mutants (supplementary material Fig. S2) (Gremski et al., 2007), suggesting that HEC regulators can potently interfere with auxin homeostasis, transport or response. Third, genetic reduction of auxin levels in yucca 1 (yuc1) yuc2 yuc6 triple mutants and yuc1 yuc4 double mutants causes fruit phenotypes very similar to hec1,2,3 (Cheng et al., 2006; Gremski et al., 2007). As the YUC genes code for flavin monooxygenases and are central components of the auxin biosynthesis pathway, decreased auxin levels might thus underlie the hec1,2,3 phenotype.
Fig. 3.
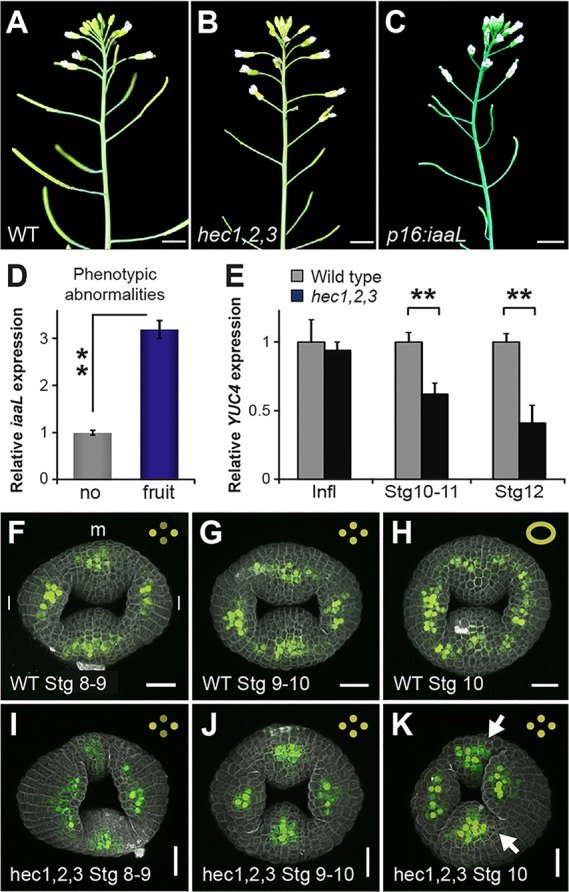
Loss of HEC function leads to impaired auxin signalling. (A-D) Wild-type (A), hec1,2,3 (B) and p16:iaaL (C) plants. Overexpressing the bacterial iaaL gene mimics the hec1,2,3 fruit phenotype. The overexpression phenotype correlates with the expression level of the iaaL transgene; plants with strong iaaL expression show distinct fruit phenotypes, whereas weak transgene expression does not cause any phenotypic alterations (D). (E) mRNA expression levels of YUC4 in dissected inflorescences (Infl) and gynoecia at stage 10-11 and stage 12 of wild type and hec1,2,3. **P<0.01. Error bars: s.d. of two (D) or three (E) biological replicates. (F-K) pDR5:3xYFP-NLS expression in the developing gynoecium of wild type (F-H) and hec1,2,3 mutants (I-K) at stage 8-9 (F,I), stage 9-10 (G,J), and stage 10 (H,K). In wild type, four regions of local auxin response are connected to a ring-like structure at the onset of stage 10 (F-H, insets), but hec1,2,3 mutants fail to establish this radial symmetry (I-K). l: lateral regions; m: medial regions. Arrows in K indicate retarded growth in the medial region. F-K, n≥9. Scale bars: 0.5 cm (A-C) and 20 µm (F-K). See also supplementary material Figs S2 and S3.
To test the connection between HEC genes and the auxin pathway experimentally, we first reduced the pool of active auxin by overexpressing the auxin conjugating enzyme iaaL and observed defects that closely resembled hec1,2,3 mutant fruit phenotypes (Fig. 3A-D). Building on this result, we analysed expression levels of YUC transcripts in wild-type and hec1,2,3 mutant inflorescences and gynoecia at multiple developmental stages to identify potential regulatory interaction between the HEC factors and YUC genes. Whereas the mRNA levels of YUC1, YUC2 and YUC6 were unaffected, YUC4 abundance was strongly reduced in hec1,2,3 throughout gynoecium development (Fig. 3E). Consistently, similar to HEC1, YUC4 is expressed at the apex of wild-type gynoecia (Fig. 4J; supplementary material Fig. S1) (Cheng et al., 2006). The specific effect on YUC4 expression indicated that the hec1,2,3 triple mutant phenotype might at least partially be caused by decreased auxin biosynthesis levels.
Fig. 4.
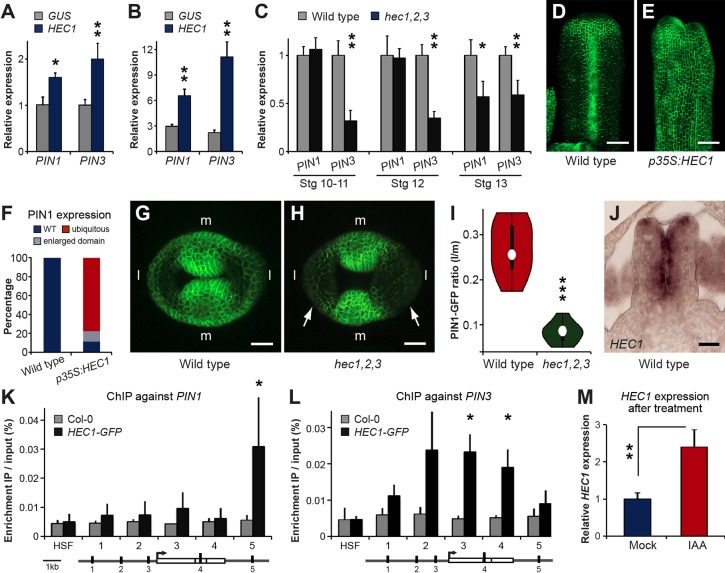
HEC1 controls PIN expression. (A-B) mRNA expression levels of PIN1 and PIN3 in inflorescences of pAlcA:GUS and pAlcA:HEC1 plants after ethanol induction (A) or ethanol induction and auxin (IAA) treatment (B) measured by qRT-PCR. (C) Expression of PIN1 and PIN3 in gynoecia of wild type and hec1,2,3 at multiple developmental stages measured by qRT-PCR. (D-F) pPIN1:PIN1-GFP activity in stage 10 fruits of wild type (D) and p35S:HEC1 (E). In contrast to wild type (D and F; n=23), p35S:HEC1 show ubiquitous PIN1-GFP expression (E and F; n=27). (G-I) Reduction of pPIN1:PIN1-GFP expression at stage 9-10 in the lateral part (l) of hec1,2,3 gynoecia (H and I, n=9 plants with 3 gynoecia imaged) compared with wild type (G and I, same sample size as hec1,2,3). Image analysis revealed a significant difference in the lateral:medial (l/m) PIN1-GFP intensity ratio (***P=3.5×10−6, GFP signal threshold=4σ). (J) HEC1 mRNA expression in wild type stage 8. (K,L) ChIP experiment against PIN1 (K) and PIN3 (L) using a stable p35S:HEC1-GFP line. HSF1 served as negative control. (M) HEC1 mRNA expression in mock and IAA-treated wild-type inflorescences. Error bars: s.d. of three (C,K-M) or four (A,B) biological replicates. *P<0.05; **P<0.01. Scale bars: 50 µm (D,E) and 20 µm (G,H,J). See also supplementary material Fig. S4.
Recently, IND, another bHLH transcription factor closely related to the HEC factors, has been shown to directly regulate auxin distribution in the fruit (Sorefan et al., 2009). Therefore, we next followed auxin responses during gynoecium and fruit development in wild type and hec1,2,3 mutants by using the pDR5:3xYFP-NLS reporter (Benková et al., 2003) to investigate defects in the spatial organisation of auxin signalling. Wild-type gynoecia showed the previously described characteristic transition from a bilateral symmetry of auxin signalling output with two medial and two lateral foci at stage 8 to a radial symmetry at stage 10, which is required for the radialisation of the style (Fig. 3F-H) (Moubayidin and Østergaard, 2014). In contrast, hec1,2,3 mutant gynoecia failed to form this ring-like DR5 expression pattern and retained a pattern with four prominent and isolated auxin signalling foci (Fig. 3I-K). This illustrated that HEC gene function is necessary for the auxin radialisation process, and provided evidence that spatially disturbed auxin signalling might underlie the observed carpel fusion, split-style and stigma developmental defects of hec1,2,3 and hec1,2,3 spt mutants. In addition, we observed a strong difference in the pDR5:3xYFP-NLS signal between wild type and hec1,2,3 in the replum of stage 17 fruits, a later stage of reproductive development at which the hec1,2,3 fruit phenotype became more prominent. In wild type, DR5:3xYFP-NLS signal was present in the replum region, but not in the separation layer at the flanks of the replum. A local auxin minimum is required for valve margin specification, and a failure in defining the valve margin separation layer leads to reduced seed dispersal, as observed in ind and spt mutants (Girin et al., 2011; Sorefan et al., 2009). In contrast to wild type, fruits of hec1,2,3, however, did not show consistent YFP signal in the central replum, but rather a moderate signal was found at the valve margin crease (supplementary material Fig. S3). Consistently, the pHEC1:GUS reporter indicated that HEC1 is expressed in the replum and septum of fruits (supplementary material Fig. S3), suggesting that HEC1 might also have a function in regulating auxin distribution or response during post-fertilisation development.
HEC1 controls PIN1 expression
Our data indicated that HEC1 activity is essential for proper spatio-temporal auxin signalling during gynoecium development. But what are the mechanisms that possibly translate HEC1 activity into specific auxin outputs apart from modulation of biosynthesis? To explore a potential function of HEC1 in the regulation of auxin transport, we performed real-time qRT-PCR analyses on inflorescences and gynoecia at multiple stages of development using the hec1,2,3 mutant as well as an inducible HEC1 allele (p35S:AlcR; AlcA:HEC1) and analysed the transcriptional response of major components of the polar auxin transport machinery. After overnight induction, PIN1 and PIN3 abundance was significantly increased in inflorescences of the pAlcA:HEC1 line compared with the respective GUS control and this effect was further enhanced by auxin co-treatment (Fig. 4A,B). Conversely, PIN3 expression was strongly reduced in hec1,2,3 compared with wild type throughout gynoecium development, and lower PIN1 levels were found from stage 13 (Fig. 4C). Interestingly, we did not observe any changes in the transcript levels of the PID and WAG2 kinases, which facilitate PIN polarisation and are directly regulated by IND, illustrating a specific regulation of different components of the auxin transport machinery by related bHLH transcription factors (supplementary material Fig. S4) (Sorefan et al., 2009). To assess the effect of the HEC regulators on PIN expression in the developing gynoecium with cellular resolution, we transformed a constitutive p35S:HEC1 construct into a stable pPIN1:PIN1-GFP reporter line (Heisler et al., 2005) and analysed stage 9/10 gynoecia where morphology was still relatively unaffected. In wild type, the PIN1-GFP fusion protein specifically accumulated in cells at the top of the gynoecium and in the presumptive replum. Gynoecia of p35S:HEC1 plants instead displayed widespread and ectopic PIN1-GFP expression, consistent with the results of the real-time qRT-PCR analysis (Fig. 4D-F). Conversely, we found a reduction of PIN1-GFP protein abundance in the apical lateral part of stage 9-10 gynoecia from hec1,2,3 mutants crossed with the pPIN1:PIN1-GFP reporter line compared with wild type, whereas medial PIN1-GFP expression was unchanged (Fig. 4G-H). Using quantitative image analysis, we could confirm that the differences in the lateral/medial PIN1-GFP accumulation ratio between wild type and hec1,2,3 were statistically significant (Fig. 4I). These findings were also consistent with the expression of HEC1 in lateral spots at the apical part of the early gynoecium (Fig. 4J).
Taken together, our results indicate that HEC activity in the developing gynoecium is necessary and sufficient to drive apical PIN1 expression. To test the directness of the HEC1-PIN1/3 regulatory interaction, we next performed chromatin immunoprecipitation (ChIP) experiments followed by qPCR using p35S:HEC1-GFP and wild-type control seedlings. In the HEC1-GFP line we found a significant enrichment of a fragment downstream of PIN1 as well as of fragments from the PIN3 promoter and 3rd intron (Fig. 4K,L). In summary, these results show that HEC1 promotes auxin transport by directly activating the expression of PIN1 and PIN3 efflux carriers. Lastly, we wondered whether HEC1 expression itself might be under control of auxin. To this end, we analysed HEC1 transcript levels in inflorescences of wild-type plants that had been treated with auxin (50 µM IAA) for 2 h. We found that HEC1 mRNA expression was elevated (Fig. 4M), demonstrating that HEC1 is tightly integrated into the auxin signalling network both at the input and output level.
Dual mode of HEC1 function in SAM maintenance and gynoecium development
Having shown on a regulatory basis that HEC1 impinges on auxin biosynthesis and transport during gynoecium development, we asked whether this mechanism is also important for the stem cell control activities of HEC1 in the SAM (Schuster et al., 2014). Because the balance between auxin and cytokinin in the centre of the meristem is essential for stem cell maintenance (Zhao et al., 2010), we tested the effects of HEC1 in a setting with greatly reduced polar auxin transport. Not only should a pin1 mutant background provide a sensitised environment for testing the effects of elevated or decreased auxin levels in stem cells, but also reveal whether HEC regulators control stem cell behaviour by modulating PIN1 expression, as in this scenario their effect should be fully supressed. Expression of HEC1 from the pCLV3 promoter in a pin1 mutant background caused massive stem cell over-proliferation, just as in wild type (Fig. 5A,D,E), demonstrating that HEC1 function in shoot stem cells is independent of PIN1 activity. Consistently, we did not observe obvious changes in PIN1 expression or localisation in SAMs of a p35S:HEC1 line that carried a pPIN1:PIN1-GFP reporter construct and showed a significantly enlarged meristem (supplementary material Fig. S5).
Fig. 5.
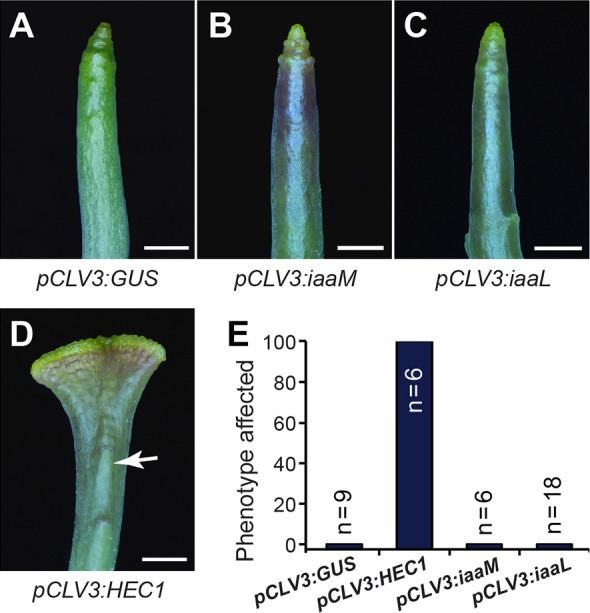
HEC1-mediated stem cell over-proliferation is independent of auxin transport and concentration. (A-D) Transgenic plants expressing GUS (A), iaaM (B), iaaL (C) or HEC1 (D) under the control of the CLV3 promoter in a pin1 mutant background. All pCLV3:HEC1-pin1 T1 lines showed enlarged meristems, while pCLV3:GUS-pin1 controls, pCLV3:iaaM-pin1 and pCLV3:iaaL-pin1 T1 lines did not (E). Scale bars: 1 mm. See also supplementary material Fig. S5.
Next, we extended our study and tested if the effects of locally reduced or enhanced auxin levels in general could influence stem cell proliferation in the shoot apex as they do affect morphogenesis of the gynoecium. To this end, we introduced pCLV3:iaaL or pCLV3:iaaM transgenes, which locally reduced or enhanced auxin content in the stem cell domain, respectively, into the pin1 mutant background (Romano et al., 1991, 1995). Neither of these constructs provoked an over-proliferation phenotype (Fig. 5B,C,E). Based on these findings, we conclude that the regulation of auxin signalling is likely to be important for the roles of HEC1 in gynoecium development, while in stem cell regulation HEC1 acts through a diverse set of transcriptional targets.
HEC1 and SPT mutants are hypersensitive to cytokinin
A major function of HEC1 in the context of the SAM is the regulation of cytokinin response by activating the expression of several type-A ARRs (Schuster et al., 2014). Interestingly, recent observations indicate that the phytohormone cytokinin is also important for ovule development, gynoecium as well as fruit patterning and morphogenesis (Bartrina et al., 2011; Marsch-Martínez et al., 2012a,b). We therefore tested whether HEC genes play a role in regulating cytokinin signalling in the gynoecium and fruit. To this end, we used a pharmacological approach and treated developing flowers of hec1,2, hec1,2,3 and spt mutants with cytokinin at levels that do not cause any phenotypic alterations in wild type (50 µM BA). While fruits of wild-type plants did not show any morphological changes (Fig. 6A,E), both hec and spt mutants were hypersensitive to cytokinin: the fruit was apically unfused and displayed extensive tissue proliferation at the top (Fig. 6B-D versus F-H). This phenotype was already present in gynoecia of cytokinin treated hec1,2,3 mutants at earlier stages of development (Fig. 6I-L). It is important to mention that extensive cytokinin treatment of wild-type plants led to massive over-proliferation at the external medial region along the entire fruit, but never led to unfused carpels and apically restricted tissue proliferation as observed in hec and spt mutants (supplementary material Fig. S6) (Marsch-Martínez et al., 2012a). Taken together, the over-proliferation phenotype of cytokinin-treated hec and spt mutants points towards a function of the HEC1-SPT module in negatively modulating cytokinin signalling during gynoecium development, in line with the reported activation of negative cytokinin signalling components by HEC1.
Fig. 6.
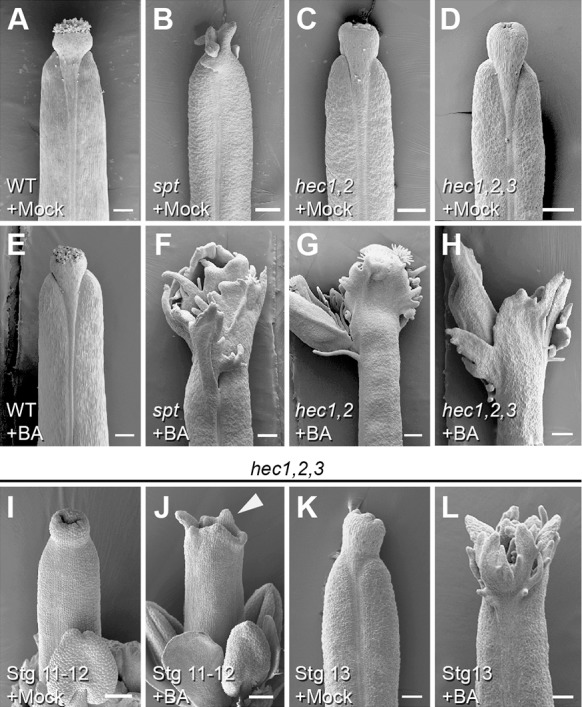
hec and spt mutants are hypersensitive to cytokinin. (A-H) Scanning electron microscopy of wild-type (A,E), spt (B,F), hec1,2 (C,G) and hec1,2,3 (D,H) fruits after mock (A-D) or cytokinin (50 µM BA) (E-H) treatment. Cytokinin treatment of spt, hec1,2 and hec1,2,3 mutants lead to apically unfused fruits showing ectopic tissue proliferation (F-H), whereas fruits of wild-type plants do not display any phenotypic alterations after treatment compared with mock controls (A,E). SEM images show stage 17b fruits, except panel B (stage 15). (I-L) hec1,2,3 mutant gynoecia stage 11-12 (I,J) and stage 13 (K,L) after mock (I,K) and cytokinin (J,L) treatment. The arrowhead in J indicates the extensions at the top of the gynoecium. Scale bars: 200 µm (A-H) and 100 µm (I-L). See also supplementary material Fig. S6.
DISCUSSION
HEC1 and SPT interact to control gynoecium development
Our phenotypic analysis of the hec1,2,3 spt quadruple mutant showed that HEC1 and SPT genetically interact during gynoecium development. Combined with the significant overlap of their spatio-temporal expression pattern and the previously reported protein interaction both in yeast and in planta (Gremski et al., 2007; Schuster et al., 2014), this clearly points towards a role for the HEC1-SPT complex during female reproductive development. It should be pointed out that hec and spt mutants show shared as well as clearly distinct phenotypes. Whereas both hec and spt mutants exhibit short fruits and reduced fertility as a result of disturbed gynoecium development (Alvarez and Smyth, 1999; Gremski et al., 2007; Heisler et al., 2001), the most obvious difference is that hec1,2,3 plants, in contrast to spt mutants, have only minor defects related to gynoecium fusion. The combination of hec1,2,3 and spt, however, severely enhanced this aspect of the spt mutant phenotype. A comparable phenomenon was observed in alcatraz-1 (alc-1) spt-2 double mutants. Here, similar to hec1,2,3, alc-1 alone did not show any carpel fusion defects, but the double mutant exhibited a substantial enhancement of this particular phenotype (Groszmann et al., 2011). One possible scenario to explain this would involve the functional redundancy of HEC genes with other bHLH transcription factors, including SPT, during female reproductive development. Interestingly, we previously showed that in the context of the shoot apical meristem, HEC function is the restricting component of the HEC1-SPT module, but during gynoecium patterning, SPT seems to be limiting. It will be important to analyse the role of ALC in this regulatory system in future studies. As the margins of the two fused carpels display meristematic characteristics, the role of HEC1 in carpel fusion presented in this study fits well with its function as a regulator of cell proliferation in the SAM. Taken together, we have demonstrated the importance of the HEC-SPT module in the developing gynoecium and provide evidence for a yet unknown role of the HEC genes during carpel fusion.
HEC1 regulates phytohormone responses in the developing gynoecium
Phytohormones are known to play key roles during flower and fruit development. Whereas the importance of auxin and gibberellin is well established, the role of cytokinin function in reproductive tissue development is less well understood (Arnaud et al., 2010; Daviere and Achard, 2013; Østergaard, 2009). Recent studies demonstrate that cytokinin promotes cell proliferation in early reproductive tract development and regulates valve margin morphogenesis at later stages (Bartrina et al., 2011; Marsch-Martínez et al., 2012a,b). Here, we found that both hec and spt mutants are hypersensitive to cytokinin treatment. As HEC1/2/3 can activate type-A ARR genes, which are negative regulators of cytokinin signalling, this observation suggests that HEC transcription factors function by restricting cytokinin responses during gynoecium development, as supported by the massive tissue proliferation at the apex of the gynoecium in hec1,2,3 upon cytokinin treatment.
In addition to the regulation of cytokinin responses, we also found that HEC1 modulates auxin biosynthesis and distribution in the gynoecium by activating the expression of YUC4, as well as PIN1 and PIN3 genes, respectively. Interestingly, a previous study demonstrated that SPT interacts with IND to regulate the expression of PID and thus ultimately controls polar localisation of PIN proteins (Fig. 7) (Girin et al., 2011). This nicely demonstrates how related bHLH transcription factors can control different components of the same signalling pathway. We propose that the lack of PIN1 expression in the apical lateral part of early gynoecia observed in hec1,2,3 mutants might prevent the establishment of the auxin radial symmetry, which is required for the radialisation of the style (Moubayidin and Østergaard, 2014). In addition to the role of HEC genes in regulating auxin signalling during gynoecium development, we also found evidence for an auxin-mediated function of HECs during later stages of female reproductive development. Future experiments should further analyse the in vivo relevance of the auxin-HEC regulatory interaction. This could be done by artificially expressing the auxin synthesis gene iaaM under the control of the HEC1 promoter and searching for a hec1,2,3 mutant fruit phenotype rescue, analogous to previous work done on yuc1,2,6 (Cheng et al., 2006). Interestingly, in contrast to the regulation of cytokinin signalling, which represents a common feature of HEC function in both SAM and gynoecium, the control of auxin distribution seems to be specific for reproductive development.
Fig. 7.
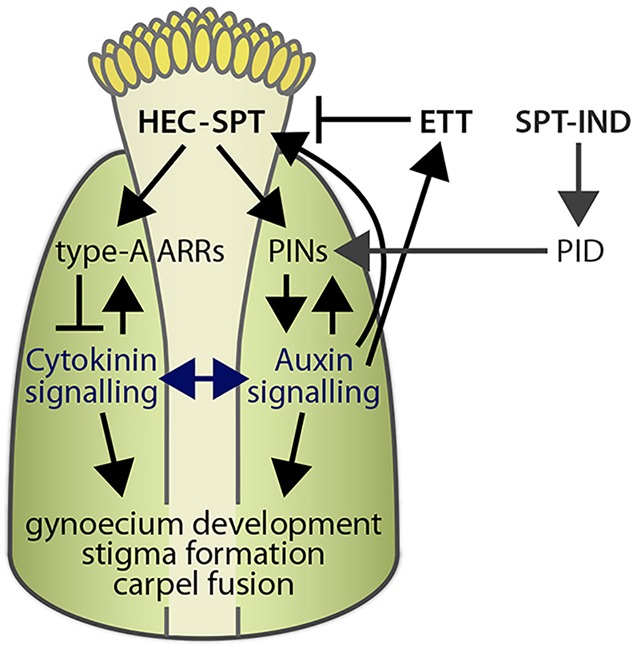
Hypothetical model of HEC gene function during gynoecium development. HEC1 interacts with SPT to control carpel fusion, and both transcription factors buffer auxin and cytokinin signals during gynoecium development. This might involve type-A ARRs, which antagonise cytokinin function. HEC1 stimulates auxin biosynthesis and directly activates the expression of PIN1 and PIN3 auxin efflux transporters and thus ultimately regulates auxin distribution during early stages of gynoecium development. Interestingly, the SPT-IND complex binds to the promoter of the PID gene that modulates PIN polarisation. This highlights how combinatorial effects of related bHLH transcription factors regulate distinct components of the auxin signalling machinery. Finally, HEC1 itself is tightly integrated into the auxin signalling network, and its spatial expression seems to be partly controlled by auxin-dependent activation and ETT mediated repression. Cross-talk between auxin and cytokinin pathways is an important feature of shoot meristem control and might also play a role in the developing gynoecium and fruit.
Cross-talk between auxin and cytokinin is important for the control of both root and shoot stem cell systems, and has also been proposed to play a role in gynoecium morphogenesis (Marsch-Martínez et al., 2012b; Müller and Sheen, 2008; Zhao et al., 2010). Besides the apical-basal gradient of auxin with high levels of auxin at the top of the gynoecium and low levels at the bottom (Nemhauser et al., 2000), a reverse gradient could exist for cytokinin with a maximum concentration at the basal and a minimum concentration at the apical end (Østergaard, 2009; Sundberg and Østergaard, 2009). Because HEC1 can impinge on both auxin and cytokinin signalling, it needs to be further elucidated whether HEC1 functions as a central hub to balance the local response ratio between both hormones.
In summary, based on the results presented in this study and previous work, we suggest the following working model (Fig. 7): HEC1/2/3 transcription factors control gynoecium development in conjunction with SPT by balancing phytohormone responses, most notably auxin and cytokinin. First, these factors act together, at least in part, by modulating cytokinin action, potentially through the activation of type-A ARR genes. Second, HEC1/2/3 control YUC4 and PIN expression and thus ultimately local auxin signalling; we propose that this function is important for gynoecium development, but might also play a role in the development of the fruit. Strikingly, HEC genes themselves are tightly integrated into the auxin signalling network: auxin stimulates the expression of HEC1, but also limits HEC and SPT activity through ETT function (Gremski et al., 2007; Heisler et al., 2001). The specific mode of each hormone action influenced by HEC transcription factors needs to be elucidated in future studies.
MATERIALS AND METHODS
Plant material and treatments
Plants of Columbia background were grown at 23°C in long days. Ethanol vapour inductions were performed overnight by placing a tip-box filled with 95% ethanol into the plant tray. For inductions with IAA, inflorescences were incubated for 2 h in ½ MS medium containing 50 µM IAA and 0.015% Silwet L-77; 0.1% dimethylsulfoxide (DMSO). 0.015% Silwet L-77 in ½ MS was used as control. Cytokinin treatments were performed by spraying 50 µM 6-benzyladenine (BA) once a week on inflorescences during a 3-week period after bolting.
The spt allele corresponds to spt-12 (Ichihashi et al., 2010), the pSPT:GUS reporter line is pSPT:6253-GUS (Groszmann et al., 2010) and the hec1,2,3 triple mutant was previously described (Schuster et al., 2014).
Transgenes
The HEC1 coding sequence was amplified using Gateway tailed primers and cloned into pGEM-T Easy (Promega) for sequencing. For generating constitutive overexpression constructs it was then recombined into pDONR221 using Gateway Technology (Invitrogen) and further recombined into pGREEN II destination vectors carrying tissue-specific promoters. The same procedures were used for making constructs of GUS control, iaaL and iaaM expression. The ethanol-inducible HEC1 line and the p16 promoter are described in Schuster et al. (2014). To assess auxin signalling activity, the DR5 reporter driving the expression of 3xYFP-NLS was transformed into wild type and hec1,2,3. For monitoring PIN1 expression upon alterations of HEC1 activity, p35S:HEC1 was transformed into a stable pPIN1:PIN1-GFP line [provided by Marcus Heisler (Heisler et al., 2005)], and hec1,2,3 triple mutants were crossed with the same PIN1 reporter line.
Quantitative real-time RT-PCR
qRT-PCR was performed on dissected inflorescence apices and on gynoecia at multiple developmental stages. Tissue was collected in microcentrifuge tubes floating on liquid nitrogen. Twenty plant samples were pooled for each replicate, and RNA was prepared using the RNeasy Plant Mini Kit (Qiagen). After DNase treatment, cDNA was prepared from 1 µg total RNA using the RevertAid First Strand cDNA Synthesis Kit (Fermentas). Quantitative real-time RT-PCR was carried out using SYBR Green and amplification of TUBULIN served as control. Sequences for all primers are listed in supplementary material Table S1.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed according to (Gendrel et al., 2005) with minor modifications. Col-0 and p35S:HEC1-GFP seedlings were harvested 12 days after germination. Overnight immunoprecipitation was performed using GFP-trap (Chromotek) and DNA isolation was conducted using MinElute Reaction Cleanup Kit (Qiagen). For each individual biological replicate, technical duplicates were obtained by splitting the samples after sonication, and by processing them separately in the subsequent steps.
Microscopy
Confocal laser scanning microscopy, scanning electron microscopy and GUS staining were performed in accordance with standard protocols. The lateral:medial expression ratio of the pPIN1:PIN1-GFP reporter in gynoecia of wild type and hec1,2,3 mutant was determined by thresholding the GFP signal intensity to the tissue background intensity of the same image. The threshold was determined by the mean of the background intensity plus four standard deviations.
Statistical analysis
For statistical analysis, data was first tested for normality using the Shapiro–Wilk test. Then, means were compared pair-wise using either Welch's t-test or the Wilcoxon rank sum test. All calculations were performed in R.
Acknowledgements
We thank Remko Offringa, University of Leiden, Netherlands, for providing the iaaL transgene, Marcus Heisler, EMBL Heidelberg, Germany, for the pPIN1:PIN1-GFP construct and seeds, Marty Yanofsky, University of California San Diego, USA, for hec1,3 seeds, and David Smyth, Monash University, Australia, for the pSPT:GUS line. We also thank Jeremy Skepper from the Cambridge Advanced Imaging Centre, Raymond Wightman and the Microscopy Core Facility at the Sainsbury Laboratory, University of Cambridge, for help with SEM, Elliot Meyerowitz for generous support at the Sainsbury Laboratory, and the members of the Lohmann lab for critically reading the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
C.S., C.G. and J.U.L. designed the research and analysed the data; C.G. performed ChIP assays, cLSM analyses, generated the hec1,2,3 spt quadruple mutant and commented on the manuscript; C.S. performed all other experiments, initiated this study, and prepared and revised the manuscript together with J.U.L.
Funding
This research was supported by Deutsche Forschungsgemeinschaft (DFG) grant [SFB873] and European Research Council (ERC) grant [282139] ‘StemCellAdapt awarded to J.U.L., a PhD fellowship from the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (C.G.), and Gatsby Charitable Trust grants [GAT3272/C and GAT3273-PR1] awarded to E.M.M. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.120444/-/DC1
References
- Alvarez J. and Smyth D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377-2386. [DOI] [PubMed] [Google Scholar]
- Alvarez J. and Smyth D. R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163, 17-41. 10.1086/324178 [DOI] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T. A., Lawrenson T., Sablowski R. and Ostergaard L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24, 2127-2132. 10.1101/gad.593410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I., Otto E., Strnad M., Werner T. and Schmulling T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69-80. 10.1105/tpc.110.079079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G. and Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591-602. 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- Bennett S. R. M., Alvarez J., Bossinger G. and Smyth D. R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505-520. 10.1046/j.1365-313X.1995.8040505.x [DOI] [Google Scholar]
- Cheng Y., Dai X. and Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790-1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviere J.-M. and Achard P. (2013). Gibberellin signaling in plants. Development 140, 1147-1151. 10.1242/dev.087650 [DOI] [PubMed] [Google Scholar]
- Edlund A. F., Swanson R. and Preuss D. (2004). Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16 Suppl., S84-S97. 10.1105/tpc.015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund D. M., Staldal V., Valsecchi I., Cierlik I., Eriksson C., Hiratsu K., Ohme-Takagi M., Sundstrom J. F., Thelander M., Ezcurra I. et al. (2010). The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22, 349-363. 10.1105/tpc.108.064816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Pelaz S. and Yanofsky M. F. (1999). Control of carpel and fruit development in arabidopsis. Annu. Rev. Biochem. 68, 321-354. 10.1146/annurev.biochem.68.1.321 [DOI] [PubMed] [Google Scholar]
- Gendrel A.-V., Lippman Z., Martienssen R. and Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213-218. 10.1038/nmeth0305-213 [DOI] [PubMed] [Google Scholar]
- Girin T., Paicu T., Stephenson P., Fuentes S., Korner E., O'Brien M., Sorefan K., Wood T. A., Balanza V., Ferrandiz C. et al. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23, 3641-3653. 10.1105/tpc.111.090944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K., Ditta G. and Yanofsky M. F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134, 3593-3601. 10.1242/dev.011510 [DOI] [PubMed] [Google Scholar]
- Groszmann M., Bylstra Y., Lampugnani E. R. and Smyth D. R. (2010). Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 61, 1495-1508. 10.1093/jxb/erq015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M., Paicu T., Alvarez J. P., Swain S. M. and Smyth D. R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68, 816-829. 10.1111/j.1365-313X.2011.04732.x [DOI] [PubMed] [Google Scholar]
- Heisler M. G., Atkinson A., Bylstra Y. H., Walsh R. and Smyth D. R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089-1098. [DOI] [PubMed] [Google Scholar]
- Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., Long J. A. and Meyerowitz E. M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899-1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Ichihashi Y., Horiguchi G., Gleissberg S. and Tsukaya H. (2010). The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51, 252-261. 10.1093/pcp/pcp184 [DOI] [PubMed] [Google Scholar]
- Jones B., Gunneras S. A., Petersson S. V., Tarkowski P., Graham N., May S., Dolezal K., Sandberg G. and Ljung K. (2010). Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22, 2956-2969. 10.1105/tpc.110.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martínez N., Ramos-Cruz D., Irepan Reyes-Olalde J., Lozano-Sotomayor P., Zúñiga-Mayo V. M. and de Folter S. (2012a). The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 72, 222-234. 10.1111/j.1365-313X.2012.05062.x [DOI] [PubMed] [Google Scholar]
- Marsch-Martínez N., Reyes-Olalde J. I., Ramos-Cruz D., Lozano-Sotomayor P., Zúñiga-Mayo V. M. and de Folter S. (2012b). Hormones talking: does hormonal cross-talk shape the Arabidopsis gynoecium? Plant Signal. Behav. 7, 1698-1701. 10.4161/psb.22422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L. and Østergaard L. (2014). Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr. Biol. 24, 2743-2748. 10.1016/j.cub.2014.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B. and Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094-1097. 10.1038/nature06943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J. L., Feldman L. J. and Zambryski P. C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877-3888. [DOI] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M. K., Bell C. J. and Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of arabidopsis floral bud formation. Plant Cell 3, 677-684. 10.1105/tpc.3.7.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard L. (2009). Don't ‘leaf’ now. The making of a fruit. Curr. Opin. Plant Biol. 12, 36-41. 10.1016/j.pbi.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Romano C. P., Hein M. B. and Klee H. J. (1991). Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev. 5, 438-446. 10.1101/gad.5.3.438 [DOI] [PubMed] [Google Scholar]
- Romano C. P., Robson P. R. H., Smith H., Estelle M. and Klee H. J. (1995). Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27, 1071-1083. 10.1007/BF00020881 [DOI] [PubMed] [Google Scholar]
- Schuster C., Gaillochet C., Medzihradszky A., Busch W., Daum G., Krebs M., Kehle A. and Lohmann J. U. (2014). A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28, 438-449. 10.1016/j.devcel.2014.01.013 [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L. and Meyerowitz E. M. (1990). Early flower development in arabidopsis. Plant Cell 2, 755-767. 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg J. J., Myrenås M., Kuusk S., Lagercrantz U., Kowalczyk M., Sandberg G. and Sundberg E. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 47, 112-123. 10.1111/j.1365-313X.2006.02775.x [DOI] [PubMed] [Google Scholar]
- Sorefan K., Girin T., Liljegren S. J., Ljung K., Robles P., Galván-Ampudia C. S., Offringa R., Friml J., Yanofsky M. F. and Østergaard L. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459, 583-586. 10.1038/nature07875 [DOI] [PubMed] [Google Scholar]
- Ståldal V., Sohlberg J. J., Eklund D. M., Ljung K. and Sundberg E. (2008). Auxin can act independently of CRC, LUG, SEU, SPT and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytol. 180, 798-808. 10.1111/j.1469-8137.2008.02625.x [DOI] [PubMed] [Google Scholar]
- Sundberg E. and Østergaard L. (2009). Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. 1, a001628 10.1101/cshperspect.a001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaud E., Brioudes F., Szecsi J., Leroux J., Brown S., Perrot-Rechenmann C. and Bendahmane M. (2011). AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23, 973-983. 10.1105/tpc.110.081653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Andersen S. U., Ljung K., Dolezal K., Miotk A., Schultheiss S. J. and Lohmann J. U. (2010). Hormonal control of the shoot stem-cell niche. Nature 465, 1089-1092. 10.1038/nature09126 [DOI] [PubMed] [Google Scholar]