Abstract
The unique cell cycle dynamics of meiosis are controlled by layers of regulation imposed on core mitotic cell cycle machinery components by the program of germ cell development. Although the mechanisms that regulate Cdk1/Cyclin B activity in meiosis in oocytes have been well studied, little is known about the trans-acting factors responsible for developmental control of these factors in male gametogenesis. During meiotic prophase in Drosophila males, transcript for the core cell cycle protein Cyclin B1 (CycB) is expressed in spermatocytes, but the protein does not accumulate in spermatocytes until just before the meiotic divisions. Here, we show that two interacting proteins, Rbp4 and Fest, expressed at the onset of spermatocyte differentiation under control of the developmental program of male gametogenesis, function to direct cell type- and stage-specific repression of translation of the core G2/M cell cycle component cycB during the specialized cell cycle of male meiosis. Binding of Fest to Rbp4 requires a 31-amino acid region within Rbp4. Rbp4 and Fest are required for translational repression of cycB in immature spermatocytes, with Rbp4 binding sequences in a cell type-specific shortened form of the cycB 3′ UTR. Finally, we show that Fest is required for proper execution of meiosis I.
KEY WORDS: Drosophila, Spermatogenesis, Meiosis, Cyclin B, Translational control
Summary: The translation timing of Cyclin B1 RNA during meiotic prophase in the male Drosophila germline is regulated by two interacting proteins, Rbp4 and Fest.
INTRODUCTION
Meiosis, the pair of specialized cell divisions required to convert germline diploid progenitor cells into haploid gametes, is an essential process for sexual reproduction in eukaryotes. After pre-meiotic DNA replication, germ cells enter an extended G2 cell cycle phase, termed meiotic prophase, during which homologous chromosomes pair and interact, and an extensive, cell type-specific transcription program turns on to set up gamete differentiation. The homologs then segregate to different daughter cells, commonly during the first meiotic division, followed by segregation of sister chromatids during meiosis II without an intervening S phase. As in mitosis, the timing of key cell cycle events is choreographed by regulated activation and deactivation of cyclin-dependent kinase (Cdk) complexes, in which cyclins play key roles in regulating the timing and targets of Cdk activity. B-type cyclins in particular are instrumental to negotiating the G2/M transition in both mitosis and meiosis.
The developmental program that specifies germ cell differentiation imposes additional layers of regulation on core cell cycle regulatory circuitry components such as the cyclins to produce the specialized cell cycles of meiosis. Although the events that regulate progression from G2 to M phase of the first meiotic division during oocyte maturation and activation have been studied extensively, much less is known about how the meiotic cell cycle is controlled in male germ cells [Lamitina and L'Hernault, 2002; Sun et al., 2010; and reviewed by Wolgemuth et al. (2013)]. Sporulation in yeast may arguably be compared to spermatogenesis in animals: in both cases the meiotic divisions take place prior to terminal differentiation. Recent studies in yeast have revealed stage-specific translational repression followed by activation of expression of Cyclin B3 (Clb3) during meiosis (Carlile and Amon, 2008), although the trans-acting factors acting on the CLB3 RNA to regulate translation are not known.
Here, we show that expression of Cyclin B1 (CycB) is under translational repression by action of two interacting factors expressed under control of the germ cell developmental program during meiosis I in male germ cells in Drosophila. In the Drosophila testis, male germline stem cells at the apical tip produce new germline stem cells and daughter cells termed gonialblasts that initiate differentiation. Each gonialblast becomes enclosed in two somatic cyst cells, then embarks on four rounds of mitosis, producing a cyst of 16 interconnected germ cells, which then undergo premeiotic DNA synthesis and together enter meiotic prophase. During the 3.5 days of meiotic prophase, the resulting spermatocytes grow 25-fold in volume and turn on an extensive transcription program in preparation for spermatid differentiation (Fuller, 1993; Kierszenbaum and Tres, 1978). At the completion of meiotic prophase, the spermatocytes undergo in rapid succession the two meiotic divisions, producing a cyst of 64 haploid round spermatids (Fig. 1A). The male germ cells up through the meiotic divisions are displayed in rough developmental order along the first third of the testis, with bundled flagella of elongating spermatid cysts extending up through the testis lumen.
Fig. 1.
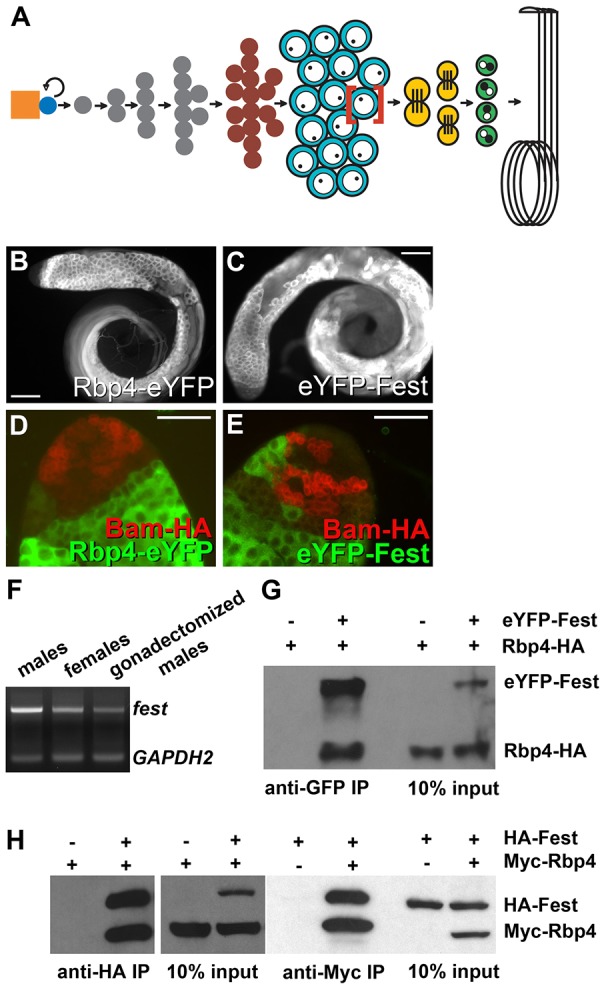
Rbp4 and Fest are expressed shortly after the onset of spermatocyte development and interact physically. (A) Schematic of germ cell development in the Drosophila male. Due to space constraints, only one spermatocyte (red brackets) is shown advancing through the meiotic and post-meiotic stages. Orange, hub; blue, germline stem cell; gray, spermatogonia; dark red, germ cells undergoing premeiotic S DNA replication; turquoise, spermatocytes; yellow, meiotic divisions; green, round spermatids. (B,C) Anti-GFP immunofluorescence of testes from flies expressing (B) Rbp4-eYFP or (C) eYFP-Fest. Scale bars: 100 µm. (D,E) Anti-GFP (green) and anti-HA (red) immunofluorescence on testes from flies expressing (D) Rbp4-eYFP, Bam-HA or (E) eYFP-Fest, Bam-HA. Scale bars: 50 µm. (F) RT-PCR on fest transcript from RNA from males, females and gonadectomized males. Control: GAPDH2. (G) Anti-GFP, anti-HA western blot of anti-GFP immunoprecipitations from testes of Rbp4-HA flies and eYFP-Fest, Rbp4-HA flies. (H) Anti-HA, anti-Myc western blot of anti-HA immunoprecipitations (left panels) or anti-Myc immunoprecipitations (right panels) from S2 cells transfected with either HA-Fest, Myc-Rbp4 or both, as indicated.
CycB protein is expressed in spermatogonia in the mitotic region at the apical tip, but is downregulated in immature and growing spermatocytes by translational repression, as we show in this study. The cycB RNA had previously been shown to be expressed at low levels at the tip of the testis, where spermatogonial cells are undergoing mitotic proliferation, downregulated after completion of the mitotic divisions, then re-expressed at high levels in primary spermatocytes, during the spermatocyte growth period (White-Cooper et al., 1998). By contrast, the level of CycB protein in spermatocytes remained low until just before spermatocytes entered the G2/MI transition, a nearly three-day delay. CycB protein began to accumulate in the spermatocyte cytoplasm just before chromatin condensation initiated, was high at prometaphase, then entered the nucleus and was almost immediately degraded at metaphase (White-Cooper et al., 1998).
Here, we identify the predicted RNA binding protein Rbp4 and the novel protein Fest as cell type-specific repressors of cycB translation during meiotic prophase in growing spermatocytes. In flies mutant for either rbp4 or fest, CycB protein accumulates prematurely, in immature spermatocytes. Loss of function of Rbp4 [formerly published under the name Testis-specific RRM protein, or Tsr, but now termed Rbp4 (FlyBase)] was previously shown to result in male sterility and abnormally high levels of several proteins in the Drosophila testis (Haynes et al., 1997), but the proteins/RNA targets were not identified. We show that Rbp4 binds a short form of the cycB 3′ UTR expressed during male meiosis, and that conserved sequences within the short 3′ UTR are required for translational repression of an epitope-tagged CycB reporter in early spermatocytes in vivo. In addition, we show that Fest is required for spermatocytes to progress correctly into metaphase of meiosis I, and that a 31-amino acid region of Rbp4 is required for binding of Fest. Both Rbp4 and Fest are expressed at the onset of the spermatocyte transcription program, so that by the time the cycB RNA is transcribed in spermatocytes, it enters a cytoplasm prepared by the developmental program for proper regulation of this key component of the cell cycle machinery for meiosis I.
RESULTS
Two interacting proteins are expressed soon after the onset of spermatocyte differentiation
The RRM class predicted RNA-binding protein Rbp4 is expressed cell type-specifically under control of the developmental program for male gametogenesis. Examination of FlyAtlas, a survey of expression data from 25 adult and larval Drosophila tissues, indicated that rbp4 transcript was significantly expressed only in testis (Chintapalli et al., 2007), consistent with the northern blot data published by Haynes and colleagues (Haynes et al., 1997). Within the testis, analysis of a C-terminal-tagged Rbp4 in vivo reporter expressed under control of rbp4 genomic regulatory sequences (Materials and Methods) revealed expression of Rbp4-eYFP in very early spermatocytes and subsequent germ cell types through early elongating spermatid stages (Fig. 1B). In all cases the Rbp4-eYFP fusion protein was cytoplasmic. The stages of expression and subcellular localization of the Rbp4-eYFP reporter were similar to the distribution of Rbp4 (Tsr) protein described by Haynes and colleagues, based on immunohistochemical staining with an anti-Rbp4 antibody (anti-Tsr, no longer available, Haynes et al., 1997).
The novel protein Fest, encoded by CG9975, implicated as a putative binding partner of Rbp4 (BioGRID, Giot et al., 2003), is also expressed starting in early spermatocytes and continuing into later stages of germ cell differentiation, similar to the expression pattern of Rbp4. An eYFP-Fest in vivo reporter expressed from the fest promoter (Materials and Methods) drove expression of cytoplasmic eYFP-Fest, starting in immature spermatocytes, with the reporter protein remaining detectable in mature spermatocytes and later stages (Fig. 1C).
Double-staining for either Rbp4-eYFP or eYFP-Fest and the spermatogonial differentiation marker Bam revealed that expression of Rbp4-eYFP and eYFP-Fest initiated in young primary spermatocytes, soon after completion of pre-meiotic S phase. Bam protein is expressed in 4-, 8- and early 16-cell germline cysts, and is downregulated upon completion of pre-meiotic S phase (Insco et al., 2009). Immunostaining of testes from flies carrying a Bam-HA genomic transgene and either the Rbp4-eYFP or eYFP-Fest reporter with anti-HA and anti-GFP revealed a small gap between downregulation of Bam-HA in young 16-cell cysts and the onset of Rbp4-eYFP or eYFP-Fest expression (Fig. 1D,E), indicating that expression of the Rbp4 and Fest proteins began after completion of pre-meiotic DNA replication.
FlyAtlas indicated robust expression of the fest/CG9975 RNA in testis (Chintapalli et al., 2007). RT-PCR of fest from males, females and gonadectomized males showed that fest transcript expression was enriched in, but not exclusive to, the testis (Fig. 1F). The Fest protein had no currently recognized domains, based on PROSITE analysis. However, the C-terminal half (aa239-494) of Fest was conserved in other insects and in nematodes, with 22% identity and 39% homology to C. elegans hypothetical protein C44B9.2.
Co-immunoprecipitation studies confirmed that the Rbp4 and Fest proteins physically interact. Immunoprecipitation of eYFP-Fest with anti-GFP brought down Rbp4-HA (identical to Rbp4-eYFP except for the epitope tag) from testis extracts from flies carrying both transgenes. Rbp4-HA was not detected in control immunoprecipitations with anti-GFP from testis extracts from flies carrying Rbp4-HA alone (Fig. 1G). In addition, Myc-tagged Rbp4 and HA-tagged Fest co-immunoprecipitated from extracts of Drosophila S2 cells transiently transfected with Myc-Rbp4 and HA-Fest, indicating that the interaction did not require additional testis-specific proteins. Immunoprecipitation with anti-HA brought down Myc-Rbp4 from cells transfected with both Myc-Rbp4 and HA-Fest but not from extracts of control cells transfected with Myc-Rbp4 alone (Fig. 1H, left panels). The reverse was also true: immunoprecipitation with anti-Myc antibody brought down HA-Fest from extracts co-transfected with HA-Fest and Myc-Rbp4 but not from control cells transfected with HA-Fest alone (Fig. 1H, right panel).
Rbp4 and Fest block accumulation of CycB protein in early spermatocytes
Function of Rbp4 and Fest is required in spermatocytes for the normal delay in CycB protein accumulation during meiotic prophase in males. Loss of function of rbp4 was generated by making flies trans-heterozygous for the deficiency Df(3R)Exel6169 and an rbp4 allele (rbp4LL06910) caused by the PBac{SAstopDsRed}LL06910 transposon insertion (for details see Materials and Methods). Loss of function of fest was generated by making flies trans-heterozygous for the deficiency Df(2R)BSC26 and a CRISPR-generated 8-nucleotide deletion (Materials and Methods) early in the fest coding region that results in truncation of the Fest protein after only 8 amino acids. In wild-type testes, CycB protein was low in immature spermatocytes (Fig. 2A, yellow lines), then accumulated to high levels in late spermatocytes just before they enter the meiotic divisions (Fig. 2A, arrowheads), as described by White-Cooper et al. (1998). In either rbp4 or fest mutant testes, however, CycB protein was detected even in immature spermatocytes near the spermatogonial region of the testis (Fig. 2B,C, yellow lines). Similar premature expression of CycB in immature spermatocytes was also observed when either rbp4 or fest was knocked down by RNAi in late spermatogonia and spermatocytes under control of a bamGal4 driver (supplementary material Fig. S1 and supplementary Materials and Methods), indicating that the requirement for Rbp4 and Fest function is cell-autonomous to the germline. The small gap between mitotic CycB expression and the premature CycB expression in immature spermatocytes in rbp4 and fest is probably due to the timing of re-initiation of cycB transcription in spermatocytes. Expression of CycB protein in immature spermatocytes in testes from rbp4 or fest mutant males was not due to a noticeable change in cycB RNA levels, as assayed by in situ hybridization (Fig. 2E,F versus D).
Fig. 2.

Rbp4 and Fest repress CycB accumulation in immature spermatocytes. (A-C) Anti-CycB immunofluorescence on wild-type (A), rbp4 mutant (B) and fest mutant testes (C). Yellow lines mark the region containing immature spermatocytes. Scale bar: 100 µm. (D-F) In situ hybridization on wild-type (D), rbp4 mutant (E) and fest mutant (F) testes with antisense cycB probe. Scale bar: 100 µm. (G-I) Anti-PH3Thr3 immunofluorescence on wild-type (G), rbp4 mutant (H) and fest mutant (I) testes. Scale bars: 100 µm. (G′,H′) Higher magnification of boxed areas in G,H. Scale bars: 20 µm.
Surprisingly, premature expression of CycB protein in rbp4 mutant males did not dramatically alter the timing of entry into the meiotic divisions. Immunofluorescence with antibody against phospho-histone3-Thr3 (PH3Thr3), a marker of dividing cells, showed signal in wild-type (Fig. 2G,G′) and rbp4 mutant (Fig. 2H,H′) testes, without a dramatic difference in positioning of the PH3Thr3+ cysts relative to the apical tip of the testis. No PH3Thr3 signal was observed in the fest mutant spermatocytes (Fig. 2I), suggesting that these cells not only do not undergo metaphase of meiosis I prematurely but do not undergo it at all.
Fest is required to set up a metaphase spindle in meiosis I
Loss of fest function resulted in a distinctive germ cell arrest phenotype, with no sperm or even elongating spermatids observed. Male germ cells from flies bearing a tubulin-GFP transgene were viewed by phase-contrast microscopy in unfixed testis squash preparations combined with fluorescence microscopy to visualize tubulin (GFP) and chromatin (Hoechst dye). In wild-type control testes, cells that had advanced to prometaphase of meiosis I showed condensed chromosomes, a rounded nucleus and asters appearing at the cell poles (Fig. 3A-A″). Wild-type cells in metaphase had condensed bivalent chromosomes bunched tightly together in a single knot (Fig. 3B″), whereas wild-type cells in anaphase had separated their chromosomes to opposite poles (Fig. 3C″). Metaphase and anaphase spindles from wild-type males were well organized, barrel-shaped and clearly nucleated at the poles (Fig. 3B′,C′). By contrast, spermatocytes from fest mutant flies did not appear to approach metaphase normally. Instead, the fest mutant spermatocytes appeared to arrest with bivalent chromosomes paired, tightly condensed and congressed near the center of the cell (Fig. 3D″). Although the condensed bivalents appeared very rarely in a tight knot (Fig. 3E″), the same cells never showed prometaphase asters or a metaphase spindle. Instead of forming a barrel-shaped spindle, fest spermatocytes had a diffuse microtubule array (Fig. 3D′,E′) that eventually overwhelmed the fest germ cells (Fig. 3F′,G′) and their still-arrested chromosomes (Fig. 3F″,G″). As a result of this arrest in an aberrant stage, fest testes were half-filled with small, sausage-shaped cells, earning the gene its full moniker: wurstfest. Knockdown of fest in late spermatogonia and spermatocytes by RNAi gave a similar phenotype (supplementary material Fig. S2), consistent with a cell-autonomous function for Fest in the germline.
Fig. 3.
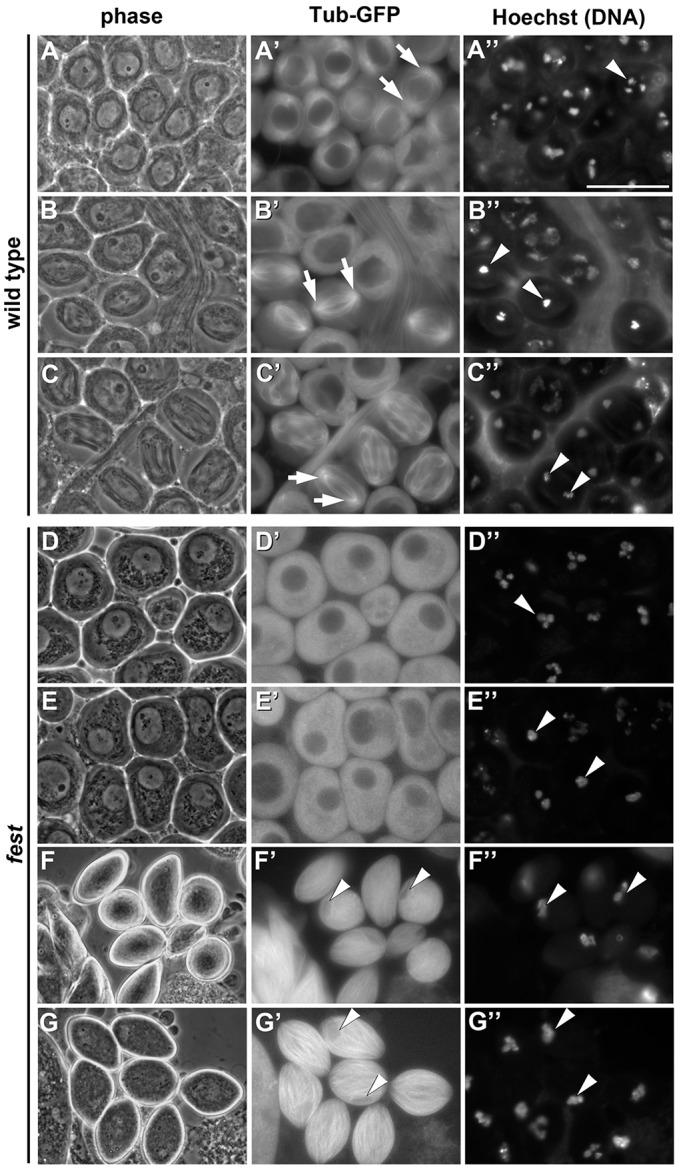
Fest is required for proper entry into metaphase of meiosis I. (A-G″) Live squashes of wild-type (A-C″) and fest mutant (D-G″) testes, both expressing Tubulin-GFP. (A-G) Phase images. (A′-G′) Tub-GFP. Arrows in A′,B′,C′ indicate spindle poles. (A″-G″) Hoechst (DNA). Arrowheads indicate condensed chromosomes. (A-A″) Wild-type prometaphase of meiosis I. (B-B″) Wild-type metaphase I. (C-C″) Wild-type anaphase I. (D-D″) fest mutant spermatocytes showing condensed chromosomes (D″) but no evidence of microtubule asters (D′, compare with A′). (E-E″) A rare class of fest mutant spermatocytes with chromosomes condensed and congressed to the center of the cell (E″) but lacking a metaphase spindle (E′, compare with B′). (F-F″) fest spermatocytes with dense bundles of microtubules and nuclei in the process of getting squashed (arrowheads, F′). (G-G″) The terminal phenotype of fest mutant germ cells. Microtubules are dramatically dense and disorganized, compressing the still-arrested nucleus (arrowheads, G′). Scale bar: 50 µm in A″.
By contrast, the rbp4 loss-of-function meiotic division phenotype was much milder, as described (Haynes et al., 1997). Spermatocytes in an rbp4 mutant were able to enter and progress through the meiotic divisions, but showed occasional cytokinesis defects, as indicated by the presence of round spermatids containing two or more nuclei and one large mitochondrial derivative (supplementary material Fig. S3).
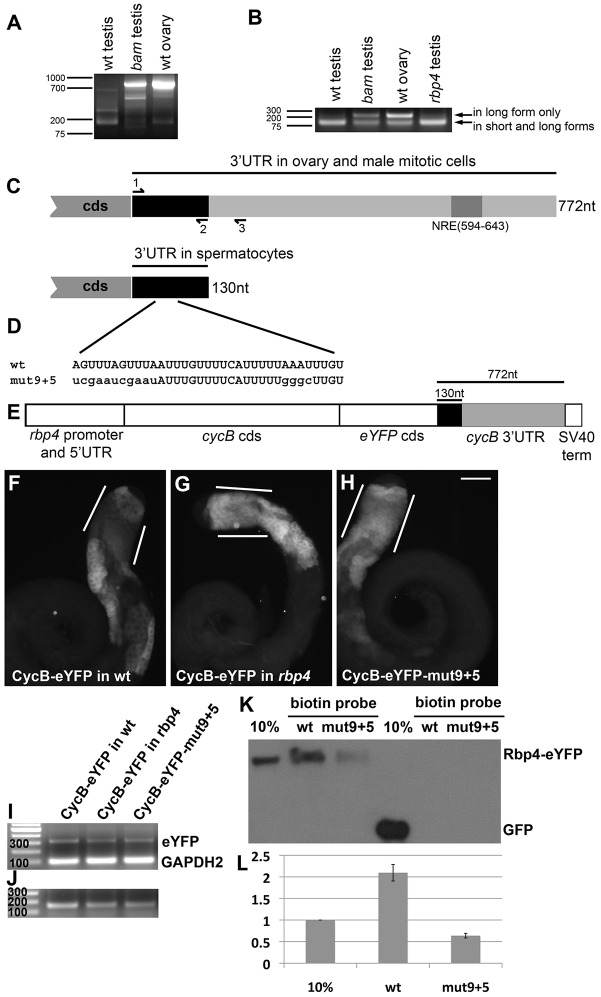
A cell type-specific short cycB 3′ UTR is bound by Rbp4 and contains conserved sequences required for translational repression
The cycB transcript expressed in meiotic and post-meiotic male germ cells has a short 3′ UTR, probably generated by cell type-specific 3′-end formation. 3′ RACE (Fig. 4A) with a primer in the cycB open reading frame and RT-PCR (Fig. 4B) with a trio of primers in the cycB 3′ UTR (Fig. 4C, primers 1-3) on RNA from wild-type ovaries and testes from wild-type, bam or rbp4 mutant males revealed that a short form of the cycB 3′ UTR predominated in wild-type and rbp4 testes. By contrast, the dominant form in bam mutant testes, which contain mitotic spermatogonia but not spermatocytes or spermatids, was the full-length 772-nt 3′ UTR also found in wild-type ovaries (Fig. 4A,B). Sequencing of the short 3′ RACE product expressed in wild-type testes revealed that the short form extended 130 nt from the stop codon, without other differences from the long 3′ UTR, suggesting that the 130-nt 3′ UTR resulted from alternative 3′ end processing rather than alternative splicing. The shortening of the cycB 3′ UTR to 130 nt removed the Nanos response element (Fig. 4C, NRE), which targets the cycB RNA for translational repression by Nanos in early germ cells in embryos (Kadyrova et al., 2007, Asaoka-Taguchi et al., 1999). The 130-nt short form of the cycB 3′ UTR predominated in rbp4 mutant testes, indicating that wild-type function of Rbp4 is not required for generating the 130-nt 3′ UTR.
Fig. 4.
Sequences in the cycB 3′ UTR are required for translational repression. (A) 3′ RACE PCR on the cycB transcript from RNA collected from wild-type testis, wild-type ovary, bam mutant testis (spermatogonia accumulate, and spermatocytes and spermatids are absent). Forward primer for 3′ RACE was nearly identical to primer #1 in C, just 4 bases longer. (B) RT-PCR from RNA collected from wild-type testis, wild-type ovary, bam mutant testis and rbp4 mutant testis, using primers #1-3, shown in C. Predicted products: 124 bp from short and long form; 227 bp from long form only. (C) Schematic of the cycB 3′ UTR as detected in ovary and spermatogonia versus in spermatocytes. NRE, Nanos response element. (D) A conserved proportion of the short cycB 3′ UTR, with mut9+5 variation created by site-directed mutagenesis (mutated nucleotides shown in lowercase). (E) Schematic of CycB-eYFP in vivo reporter. (F-J) Anti-GFP immunofluorescence on: (F) CycB-eYFP in wild-type testis, (G) CycB-eYFP in rbp4 mutant testis and (H) CycB-eYFP-mut9+5 in wild-type testis. White lines indicate early spermatocytes. Scale bar: 100 µm in H. (I,J) RT-PCR on CycB-eYFP, CycB-eYFP in rbp4 and CycB-eYFP-mut9+5 reporters. (I) Amplifying eYFP (313-bp expected product) and (control) GAPDH2 (100 bp) to assay reporter transcript levels. (J) Using an eYFP forward primer and cycB 3′ UTR reverse primers 2 and 3. Predicted products: 157 bp from short and long form; 260 bp from long form only – not detected. (K) Anti-GFP western blot of a biotin RNA pull-down from Rbp4-eYFP or Ubi-GFP testis extract. Wild-type and mutant biotin probes as indicated. (L) Quantification via ImageJ of three independent biotin RNA pull-downs from Rbp4-eYFP. The mean of the 10% input bands was set to 1; a value of 2 for the wild-type probe indicates that that probe pulled down ∼20% of the Rbp4-eYFP input. Error bars indicate s.e.m.
Conserved sequences in the cycB 3′ UTR were required for translational repression of an in vivo CycB-eYFP reporter in immature spermatocytes. A hybrid transgene reporter containing 0.7 kb upstream of the rbp4 transcription start site, the rbp4 5′ UTR, the cycB protein coding sequence fused in frame to eYFP and 772 nt of cycB 3′ sequence (large enough to encode either the 130-nt 3′ UTR or the 772-nt 3′ UTR), followed by an SV40 terminator (Fig. 4E), drove expression of CycB-eYFP protein in a corona of very early spermatocytes, probably due to the early onset of transcription from the rbp4 promoter. However, after this corona region, expression of CycB-eYFP protein was repressed in early spermatocytes, with CycB-eYFP expressed again in mature spermatocytes (Fig. 4F, lines), similar to endogenous CycB (Fig. 2A). Also, similar to endogenous CycB, loss of function of rbp4 resulted in expression of the CycB-eYFP reporter in early spermatocytes (Fig. 4G, lines), indicating that the reporter transgene contained sequences sufficient for translational repression in immature spermatocytes dependent on Rbp4. A milder de-repression effect was observed in the fest mutant background (supplementary material Fig. S4). When a 35-nt stretch within the 130-nt cycB short 3′ UTR conserved within Drosophilids was mutated in two patches (mut9+5, Fig. 4D) in the CycB-eYFP reporter, CycB-eYFP was expressed in immature spermatocytes (Fig. 4H, lines), indicating that sequences within the conserved region were required for translational repression of CycB in immature spermatocytes. High early expression of the wild-type CycB-eYFP reporter expressed in the rbp4 mutant, or the mutated CycB-eYFP-mut9+5 reporter in an otherwise wild-type testis, was not due to increased expression of the reporter RNA, based on results from RT-PCR against eYFP and a GAPDH2 control (Fig. 4I). Furthermore, the alternative polyadenylation of the reporters in testis recapitulated that of the endogenous cycB RNA. In RT-PCR with an eYFP forward primer and reverse primers 2 and 3 in the cycB 3′ UTR (Fig. 4C), a 260-bp product corresponding exclusively to the long form of the 3′ UTR was not detected (Fig. 4J, versus 157 bp for short and long).
The Rbp4 protein associated with the cycB 130-nt 3′ UTR in biotin pull-down assays. When a biotinylated form of the 130-nt testis-specific 3′ UTR synthesized in vitro was incubated with testis extract from Rbp4-eYFP-expressing flies, then recovered with streptavidin beads, Rbp4-eYFP protein was pulled down with the RNA (Fig. 4K, wt). Binding of Rbp4 was noticeably reduced when a probe mutated for the 35-nt conserved region was used (Fig. 4K, mut9+5, quantified in Fig. 4L). GFP driven by the ubiquitin promoter did not bind either wild-type or mutated cycB 3′ UTR (Fig. 4K, right lanes).
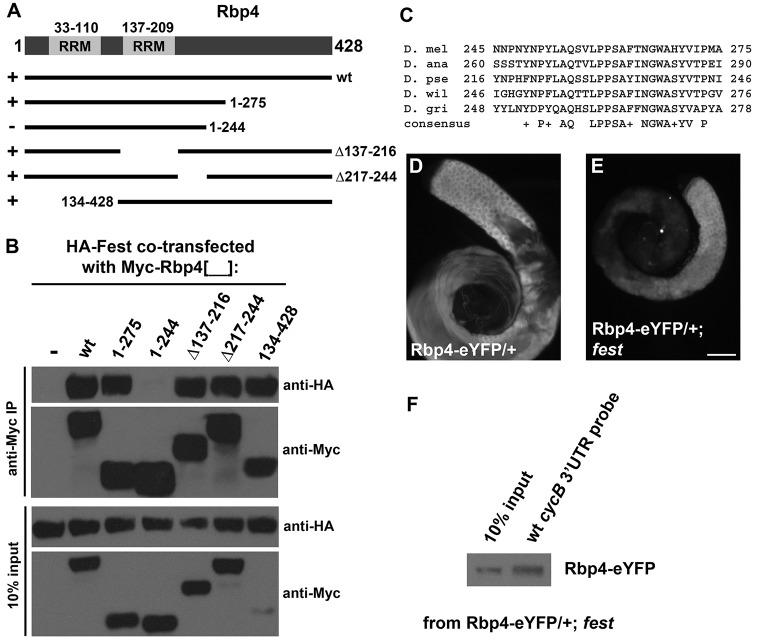
A 31-amino acid segment of Rbp4 is required for binding of Fest
Mapping of the parts of Rbp4 required for binding of Fest revealed that a 31-amino acid region in the C-terminal half of the Rbp4 protein, close to, but not abutting, the second RRM domain, was required for the Rbp4-Fest interaction. Drosophila S2 cells were transfected with either HA-Fest alone or HA-Fest in combination with full-length (wt) or deleted versions of Myc-Rbp4, as diagrammed (Fig. 5A). Immunoprecipitation with anti-Myc followed by western blots probed with anti-Myc and anti-HA showed that Rbp4-1-275 was able to immunoprecipitate HA-Fest, but the next truncation in the series, Rbp4-1-244, was not (Fig. 5B), even though the truncated Rbp4 containing amino acids 1-244 was expressed at normal levels and present in the extract. All other Rbp4 deletions/truncations were able to bind Fest, indicating that the region of Rbp4 between residues 244 and 275 was particularly essential for Fest binding. This region is ∼28 aa from the C-terminal end of the second RRM domain (Fig. 5A), includes several aromatic residues and is moderately conserved among Rbp4 homologs throughout the order Diptera (Fig. 5C).
Fig. 5.
A 31-amino acid domain in Rbp4 is required for binding to Fest. (A) Diagram of Rbp4 protein structure and truncated Rbp4 proteins tested in S2 cells for binding to Fest. RRM, RNA recognition motif. (B) Anti-HA, anti-Myc western blot of anti-Myc immunoprecipitations (top panels) and 10% input (bottom panels) from S2 cells transfected with HA-Fest alone or HA-Fest with various Myc-Rbp4 truncations, as indicated. (C) Alignment of Rbp4 residues 245-275 with Rbp4 homologs in other species. (D,E) Anti-GFP immunostaining of Rbp4-eYFP/+ testis (D) and Rbp4-eYFP/+; fest testis (E). Scale bar: 100 µm in E for D and E. (F) Biotin RNA pull-down from Rbp4-eYFP/+, fest testis, using the wild-type 130 nt cycB 3′ UTR probe.
Activity of Fest is neither required for stability of Rbp4 protein nor required for pull-down of Rbp4 by biotinylated 130-nt cycB 3′ UTR. Anti-GFP immunostaining of Rbp4-eYFP in wild-type and fest mutant testes revealed similar expression of the Rbp4-eYFP reporter protein (Fig. 5D,E). When a biotin RNA pull-down from testis extract from Rbp4-eYFP/+; fest flies was performed with the cycB 130 nt 3′ UTR as probe, ∼20% of the Rbp4-eYFP in the sample was brought down by the biotin-labeled probe (Fig. 5F), comparable to that seen for Rbp4-eYFP in a wild-type background (Fig. 4K).
DISCUSSION
Our findings show that the developmental program of male gametogenesis imposes several levels of cell type- and stage-specific post-transcriptional control on expression of the key G2/M cell cycle regulatory component CycB during meiotic prophase and identify two key developmentally regulated trans-acting factors involved. First, the cycB RNA expressed in spermatocytes has a short 3′ UTR, only 130 nt long and missing previously identified translational regulatory sequences used in other cell types. Second, the RNA-binding protein Rbp4, expressed starting early in meiotic prophase soon after completion of pre-meiotic DNA synthesis, binds the short 3′ UTR and blocks translation of cycB in immature spermatocytes. Third, the Rbp4-interacting protein Fest, also upregulated early in the spermatocyte period, is also required for blocking CycB expression in immature spermatocytes.
Rbp4 and Fest RNA and protein are expressed in very early spermatocytes prior to onset of transcription of cycB, which depends on action of the tMAC complex (White-Cooper et al., 1998). As a result, when the cycB RNA is expressed, it arrives in a cytoplasm already primed for its proper cell type- and stage-specific translational repression. Expression of Cyclin B3 (Clb3) protein in budding yeast has been shown to be restricted to meiosis II via sequences in the CLB3 3′ UTR that block translation during meiosis I. Although translational repression of CLB3 in meiosis I was important to prevent premature separation of sister chromatids, an event appropriate for meiosis II rather than meiosis I (Carlile and Amon, 2008), trans-acting factors responsible for the stage-specific translational repression have yet to be identified. Our data reveal that translational repression of a cyclin (in this case CycB) is also a key feature of meiotic prophase during spermatogenesis in a metazoan animal. Surprisingly, expression of CycB in immature spermatocytes – either in rbp4 or fest mutants or by a mutated CycB-eYFP reporter – was insufficient to drive those cells immediately into meiotic division. This might be because action of the Cdc25 cell cycle phosphatase encoded by twine, which is also translationally repressed in immature spermatocytes and becomes translationally activated by the RNA-binding protein Boule only in mature spermatocytes (Alphey et al., 1992; Maines and Wasserman, 1999), is also required to generate active Cdk1/CycB.
One general model for Fest function invokes the possibility that Rbp4 recruits Fest to the cycB 3′ UTR, where Fest is able to interfere with cycB translation. However, we found no compelling evidence of specific binding of Fest to the cycB 3′ UTR in biotin pull-down experiments from testis extracts from flies expressing eYFP-Fest either with or without functional Rbp4 (data not shown), which suggests that either Fest is not recruited to the cycB 3′ UTR, or that our biotin pull-down assay has limitations in detecting indirect RNA-protein interactions. As a result, it is important to consider other mechanisms for Fest function, including the possibility that binding of Fest to Rbp4 is needed only briefly to enact a post-translational modification of or conformational change within Rbp4 to promote its ability to recruit partners and/or repress translation. It is also technically possible that Fest and Rbp4 act in parallel pathways to regulate CycB. Finally, as the fest germ cell phenotype is dramatically stronger than that of rbp4, it is likely that Fest regulates other proteins in addition to Rbp4.
It is not yet known how information about spermatocyte maturation is communicated to Rbp4 or Fest to allow translation of cycB in mature spermatocytes. One or more proteins could respond to input regarding cell size, given that spermatocytes grow 25-fold in volume during meiotic G2. Alternatively, given that translation of cycB in mature spermatocytes requires function of the testis TAF proteins, signals indicating the completion of the spermatocyte transcription program (not merely its onset) could trigger the reprieve from translational repression. Another possibility might be a meiotic arrest checkpoint mechanism triggered by transcriptional activity from unpaired chromatin, as seen in mammalian spermatocytes (Odorisio et al., 1998). Whatever the stimulus, it is clear that through stage-specific expression of the translational regulators Rbp4 and Fest in very early spermatocytes, the developmental program of male germ cell differentiation exerts additional layers of control over the core cell cycle machinery.
MATERIALS AND METHODS
Fly husbandry and stocks
Flies were maintained on dextrose/cornmeal at 21°C (stocks) and 25°C (crosses). Flies carrying Df(3R)Exel6169 or Df(2R)BSC26 were obtained from the Bloomington Stock Center (#7648 and #6866, respectively). Flies carrying PBac{SAstopDsRed}LL06910 (rbp4LL06910) were obtained from the Drosophila Genomics Resource Center (#141934). Tub-GFP flies were obtained from the Glover lab (Inoue et al., 2004).
The PBac{SAstopDsRed}LL06910 transposon insertion is located between the third and fourth exons of rbp4 (FlyBase) and predicted to truncate the Rbp4 protein at residue 194, 15 amino acids before the end of the second of two RRM domains. Levels of rbp4 transcript from rbp4LL06910/Df testes were much lower than in wild type, as assayed by RT-PCR against the part of the rbp4-coding sequence 5′ of the P element insertion site (supplementary material Fig. S5). Any low levels of protein generated from this allele of rbp4 might have residual function, particularly RNA-binding capability.
The loss-of-function allele of fest was generated using the CRISPR approach (Bassett et al., 2013). A G(N19)NGG sequence running 3′ to 5′ early in the fest coding sequence was selected, with GGTGCTGGTCTTAGGTGCAA added to the gene-specific oligonucleotide. PCR and in vitro transcription were performed as described (Bassett et al., 2013). The resulting CRISPR RNA was purified using Trizol and injected into embryos of Act5-Cas9 flies (Port et al., 2014). Surviving adults were crossed individually to Df(2R)BSC26/TM6B, and non-TM6B male progeny were scored for germ cell phenotypes. TM6B siblings from vials with a high incidence of hits were crossed again to Df(2R)BSC26/TM6B to identify and recover loss-of-function alleles. About 300 bp of sequence surrounding the CRISPR target site was PCR-amplified and sequenced in independent lines. Two alleles resulting in the same phenotype were recovered: a 1-nt deletion (29 nt into the coding sequence) and an 8-nt deletion (starting 25 nt into the coding sequence). The latter allele was the one used for the experiments described here.
Plasmids and transgenic flies
For the S2 cell experiments, tagged proteins were created by cloning the relevant coding sequence into copper-inducible pMT-Myc or pMT-HA (Bunch et al., 1988). For Myc-Rbp4 and the Myc-Rbp4 truncations, the rbp4 coding sequence (full-length or partial) was cloned into the KpnI/SacI sites of pMT-Myc; for HA-Fest, the fest coding sequence was cloned into the SacI/SacII sites of pMT-HA.
Rbp4-eYFP was built in pBluescript (pBS) and then moved into the NotI/EcoRI sites of pCaSpeR4 (Pirrotta, 1988): NotI -rbp4 promoter (716 bp, directly 5′ of annotated transcription start site) and 5′ UTR (90 bp) -SpeI -rbp4 coding sequence (1287 bp) -SmaI/EcoRV [non-recleavable] -eYFP coding sequence (717 bp) -SmaI -rbp4 3′ UTR (141 bp) and 3′ genomic sequence (148 bp) -EcoRI.
eYFP-Fest was built in pBS and then moved into the XbaI/XhoI sites of pCaSpeR4: XbaI -fest promoter (590 bp, directly 5′ of annotated transcription start site) and 5′ UTR (275 bp) -SpeI -eYFP coding sequence (717 bp) -SmaI -fest coding sequence (1542 bp) -EcoRI -fest 3′ UTR and genomic sequence (1255 bp total) -EcoRI-XhoI.
The CycB-eYFP reporter was built in pBS and then moved into the NotI/BamHI sites of pCaSpeR4 containing an SV40 terminator: NotI -rbp4 promoter (716 bp) and 5′ UTR (90 bp) -SpeI/XbaI [non-recleavable] -cycB coding sequence (1590 bp) -SpeI/XbaI [non-recleavable] -eYFP coding sequence (717 bp) -SpeI -cycB 3′ UTR and 3′ genomic sequence (772 bp) -BamHI. For testing the mutated version of the cycB 3′ UTR, the 3′ UTR was subjected to site-directed mutagenesis and then swapped into the SpeI and BamHI sites of the reporter within pBS and then re-transferred to pCaSpeR4/SV40. For both wild-type and mutated reporters, at least two independent transgenic lines were characterized and showed consistent expression patterns. An eYFP reporter without the cycB coding sequence, which promotes rapid clearance of the fusion protein, showed perdurance of the eYFP from the ‘corona,’ obscuring the translational ‘off’ state in immature spermatocytes.
All plasmids were injected into yw embryos as per Rubin and Spradling (1982).
3′ RACE and RT-PCR
For 3′ RACE on the cycB RNA, a forward primer within the cycB-coding sequence (5′-TGCGGTCCAAGGCGGACTGGAAG-3′) was used together with a 3′ RACE primer (5′-GCACGGTCACGGTCCAGCT12-3′) for 35 cycles at 60°C annealing temperature. For RT-PCR on the cycB 3′ UTR, a forward primer (#1 in Fig. 2C) was used in combination with a pair of reverse primers (#2 and #3) designed to amplify from (#2) either both long and short forms of the 3′ UTR or (#3) just the long form. Expected products were 124 bp (from both short and long 3′ UTRs) versus 227 bp (long 3′ UTR only). The 3′ UTR length of the CycB-eYFP reporters was assayed by RT-PCR with a forward primer from the GFP-coding sequence and the same pair of reverse primers as above (#2 and #3). The expected products were 157 bp (short and long 3′ UTRs) and 260 bp (long 3′ UTR only). For RT-PCR to assay the relative transcript abundance of the CycB-eYFP reporters, primers to the eYFP-coding sequence (313-bp expected product) and to GAPDH2 (100 bp) were used in the same reaction. The RT-PCR primers for fest amplified a predicted 514-bp product from the 3′ end of the fest-coding sequence. In all cases, RNA samples were prepared using Trizol, and cDNA was generated using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare).
Histology
All testis dissections were carried out in 1× PBS in an 8-well dish unless otherwise noted. For CycB whole-mount immunostaining, testes were processed in 1.5-ml tubes through ice-cold methanol (5 min) and ice-cold acetone (2 min) before continuing. For GFP and PH3Thr3 immunostaining experiments, testes were fixed in 1.5-ml tubes in 4% formaldehyde (1 h) and permeabilized in 0.3% deoxycholate/0.3% Triton-X/PBS (30 min) before continuing. For the HA/GFP immunostaining, dissected testes were placed on a drop of PBS on a microscope slide, flattened under a coverslip and frozen in liquid nitrogen. The coverslip was removed with a razor blade, and slides were processed through 4% formaldehyde and 0.3% DOC, as above. All samples were subsequently washed in PBS/0.1% Triton X (PBSTr) and blocked in PBSTr/3% BSA (30 min) before incubating with primary antibody at 4°C (overnight). Samples were then washed in PBSTr/3% BSA (30 min), incubated with secondary antibody at room temperature (2 h) in the dark, washed twice with PBSTr and mounted using Vectashield with DAPI (Vector Labs). Antibody sources and dilutions: anti-CycB (1:30; F2F4, mouse, BD Biosciences), anti-HA (1:1000; 16B12, mouse, Covance), anti-GFP (1:3000; #A11122, rabbit, Invitrogen/Life Technologies), anti-PH3Thr3 (1:200; #07-424, rabbit, Millipore). Secondary antibodies were all Alexa Fluor-conjugated and used at a 1:200 dilution: donkey anti-rabbit 488, goat anti-mouse 488 and goat anti-mouse 568 (Molecular Probes). In situ hybridization with probe generated from the cycB coding sequence was performed as in White-Cooper et al. (1998).
Unfixed squashes were performed by placing dissected testes on a drop of PBS on a slide, opening the testis sheath, gently placing a glass coverslip on top and wicking away excess PBS with the edge of a Kimwipe until germ cells were in a single layer. For Hoechst staining, Hoechst 33342 was added to PBS to a final concentration of 10 µg/ml.
Images from immunostaining and phase-Hoechst staining were captured by a Photometrics CoolSNAP CCD camera connected to a Zeiss Axioskop microscope, with fluorescence illumination provided by an X-Cite 120 excitation light source. Phase image panels in supplementary material Fig. S1 and images of in situ hybridizations (Fig. 2) were captured by a Spot RT3 CCD camera affixed to a Zeiss Axioskop microscope.
Tissue culture
Drosophila S2 cells were maintained in Schneider's S2 cell medium (Gibco) plus 12.5% fetal bovine calf serum and 0.1 mg/ml gentamycin. Cells were transiently transfected using FuGENE HD (Roche/Promega) or Effectene (Qiagen) in a 6-well plate format, following package instructions. After 48 h, transfected cells were transferred to T-75 tissue culture flasks (+13 ml fresh medium), and pMT promoter expression was induced with copper sulfate added to a final concentration of 0.7 mM. Cells were harvested for immunoprecipitation 24 h later.
Co-immunoprecipitations
Anti-Myc (4A6, Millipore/Upstate) and anti-HA (16B12, Covance) were individually conjugated to pan-mouse IgG Dynabeads (Invitrogen). Sufficient beads for 50 µl beads/IP were blocked briefly in 3% BSA in phosphate-buffered saline +0.1% Tween (PBST) and then incubated for 1 h at RT with lysis buffer (20 mM Tris, 135 mM NaCl, 10% glycerol, 1% NP40, 10 mM EDTA) plus antibody (1:100), with 2 µl antibody/200 µl lysis buffer per IP. Beads were split into individual tubes, with 50 µl beads per tube, and washed three times for 5 min with 1 ml 0.2 M triethanolamine, pH 8.2, then incubated with 1 ml of 29 mM dimethyl pimelimidate, 0.2 M triethanolamine for 30 min at RT. Following a 15-min wash with 50 mM Tris and three 5-min washes with PBST, beads were stored overnight or used directly in the next step. After beads were washed twice with 100 mM glycine, pH 2.5, to remove un-crosslinked antibody, they were incubated in 10% BSA, 50 mM Tris for 1 h to block non-specific binding.
For IPs from testis extract (Fig. 1G), 70+7 testis pairs were dissected per IP+input. The dissected testes were mechanically disrupted in lysis buffer (220 µl each) via a 1-cc syringe and 25×5/8-gauge needle. For IPs from S2 cells, the transfected cells were spun down, washed once in PBS and spun again before lysis buffer (220 µl each) was added. Lysis buffer contained 20 mM Tris, 135 mM NaCl, 10% glycerol, 1% NP40, 10 mM EDTA and 1× complete protease inhibitor (Roche). Lysis was allowed to proceed for 30 min at 4°C with rocking, then the insoluble fraction was pelleted and discarded. A 20 µl portion of lysate was saved as 10% input for each sample. Lysates were precleared by incubating with 20 µl mouse IgG Dynabeads (blocked with 3% BSA/PBST but not conjugated to antibody) at 4°C for 45 min. The lysates were then incubated with antibody-conjugated beads for 3-4 h at 4°C with rocking. Beads were then washed twice with 1 ml lysis buffer (5 min, 4°C), and bound proteins were eluted with 40 µl elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris, 1× complete protease inhibitor) at 70°C for 30 min with frequent vortexing. Laemmli sample buffer was added, and samples were boiled for 10 min. Eluted proteins were analyzed by western blot.
Biotin pull-downs
Biotin-labeled probes: wild-type and mutant versions of the 130-nt cycB 3′ UTR were cloned into the SpeI/BamHI sites of pBluescript. Plasmids were linearized with BamHI to generate templates for sense probes. Biotin-labeled probes were generated via in vitro transcription using biotin RNA labeling mix and T7 RNA polymerase (Roche), treated with DNAse and purified using NucAway Spin Columns (Ambion). Probe concentration was determined by Nanodrop and probe integrity was verified by agarose gel.
For each pull-down, 50 testis pairs (from Rbp4-eYFP flies) or 80 pairs (from Rbp4-eYFP/+; fest flies) were dissected and homogenized in 100 µl lysis buffer [100 mM NaCl, 50 mM Tris, 0.4 mM EDTA, 1% NP40, 1× complete protease inhibitor (Roche), 1 µl/ml SUPERaseIN (Applied Biosystems)] with a 1-ml syringe and a 25×5/8-gauge needle, then lysed at 4°C for 30 min. Lysate was centrifuged for 5 min, then 10 µl supernatant was set aside for input (10%). The remaining supernatant was pre-cleared with streptavidin beads (streptavidin magnesphere paramagnetic particles, Promega) for 30 min at room temperature, then split into four tubes (100 µl each) and incubated with 10 pmol of biotin-labeled probe for 30 min at RT. Each sample was then incubated with fresh streptavidin beads (30 min, RT) and washed five times for 10 min. Laemmli sample buffer was added, and samples were boiled for 10 min and analyzed by western blot.
Western blots
Samples were run on a SDS-PAGE gel (TGX, BioRad) and proteins were blotted onto a PVDF membrane overnight in 0.025 M Tris, 0.192 M glycine. The blot was blocked in 5% milk in Tris-buffered saline (TBS), incubated with primary antibody (mouse anti-GFP, 1:2000, Roche; mouse anti-HA, 1:5000, Covance; mouse anti-Myc, 1:5000, Millipore/Upstate) in 5% milk/TBS for 1 h, rinsed briefly with 5% milk/TBS, then incubated with HRP-conjugated anti-mouse IgG (Promega) at 1:10,000 for 1 h. After >2 h of washes in TBS, signals were detected using Western Lightning Plus-ECL reagents and exposed to autoradiography film (BioMax XAR, Kodak).
Acknowledgements
We would like to thank the Vienna Drosophila RNAi Center and Bloomington Drosophila Stock Center for fly stocks, FlyAtlas and FlyBase for invaluable online resources, the Bullock lab for Act5-Cas9 flies, the Glover lab for Tub-GFP flies and the members of the Fuller lab for their helpful discussions and feedback.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
B.S.G. performed the initial experiments, including identifying Rbp4 as a repressor of cycB translation and mapping cis-acting regulation to the cycB 3′ UTR. C.C.B. carried out the remaining experiments. C.C.B. and M.T.F. designed the study and prepared the manuscript.
Funding
This research was supported by funds from the National Institutes of Health (NIH) Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) [U54 HD068158] as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.122341/-/DC1
References
- Alphey L., Jimenez J., White-Cooper H., Dawson I., Nurse P. and Glover D. M. (1992). Twine, a Cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69, 977-988. 10.1016/0092-8674(92)90616-K [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K. and Kobayashi S. (1999). Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1, 431-437. 10.1038/15666 [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P. and Liu J.-L. (2013). Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220-228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch T. A., Grinblat Y. and Goldstein L. S. B. (1988). Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 16, 1043-1061. 10.1093/nar/16.3.1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T. M. and Amon A. (2008). Meiosis I is established through a division-specific translational control of a cyclin. Cell 133, 280-291. 10.1016/j.cell.2008.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J. and Dow J. A. T. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715-720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Fuller M. T. (1993). Spermatogenesis. In The Development of Drosophila Melanogaster, Vol. I (ed. Bate M. and Arias A. M.), pp. 71-147. Plainview, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E. et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727-1736. 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Cooper M. T., Pype S. and Stolow D. T. (1997). Involvement of a tissue-specific RNA recognition motif protein in Drosophila spermatogenesis. Mol. Cell. Biol. 17, 2708-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y. H., Savoian M. S., Suzuki T., Mathe E., Yamamoto M.-T. and Glover D. M. (2004). Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166, 49-60. 10.1083/jcb.200402052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco M. L., Leon A., Tam C. H., McKearin D. M. and Fuller M. T. (2009). Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc. Natl. Acad. Sci. USA 106, 22311-22316. 10.1073/pnas.0912454106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H. and Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519-1527. 10.1242/dev.002212 [DOI] [PubMed] [Google Scholar]
- Kierszenbaum A. L. and Tres L. L. (1978). RNA transcription and chromatin structure during meiotic and postmeiotic stages of spermatogenesis. Fed. Proc. 37, 2512-2516. [PubMed] [Google Scholar]
- Lamitina S. T. and L'Hernault S. W. (2002). Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development 129, 5009-5018. [DOI] [PubMed] [Google Scholar]
- Maines J. Z. and Wasserman S. A. (1999). Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell Biol. 1, 171-174. 10.1038/11091 [DOI] [PubMed] [Google Scholar]
- Odorisio T., Rodriguez T. A., Evans E. P., Clarke A. R. and Burgoyne P. S. (1998). The meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nat. Genet. 18, 257-261. 10.1038/ng0398-257 [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1988). Vectors for P-mediated transformation in Drosophila. Biotechnology 10, 437-456. 10.1016/b978-0-409-90042-2.50028-3 [DOI] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T. and Bullock S. L. (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. 111, E2967-E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. and Spradling A. C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348-353. 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- Sun S. C., Lee S. E., Xu Y. N. and Kim N. H. (2010). Perturbation of Spc25 expression affects meiotic spindle organization, chromosome alignment and spindle assembly checkpoint in mouse oocytes. Cell Cycle 9, 4552-4559. 10.4161/cc.9.22.13815 [DOI] [PubMed] [Google Scholar]
- White-Cooper W., Schäfer M. A., Alphey L. S. and Fuller M. T. (1998). Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 125, 125-134. [DOI] [PubMed] [Google Scholar]
- Wolgemuth D. J., Manterola M. and Vasileva A. (2013). Role of cyclins in controlling progression of mammalian spermatogenesis. Int. J. Dev. Biol. 57, 159-168. 10.1387/ijdb.130047av [DOI] [PMC free article] [PubMed] [Google Scholar]