Abstract
The ability to induce targeted mutations in somatic cells in developing organisms and then track the fates of those cells is a powerful approach both for studying neural development and for modeling human disease. The CRISPR/Cas9 system allows for such targeted mutagenesis, and we therefore tested it in combination with a piggyBac transposase lineage labeling system to track the development of neocortical neural progenitors with targeted mutations in genes linked to neurodevelopmental disruptions and tumor formation. We show that sgRNAs designed to target PTEN successfully decreased PTEN expression, and led to neuronal hypertrophy and altered neuronal excitability. Targeting NF1, by contrast, caused increased astrocytogenesis at the expense of neurogenesis, and combined targeting of three tumor suppressors (PTEN, NF1 and P53) resulted in formation of glioblastoma tumors. Our results demonstrate that CRISPR/Cas9 combined with piggyBac transposase lineage labeling can produce unique models of neurodevelopmental disruption and tumors caused by somatic mutation in neural progenitors.
KEY WORDS: CRISPR/Cas9, Neural progenitors, Lineage, piggyBac, In utero electroporation, Glioblastoma multiforme
Summary: The CRISPR/Cas9 system is used to target three tumor suppressor genes (PTEN, NF1 and P53), alone and in combination with piggyBac transposase lineage marking, to analyze their function in the developing mouse brain.
INTRODUCTION
A general approach to studying developmental mechanisms is to mutate genes of interest, and then evaluate the resulting phenotypes. Recent breakthroughs in genome engineering technology adapted from clustered regularly interspaced short palindromic repeats (CRISPR) and Cas9 systems in prokaryotes (Cong et al., 2013; Mali et al., 2013) have now been applied with unprecedented speed and precision to a wide variety of approaches. The type II CRISPR/Cas9 system has been used to generate transgenic animals in multiple species (Cho et al., 2013; Gratz et al., 2013; Yang et al., 2013; Hai et al., 2014), to induce or correct genetic mutations in vivo (Ding et al., 2014), to screen for genes involved in specific biological processes (Shalem et al., 2014; Wang et al., 2014), to activate or repress gene expression (Cheng et al., 2013; Gilbert et al., 2013; Qi et al., 2013), to isolate (Fujita and Fujii, 2013; Chen et al., 2013) specific regions of interest within the genome, and to knock out genes involved in synaptic transmission (Incontro et al., 2014; Straub et al., 2014).
In utero electroporation (IUE) is an efficient method to deliver multiple transgenes into rodent neural progenitors in the developing forebrain. It has been used in studies of neocortical development (LoTurco et al., 2009), and when combined with a binary piggyBac transposon plasmid system, IUE can be used as a lineage and fate-mapping tool for neural progenitors (Chen and LoTurco, 2012; Chen et al., 2014b; Siddiqi et al., 2014). The system has also been used to generate animal models of CNS tumors (Chen et al., 2014a; Glasgow et al., 2014). In this report, we combine the CRISPR/Cas9 system with IUE and piggyBac transposase lineage labeling to induce somatic mutations in cells in neural progenitor lineages and to investigate the effects on cells in the lineage. We tested two short guide sequences (sgRNAs) targeting rat PTEN, and these successfully abolished PTEN expression in neurons. Moreover, PTEN null neurons demonstrated phenotypes that were similar to PTEN knockout or RNAi knockdown neurons previously reported (Kwon et al., 2001; Fraser et al., 2004; Luikart et al., 2011) including hypertrophy and altered neuronal excitability. We also found that sgRNA designed against NF1 leads to loss of NF1 expression and to a neural progenitor fate switch that increased gliogenesis. Finally we show that targeting three tumor suppressors [PTEN, NF1 and P53 (TRP53 – Mouse Genome Informatics)] by multiplex CRISPR/Cas9, leads to the formation of glioblastoma multiforme (GBM) tumors. Our results demonstrate that the CRISPR/Cas9 system can be used to efficiently target genes in developing brain in vivo and to generate novel models of glioblastoma and neurodevelopmental disruption.
RESULTS
Knocking out PTEN expression using CRISPR-induced somatic mutation
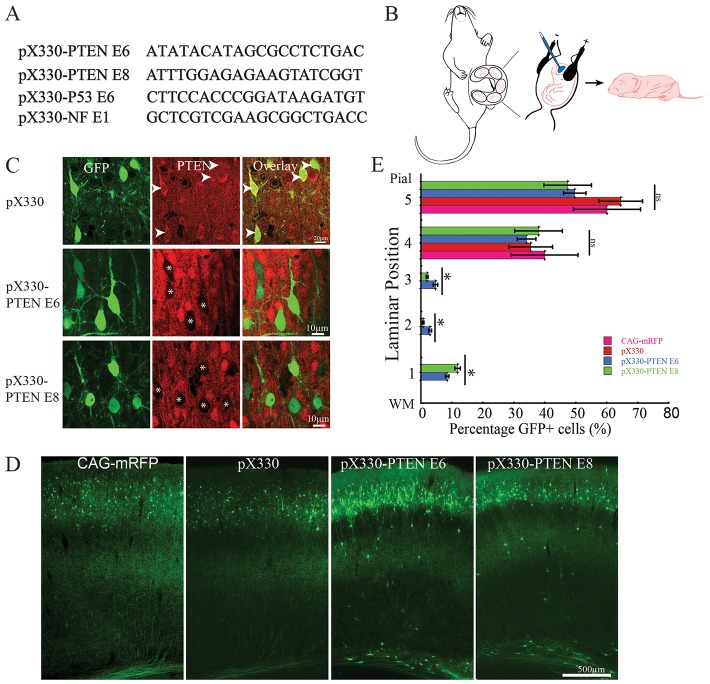
To investigate the application of CRISPR to the central nervous system, we designed two short guide RNAs (sgRNAs) to target either of two exons of the rat phosphatase and tensin homolog (PTEN; exon 6 or exon 8) (Fig. 1A). We cloned these two sgRNAs into pX330 vector containing the wild-type human codon optimized SpCas9 (Cong et al., 2013). In vitro SURVEYOR assay in rat neuroblastoma B104 cells showed that both sgRNAs were able to introduce mutations at targeted PTEN genomic loci in rat neuroblastoma cells (Fig. S1A). To test whether the sgRNAs targeting PTEN were able to introduce mutation in vivo, we used IUE to deliver pX330-PTEN E6 and pX330-PTEN E8 into embryonic day (E) 14 neocortical neural progenitors in rats (Fig. 1B). Empty pX330 vector containing wild-type Cas9 but no sgRNA was used to control for Cas9 activity, and pCAG-mRFP was used as IUE control. All plasmids were used at the concentration of 1.5 µg/μl. pCAG-eGFP (1 µg/µl) was co-electroporated with CRISPR constructs, empty pX330 and pCAG-mRFP to visualize transfected cells. To further test whether sgRNA directed Cas9 targeting of PTEN can induce somatic mutation in vivo, we performed an in vivo SURVEYOR assay on genomic DNAs isolated from acute dissected transfected tissue from a postnatal day (P) 19 pX330-PTEN E6-transfected brain. As shown in Fig. S1B, sgRNA targeting PTEN exon 6 successfully induced mutations at the PTEN locus in vivo.
Fig. 1.
CRISPR/Cas9 knocks out PTEN expression in vivo. (A) sgRNA sequences used in this study. (B) Schematic of IUE. Plasmids DNA were injected into lateral ventricle of E14-15 rat embryos followed by electroporation. Analyses were performed at various postnatal times. (C) Representative images of PTEN antibody staining on brains sections from pX330-, pX330-PTEN E6- and pX330-PTEN E8-transfected brains. CRISPR constructs targeting PTEN exon 6 and 8 abolished endogenous PTEN expression in postmitotic neurons. IUE was performed at E14 and brains were analyzed at P19. Arrowheads indicate GFP-labeled PTEN positive neurons. Asterisks indicate GFP-labeled PTEN-negative neurons. (D) Representative images from P19 brains transfected with CAG-mRFP, pX330, pX330-PTEN E6 and pX330-PTEN E8. A small fraction neurons transfected with PTEN-targeting CRISPR constructs showed disrupted migration. IUE was performed at E14 and brains were analyzed at P19. (E) Quantification results of neuronal migration. Cerebral cortex was divided into five bins with bin 1 close to white matter and bin 5 close to pial surface. About 10% of PTEN-targeting CRISPR-transfected neurons failed to migrate to their normal laminar position (n=3, one-way ANOVA; *P<0.05; ns, no significant difference; data are presented as means±s.e.m.). WM, white matter.
In immunofluorescence histochemistry for PTEN we found that 86±3.8% (n=3 animals) of pX330-PTEN E6 and 82±5.2% (n=3 animals) pX330-PTEN E8-transfected neurons (GFP positive) were negative for PTEN (Fig. 1C). By contrast, in control transfected brains (n=3) 1.6±0.8% of neurons (5/314) were found to be PTEN negative by pX330-transfected animals (one-way ANOVA: F=88.05, P=1.61×10−6; followed by post hoc Tukey HSD test: pX330-PTEN E6 versus pX330, P=0.001; pX330-PTEN E8 versus pX330, P=0.001). These results indicate that both sgRNAs targeting PTEN were able to introduce mutations in vivo at PTEN genomic loci that resulted in loss of PTEN protein expression with high efficacy.
PTEN mutated neurons were hypertrophic and showed altered neurophysiology
We assessed the phenotypes of neurons generated from E14 neural progenitors transfected with pX330-PTEN E6, pX330-PTEN E8, or the two controls (n=3 for each group). In all four conditions, the transfected cells migrated normally to their correct laminar positions, suggesting that they had acquired an appropriate neuronal identity. In both PTEN CRISPR conditions ∼10% of transfected neurons were ectopically located in white matter or deep cortical lamina (Fig. 1D,E), a significantly greater proportion than were in those ectopic positions in control transfections (one-way ANOVA: F=32.91, P=7.53×10−5; followed by post hoc Tukey HSD test: pX330-PTEN E6 versus pX330, P=0.001; pX330-PTEN E6 versus CAG-mRFP, P=0.001; pX330-PTEN E6 versus pX330, P=0.025; pX330-PTEN E6 versus CAG-mRFP, P=0.025). This result is consistent with previous studies showing that neuronal migration defects occur in a subset of PTEN null neurons (Backman et al., 2001; Kwon et al., 2001; Zhu et al., 2012).
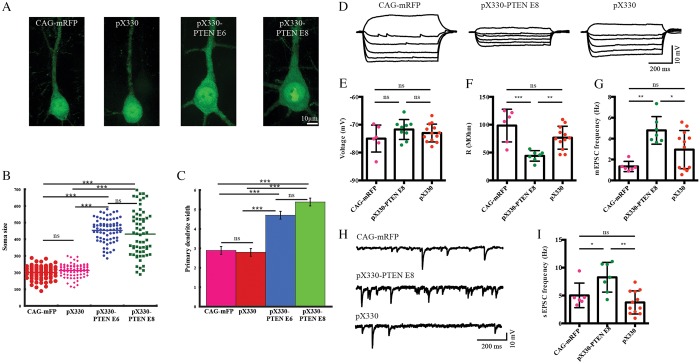
We also quantified the area of neuronal somas in PTEN CRISPR-targeted neurons in layer II/III, and consistent with previous reports of hypertrophy in cases of PTEN deletion or knockdown (Kwon et al., 2001; Fraser et al., 2004; Luikart et al., 2011), found that neurons transfected with PTEN-targeting sgRNAs showed substantially increased somal areas (Fig. 2A). At P19, pX330-transfected neurons (212.7±5.7 µm2, n=85) and CAG-mRFP-transfected neurons (201.2±5 µm2, n=74) had areas that did not significantly differ (one way ANONVA: F=3.14, P=0.08) (Fig. 2B). Neurons transfected with pX330-PTEN E6, by contrast, had significantly increased soma size (453.8±5.2 µm2, n=72) compared with neurons transfected with pX330 or CAG-mRFP (one-way ANOVA: F=559.28, P=1.11×10−16; followed by post hoc Tukey HSD test: pX330-PTEN E6 versus pX330, P=0.001; pX330-PTEN E6 versus CAG-mRFP, P=0.001) (Fig. 2B). Similarly, pX330-PTEN E8-transfected neurons had an average somal size of 428.5±12 µm2 (n=88); a value significantly greater than those of neurons transfected with pX330 or CAG-mRFP (one-way ANOVA: F=241.83, P=1.11×10−16; followed by post hoc Tukey HSD test: pX330-PTEN E8 versus pX330, P=0.001; pX330-PTEN E8 versus CAG-mRFP, P=0.001), and the soma sizes of pX330-PTEN E6 and pX330-PTEN E8 transfected neurons did not significantly differ (one-way ANOVA: F=2.77, P=0.98) (Fig. 2B). To quantify and compare another measure of neuronal hypertrophy, we measured the width of the primary dendrites at 50 µm from the center of the soma. Neurons transfected with the two control plasmids, CAG-mRFP and pX330, had similar primary dendritic widths of 2.9±0.2 µm (n=14), and 2.8±0.2 µm (n=25), respectively (one-way ANOVA: F=0.13, P=0.72) (Fig. 2C). In contrast, the primary dendritic widths in pX330-PTEN E6-transfected neurons were 4.7±0.2 µm (n=24), which was significantly larger than CAG-mRFP or pX330-transfected neurons (one-way ANOVA: F=30.54, P=7.13×10−10; followed by post hoc Tukey HSD test: pX330-PTEN E6 versus pX330, P=0.001; pX330-PTEN E6 versus CAG-mRFP, P=0.001). Similarly, pX330-PTEN E8-transfected neurons had significantly thicker primary dendrites (5.4±0.2 µm, n=18) than CAG-mRFP or pX330-transfected neurons (Fig. 2C) (one-way ANOVA: F=55.92, P=6.96×10−14; followed by post hoc Tukey HSD test: pX330-PTEN E8 versus pX330, P=0.001; pX330-PTEN E8 versus CAG-mRFP, P=0.001).
Fig. 2.
PTEN null neurons were hypertrophic and exhibited altered membrane properties. (A) Representative images of P19 neurons transfected at E14 with CAG-mRFP, pX330, pX330-PTEN E6 and pX330-PTEN E8. (B,C) Neurons transfected with PTEN-targeting CRISPR were hypertrophic. PTEN-targeting CRISPR-transfected neurons displayed increased soma size (B) and thickened primary dendrite (C). (D-I) PTEN-targeting CRISPR-transfected neurons showed altered neurophysiological properties. (D) Representative traces from CAG-mRFP-, pX330- and PTEN-targeting CRISPR-transfected neurons representing voltage responses to −300, −250, −200, −150, −100, −50, −20 and 50 pA step current injections. (E) CAG-mRFP-, empty CRISPR construct pX330- and PTEN-targeting CRISPR-transfected neurons showed similar resting membrane potential. However, PTEN-targeting CRISPR-transfected neurons demonstrated markedly decreased input resistance (F), increased miniature EPSCs (G) and spontaneous EPSCs (I). Representative traces of spontaneous EPSCs are shown in H. One-way ANOVA followed by Turkey's multiple comparison test, *P<0.05; **P<0.01; ***P<0.001; ns, no significant difference; data are presented as means±s.e.m.
We next examined the electrophysiological properties of transfected neurons. Neurons transfected with CAG-mRFP, pX330 and pX330-PTEN E8 had similar resting membrane potentials (CAG-mRFP, −74.99±1.97 mV, n=13; pX330, −72.98±0.89 mV, n=6; pX330-PTEN E8, −71.71±1.19 mV, n=9. One-way ANOVA: F2,25=1.43, P=0.26; followed by Tukey's multiple comparisons test: difference not significant between any two conditions; Fig. 2E). However, consistent with the increased soma size in pX330-PTEN E8-transfected cortical pyramidal neurons, we found significantly lower input resistance in this condition when compared with neurons transfected with the other two constructs (pX330-PTEN E8, 44.03±3.57 MΩ, n=12; CAG-mRFP, 98.42±11.98 MΩ, pX330: 76.66±5.941 MΩ. One-way ANOVA, F2,22=11.48, P=0.0004; followed by Tukey's multiple comparisons test: pX330-PTEN E8 versus pX330, P=0.009; pX330-PTEN E8 versus CAG-mRFP, P=0.0003; pX330 versus CAG-mRFP, P=0.11; Fig. 2A,D,F). We also measured the frequencies of spontaneous synaptic events (sEPSC) in each condition. Neurons transfected with pX330-PTEN E8 have markedly higher sEPSC frequencies compared with ones transfected with pX330 or CAG-mRFP (pX330-PTEN E8, 8.27±1.01 Hz, n=7; pX330, 3.75±0.60 Hz, n=12; CAG-mRFP, 5.03±0.90 Hz, n=6. One-way ANOVA: F2,22=8.74, P=0.002; followed by Tukey's multiple comparisons test: pX330-PTEN E8 versus pX330, P=0.001; pX330-PTEN E8 versus CAG-mRFP, P=0.04; pX330 versus CAG-mRFP, P=0.52; Fig. 2H,I). Similarly, in the presence of tetrodotoxin, no difference in miniature EPSC (mEPSC) frequencies were observed between CAG-mRFP and pX330-transfected neurons, whereas mEPSC frequencies of pX330-PTEN E8-transfected neurons remained significantly higher (pX330-PTEN E8, 4.79±0.50 Hz, n=7; pX330, 2.95±0.53 Hz, n=12; CAG-mRFP, 1.33±0.19 Hz, n=6. One-way ANOVA: F2,22=8.85, P=0.001; followed by Tukey's multiple comparisons test: pX330-PTEN E8 versus pX330, P=0.04; pX330-PTEN E8 versus CAG-mRFP, P=0.001; pX330 versus CAG-mRFP, P=0.10; Fig. 2G). Taken together, the phenotypes from the PTEN CRISPR-transfected neurons were remarkably consistent with the loss-of-function PTEN phenotypes previously described in mouse knockouts, and RNAi-treated neurons (Kwon et al., 2001; Fraser et al., 2004; Luikart et al., 2011).
NF1 and P53 CRISPR
To explore further the application of CRISPR/Cas9 and to study the effects of somatic mutations on the lineage of neural progenitors, we designed and tested sgRNAs targeting rat neurofibromin 1 (NF1) exon 1 (pX330-NF1 E1) and P53 exon 6 (pX330-P53 E6). Both sgRNAs were able to introduce mutations at the targeted genomic locus in rat neuroblastoma B104 cells as determined by SURVEYOR assays (Fig. S1C). To test the efficacy of sgRNAs targeting NF1 and P53 on protein expression in vivo, we initially tested for changes in expression of both proteins in neurons labeled without the piggyBac transposon lineage label, thereby only labeling the immediately generated cohort of neurons and not subsequently generated cells in the progenitor lineage (e.g. astrocytes and oligodendrocytes). Immunohistochemistry showed that sgRNAs targeting NF1 and P53 reduced the percentage of cells expressing NF1 and P53 expression in post mitotic neurons at P19 (Fig. S1D-G). For NF1 CRISPR-transfected brains 22±0.2% (132/564, n=3 brains) were NF1 negative, whereas in three brains transfected with pX330, 1.78±0.06% (10/514) of transfected neurons were negative for NF1 (Fig. S1D,E) (one-way ANOVA: F=71.40, P=1.22×10−6). Similarly, 23.5±1.8% of pX330-P53 E6-transfected neurons (43/182, n=3, at least 3 sections from each brain) were negative for P53 (Fig. S1G) and 0.5±0.3% (4/420, n=3, at least 3 sections from each brain) transfected neurons were negative for P53 in pX330-transfected brains (Fig. S1F) (n=3, 3 sections from each brain; one-way ANOVA: F=107.15, P=1.12×10−6).
Tracking CRISPR-targeted neocortical progenitor lineages using piggyBac lineage mapping
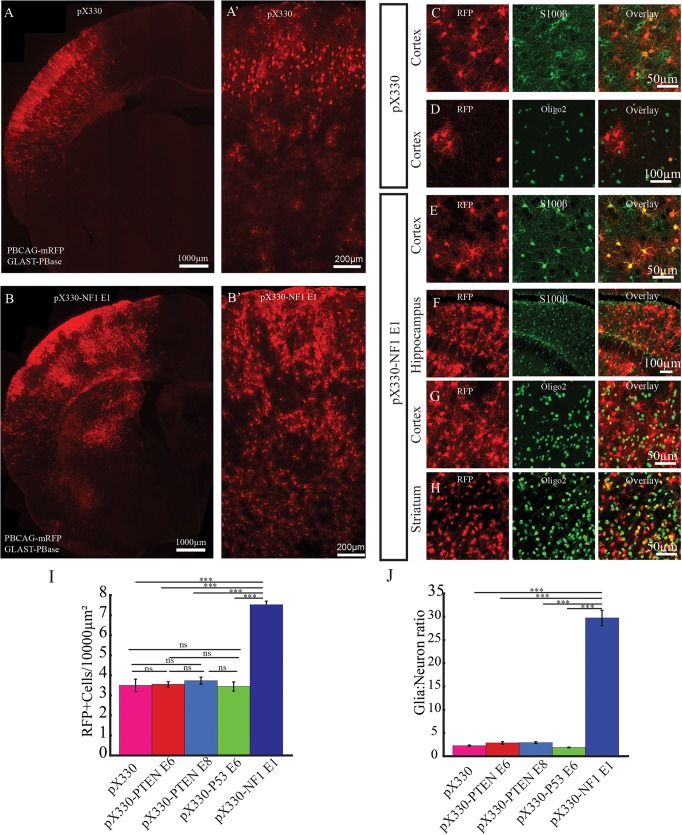
Standard IUE with episomal plasmids fails to label the entire lineage of neocortical progenitors because episomal plasmids are lost with cell division (Chen and LoTurco, 2012; Chen et al., 2014b; Siddiqi et al., 2014). In order to label the lineage of CRISPR-targeted progenitors in vivo we used a binary piggyBac transposase plasmid system that, when transfected by IUE, produces stable integration of donor transgenes into the entire progenitor lineage. For these experiments a piggyBac donor plasmid (PBCAG-eGFP or PBCAG-mRFP) was co-transfected with a helper plasmid encoding piggyBac transposase (GLAST-PBase) and CRISPR/Cas9 constructs containing sgRNAs targeting NF1, PTEN and P53, alone and in combination. A comparison of the total number of cells in the lineages resulting from transfection of the different CRISPR/Cas9 vectors (see pX330: Fig. 3A,A′,C,D; pX330-P53 E6: Fig. S2A,A′; pX330-PTEN E6: Fig. S2B,B′; pX330-PTEN E8: Fig. S2C,C′ and pX330-NF1 E1: Fig. 3B,B′,E-H), indicated that pX330-NF1 E1 caused an increase in the numbers of glial cells (Fig. 3B,I,J). As shown in Fig. 3B, patches of labeled glia cells containing cells positive for either S100β (Fig. 3E,F) or olig2 (Fig. 3G,H) were evident in the NF1 CRISPR transfection condition in neocortex (Fig. 3E,G), hippocampus (Fig. 3F) and striatum (Fig. 3H). The density of transfected cells in neocortex was 3.5±0.3, 3.6±0.1, 3.7±0.2, 3.4±0.2 and 7.5±1.8 cells/104 µm2 in sections containing neurons transfected with pX330, pX330-PTEN E6, pX330-PTEN E8, pX330-P53 E6 and pX330-NF1 E1, respectively (Fig. 3I) (n=3 brains, 3 sections quantified in each brain). Thus, there was an approximately twofold higher density of transfected cells in the pX330-NF1 E1 conditions that was significantly higher than in the other four conditions) (one-way ANOVA: F=138.53, P=1.19×10−11; followed by post hoc Tukey HSD test: pX330 versus pX330-NF1 E1, P=0.001; pX330-PTEN E8 versus pX330-NF1 E1, P=0.001; pX330-PTEN E6 versus pX330-NF1 E1, P=0.001; pX330-P53 E6 versus pX330-NF1 E1, P=0.001). The increased number of transfected cells in the NF1-CRISPR condition did not appear to be as a result of an increase in ongoing proliferation as assays of Ki67 labeling showed that there was no significant difference in the percentage of transfected cells that were positive for Ki67 (Fig. S3A-D,F-G). (pX330-labeled glia=1065, Ki67+=9, 0.8±0.2%, n=3 brains; pX330-PTEN E8-labeled glia=1538, Ki 67+=17, 1.1±0.3%, n=3 brains; pX330-P53 E6-labeled glia=1254, Ki67+=18, n=3 brains; pX330-NF1 E1-labeled glia=1564, Ki67+=25, 1.4±0.2%, n=3 brains. One-way ANOVA: F=1.63, P=0.20; followed by post hoc Tukey HSD test: pX330 versus pX330-NF1 E1, P=0.18; pX330-PTEN E6 versus pX330-NF1 E1, P=0.90; pX330-PTEN E8 versus pX330-NF1 E1, P=0.67; pX330-P53 E6 versus pX330-NF1 E1, P=0.90).
Fig. 3.
Loss of NF1 expression and increased gliogenesis. (A-B′) Representative images of P19 rat brains transfected with either control empty pX330 (A,A′) or pX330-NF1 E1 (B,B′). IUE was performed at E14 with the piggyBac transposon system (GLAST-PBase and PBCAG-mRFP) to track the lineage of transfected neural progenitors. A and B are representative images of transfected hemispheres. A′ and B′ are representative images of transfected neocortex. (C-H) In brains transfected with pX330, astrocytes (C) and oligodendrocytes (D) were labeled in neocortex. However, in brains transfected with pX330-NF1 E1, mRFP-labeled astrocytes (E,F) and oligodendrocytes (G,H) were frequently found in neocortex (E,G), hippocampus (F) and striatum (H). (I,J) The density of mRFP-labeled cells in neocortex (I) as well as the glia:neuron ratio of mRFP-labeled cells (J) were significantly higher in pX330-NF1 E1-transfected brains (n=3, one-way ANOVA, followed by post hoc Tukey HSD; ***P<0.001; ns, no significant difference; data are presented as means±s.e.m.).
The increased number of cells in the NF1 CRISPR-treated lineages appeared to be due primarily to an increase in glial cells. We therefore determined the percentage of glia relative to neurons in the lineages generated in the five transfection conditions. Transfected neurons and glia show distinct morphologies (Fig. S4) and could be quantified separately. Their identity as neurons or glia (both astrocytes and oligodendrocytes) was confirmed by S100β and Olig2 immunopositivity for the glial cell populations or uniform negativity for those markers as well as the presence of axons and dendrites on labeled neurons. Quantification showed that the percentage of transfected cells identified as glia in transfected brains were 96.7±0.1% for pX330-NF1 E1 (n=3 brains, 3 sections from each brain; total: 8835 glia, 309 neurons), 69.3±1.1% for pX330 (total: 2321 glia, 1028 neurons), 75.8±2.7% for pX330-PTEN E8 (total: 2142 glia, 673 neurons), 75.3±3.0% for pX330-PTEN E6 (total: 2327 glia, 756 neurons) and 66.6±0.4% for pX330-P53 E6 (total: 2321 glia, 1028 neurons) (n=3 brains, 3 sections from each brain). Statistical analysis revealed that there were significantly more labeled glia in pX330-NF1 E1-transfected brains than in brains transfected with any of the other CRISPR constructs (one-way ANOVA: F=79.57, P=6.48×10−10; followed by post hoc Tukey HSD test: pX330 versus pX330-NF1 E1, P=0.001; pX330-PTEN E8 versus pX330-NF1 E1, P=0.001; pX330-PTEN E6 versus pX330-NF1 E1, P=0.001; pX330-P53 E6 versus pX330-NF1 E1, P=0.001). No significant difference was observed between pX330-transfected brains and brains transfected with P53 targeting CRISPR constructs (one-way ANOVA: F=15.68, P=0.07). However, compared with pX330- and pX330-P53 E6-transfected brains, there was increased labeling of glia in pX330-PTEN E6- and pX330-PTEN E8-transfected brains (one-way ANOVA: F=11.60, P=0.0003; followed by post hoc Tukey HSD test: pX330-PTEN E6 versus pX330, P=0.042; pX330-PTEN E6 versus pX330-P53 E6, P=0.0017; pX330-PTEN E8 versus pX330, P=0.0025; pX330-PTEN E8 versus pX330-P53 E6, P=0.048; pX330 versus pX330-P53 E6, P=0.30). The increased percentage of glial cells in the lineages targeted by pX330-NF1 E1corresponded to increases in the ratio of glia to neurons: glia:neuron=29.8±1.7 for pX330-NF1 E1, 2.3±0.1 for pX330, 2.9±0.2, for pX330-PTEN E6, 2.9±0.2 for pX330-PTEN E8 and 1.9±0.1 for pX330-P53 E6 (Fig. 3J) (one-way ANOVA: F=403.56, P=4.55×10−15; followed by post hoc Tukey HSD test: pX330 versus pX330-NF1 E1, P=0.001; pX330-PTEN E6 versus pX330-NF1 E1, P=0.001; pX330-PTEN E8 versus pX330-NF1 E1, P=0.001; pX330-P53 E6 versus pX330-NF1 E1, P=0.001). The shift in glia:neuron ratio is consistent with previous experiments using conditional knockout alleles showing a role for NF1 in glial and neuronal fates from progenitors in the subventricular zone (SVZ) of postnatal forebrain (Wang et al., 2012).
Multiplex CRISPR/Cas9 of PTEN, NF1 and P53 in neocortical progenitors leads to formation of glioblastoma multiforme tumors
Somatic mutation of tumor suppressor genes has been implicated in several cancers (Watson et al., 2013), including tumors of the developing central nervous system. However, it is not entirely certain which combinations of mutations in different progenitor types are required for tumor formation (Jacques et al., 2010; Visvader, 2011). We therefore tested whether CRISPRs targeting tumor suppressors alone or in combination in neocortical progenitors would lead to formation of tumors. CRISPR/Cas9 targeting P53 (Figs S2A and S3C), NF1 (Fig. 3; Fig. S3B) or PTEN (Fig. 1; Figs S2B,C and S3D) alone appeared insufficient to induce tumor formation by 19 days after birth (n=5 animals in each condition). In addition, we found no significant increase in the Ki67 mitotic index for transfected cells in the conditions targeting NF1, PTEN or P53 alone relative to controls (Fig. S3F) (Ki67+ cells/mm2: pX330=38.46±6.2, n=3 brains; pX330-PTEN E8=18.37±6.7, n=3 brains; pX330-NF1 E1=48.22±10.50, n=3 brains; pX330-P53 E6=24.06±4.51, n=3. One-way ANOVA: F=3.06, P=0.056; followed by post hoc Tukey HSD test: pX330 versus pX330-PTEN E8, P=0.25; pX330 versus pX330-NF1 E1, P=0.90; pX330 versus pX330- P53 E6, P=0.55). In contrast, when we transfected a combination of three tumor suppressor CRISPR constructs, large tumors were found in every brain examined (n=11). The induced tumors were aggressive and fatal in all animals transfected with combinations and allowed to survive past P60 (n=8) with all animals dying by P75. Upon postmortem analysis all such animals were found to have large cerebral tumors. A clinical histopathological analysis of paraffin-sectioned tumor tissue revealed that tumors displayed histopathological features characteristic of human GBM that included cells diffusely infiltrating into neighboring tissue (Fig. S5A,B) and glomeruloid vascular proliferation (Fig. S5B). Some regions of tumors showed large multinucleated cells, suggesting the possible differential diagnosis of giant cell glioblastoma (Fig. S5C, black arrow), but these multinucleated cells were infrequent. Tumor cells expressed a combination of markers typical of glioblastoma tumor cells including strong positivity for vimentin (Fig. S5D), GFAP (Fig. S5E) and MAP2 (Fig. S5F). By contrast, in normal brain tissue, antibodies against GFAP (Fig. S5G) and vimentin (Fig. S5H) marked astrocytes, whereas MAP2 antibodies specifically labeled neurons (Fig. S5I). Taken together, the histopathological features of CRISPR-induced tumors are most similar to human GBM (WHO grade IV). This highly malignant and aggressive type of tumor is consistent with the 100% lethality we observed.
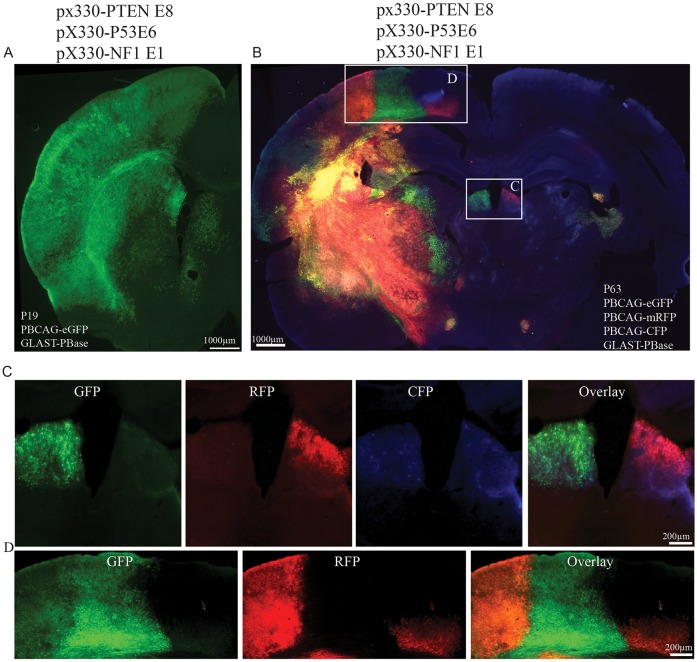
We also applied a multicolor clonal labeling approach based on the piggyBac transposon plasmid system (Chen et al., 2014a; Siddiqi et al., 2014) to visualize tumor clonal boundaries. This system works by the stochastic integration and varied expression of three different fluorescent protein reporters delivered by a mixture of fluorescent protein encoding donor plasmids (Chen et al., 2014a; Siddiqi et al., 2014). Clonally related cells inherit the same set of integrated fluorescent reporters, and thus will be labeled with the fluorescent proteins (Chen et al., 2014a). By P19 large masses of uniformly colored cells were frequently observed (n=8 brains) (Fig. 4A,B) in brains transfected with the mixture of CRISPR constructs and the multicolor labeling plasmids at E14. We found a significant increase in the Ki67-positive index in these unicolor masses, indicating increased proliferation in the tumors (Fig. S3E-G) (Ki67+ cell/mm2 in brains transfected with pX330-PTEN E8, pX330-P53 E6 and pX330-NF1 E1: 1112.36±289.79. One-way ANOVA: F=6.93, P=0.0023; followed by post hoc Tukey HSD test: pX330 versus pX330-PTEN E8/P53 E6/NF1 E1, P=0.018; pX330-PTEN E8 versus pX330-PTEN E8/P53 E6/NF1 E1, P=0.008; pX330-P53 E6 versus pX330-PTEN E8/P53 E6/NF1 E1, P=0.008; pX330-NF1 E1 versus pX330-PTEN E8/P53 E6/NF1 E1, P=0.019). Transfections by IUE are unilateral, and yet in six out of six brains examined, tumor cells and masses were found in contralateral cerebral hemispheres, suggesting that transformed cells were migratory and invasive. Tumors contralateral to the transfection location were either in single colored clusters (Fig. 4C), or in mixed color clusters (Fig. 4D) indicating further that invasive tumors could be derived from single or multiple clones. Large singly colored masses of cells were never observed in brains transfected with the multicolor piggyBac system and CRISPR targeting tyrosinase (pX330-Tyr1) (Fig. S6A,B) (n=3) or pX330-NF1 E1 (Fig. S6C,D) (n=3) suggesting that the large clonal clusters are dependent on the formation of tumors.
Fig. 4.
Multiplex CRISPR-induced tumors. (A) Representative images of P19 brain transfected with CRISPR targeting PTEN, P53 and NF1. Densely packed GFP-labeled cells were frequently found throughout the transfected cerebral hemisphere. (B) Representative image from postmortem P63 rat brains transfected with CRISPR targeting PTEN, P53 and NF1 together with multicolor piggyBac system. (C) Magnified view of boxed area in B shows two different colored tumor clusters, suggesting clonal separation. (D) Magnified view of boxed area in B shows mixing of three different colored tumor clusters.
Mutagenesis and off-targets in CRISPR-induced tumors
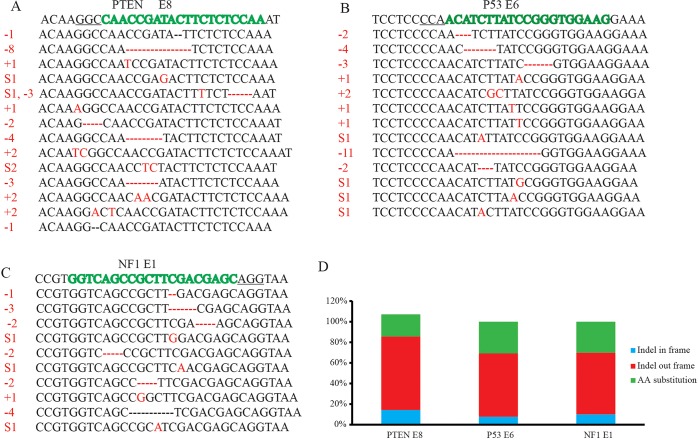
In order to identify the CRISPR/Cas9-induced mutations in the targeted regions of the tumor suppressor genes, we sorted GFP-labeled cells by FACS, amplified and subcloned genomic regions of PTEN, NF1 and P53 that were targeted by sgRNAs, and then sequenced the resulting clones by Sanger sequencing. We sequenced a total of 25 bacterial clones containing plasmids with inserts amplified from exon 8 of PTEN (Fig. 5A), 28 amplified from exon 6 of P53 (Fig. 5B), and 18 amplified from exon 1 of NF1 (Fig. 5C), and found a total of 37 with mutations in the targeted regions. The mutations included insertions, deletions and substitutions in the vicinity of the predicted Cas9 cleavage site (Fig. 5A-C). Of the 37 mutations identified, 23 were out-of-frame mutations creating premature stop codons (Fig. 5D), and 14 were in-frame missense mutations. Interestingly, no mutation occurred twice in this data set, suggesting an even larger diversity of existing mutations in the tissue. For this analysis we isolated cells from the entire population of transfected cells at P30 from four brains, a time prior to the formation of clearly developed and easily separable tumor masses. Given the diversity of mutations identified and the relatively early time in oncogenesis that we isolated cells for this assay, it could be interesting in future studies to determine whether there is a selection for different mutations over time and tumor maturation.
Fig. 5.
Induced mutations at targeted loci in sorted tumor cells. (A-C) Indels at PTEN (A), P53 (B) and NF1 (C) locus in induced GBM. Wild-type (WT) sequences are listed at the top of each figure. sgRNA sequence is marked in green and protospacer adjacent motif (PAM) sequence is underlined. Identified mutations are shown in red. −, deletion; +, insertion; S, substitution; numbers in red indicate number of nucleotides that have changed. (D) Summary of the mutation type (indel in frame, indel out frame and amino acids substitution) introduced at PTEN, P53 and NF1 loci.
To evaluate mutation efficiency and potential off-targets in tumors induced by TRIPLEX CRISPR, we performed SURVEYOR assays using genomic DNA isolated and amplified from acutely dissected tumor tissue. As shown in Fig. S7A, PTEN exon 8, P53 exon 6 and NF1 exon 1 showed an indel percentage of 47%, 81% and 24%, respectively. To investigate the specificity of the CRISPR/Cas9 system, we evaluated seven to eight potential off-targets for each target by SURVEYOR. The potential off-targets were predicted using a CRISPR design algorithm (http://crispr.mit.edu/) (Hsu et al., 2013) and based on sequence similarity to designed sgRNAs, of between one and four mismatches (Table S1). Unlike the targeted genomic regions that matched sgRNAs precisely, SURVEYOR detected no mutations at the evaluated potential off-target sites with one to four mismatches (Fig. S7B-F). Although we did not perform a genome-wide assessment of potential off-targets, our evaluation of 22 potential off-targets and failure to detect mutation at these sites is consistent with previous reports of low off-target rates of CRISPR/Cas9 in rats (Ma et al., 2014) and zebrafish (Hruscha et al., 2013).
DISCUSSION
The CRISPR/Cas9 system can introduce mutations in vivo at targeted genomic loci and abolishes gene expression with remarkably high efficiency (Incontro et al., 2014; Straub et al., 2014). Our results showed that sgRNAs targeting two different exons of PTEN (exon 6 and 8) reduced PTEN expression at similar levels; 86±3.8% and 82±5.2%, respectively, of labeled neurons transfected at E14-15 and assessed at P19 were immunonegative for PTEN (Fig. 1C,D). This level of lost expression is consistent with recent reports that more than 90% of CRISPR/Cas9-induced mutations were out-of-frame insertion or deletions (Incontro et al., 2014) in postmitotic neurons. The high out-of-frame mutation rate might be as a result of the fact that the persistent Cas9/sgRNA complex in postmitotic neurons could continue to generate double-strand breaks until non-homologous end joining repair abolishes the sgRNA binding site (Incontro et al., 2014). In contrast to postmitotic neurons, we found that the out-of-frame mutation rate in highly dividing tumor cells was 62% (Fig. 5), and similarly, we found that the efficiency of sgRNAs targeting NF1 and P53 in neurons was lower compared with PTEN mutation rates. The differences might be as a result of both the loss of Cas9/sgRNA-expressing plasmids in mitotic tumor cells, and to differences in the efficiency of targeting different genomic loci. For inducing higher rates of loss of function, it might be desirable to increase the out-of-frame mutation rate. A recent study has suggested that selecting target sequences near microhomology domains can increase the rate of indels that cause out-of-frame mutations (Bae et al., 2014). When we examined the three target regions we have sequenced in this study (PTEN E8, NF1 E1 and P53 E6) we found that sgRNA for PTEN E8, whose targeted region contained these microhomology domains, had an out-of-frame indel frequency of 71%, whereas sgRNAs whose targeted regions did not contain microhomology domains had out-of-frame indel frequencies of 61% (P53 E6) and 60% (NF1 E1), respectively.
Genome-wide binding assays of Cas9 in mammalian cells have shown that dCas9 (mutant Cas9) is able to bind to genomic DNA even without the presence of sgRNA (Wu et al., 2014). It is possible that Cas9 interaction with genomic DNA could cause experimental artifacts without a sgRNA. Although we have not performed a genome-wide study, we measured several properties of neuronal development in cells that were induced to overexpress Cas9 without sgRNA, i.e., pX330 blank vector transfection. We found that several sensitive electrophysiological and developmental outcomes were unaffected by expression of wild-type Cas9 by pX330. These unaffected outcomes, relative to fluorescent protein expression alone, included resting membrane potential and input resistance, frequency of synaptic events (Fig. 2), neuronal migration (Fig. 1D,E) and ratio of neurons to glia (Fig. 3J). These results suggest that expression of wild-type Cas9 without sgRNA fails to cause functional developmental changes in developing neurons.
The system of plasmids described here extends the application of the CRISPR/Cas9 system to assessments of modified lineages of mutated neural progenitors. We showed that loss of NF1 function caused an apparent neural progenitor fate switch and resulted in increased gliogenesis. We further demonstrated that multiplex CRISPR/Cas9, via transfection with sgRNAs targeting multiple tumor suppressors, results in GBM formation. Our results are entirely consistent with, and extend, the recent work by Gronych and colleagues (Zuckermann et al., 2015) who showed that simultaneously targeting PTEN, P53 and NF1 with CRISPR/Cas9 delivered via IUE induced GBM formation in mice. In our study, we combined the tumor-inducing CRISPR/Cas9 constructs with the multicolor piggyBac transposon system to label both the lineage of transformed cells and to distinguish clonal boundaries (Fig. 5). This approach should be useful for molecular profiling experiments and to determine whether tumor clones with different mutations contribute to tumor heterogeneity. Our study was also performed in rat and with different sgRNA sequences than those used by Zukermann et al. (2015), demonstrating both the utility of CRISPR for modeling human disease in multiple species and also the potent effect of triple mutations in PTEN, P53 and NF1. We also showed, similar to Zukermann et al. (2015), that although P53, NF1 and PTEN targeting alone do not result in tumors, distinct neuropathologies do result after NF1 or PTEN alone are targeted.
These results are consistent with the ‘multiple hit’ theory of tumorigenesis; accumulation of multiple mutations is required for tumorigenic transformation (Knudson, 1971). In many cases, tumor suppressor genes act in a cooperative manner to prevent development of tumors (Zhu et al., 2005; Kwon et al., 2008; Gregorian et al., 2009). As we showed here, loss of a single tumor suppressor was insufficient for tumor formation. The ease with which CRISPR/Cas9 can be multiplexed should be particularly valuable in studies of the formation of glial tumors in the developing brain.
Off-target effects of CRISPR/Cas9 remains a concern. We used two sgRNAs targeting different exons of PTEN and showed that neurons transfected with the two different sgRNAs exhibited the same phenotype, suggesting that the resultant phenotype is not a result of an off-target effect. We also showed that PTEN null neurons exhibited phenotypes similar to those generated in PTEN knockout or RNAi knockdown neurons (Kwon et al., 2001; Fraser et al., 2004; Luikart et al., 2011). Though we tested only one sgRNA-targeting NF1, we showed that the specific phenotype caused by NF1 sgRNA was similar to that observed in NF1 knockout studies (Wang et al., 2012). We further evaluated via SURVERYOR assay the top potential off-target sites of sgRNAs used to induce GBM, and found no mutations in those 22 evaluated sites. Limitations of using the SURVEYOR assay to evaluate potential off-targets should, however, be considered. Because of the relatively low sensitivity of the SURVERYOR assay compared with NGS approaches, not all off-target mutations, including those that might occur infrequently, would be detected (Wang et al., 2015). Recent studies using whole genome sequencing approaches to detect CRISPR off-targets revealed off-target sites not predicted by sequence similarity to target or by ChIP (Ran et al., 2015; Tsai et al., 2015). These off-targets sites include those containing mismatches to the guide as well as insertion and deletion mismatches in the guide target heteroduplex (Ran et al., 2013; Bae et al., 2014; Lin et al., 2014). Using integrase-defective lentiviral vectors, Wang and colleagues detected Cas9 off-target sites with 1 to 13 base pair mismatches between single sgRNA and its genomic targets (Wang et al., 2015). These results underline the need to better understand the mechanism of Cas9 activity, and for the importance of adequate controls. Whole genome sequencing also confirmed that using either truncated sgRNA (Tsai et al., 2015), nickase paired design (Frock et al., 2015) and modified sgRNA design might greatly reduce off-targets (Cho et al., 2014; Kim et al., 2015).
Despite the concern of off-target effect, for most experimental purposes CRISPR/Cas9 is sufficiently specific, and is likely much less prone to off-targets than RNAi or shRNA treatments that can interfere with endogenous miRNA and siRNA systems. Kim and colleagues used Digenome-seq to profile CRISPR/Cas9 off-target effects genome wide in human cells and found that Cas9 nuclease was highly specific, inducing off-target mutations at merely several rather than thousands of sites in the genome (Kim et al., 2015). Hruscha et al. (2013) reported that the mutation rates at potential off-target sites were between 1.11% and 2.5% in zebrafish (Hruscha et al., 2013), and Ma et al. (2014) found only one off-target mutation in 39 transgenic rats created using CRISPR/Cas9 (Ma et al., 2014). Most recently, Gronych and colleagues showed, by whole genome sequencing of medulloblastoma induced by CRISPR targeting of Ptch1, that there were no detectable off-target effects (Zuckermann et al., 2015). Such high specificity makes CRISPR/Cas9 a valuable tool for inducing somatic mutations in developing cells. When combined with lineage labeling, the high efficiency of gene silencing, specificity and multiplex possibilities make this approach a particularly valuable tool for studying mechanisms of normal and abnormal development of the neocortical neural progenitors.
MATERIALS AND METHODS
Plasmids and sgRNA sequences
Guide RNA sequences were designed using an online program (http://crispr.mit.edu) as described by Hsu et al. (2013). Guide sequences were cloned into pX330 vector (Addgene, plasmid 42230) (Cong et al., 2013) following normal cloning procedure. The guide sequences are as follows; PTEN exon 6 forward: 5′-GATATACATAGCGCCTCTGAC-3′; PTEN exon 6 reverse: 5′-GTCAGAGGCGCTATGTATATC-3′; PTEN exon 8 forward: 5′-ATTTGGAGAGAAGTATCGGT-3′; PTEN exon 8 reverse: 5′-ACCGATACTTCTCTCCAAAT-3′; NF1 exon 1 forward: 5′-GGTCAGCCGCTTCGACGAGC-3′; NF1 exon 1 reverse: 5′-GCTCGTCGAAGCGGCTGACC-3′; P53 exon 6 forward: 5′-CTTCCACCCGGATAAGATGT-3′; P53 exon 6 reverse: 5′-ACATCTTATCCGGGTGGAAGC-3′; Tyr1 forward: 5′-GTGCGGCCAGCTTTCAGGCAG-3′; Tyr1 reverse: 5′-CTGCCTGAAAGCTGGCCGCAC-3′.
Animals
Pregnant Wistar rats were obtained from Charles River Laboratories (Wilmington, MA, USA) and maintained at the University of Connecticut vivarium on a 12 h light cycle and fed ad libitum. Animal gestational ages were determined via palpation prior to and confirmed during surgery. Both male and female subjects were used for tumor induction. All procedures and experimental approaches were approved by the University of Connecticut IACUC.
In utero electroporation
In utero electroporation was performed as previously described (Chen et al., 2014b). Briefly, electroporation was performed at E14 or E15 and gestation age was confirmed during surgery.
Image acquisition
Multi-color imaging was performed as previously described (Chen et al., 2014a) using Zeiss Axio imager M2 microscope with Apotome with 488/546/350 nm filter cubes and the X-Cite series 120Q light source and Leica SP5 confocal microscope. All the images were further processed in Adobe Photoshop CS3 software.
Immunohistochemistry
Animals were deeply anesthetized with isoflurane and perfused transcardially with 4% paraformaldehyde/PBS (4% PFA). Samples were post-fixed overnight in 4% PFA. For immunofluorescence, brains were sectioned at 65 µm thickness on a vibratome (Leica VT 1000S). Sections were processed as free-floating sections and stained with PTEN (Cell Signaling, 9559, 1:50), NF1 (Santa Cruz, Sc-30918, 1:50), Oligo2 (Millipore, MABN 50, 1:500) P53 (Cell Signaling, 2524, 1:50), S100B (Dako, Z0311, 1:500) or Ki67 (Abcam, ab16667, 1:1000) antibody. Images were acquired and processed as previously described (Chen et al., 2014a; Siddiqi et al., 2014).
For histological analysis, immunohistochemistry and Hematoxylin and Eosin staining were carried out on paraffin-embedded 4 µm sections as described by Schick et al. (2007a,b), using GFAP (Dako, Z0334, 1:1000), MAP2c (Sigma, M4403, 1:2000), and Vimentin (DAKO, M0725, 1:1000) antibody.
SURVEYOR assay
For in vitro SURVEYOR assay, rat neuroblastoma B104 cells were transfected with CRISPR constructs using lipofectamine 2000 (Invitrogen) according to manufacturer's instruction. Two days post transfection, cells were harvested, and genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen). Genomic region flanking targeting sites were PCR amplified and SUVEYOR assay was performed using a SURVEYOR assay kit (Transgenomics) according to the manufacturer's instruction. For in vivo SURVEYOR assay, we carefully dissected out transfected cortical region from freshly removed brain under fluorescent dissection microscope. Brain tissues were chopped into small pieces and genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen). Primers for SURVEYOR assays are; PTEN exon 6 forward: 5′-CATCCATGTGAGGAACCCTGGGTG-3′; PTEN exon 6 reverse: 5′-CACTTACTGCAAGTTCCGCC-3′; PTEN exon 8 forward: 5′-CCACAAGGTGTTTGCCTTCA-3′; PTEN exon 8 reverse: 5′-CTCGAGATCCAAAGGCATTG-3′; NF1 exon 1 forward: 5′-GACGTCACCTCCAGGAGGAC-3′; NF1 exon 1 reverse: 5′-TGGGATAAAGGGGATGGAGGG-3′; P53 exon 6 forward: 5′-CCCACTTTGACCCTTGATCC-3′; P53 exon 6 reverse: 5′-CAGGCACAAACACGAACCTC-3′. Indel percentage was calculated as previously described (Cong et al., 2013).
Statistical analysis
Statistical analysis was performed using KaleidaGraph version 4.0 (Synergy Software, 2006). A confidence interval of 95% (P<0.05) was required for values to be considered statistically significant. All data are presented as means±s.e.m.
FACS and sequencing
Transfected areas from four animals (P30, no obvious presence of tumor) transfected with PBCAG-eGFP, GLAST-PBase together with multiplex CRISPRs targeting PTEN, P53 and NF1 were acutely dissected under fluorescent dissection microscope and enzymatically dissociated into a single cell suspension. FACS was performed on a Becton Dickinson FACSAria Flow Cytometer. Sorted cells were pelleted by centrifugation, and genomic DNA of sorted cells was extracted using DNeasy Blood and Tissue Kit (Qiagen). Targeted genomic areas flanking sgRNA binding sites were PCR amplified using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, 11304-011). PCR products were ligated into pCR4-TOPO vector (Invitrogen, K4530-20) for sequencing. Primers used for targeted sequencing were as follows; PTEN exon 8 forward: 5′-CCACAAGGTGTTTGCCTTCA-3′; PTEN exon 8 reverse: 5′-CTCGAGATCCAAAGGCATTG-3′; NF1 exon 1 forward: 5′-GGGGAGGACATGGCCGCACACAGGC-3′; NF1 exon 1 reverse: 5′-GCTCCTTCCTCCCCTCACCCACCGCGTC-3′; P53 exon 6 forward: 5′-CCCACTTTGACCCTTGATCC-3′; P53 exon 6 reverse: 5′-CAGGCACAAACACGAACCTC-3′.
Acknowledgements
The authors thank Dr Feng Zhang (MIT) for sharing the pX330 construct and Dr Akiko Nyshiyama (University of Connecticut) for providing the Oligo2 antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
F.C. and J.L. designed the experiments. F.C., J.R., A.C. and A.B. performed the experiments. F.C., J.R., A.C., A.B. and J.L. analyzed the data. F.C. and J.L. wrote the manuscript.
Funding
This work is supported by grants from the National Institutes of Health [RO1HD055655 and R01MH056524 to J.L.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.118836/-/DC1
References
- Backman S. A., Stambolic V., Suzuki A., Haight J., Elia A., Pretorius J., Tsao M.-S., Shannon P., Bolon B., Ivy G. O. et al. (2001). Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29, 396-403. 10.1038/ng782 [DOI] [PubMed] [Google Scholar]
- Bae S., Kweon J., Kim H. S. and Kim J.-S. (2014). Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705-706. 10.1038/nmeth.3015 [DOI] [PubMed] [Google Scholar]
- Chen F. and LoTurco J. (2012). A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods 207, 172-180. 10.1016/j.jneumeth.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E. H., Weissman J. S., Qi L. S. et al. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479-1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Becker A. J. and LoTurco J. J. (2014a). Contribution of tumor heterogeneity in a new animal model of CNS tumors. Mol. Cancer Res. 12, 742-753. 10.1158/1541-7786.MCR-13-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Maher B. J. and LoTurco J. J. (2014b). PiggyBac transposon-mediated cellular transgenesis in mammalian forebrain by in utero electroporation. Cold Spring Harb. Protoc. 2014, pdb.prot073650 10.1101/pdb.prot073650 [DOI] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., Theunissen T. W., Rangarajan S., Shivalila C. S., Dadon D. B. and Jaenisch R. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163-1171. 10.1038/cr.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S. and Lee J. (2013). Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195, 1177-1180. 10.1534/genetics.113.155853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., Bae S. and Kim J.-S. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132-141. 10.1101/gr.162339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Strong A., Patel K. M., Ng S.-L., Gosis B. S., Regan S. N., Cowan C. A., Rader D. J. and Musunuru K. (2014). Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 115, 488-492. 10.1161/CIRCRESAHA.115.304351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M. M., Zhu X., Kwon C.-H., Uhlmann E. J., Gutmann D. H. and Baker S. J. (2004). Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 64, 7773-7779. 10.1158/0008-5472.CAN-04-2487 [DOI] [PubMed] [Google Scholar]
- Frock R. L., Hu J., Meyers R. M., Ho Y.-J., Kii E. and Alt F. W. (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179-186. 10.1038/nbt.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T. and Fujii H. (2013). Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 439, 132-136. 10.1016/j.bbrc.2013.08.013 [DOI] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A. et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442-451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow S. M., Zhu W., Stolt C. C., Huang T.-W., Chen F., LoTurco J. J., Neul J. L., Wegner M., Mohila C. and Deneen B. (2014). Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat. Neurosci. 17, 1322-1329. 10.1038/nn.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., Harrison M. M., Wildonger J. and O'Connor-Giles K. M. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029-1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorian C., Nakashima J., Dry S. M., Nghiemphu P. L., Smith K. B., Ao Y., Dang J., Lawson G., Mellinghoff I. K., Mischel P. S. et al. (2009). PTEN dosage is essential for neurofibroma development and malignant transformation. Proc. Natl. Acad. Sci. USA 106, 19479-19484. 10.1073/pnas.0910398106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T., Teng F., Guo R., Li W. and Zhou Q. (2014). One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24, 372-375. 10.1038/cr.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C. and Schmid B. (2013). Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982-4987. 10.1242/dev.099085 [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O. et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827-832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S., Asensio C. S., Edwards R. H. and Nicoll R. A. (2014). Efficient, complete deletion of synaptic proteins using CRISPR. Neuron 83, 1051-1057. 10.1016/j.neuron.2014.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques T. S., Swales A., Brzozowski M. J., Henriquez N. V., Linehan J. M., Mirzadeh Z., O’ Malley C., Naumann H., Alvarez-Buylla A. and Brandner S. (2010). Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 29, 222-235. 10.1038/emboj.2009.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Bae S., Park J., Kim E., Kim S., Yu H. R., Hwang J., Kim J.-I. and Kim J.-S. (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237-243, 1 p following 243 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- Knudson A. G., Jr. (1971). Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 68, 820-823. 10.1073/pnas.68.4.820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.-H., Zhu X., Zhang J., Knoop L. L., Tharp R., Smeyne R. J., Eberhart C. G., Burger P. C. and Baker S. J. (2001). Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat. Genet. 29, 404-411. 10.1038/ng781 [DOI] [PubMed] [Google Scholar]
- Kwon C.-H., Zhao D., Chen J., Alcantara S., Li Y., Burns D. K., Mason R. P., Lee E. Y.-H. P., Wu H. and Parada L. F. (2008). Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 68, 3286-3294. 10.1158/0008-5472.CAN-07-6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cradick T. J., Brown M. T., Deshmukh H., Ranjan P., Sarode N., Wile B. M., Vertino P. M., Stewart F. J. and Bao G. (2014). CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 42, 7473-7485. 10.1093/nar/gku402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco J., Manent J.-B. and Sidiqi F. (2009). New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb. Cortex 19 Suppl 1, i120-i125. 10.1093/cercor/bhp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart B. W., Schnell E., Washburn E. K., Bensen A. L., Tovar K. R. and Westbrook G. L. (2011). Pten knockdown in vivo increases excitatory drive onto dentate granule cells. J. Neurosci. 31, 4345-4354. 10.1523/JNEUROSCI.0061-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Ma J., Zhang X., Chen W., Yu L., Lu Y., Bai L., Shen B., Huang X. and Zhang L. (2014). Generation of eGFP and Cre knockin rats by CRISPR/Cas9. FEBS J. 281, 3779-3790. 10.1111/febs.12935 [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E. and Church G. M. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823-826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P. and Lim W. A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173-1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., Zetsche B., Shalem O., Wu X., Makarova K. S. et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186-191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick V., Majores M., Engels G., Hartmann W., Elger C. E., Schramm J., Schoch S. and Becker A. J. (2007a). Differential Pi3K-pathway activation in cortical tubers and focal cortical dysplasias with balloon cells. Brain Pathol. 17, 165-173. 10.1111/j.1750-3639.2007.00059.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick V., Majores M., Koch A., Elger C. E., Schramm J., Urbach H. and Becker A. J. (2007b). Alterations of phosphatidylinositol 3-kinase pathway components in epilepsy-associated glioneuronal lesions. Epilepsia 48 Suppl 5, 65-73. 10.1111/j.1528-1167.2007.01291.x [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G. et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84-87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi F., Chen F., Aron A. W., Fiondella C. G., Patel K. and LoTurco J. J. (2014). Fate mapping by PiggyBac transposase reveals that neocortical GLAST+ progenitors generate more astrocytes than Nestin+ progenitors in rat neocortex. Cereb. Cortex 24, 508-520. 10.1093/cercor/bhs332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C., Granger A. J., Saulnier J. L. and Sabatini B. L. (2014). CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS ONE 9, e105584 10.1371/journal.pone.0105584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., Thapar V., Wyvekens N., Khayter C., Iafrate A. J., Le L. P. et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187-197. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J. E. (2011). Cells of origin in cancer. Nature 469, 314-322. 10.1038/nature09781 [DOI] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M. and Lander E. S. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80-84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kim E., Wang X., Novitch B. G., Yoshikawa K., Chang L.-S. and Zhu Y. (2012). ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell 150, 816-830. 10.1016/j.cell.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Y., Wu X., Wang J., Qiu Z., Chang T., Huang H., Lin R.-J. and Yee J.-K. (2015). Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 33, 175-178. 10.1038/nbt.3127 [DOI] [PubMed] [Google Scholar]
- Watson I. R., Takahashi K., Futreal P. A. and Chin L. (2013). Emerging patterns of somatic mutations in cancer. Nat. Rev. Genet. 14, 703-718. 10.1038/nrg3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., Dadon D. B., Cheng A. W., Trevino A. E., Konermann S., Chen S. et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670-676. 10.1038/nbt.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L. and Jaenisch R. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370-1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Guignard F., Zhao D., Liu L., Burns D. K., Mason R. P., Messing A. and Parada L. F. (2005). Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 8, 119-130. 10.1016/j.ccr.2005.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Chow L. M. L., Bayazitov I. T., Tong Y., Gilbertson R. J., Zakharenko S. S., Solecki D. J. and Baker S. J. (2012). Pten deletion causes mTorc1-dependent ectopic neuroblast differentiation without causing uniform migration defects. Development 139, 3422-3431. 10.1242/dev.083154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckermann M., Hovestadt V., Knobbe-Thomsen C. B., Zapatka M., Northcott P. A., Schramm K., Belic J., Jones D. T. W., Tschida B., Moriarity B. et al. (2015). Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 6, 7391 10.1038/ncomms8391 [DOI] [PMC free article] [PubMed] [Google Scholar]