Abstract
In the developing cochlea, sensory hair cell differentiation depends on the regulated expression of the bHLH transcription factor Atoh1. In mammals, if hair cells die they do not regenerate, leading to permanent deafness. By contrast, in non-mammalian vertebrates robust regeneration occurs through upregulation of Atoh1 in the surviving supporting cells that surround hair cells, leading to functional recovery. Investigation of crucial transcriptional events in the developing organ of Corti, including those involving Atoh1, has been hampered by limited accessibility to purified populations of the small number of cells present in the inner ear. We used µChIP and qPCR assays of FACS-purified cells to track changes in the epigenetic status of the Atoh1 locus during sensory epithelia development in the mouse. Dynamic changes in the histone modifications H3K4me3/H3K27me3, H3K9ac and H3K9me3 reveal a progression from poised, to active, to repressive marks, correlating with the onset of Atoh1 expression and its subsequent silencing during the perinatal (P1 to P6) period. Inhibition of acetylation blocked the increase in Atoh1 mRNA in nascent hair cells, as well as ongoing hair cell differentiation during embryonic organ of Corti development ex vivo. These results reveal an epigenetic mechanism of Atoh1 regulation underlying hair cell differentiation and subsequent maturation. Interestingly, the H3K4me3/H3K27me3 bivalent chromatin structure observed in progenitors persists at the Atoh1 locus in perinatal supporting cells, suggesting an explanation for the latent capacity of these cells to transdifferentiate into hair cells, and highlighting their potential as therapeutic targets in hair cell regeneration.
KEY WORDS: Epigenetics of inner ear development, Sensory hair cell differentiation, Epigenetics of Atoh1 regulation, Mouse
Highlighted article: Levels of histone H3 methylation and acetylation control the expression of Atoh1, a key regulator of hair cell differentiation, during inner ear development in mice.
INTRODUCTION
Mammalian sensory hair cells in the organ of Corti do not regenerate and their loss is the most common cause of deafness (Groves, 2010). However, in non-mammalian vertebrates, these cells regenerate and restore function within weeks of loss (Stone and Cotanche, 2007). In birds, hair cell regeneration correlates with increased levels of the basic helix-loop-helix (bHLH) transcription factor Atoh1 in surviving supporting cells (Cafaro et al., 2007), followed by their subsequent proliferation and/or direct transdifferentiation. Interestingly, although hair cell loss does not lead to widespread supporting cell regeneration in mature mammals, a latent potential for direct transdifferentiation of supporting cells to hair cells persists in the newborn mouse organ of Corti (Bramhall et al., 2014; Doetzlhofer et al., 2009; Takebayashi et al., 2007; White et al., 2006), though recent reports indicate that this potential is lost during the first week after birth (Liu et al., 2012b; Maass et al., 2015).
To better understand the mechanism of Atoh1 regulation during organ of Corti differentiation and postnatal maturation, we have analyzed the changing epigenetic status of the Atoh1 locus during organ of Corti differentiation and maturation. During development, the transcriptional hierarchy that controls cell differentiation is mediated, in part, by epigenetic mechanisms facilitated by the post-translational modification of nucleosomal histones (Arney and Fisher, 2004). For instance, the simultaneous modification of histone H3 by the repressive tri-methylation of lysine 27 (H3K27me3) and the permissive tri-methylation of lysine K4 (H3K4me3) are associated with a subset of genes that are transcriptionally silent, but poised for developmentally regulated expression, and believed to be responsible for lineage-specific differentiation (Bernstein et al., 2006). This so-called ‘bivalent’ state has been observed at the Atoh1 locus in mESCs (Azuara et al., 2006), and its resolution through removal of H3K27me3 is associated with Atoh1 expression (Jørgensen et al., 2006). Another epigenetic mark present at actively transcribed genes is H3K9ac (Wang et al., 2008), which is often opposed by H3K9me3, a mark associated with gene silencing (Kouzarides, 2007; Rea et al., 2000).
The organ of Corti develops within the cochlear duct from a postmitotic prosensory domain that forms between embryonic day (E) 12.5 and E14.5 in mice (Lee et al., 2006; Ruben, 1967). This prosensory domain is subsequently patterned into a complex mosaic of sensory hair cells and nonsensory supporting cells (Kelley, 2006). Starting between E13.5 and E14.5, Atoh1 is upregulated in nascent hair cells in the mid-basal region of the cochlea, and spreads apically along the prosensory domain until patterning is complete around E17.5. Through Notch-mediated lateral inhibition, Atoh1 expression in nascent hair cells represses Atoh1 expression in surrounding progenitors, and stimulates supporting cell differentiation (Kelley, 2006; Woods et al., 2004). Although Atoh1 is required for the differentiation of hair cells, it is subsequently downregulated, starting at about E17.5 and reduced to barely detectable levels by postnatal day (P) 6 (Driver et al., 2013; Maass et al., 2015) (Fig. 1A).
Fig. 1.
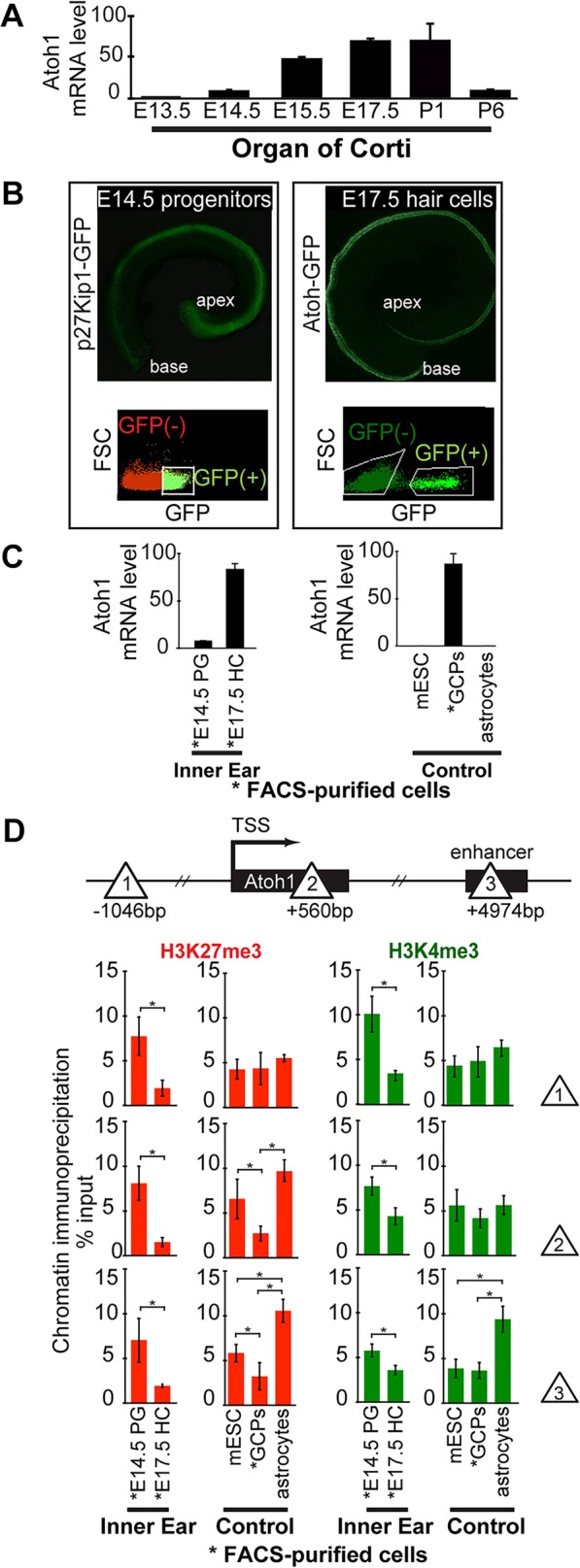
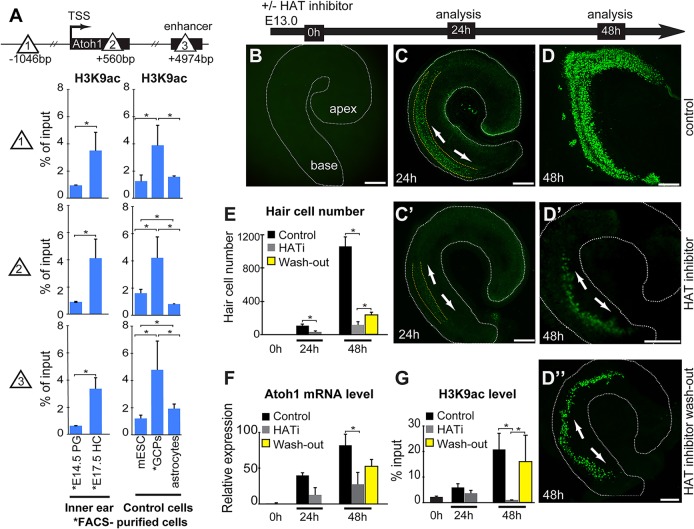
Micro-chromatin immunoprecipitation (µChIP) shows that the Atoh1 gene is bivalent (H3K27me3+ and H3K4me3+) in prosensory progenitors of the organ of Corti, and that H3K27me3 levels are strongly reduced in differentiating hair cells. (A) Relative Atoh1 mRNA levels in the cochlea increase during hair cell differentiation. Levels peak at E17.5 and then decrease during postnatal maturation. Atoh1 mRNA levels extracted from whole cochleae were analyzed at each time point by qPCR. Atoh1 expression levels are normalized to Gapdh as internal reference. Results are mean±s.e.m. (n=3). (B) Representative fluorescence images of whole-mount cochleae and FACS gating used to purify E14.5 progenitors (left, p27Kip1-GFP transgene), and nascent E17.5 hair cells (right, Atoh1-GFP transgene). (C) Relative Atoh1 mRNA levels in FACS-purified progenitors and hair cells, and in control cell types E14.5 progenitor cells (PG), E17.5 hair cells (HC), mouse embryonic stem cells (mESC) and P1 cerebellar granule cell precursors (GCPs). Atoh1 expression levels are normalized to Gapdh as internal reference. Results are mean±s.e.m. (n=3). (D) µChIP analysis of the Atoh1 locus indicates a change in relative H3K27me3 levels between Atoh1-expressing hair cells (FACS-purified from P1 Atoh1-GFP transgenic mice), compared with prosensory progenitors (FACS-purified from P1 p27Kip1-GFP transgenic mice). Schematic shows the locations across the Atoh1 locus (sites 1, 2 and 3; triangles) analyzed by µChIP qPCR for the relative abundance of H3K4me3 and H3K27me3. Results are mean±s.e.m., *P<0.05 (n=3).
Our analysis of the epigenetic status of the Atoh1 locus during organ of Corti development shows that in postmitotic prosensory domain progenitors, H3K27me3 and H3K4me3 bivalently mark the Atoh1 locus prior to Atoh1 upregulation. In nascent hair cells, a reduction of H3K27me3 and the appearance of the permissive H3K9ac accompany Atoh1 upregulation. Blocking histone acetylation in the embryonic organ of Corti blocks the basal-to-apical progression of Atoh1 upregulation and corresponding embryonic hair cell differentiation. Postnatally, hair cells downregulate Atoh1 expression, and inappropriate postnatal Atoh1 expression has been suggested to be deleterious to hair cells (Liu et al., 2012a). Downregulation of Atoh1 appears to be achieved by a reduction in histone H3 acetylation, and a simultaneous increase in H3K9me3, a marker of incipient heterochromatinization (Rea et al., 2000), providing a possible mechanism for silencing of Atoh1. Inhibiting histone deacetylation during this postnatal period blocks the downregulation of Atoh1 mRNA. Finally, we show that bivalency is maintained in early postnatal supporting cells, offering an explanation for their latent potential to transdifferentiate into hair cells, and providing a possible target for future efforts to stimulate regeneration.
RESULTS
Hair cell differentiation begins between E13.5 and E14.5 near the base of the growing cochlear duct in a prosensory domain defined by expression of the CDK inhibitor p27Kip1 (Cdkn1b – Mouse Genome Informatics) (Chen and Segil, 1999), and continues in a basal-to-apical wave before reaching the apex around E17.5 (Kelley, 2006). We used real-time quantitative PCR (qPCR) to analyze Atoh1 expression in the developing cochlear duct starting at E13.5, and observed a low level of Atoh1. Over the next several days, Atoh1 mRNA levels rise to peak expression around E17.5, before again falling to low levels by P6 (Fig. 1A) (Driver et al., 2013; Maass et al., 2015). To correlate changes in epigenetic status with changes in Atoh1 expression in specific cell types and at different ages, we used fluorescence-activated cell sorting (FACS) to separate prosensory cells prior to the onset of hair cell differentiation, from nascent hair cells that have begun to upregulate Atoh1. Prosensory cells were FACS-purified from E14.5 transgenic mice expressing GFP under the control of the prosensory marker p27Kip1 (White et al., 2006), whereas embryonic and perinatal hair cells were purified in separate experiments from a reporter strain expressing GFP under the control of an Atoh1 3′ enhancer (Chen et al., 2002; Lumpkin et al., 2003) (Fig. 1B). FACS-purified E14.5 progenitors displayed extremely low Atoh1 mRNA levels, comparable to two negative controls, mouse embryonic stem cells (mESCs) (Azuara et al., 2006) and astrocytes from prefrontal cortex of P1 mice (Jhas et al., 2006) (Fig. 1C). By contrast, nascent and maturing hair cells purified at E17.5, the peak of cochlear Atoh1 expression, showed Atoh1 levels 50 to 100 times higher – comparable to cerebellar granule cell precursors (GCPs), a positive control cell population known to be dependent on Atoh1 expression for their differentiation (Ben-Arie et al., 1997). As Atoh1 levels in nascent hair cells rise, levels of Atoh1 in the surrounding progenitors are repressed by Notch-mediated lateral inhibition as they differentiate as supporting cells (Kelley, 2006).
The Atoh1 locus is bivalent in prosensory progenitors, and reduction in H3K27 tri-methylation correlates with the onset of hair cell differentiation
H3K27me3 and H3K4me3 bivalently modify many developmentally regulated genes that are maintained in a non-expressing, but ‘poised’ state in mESCs, and during early embryonic development (Bernstein et al., 2006). Many of these genes undergo a loss of the inhibitory H3K27me3 mark at the time of transcriptional activation. To assay bivalency in E14.5 progenitors and E17.5 hair cells, we used µChIP (Dahl and Collas, 2008) from ∼25,000 FACS-purified cells and assayed the results by qPCR (Fig. 1D). We chose three regions, based on the bivalency of Atoh1 in mESCs (Fig. 1D); position −1046 bp relative to the TSS (Site 1), a region at +560 bp within the Atoh1 coding region (Site 2), and the previously characterized Atoh1 3′ autoregulatory enhancer (Helms et al., 2000) at +4974 bp (Site 3) (Fig. S1; USCS Browser, mm9, ENCODE data).
In E14.5 prosensory cells FACS-purified from p27kip1-GFP transgenic mice, and mESCs, H3K27me3 and H3K4me3 are present across the Atoh1 gene and enhancer regions (Fig. 1D, sites 1, 2 and 3), indicative of a bivalent state and consistent with the low level of Atoh1 gene expression (Fig. 1C). By contrast, in E17.5 hair cells, we observed a significant reduction in H3K27me3 levels relative to the progenitors, and this was mirrored in our positive control GCPs. H3K4me3 was also significantly elevated in progenitors across the Atoh1 locus, and maintained at a lower level in E17.5 hair cells. It did not vary significantly between mESCs and GCPs, although its level was significantly higher at the Atoh1 enhancer in non-expressing control astrocytes. Thus, loss of bivalency correlates strongly with Atoh1 transcriptional upregulation.
Atoh1 expression during hair cell differentiation is dependent on H3K9 histone acetylation
To better understand the mechanisms regulating Atoh1 expression during hair cell differentiation, we analyzed H3K9ac, typically associated with the activation of gene expression (Karmodiya et al., 2012). H3K9ac levels increased significantly at the Atoh1 locus between E14.5 progenitors and E17.5 hair cells (Fig. 2A). This was consistent with similarly high levels in positive control GCPs, and low levels in negative control mESCs and astrocytes.
Fig. 2.
Atoh1 expression and hair cell differentiation is dependent on Atoh1-associated histone acetylation. (A) Schematic shows the locations across the Atoh1 locus (sites 1, 2 and 3; triangles) analyzed by µChIP qPCR for the presence of H3K9ac in FACS-purified E14.5 progenitors (PG) and E17.5 hair cells (HC), as well as control mouse embryonic stem cells, (mESC), cerebellar granule cell precursors (GCPs), and astrocytes. H3K9ac increases upon hair cell differentiation across the Atoh1 locus (sites 1, 2 and 3). Results are mean±s.e.m., *P<0.05 (n=3). (B-D″) HATi blocks Atoh1 upregulation and hair cell differentiation in embryonic organ of Corti explant cultures. Representative images of whole-mount cochlear epithelia from Atoh1-GFP transgenic organ of Corti. Time-mated E13.0 cochlear ducts (B) were cultured in the absence (C,D) or presence (C′,D′) of HAT inhibitor (curcumin) for 24 or 48 h as indicated. Alternatively, organs were cultured in HATi for 24 h, after which the inhibitor was washed out, and organs analyzed after another 24 h (D″). Scale bars: 100 µm. Arrows indicate the approximate origin and direction of progressive Atoh1-GFP expression. (E) Quantification of hair cell number as shown in B-D″. At time point 0 h there were no Atoh1-GFP+ cells. Control organs (black bars) have on average 105±42 Atoh1-GFP+ hair cells by 24 h and 1063±195 cells by 48 h. The presence of HAT inhibitor decreases the number of Atoh1-GFP+ cells (gray bars) to 32±30 by 24 h, and to 113±114 by 48 h of treatment relative to control. Washing out the inhibitor at 24 h restores the number of hair cells to 235±52 (yellow bars) by 48 h. Results are mean±s.e.m., *P<0.05 (n=3). (F) Atoh1 mRNA levels at the 0 h, 24 h, and 48 h time points, and following inhibitor washout, in control and HATi cultures. HATi decreases the relative Atoh1 mRNA levels 95% (from 40.0±3.4 to 12.7±10.0) at 24 h of treatment, and 66% (from 82.0±15.4 to 27.6±16.4) at 48 h compared with control organs. Washout of inhibitor at 24 h, increased Atoh1 mRNA levels 1.9-fold at 48 h compared with time-matched control (from 27.6±16.4 to 52.7±9.6). Atoh1 gene expression is normalized to Gapdh as internal reference. Results are mean±s.e.m., *P<0.05 (n=3). (G) H3K9ac-µChIP qPCR assessment of the Atoh1 transcription start site (TSS) at 0 h, 24 h or 48 h in HATi-treated organotypic cultures. HAT inhibition in nascent hair cells reduced the H3K9ac levels at the Atoh1 transcription start site 37% (from 5.9±1.4% to 3.7±1.1% of input) at 24 h, and 19.8-fold (from 20.9±6.2% to 1.0±0.1%) by 48 h. Washing out the inhibitor increases the H3K9ac level 15.2-fold (from 1.0±0.1% to 16.0±1.0%), to a level almost as high as that of the control organs at 48 h. Results are mean±s.e.m., *P<0.05 (n=3).
In vivo, developmental expression of the Atoh1-GFP transgene starts in the mid-basal part of the cochlear duct between E13.5 and E14.5, and then spreads towards the apex of the cochlea (Chen et al., 2002; Kelley, 2006). In vitro, this pattern of Atoh1-GFP expression is recapitulated in embryonic organotypic cultures starting at E13.0 (Montcouquiol and Kelley, 2003), and can be assayed by the progressive expression of the Atoh1-GFP transgenic reporter over 48 h (Fig. 2B-D), as well as by qPCR of Atoh1 mRNA (Fig. S2).
To analyze whether ongoing histone acetylation is required for progressive Atoh1 expression in the developing cochlea, E13.0 Atoh1-GFP cochlear ducts were treated with the histone acetyl-transferase inhibitor (HATi) curcumin (Balasubramanyam et al., 2004). Curcumin (30 µM) was efficient in preventing Atoh1 expression (Fig. S3). Cochlear cultures treated with HATi for either 24 or 48 h (Fig. 2C′,D′) were analyzed by immunofluorescence to assess the number of Atoh1-GFP+ hair cells (Fig. 2E), by qPCR to measure the Atoh1 mRNA levels (Fig. 2F), and by µChIP to evaluate the H3K9ac levels at the Atoh1 transcription start site (Fig. 2G). At the 0 h time point (equivalent to E13.0), there are no Atoh1-GFP+ hair cells present (Fig 2B,E), whereas 24 and 48 h later, control organs have increased the total number of GFP+ cells in a basal-to-apical gradient (Fig. 2C,D,E). The presence of HATi significantly reduces the number of GFP+ cells observed at both 24 and 48 h (Fig. 2C′,D′,E). Atoh1 mRNA and H3K9ac levels were also significantly lower in HATi-treated organs compared with controls (Fig. 2F,G).
To test the contribution of ongoing acetylation to hair cell differentiation, HATi was removed from some cultures at the 24 h time point. These washout experiments demonstrated that changes brought about by HATi were reversible, and differentiation resumed upon drug removal (Fig. 2D″). Atoh1 mRNA levels in the washout samples increased compared with the 48 h treated sample (Fig. 2F), and H3K9ac levels were restored (Fig. 2G). Dependence of Atoh1 expression on H3K9 acetylation was confirmed by using a different HATi, butyrolactone 3, which inhibits GCN5-mediated H3K9 acetylation (Biel et al., 2004) (Fig. S4).
Downregulation of Atoh1 during hair cell maturation correlates with the acquisition of H3K9me3
The expression of Atoh1 decreases between E17.5 and P6 as hair cells mature (Fig. 3A). To gain insight into the molecular basis for Atoh1 downregulation, we assayed H3K9ac as well as H3K9me3, a mark associated with incipient silencing (Rea et al., 2000). Like mRNA levels, H3K9ac levels steadily decrease between E17.5 and P6 (Fig. 3B). Interestingly, this correlates with an H3K9me3 increase, which is most prominent in the region of the Atoh1 enhancer (Fig. 3B, Site 3).
Fig. 3.
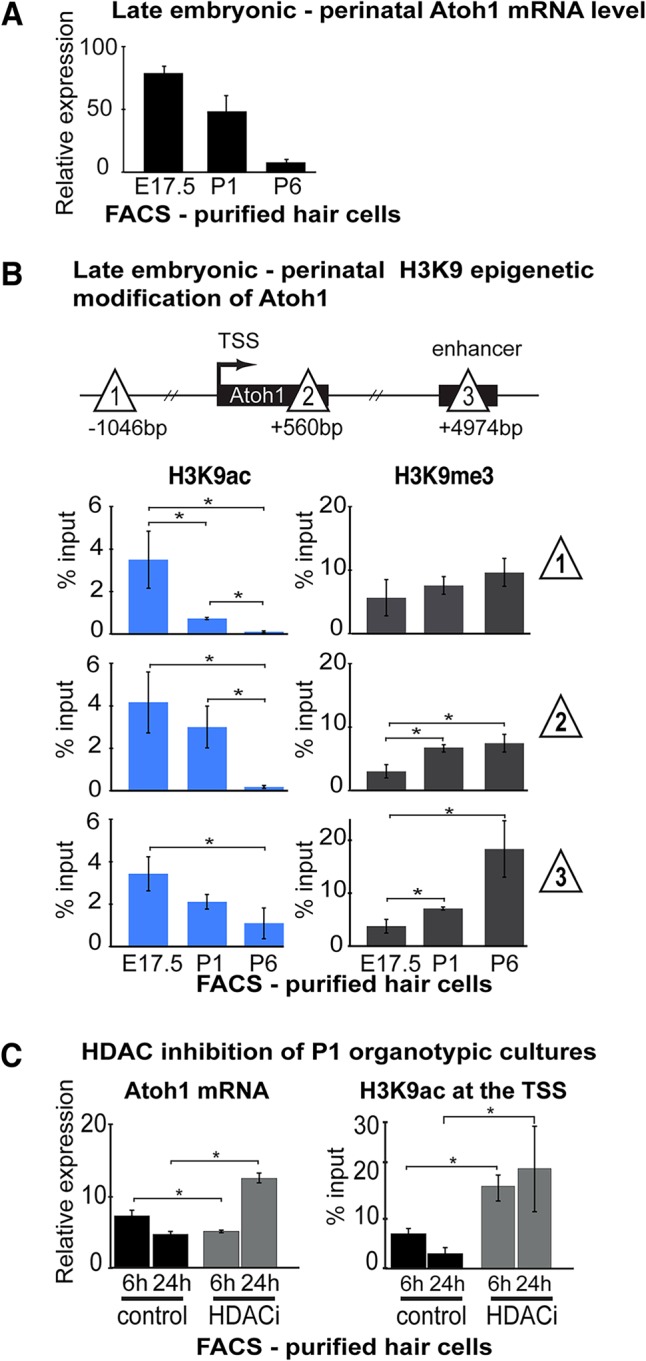
Downregulation of Atoh1 during hair cell maturation depends on H3K9 de-acetylation. Hair cells (Atoh1-GFP transgenic mice) were FACS-purified for µChIP analysis from E17.5, P1 and P6 mice. (A) Relative Atoh1 mRNA levels were measured and analyzed by qPCR. Expression levels are normalized to Gapdh internal reference. Results are mean±s.e.m. (n ≥3). (B) Schematic shows the locations across the Atoh1 locus (sites 1, 2 and 3; triangles) analyzed by µChIP qPCR for the presence of H3K9ac and H3K9me3 in FACS-purified hair cells at E17.5, P1 and P6. H3K9ac decreases from E17.5 to P6, whereas H3K9me3 levels increase over this period. Data are presented as the percent input DNA in µChIP reactions (% input). Results are mean±s.e.m., *P<0.05 (n=3). (C) HDAC inhibitor (VPA) blocks age-dependent reduction of Atoh1 mRNA (left panel) and H3K9ac levels at the Atoh1 TSS (right panel) in purified hair cells. Atoh1 mRNA relative expression increases 2.6-fold (from 4.8±0.5 to 12.6±0.7; grey bars) at 24 h of treatment compared with control (black bars). Atoh1 expression levels were normalized to Gapdh as internal reference. Results are mean±s.e.m., *P<0.05 (n=3). HDAC inhibition increases the levels of H3K9ac at the Atoh1 TSS 2.3-fold (from 7.2±1.0% to 16.5±2.6%) by 6 h, and 6.6-fold (from 3.1±1.0% to 20.0±8.5%) by 24 h of treatment. Results are mean±s.e.m., *P<0.05 (n=3).
To test the requirement for histone deacetylation of Atoh1 during postnatal downregulation, we treated P1 organ cultures for 6 or 24 h with 200 nm valproic acid (VPA), a broad-spectrum histone deacetylase inhibitor (HDACi) (Göttlicher et al., 2001) (Fig. 3C). VPA increased the Atoh1 mRNA at 24 h compared with control, correlating with the reduction of H3K9-deacetylation measured at the TSS (Fig. 3C). A similar result was obtained using a different HDACi, trichostatin A (Yoshida et al., 1990) (Fig. S5). The de-acetylation of Atoh1, along with the corresponding increase in H3K9me3, provides a potential mechanism for silencing Atoh1 expression during perinatal hair cell maturation.
Perinatal supporting cells retain a poised epigenetic state
In non-mammalian vertebrates, supporting cells undergo robust transdifferentiation in response to hair cell death (Ku et al., 2014; Stone and Cotanche, 2007). By contrast, mature mammalian supporting cells fail to transdifferentiate after hair cell loss, and deafness is permanent (Groves, 2010). However, mouse P1 supporting cells are able to transdifferentiate when Notch signaling is blocked, and this is accompanied by a sharp increase in Atoh1 mRNA expression (Doetzlhofer et al., 2009; Yamamoto et al., 2006). As it is during this same first perinatal week that the Atoh1 locus in hair cells is being silenced, we hypothesized that in P1 supporting cells Atoh1 is maintained in a transcriptionally silent, but ‘poised’ state by the presence of the repressive H3K27me3 and permissive H3K4m3 bivalency signature, previously observed in prosensory progenitors. To test this, we FACS-purified P1 supporting cells using the p27Kip1-GFP transgenic mouse (White et al., 2006) and observed that they retain high levels of H3K27me3 and H3K4me3 at the Atoh1 locus (Fig. 4). These levels are comparable to those present in E14.5 progenitors, prior to the onset of Atoh1 expression in nascent hair cells (Fig. 1D). This suggests that supporting cells retain a latent ability to activate Atoh1 expression during the perinatal period stretching from P1 to P6. By contrast, the bivalency marks are strongly reduced in hair cells, starting at P1, suggesting that the Atoh1 locus in hair cells has lost the ability to become active through the resolution of bivalency at these times.
Fig. 4.
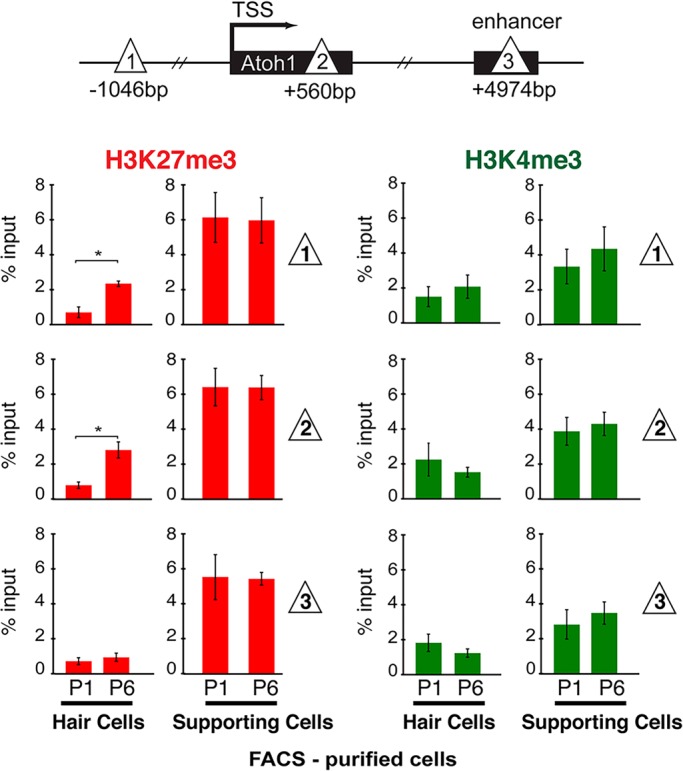
Bivalency marks H3K27me3 and H3K4me3 are maintained in perinatal supporting cells. Schematic shows the locations across the Atoh1 locus (sites 1, 2 and 3; triangles) analyzed by µChIP qPCR for the presence of H3K27me3 and H3K4me3. Hair cells (Atoh1-GFP transgenic mice) and supporting cells (p27Kip1-GFP transgenic mice) were FACS-purified from P1 and P6 mice. H3K27me3 levels remain relatively unchanged in P6 supporting cells, relative to P1. H3K4me3 is also maintained at similar levels in both P1 and P6 supporting cells. Moreover, both marks are maintained at higher levels in supporting cells at P1 and P6 compared with hair cells. Red bars, H3K27me3; green bars, H3K4me3. Data are presented as percentage of input. Results are mean±s.e.m., *P<0.05 (n=3).
DISCUSSION
As a key regulator of sensory hair cell differentiation in the inner ear, Atoh1 has become the focus of efforts to develop therapeutic strategies to overcome hearing loss. To better understand the mechanisms underlying Atoh1 transcriptional regulation, we have analyzed the epigenetic status of the Atoh1 locus during embryonic development and perinatal maturation in the organ of Corti. Our results are relevant to three important issues in organ of Corti development. First, the bivalent state at the Atoh1 locus in sensory progenitors suggests a mechanism by which prosensory cells near the apex of the cochlea are held in an undifferentiated state between E13.5 and E17.5, during which Atoh1 upregulation proceeds from the base of the cochlea. Second, the bivalent epigenetic state in perinatal supporting cells suggests a mechanism by which these cells maintain a latent potential for Atoh1 upregulation and transdifferentiation during this early postnatal phase. Third, the loss of the acetylated state of histone H3 at the Atoh1 locus in maturing hair cells, and the acquisition of H3K9me3, a mark indicative of transcriptional silencing and incipient heterochromatinization, suggest a mechanism controlling the transient nature of Atoh1 expression in hair cells.
The bivalent state of prosensory progenitors and perinatal supporting cells
We have shown that embryonic prosensory domain cells and nascent supporting cells retain a bivalent state at the Atoh1 locus (Figs 1 and 4). Bivalently marked chromatin is characteristic of many developmentally regulated genes in ESCs, and is defined by the presence of two histone post-translational modifications, H3K4me3 and H3K27me3. The presence of these two marks indicates a gene poised for expression, but repressed and waiting for an appropriate developmental stimulus (Bernstein et al., 2006). The maintenance of Atoh1 in a bivalent state in prosensory cells suggests a possible epigenetic mechanism by which Atoh1 is held in a non-expressing state during the prolonged period between initiation of Atoh1 expression in the base of the cochlea at ∼E13.5, and the final patterning of the apical regions around E17.5. Since ectopic Atoh1 is sufficient to stimulate hair cell differentiation in mice (Zheng and Gao, 2000), bivalency in prosensory cells might also underlie the limited transdifferentiation of neighboring supporting cells that is observed when hair cells are killed in the embryonic organ of Corti (Kelley et al., 1995).
Similarly, the maintained bivalency in P1 supporting cells is consistent with the retained ability to transdifferentiate into hair cell-like cells upon blockade of Notch signaling (Doetzlhofer et al., 2009; Takebayashi et al., 2007). However, the ability to transdifferentiate in response to Notch inhibition rapidly diminishes following P1 (Maass et al., 2015). In spite of this, we show that bivalency is maintained through P6, consistent with the maintenance of limited transdifferentiation potential in response to Atoh1 ectopic expression (Kelly et al., 2012; Liu et al., 2012a). At later times, ectopic Atoh1 expression does not lead to transdifferentiation (Kelly et al., 2012; Liu et al., 2012a), suggesting the hypothesis that subsequent epigenetic silencing in supporting cells underlies this failure.
Epigenetic change in nascent hair cells
Atoh1 is upregulated in nascent hair cells starting around E14, and this coincides with the repressive H3K27me3 modification at Atoh1 (Fig. 1). Removal of H3K27me3 is catalyzed by histone demethylases of the Jumanji C family (Agger et al., 2007), members of which have been reported in the P0 organ of Corti (Layman et al., 2013). It remains to be determined whether the stimulated action of these, or other, demethylases causes the selection of prosensory cells for a hair cell fate. Interestingly, astrocytes from perinatal mouse cortex, used here as a non-Atoh1-expressing control (Fig. 1C), also maintain the Atoh1 locus in a bivalent, and thus repressed, state (Fig. 1C,D), suggesting the potential for transdifferentiation to Atoh1-positive neuronal populations.
In addition to the loss of the inhibitory H3K27me3 mark, we have observed an increase in the permissive H3K9ac mark at the time of Atoh1 upregulation (Fig. 2). Our experiments with the p300/CBP histone acetyl transferase inhibitor curcumin before E14 demonstrated a need for ongoing HAT activity to maintain the basal-to-apical progression of hair cell differentiation during organ of Corti development (Fig. 2). The correlation between the increase in mRNA level and the return of histone acetylation at the locus suggests that Atoh1 chromatin state is labile and actively regulated in the service of hair cell differentiation during this time.
Following a peak of Atoh1 expression around E17.5, Atoh1 transcript levels rapidly decline to extremely low levels by P6 in hair cells (Fig. 3). This perinatal decline correlates with a rapid decline in H3K9ac, and acquisition of H3K9me3, a mark associated with incipient heterochromatinization (Rea et al., 2000). Evidence suggests that it is important to maintain Atoh1 in a silent state after the initial differentiation process is complete, as forced expression of ATOH1 in postnatal hair cells is reported to lead to cell death (Liu et al., 2012a), and an earlier report that transgenic overexpression of Atoh1 during embryogenesis results in early postnatal lethality (Helms et al., 2001). Evidence also suggests that after the perinatal period, the Atoh1 3′ enhancer is no longer able to efficiently autoregulate in response to ectopically expressed ATOH1 in supporting cells (Kelly et al., 2012; Liu et al., 2012a). We hypothesize that changes similar to these observed in hair cells at the Atoh1 locus underlie the failure of autoregulation in supporting cells at later times in maturation.
We speculate that bivalent repression of Atoh1 is the default state in much of the ectodermal lineage, which harbors several Atoh1-dependent populations (Akazawa et al., 1995; Bermingham et al., 1999; Kim et al., 2014; Lumpkin et al., 2003). This suggests that the bivalent state does not vary extensively between the progenitors of these different lineages, but rather that resolution of bivalency is regulated cell type-specifically to achieve the complex tissue-specific expression patterns observed for Atoh1. In this context, it will be interesting to chart the changes in epigenetic state of ES-derived hair cells either by directed differentiation (Koehler et al., 2013; Oshima et al., 2010) or through programming by ectopic expression of transcription factors (Costa et al., 2015).
Atoh1 is an active target for potential gene therapy studies for hair cell regeneration. Given the accessibility problems associated with transplantation into the inner ear, a targeted rewriting of the epigenetic landscape in surviving cells might provide an approach to solving the problem of hair cell regeneration. A number of small molecule epigenetic modifiers are currently under investigation. By increasing knowledge of the epigenetic landscape in surviving cell populations in the deafened ear, this study might aid in the future identification of drugs to stimulate hair cell regeneration in long-deafened individuals.
MATERIALS AND METHODS
Experimental animals
House Research Institute IACUC approved all animal procedures. For timed mating, animals were put together for 2-3 h, and the time that the vaginal plug was checked was designated as gestation day 0 (E0). For non-timed mating, animals were put together in the evening, and the plug was checked the next morning, which was designated as gestation day E0.5. Prosensory and supporting cells were isolated from transgenic mice expressing GFP under the control of p27Kip1 (White et al., 2006). Hair cells and cerebellar granule cell precursors were purified from a reporter strain expressing GFP under the control of an Atoh1 3′ enhancer (Chen et al., 2002; Lumpkin et al., 2003).
Cochlear explant cultures and drug treatment
E13 cochlear ducts were collected in PBS (Invitrogen), treated with dispase (1 mg/ml, Sigma) and collagenase (1 mg/ml, Worthington) to clear the surrounding mesenchyme and cultured on SPI Black Membrane Filters (SPI Supplies) in DMEM-F12 (Gibco) with N2 supplement (Invitrogen) and penicillin G (Sigma). Perinatal cochleae were isolated in PBS and cultured on SPI membranes in DMEM-F12/Penicillin G. All cultures were incubated in a 5% CO2/ 5% O2 humidified incubator (Forma Scientific). The explant from one ear was used as a control and the explant from the other ear was treated with a drug. Embryonic cochlear cultures were placed directly in the drug/vehicle; postnatal cultures were allowed to recover for a few hours before administering the drugs [30 µM curcumin (Enzo LifeSci, ALX-350-M010), 250 µM BA (Enzo LifeSci, ALX-778-649-18-6), 500 µM TSA (Sigma) and 200 nM VPA (Sigma)].
Cell lines
Feeder-independent mESC line E14Tg2A was generously provided by Dr Q. L. Ying (University of Southern California, USA). Cells were cultured in the presence of 1000 units ml−1 LIF on gelatin-coated tissue culture dishes (Nichols et al., 1990). Cortical astroglia cells were derived from mouse cerebral cortex as described (Kaech and Banker, 2007).
Cell purification by FACS
FACS purification of inner ear cell types was performed as described (Doetzlhofer et al., 2006). For sorting inner ear cell types, mouse cochlea were dissected, dissociated with 0.05% Trypsin (Gibco) and 1 mg/ml collagenase (Worthington) into single cell suspension and purified by FACS using a 100 µm nozzle. Only sorts with more than 96% cell purity were used for analysis of gene expression and chromatin immunoprecipitation (ChIP). Purity of the FACS sorted cells was verified by re-sorting of live cells, immunohistochemistry, and by quantitative PCR for p27Kip1 (E14.5 progenitors and supporting cells) and Atoh1 and Myo6 (hair cells) (White et al., 2006) (data not shown). For purification of cerebellar granule cell precursors, P1 cerebellum were dissected and cut into small pieces; tissue was washed several times with PBS, and incubated for 15 min at 37°C in 0.05% Trypsin (Gibco), followed by trituration in 5% fetal bovine serum using pipettes of decreasing bore size to obtain a single-cell suspension. Cells were passed through a 40 µM cell strainer to remove debris and cell clumps before proceeding with flow cytometry.
Expression analysis and quantitative real time PCR (qRT-PCR)
Total RNA was isolated using a ZR RNA MicroPrep kit (Zymo Research) including DNase I treatment (Qiagen). For RNA extraction from sorted cells, 500-2000 FACS-purified cells were sorted directly into lysis buffer; for RNA extraction from organotypic cultures, three embryonic cochlear explants were pooled for each replicate. cDNA was synthesized with iScript cDNA synthesis kit from BioRad Laboratories. qRT-PCR was performed with a SYBR Green Master Mix (Applied Biosystems) and gene-specific primers on an Applied Biosystems 7900HT Fast Real-Time PCR System. Relative quantification of gene expression was analyzed using the ΔΔCT method (Livak and Schmittgen, 2001). cDNA from postnatal day zero p27-GFPneg cochleae was used as a calibrator and expression levels were normalized to GAPDH as internal reference. The primer sequences are as follows: Gapdh: 5′-TGTGTCCGTCGTGGATCTGA-3′ (forward) and 5′-CCTGCTTCACCACCTTCTTGA-3′ (reverse), Atoh1: 5′-ATGCACGGGCTGAACCA-3′ (forward) and 5′-TCGTTGTTGAAGGACGGGATA-3′ (reverse).
Micro-chromatin immunoprecipitation (µChIP) and µChIP-qPCR
µChIP assays were performed from ∼25,000 cells per experiment, according to a previously described protocol (Dahl and Collas, 2008), with slight modifications. Briefly, FACS-purified cells were sorted into 500 µl cold PBS containing 1 mM PMSF (Sigma) and 20 mM sodium butyrate (Sigma) (for histone acetylation assays). Chromatin was cross-linked in 1% formaldehyde for 8 min (Fisher) and formaldehyde then quenched for 5 min in 125 mM glycin (Sigma) at room temperature. To remove formaldehyde, cells were centrifuged at 470 g for 10 min at 4°C in a swing-out rotor and then washed twice with ChIP-PBS. Cross-linked cells were sonicated or snap frozen in liquid nitrogen for storage at −80°C. Chromatin was sonicated to an average size of 200-300 bp using the microtip of a High Intensity Ultrasonic Processor (50 watt model; Sonics & Materials, Newtown, CT), amplitude 50 µm, power 50 (20 watts delivered to the probe), for 8×30 s, with 30 s pause. Antibody (2.4 µg) was added to 10 µl Dynabeads Protein A for 2 h, and then sonicated chromatin was added for an overnight incubation at 4°C. For each ChIP reaction, 1% chromatin was used as input DNA. Dynabeads were washed three times in RIPA, followed by 4 h chromatin de-crosslinking and elution at 68°C at 1300 rpm in an Eppendorf Thermomixer. ChIP and input DNA was purified by phenol:chloroform extraction, using 20 µg/µl glycogen as a carrier and DNA reconstituted in TE (10 mM TrisHCl,10 mM EDTA, pH 8). qPCR was performed with a SYBR Green Master Mix (Applied Biosystems) and locus-specific primers on an Applied Biosystems 7900HT Fast Real-Time PCR System with the following parameters: denaturation for 5 min at 95°C, followed by 40 cycles of 15 s at 95°C, and 60 s at 60°C. ChIP-qPCR signals were calculated as percentage of input. Antibodies used were: H3K4me3 (Active Motif 39159), H3K27me3 (Millipore 07-449), H3k9ac (Active Motif 39137), H3K9me3 (Millipore 07-442). Primers used in the ChIP-qPCR are as follows: Gapdh: 5′-GGGTTCCTATAAATACGGACTG-3′ (forward) and 5′-CTGGCACTGCACAAGAAGA-3′ (reverse), Site 1: 5′-TCAAAATGCCCAGAATCACA-3′ (forward) and 5′-CTCATAAACGCAGAGCCACAG-3′ (reverse), Site 2: 5′-CAACGACAAGAAGCTGTCCA-3′ (forward) and 5′-AACTCCGACAGAGCGTTGAT-3′ (reverse), Site 3: 5′-GGAGCATGCTTAAGCCAGAG-3′ (forward) and 5′-ATCCTTGCTGGTTCACTGCT-3′ (reverse), TSS: 5′-GGGGAGCCGGGGGAGATACAC-3′ (forward) and 5′-ACCAGGTCGCGTGCAACGAAG-3′ (reverse). It should be noted that although sonicated fragments with a mean size of 200-300 base pairs were used for µChIP analysis, fragments ranged from <200 bp to >1000 bp across the Atoh1 locus (data not shown).
Fluorescence imaging
For whole-mount fluorescence imaging, cochlear organs were dissected, placed on Superfrost Plus (Fisher) microscope slides and fixed for 1 min in 4% paraformaldehyde. Embryonic organotypic cultures were fixed and imaged directly on the SPI culture membrane following extensive washes in PBS to reduce the background coming from the membrane. Samples were mounted in Fluromount G (Southern Biotech) and analyzed with Leica CTR 6500 confocal microscope (×10 or ×20 objectives).
Statistical analysis
Data are mean±s.e.m. from at least three independent experiments. Differences between groups were tested using Student's t-tests and the null hypothesis was rejected when the P-value was <0.05. Atoh1 gene expression is normalized to Gapdh internal control. ChIP data are presented as percent of input.
Acknowledgements
We thank Sum-yan Ng for deriving the cortical astroglia cells; Juan Llamas and Welly Makmura for technical assistance; and Yassan Abdolazimi and Vincent Haoze Yu for suggestions on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Experiments were designed by all the authors, and carried out by Z.S. and T.K.; paper was written by Z.S. and N.S.
Funding
This work was supported by grants from the National Institutes of Health [#DC004189], the Hearing Health Foundation (Z.P.S., N.S.), and the generous support of the Sidgmore Family Foundation. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.126763/-/DC1
References
- Agger K., Cloos P. A. C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E. and Helin K. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731-734. 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- Akazawa C., Ishibashi M., Shimizu C., Nakanishi S. and Kageyama R. (1995). A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem. 270, 8730-8738. 10.1074/jbc.270.15.8730 [DOI] [PubMed] [Google Scholar]
- Arney K. L. and Fisher A. G. (2004). Epigenetic aspects of differentiation. J. Cell Sci. 117, 4355-4363. 10.1242/jcs.01390 [DOI] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532-538. 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K., Varier R. A., Altaf M., Swaminathan V., Siddappa N. B., Ranga U. and Kundu T. K. (2004). Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163-51171. 10.1074/jbc.M409024200 [DOI] [PubMed] [Google Scholar]
- Ben-Arie N., Bellen H. J., Armstrong D. L., McCall A. E., Gordadze P. R., Guo Q., Matzuk M. M. and Zoghbi H. Y. (1997). Math1 is essential for genesis of cerebellar granule neurons. Nature 390, 169-172. 10.1038/36579 [DOI] [PubMed] [Google Scholar]
- Bermingham N. A., Hassan B. A., Price S. D., Vollrath M. A., Ben-Arie N., Eatock R. A., Bellen H. J., Lysakowski A. and Zoghbi H. Y. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837-1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315-326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Biel M., Kretsovali A., Karatzali E., Papamatheakis J. and Giannis A. (2004). Design, synthesis, and biological evaluation of a small-molecule inhibitor of the histone acetyltransferase Gcn5. Angew. Chem. 43, 3974-3976. 10.1002/anie.200453879 [DOI] [PubMed] [Google Scholar]
- Bramhall N. F., Shi F., Arnold K., Hochedlinger K. and Edge A. S. (2014). Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2, 311-322. 10.1016/j.stemcr.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro J., Lee G. S. and Stone J. S. (2007). Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev. Dyn. 236, 156-170. 10.1002/dvdy.21023 [DOI] [PubMed] [Google Scholar]
- Chen P. and Segil N. (1999). p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581-1590. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson J. E., Zoghbi H. Y. and Segil N. (2002). The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495-2505. [DOI] [PubMed] [Google Scholar]
- Costa A., Sanchez-Guardado L., Juniat S., Gale J. E., Daudet N. and Henrique D. (2015). Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142, 1948-1959. 10.1242/dev.119149 [DOI] [PubMed] [Google Scholar]
- Dahl J. A. and Collas P. (2008). A rapid micro chromatin immunoprecipitation assay (ChIP). Nat. Protoc. 3, 1032-1045. 10.1038/nprot.2008.68 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A., White P., Lee Y.-S., Groves A. and Segil N. (2006). Prospective identification and purification of hair cell and supporting cell progenitors from the embryonic cochlea. Brain Res. 1091, 282-288. 10.1016/j.brainres.2006.02.071 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A., Basch M. L., Ohyama T., Gessler M., Groves A. K. and Segil N. (2009). Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 16, 58-69. 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver E. C., Sillers L., Coate T. M., Rose M. F. and Kelley M. W. (2013). The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol. 376, 86-98. 10.1016/j.ydbio.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlicher M., Minucci S., Zhu P., Krämer O. H., Schimpf A., Giavara S., Sleeman J. P., Lo Coco F., Nervi C., Pelicci P. G. et al. (2001). Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20, 6969-6978. 10.1093/emboj/20.24.6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves A. K. (2010). The challenge of hair cell regeneration. Exp. Biol. Med. 235, 434-446. 10.1258/ebm.2009.009281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms A. W., Abney A. L., Ben-Arie N., Zoghbi H. Y. and Johnson J. E. (2000). Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185-1196. [DOI] [PubMed] [Google Scholar]
- Helms A. W., Gowan K., Abney A., Savage T. and Johnson J. E. (2001). Overexpression of MATH1 disrupts the coordination of neural differentiation in cerebellum development. Mol. Cell Neurosci. 17, 671-682. 10.1006/mcne.2000.0969 [DOI] [PubMed] [Google Scholar]
- Jhas S., Ciura S., Belanger-Jasmin S., Dong Z., Llamosas E., Theriault F. M., Joachim K., Tang Y., Liu L., Liu J. et al. (2006). Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J. Neurosci. 26, 11061-11071. 10.1523/JNEUROSCI.1358-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen H. F., Giadrossi S., Casanova M., Endoh M., Koseki H., Brockdorff N. and Fisher A. G. (2006). Polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle 5, 1411-1414. 10.4161/cc.5.13.2927 [DOI] [PubMed] [Google Scholar]
- Kaech S. and Banker G. (2007). Culturing hippocampal neurons. Nat. Protoc. 1, 2406-2415. 10.1038/nprot.2006.356 [DOI] [PubMed] [Google Scholar]
- Karmodiya K., Krebs A. R., Oulad-Abdelghani M., Kimura H. and Tora L. (2012). H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424 10.1186/1471-2164-13-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley M. W. (2006). Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci. 7, 837-849. 10.1038/nrn1987 [DOI] [PubMed] [Google Scholar]
- Kelley M. W., Talreja D. R. and Corwin J. T. (1995). Replacement of hair cells after laser microbeam irradiation in cultured organs of corti from embryonic and neonatal mice. J. Neurosci. 15, 3013-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M. C., Chang Q., Pan A., Lin X. and Chen P. (2012). Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 32, 6699-6710. 10.1523/JNEUROSCI.5420-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-H., Li F., Ferreiro-Neira I., Ho L.-L., Luyten A., Nalapareddy K., Long H., Verzi M. and Shivdasani R. A. (2014). Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511-515. 10.1038/nature12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler K. R., Mikosz A. M., Molosh A. I., Patel D. and Hashino E. (2013). Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217-221. 10.1038/nature12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693-705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Ku Y.-C., Renaud N. A., Veile R. A., Helms C., Voelker C. C. J., Warchol M. E. and Lovett M. (2014). The transcriptome of utricle hair cell regeneration in the avian inner ear. J. Neurosci. 34, 3523-3535. 10.1523/JNEUROSCI.2606-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman W. S., Sauceda M. A. and Zuo J. (2013). Epigenetic alterations by NuRD and PRC2 in the neonatal mouse cochlea. Hear. Res. 304, 167-178. 10.1016/j.heares.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-S., Liu F. and Segil N. (2006). A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 133, 2817-2826. 10.1242/dev.02453 [DOI] [PubMed] [Google Scholar]
- Liu Z., Dearman J. A., Cox B. C., Walters B. J., Zhang L., Ayrault O., Zindy F., Gan L., Roussel M. F. and Zuo J. (2012a). Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 32, 6600-6610. 10.1523/JNEUROSCI.0818-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Owen T., Fang J., Srinivasan R. S. and Zuo J. (2012b). In vivo Notch reactivation in differentiating cochlear hair cells induces Sox2 and Prox1 expression but does not disrupt hair cell maturation. Dev. Dyn. 241, 684-696. 10.1002/dvdy.23754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lumpkin E. A., Collisson T., Parab P., Omer-Abdalla A., Haeberle H., Chen P., Doetzlhofer A., White P., Groves A., Segil N. et al. (2003). Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389-395. 10.1016/S1567-133X(03)00089-9 [DOI] [PubMed] [Google Scholar]
- Maass J. C., Gu R., Basch M. L., Waldhaus J., Lopez E. M., Xia A., Oghalai J. S., Heller S. and Groves A. K. (2015). Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front. Cell. Neurosci. 9, 110 10.3389/fncel.2015.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M. and Kelley M. W. (2003). Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J. Neurosci. 23, 9469-9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Evans E. P. and Smith A. G. (1990). Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development 110, 1341-1348. [DOI] [PubMed] [Google Scholar]
- Oshima K., Shin K., Diensthuber M., Peng A. W., Ricci A. J. and Heller S. (2010). Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell 141, 704-716. 10.1016/j.cell.2010.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z.-W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D. et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593-599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Ruben R. J. (1967). Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. Suppl. 220, 1-44. [PubMed] [Google Scholar]
- Stone J. S. and Cotanche D. A. (2007). Hair cell regeneration in the avian auditory epithelium. Int. J. Dev. Biol. 51, 633-647. 10.1387/ijdb.072408js [DOI] [PubMed] [Google Scholar]
- Takebayashi S., Yamamoto N., Yabe D., Fukuda H., Kojima K., Ito J. and Honjo T. (2007). Multiple roles of Notch signaling in cochlear development. Dev. Biol. 307, 165-178. 10.1016/j.ydbio.2007.04.035 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M. Q. et al. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897-903. 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. M., Doetzlhofer A., Lee Y. S., Groves A. K. and Segil N. (2006). Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984-987. 10.1038/nature04849 [DOI] [PubMed] [Google Scholar]
- Woods C., Montcouquiol M. and Kelley M. W. (2004). Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci. 7, 1310-1318. 10.1038/nn1349 [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Tanigaki K., Tsuji M., Yabe D., Ito J. and Honjo T. (2006). Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J. Mol. Med. (Berl) 84, 37-45. 10.1007/s00109-005-0706-9 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Kijima M., Akita M. and Beppu T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174-17179. [PubMed] [Google Scholar]
- Zheng J. L. and Gao W. Q. (2000). Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3, 580-586. 10.1038/75753 [DOI] [PubMed] [Google Scholar]