Abstract
Cell intercalation is a fundamental, coordinated cell rearrangement process that shapes tissues throughout animal development. Studies of intercalation within epithelia have focused almost exclusively on the localized constriction of specific apical junctions. Another widely deployed yet poorly understood alternative mechanism of epithelial intercalation relies on basolateral protrusive activity. Using the dorsal embryonic epidermis of Caenorhabditis elegans, we have investigated this alternative mechanism using high-resolution live cell microscopy and genetic analysis. We find that as dorsal epidermal cells migrate past one another they produce F-actin-rich protrusions polarized at their extending (medial) edges. These protrusions are controlled by the C. elegans Rac and RhoG orthologs CED-10 and MIG-2, which function redundantly to polarize actin polymerization upstream of the WAVE complex and WASP, respectively. We also identify UNC-73, the C. elegans ortholog of Trio, as a guanine nucleotide exchange factor (GEF) upstream of both CED-10 and MIG-2. Further, we identify a novel polarizing cue, CRML-1, which is the ortholog of human capping Arp2/3 myosin I linker (CARMIL), that localizes to the nonprotrusive lateral edges of dorsal cells. CRML-1 genetically suppresses UNC-73 function and, indirectly, actin polymerization. This network identifies a novel, molecularly conserved cassette that regulates epithelial intercalation via basolateral protrusive activity.
KEY WORDS: Cell intercalation, Trio, Morphogenesis, Rac GTPase, CARMIL, LRRC16A
Summary: Basolateral protrusive activity, and not apical constriction, drives cell intercalation in C. elegans embryos and is regulated by Rac/RhoG and WAVE/WASP.
INTRODUCTION
Morphogenesis requires the concurrent rearrangement of many cell types during animal development. One such coordinated cell movement, mediolateral cell intercalation, is a ubiquitous process deployed in many developmental contexts to make a tissue longer and thinner (Walck-Shannon and Hardin, 2014). Cells derived from all three germ layers can undergo mediolateral intercalation, including epithelia. The intercalation of epithelial cells is especially complex because apicobasal polarity must be maintained while cells move along the mediolateral axis. Thus far, most research on epithelial cell intercalation has focused on RhoA-mediated shrinkage of apical junctions oriented along the axis of tissue shortening (Bertet et al., 2004; Blankenship et al., 2006; Chacon-Heszele et al., 2012; Karner et al., 2009; Lienkamp et al., 2012; Nishimura et al., 2012; Simões et al., 2014).
A less well understood mechanism of epithelial intercalation involves basolateral protrusive activity. Although basolateral protrusions have been described in several systems, including the sea urchin archenteron (Hardin, 1989), the ascidian notochordal primordium (Munro and Odell, 2002a,b) and the vertebrate neural tube (Williams et al., 2014), little information is available regarding how they form. Intriguingly, recent evidence suggests that a cooperative mechanism between apical junctional constriction and basal protrusions mediates cell intercalation in the mouse neuroepithelium (Williams et al., 2014). Together, these examples suggest that basolateral protrusive activity is an important but underappreciated mechanism of epithelial cell intercalation.
A promising context for exploring basolateral protrusive activity during epithelial cell rearrangement is the dorsal epidermis of the C. elegans embryo. Four hours after fertilization, 20 dorsal epidermal cells migrate past one another in the process of dorsal intercalation (Sulston et al., 1983). These intercalating cells are bona fide, apicobasally polarized epithelia, with intact apical junctions before, during and after intercalation (McMahon et al., 2001; Patel et al., 2008; Soto et al., 2002; Williams-Masson et al., 1998; E.W.-S. and J.H., unpublished). As cells become mediolaterally polarized they adopt a wedge shape; their medial tips extend contralaterally, while their lateral edges remain rounded. Eventually, the cell body and nucleus follow as an intercalating cell makes contact with contralateral seam epidermal cells, completing intercalation (Fig. 1A).
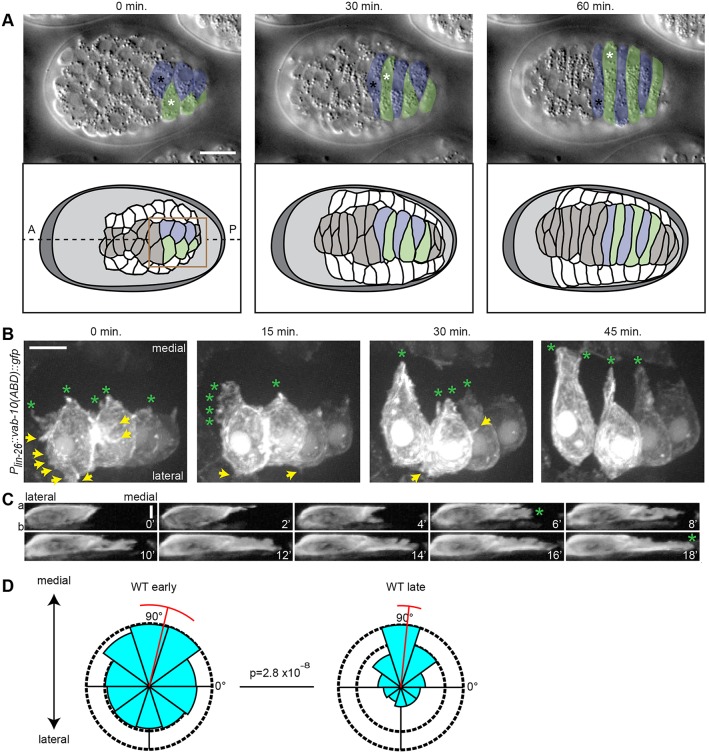
Fig. 1.
Intercalating dorsal epidermal cells exhibit basolateral protrusions. (A) (Top) DIC images of dorsal intercalation in a wild-type C. elegans embryo (dorsal view). Right nucleus, black asterisk; left nucleus, white asterisk. (Bottom) Corresponding cartoon. The box indicates the approximate area magnified in B. In this and subsequent figures anterior (A) is to the left, posterior (P) to the right; right and left cells are pseudocolored in blue and green, respectively. (B) Mosaic expression of an epidermal-specific F-actin reporter [Plin-26::vab-10(actin binding domain)::gfp] reveals dynamic protrusions during intercalation (dorsal view). Initially, small protrusions can be seen along the medial edge (green asterisks) and at positions lateral to the leading edge (yellow arrows), but the latter are eventually withdrawn as intercalation proceeds. (C) Reslices of F-actin reporter signal orthogonal to the mediolateral axis in a second embryo show that basolateral protrusions are initially extended along the apicobasal axis but consolidate into an extending medial tip (green asterisk). Apical (a) is up, basal (b) is down. Time, min. (D) Protrusions in wild-type (WT) cells become medially polarized as intercalation proceeds. Rose plots of the angle of protrusion relative to the cell centroid early (prior to and during wedging, left) and late (during tip and cell body extension, right). Red bar denotes the circular mean and deviation (Mardia-Watson-Wheeler, P=2.8×10−8). Scale bars: 10 µm in A; 5 µm in B,C.
Dorsal intercalation is marked by the formation of medially directed protrusions that extend basal to the apical junction (Williams-Masson et al., 1998). These basolateral protrusions, and not apical junction rearrangement, appear to be the major driver of dorsal intercalation, as null mutants for cadherin complex components can intercalate successfully (E.W.-S. and J.H., unpublished; Costa et al., 1998). Four-dimensional microscopy indicates that basolateral protrusions in dorsal epidermal cells bear similarities to lamellipodia in other migrating cell types (Heid et al., 2001). Pharmacological studies suggest that actin filaments are required for intercalation (Williams-Masson et al., 1998). Beyond a basic role for actin, however, the molecular basis for medial tip extension is not well understood.
Actin networks can be formed through nucleation of existing filaments via the Arp2/3 complex, which must be activated by nucleation-promoting factors of the Wiskott-Aldrich syndrome protein (WASP) family, namely WASP and WAVE (Takenawa and Suetsugu, 2007). Rho family GTPases are the predominant regulators of such F-actin networks. In its GTP-bound state, Rac can activate the highly conserved, pentameric WAVE complex, while Cdc42 can activate WASP by relieving its autoinhibition (Kim et al., 2000; Miki et al., 1998; Patel et al., 2008). The RhoG ortholog mig-2 also genetically interacts with wsp-1 (Shakir et al., 2008), although it is unclear if the activation is direct. Rather surprisingly, the main Rac homolog expressed during early embryonic development in C. elegans, ced-10, is not reported to be required for intercalation (Patel et al., 2008; Soto et al., 2002). By contrast, mutations in components of the C. elegans WAVE complex, WVE-1/WAVE, GEX-2/Sra1 and GEX-3/Nap1, lack any significant epidermal motility, indicating that they are required for epidermal intercalation (Patel et al., 2008; Soto et al., 2002). Previous analyses of WSP-1/WASP function (Withee et al., 2004) have not identified a role for WSP-1 during dorsal intercalation, although no detailed analysis of motility was performed.
Here, we analyze in detail the formation of actin-rich dynamic protrusions in intercalating dorsal cells. We use a novel tissue-specific system to show that, rather than RhoA, the Rho family GTPases CED-10/Rac and MIG-2/RhoG redundantly control these protrusions upstream of the WAVE complex and WASP, respectively. Furthermore, we show that the guanine nucleotide exchange factor (GEF) UNC-73/Trio, which is known to activate both CED-10/Rac and MIG-2/RhoG, regulates intercalation. Additionally, we show that CRML-1/CARMIL normally polarizes protrusive activity upstream of UNC-73 by inhibiting motility at the lateral edges of intercalating cells. Our results represent the first detailed analysis of actin dynamics during dorsal intercalation, and identify a novel, phylogenetically conserved molecular cassette that regulates protrusion dynamics in intercalating cells that rely on basolateral protrusive activity.
RESULTS
Intercalating dorsal epidermal cells produce highly dynamic, actin-rich protrusions
To investigate F-actin dynamics in living embryos we utilized the actin-binding domain of a C. elegans spectraplakin, VAB-10, fused to GFP and specifically expressed in the epidermis (Gally et al., 2009) as a probe, focusing on embryos that had mosaically lost the transgene in one half of intercalating cells. We found numerous protrusions throughout intercalation (Fig. 1B). Initially, protrusions formed along the entire apicobasal axis of intercalating cells, but only persisted basolaterally (Fig. 1C). As intercalation proceeded, protrusions significantly decreased in number [12.3±0.7 early, 9.4±0.8 late (mean±s.e.m.); P<0.005, two-tailed Student's t-test] and became increasingly restricted to the medial edge (Fig. 1D). Together, these data indicate that dynamic F-actin-containing protrusions are directly correlated with migration of dorsal epidermal cells, and that these protrusions rapidly become polarized along the mediolateral axis.
CED-10/Rac promotes protrusive activity in dorsal epidermal cells
Next, we investigated the functional importance of these protrusions during dorsal intercalation. Rac is the canonical Rho family GTPases responsible for tractive lamellipodia (Nobes and Hall, 1995). Given the lamellar appearance of the protrusions in dorsal epithelial cells, we reasoned that Rac would be an important regulator of their motility. The C. elegans genome encodes two Rac orthologs, CED-10 and RAC-2, and a distinct Rac- and Cdc42-like protein called MIG-2, an ortholog of mammalian RhoG and Drosophila Mtl (Lundquist, 2006). Given its predominant role in morphogenesis we focused first on CED-10/Rac.
To study CED-10 specifically in epidermal cells, we developed a novel tissue-specific, inducible, transgenic expression system. We expressed canonical dominant-negative (DN) T17N or constitutively active (CA) Q61L mutants of CED-10 (Bourne et al., 1991) driven by a variant of the lin-26 promoter that is expressed specifically in epidermal cells (see Materials and Methods) (Landmann et al., 2004). To regulate the expression of these transgenes, we exploited the endogenous nonsense-mediated mRNA decay (NMD) system, which identifies and degrades aberrant transcripts with a premature stop codon (Chang et al., 2007). The DNA encoding each fusion protein was fused to sequence encoding a long 3′UTR that contained an early stop codon, thus rendering the mRNA produced from these constructs sensitive to NMD. To make the system inducible, these tissue-specific, dominant, NMD-sensitive transgenes were put into a temperature-sensitive NMD background, smg-1(cc546ts) (Domeier et al., 2000). At the restrictive temperature (25°C) the transgenes are expressed, but at the permissive temperature (15°C) the transcripts are degraded, allowing inducible, tissue-specific protein expression (Fig. 2A, Fig. S1).
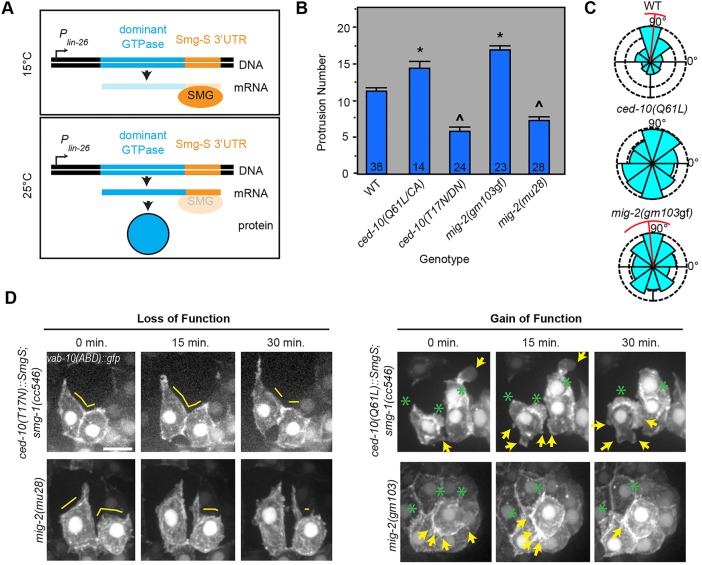
Fig. 2.
The two small GTPases CED-10/Rac and MIG-2/RhoG control protrusive activity in intercalating dorsal epidermal cells. (A) Scheme for tissue-specific conditional expression of transgenic transcripts in C. elegans. (B) The average protrusion number per cell is significantly higher in ced-10(CA) and mig-2(gm103gf) mutants compared with wild type (WT), but significantly lower in ced-10(DN) and mig-2(mu28) mutants. *P≤0.02 (ANOVA) versus wild type, ced-10(DN) and mig-2(mu28); ^P≤1×10−4 (ANOVA) versus wild type, ced-10(CA) and mig-2(gm103gf). Error bars indicate s.e.m. Sample size (number of cells) is indicated at the bottom of each bar. (C) Protrusion angles are less medially focused in ced-10(CA) and mig-2(gm103gf) during tip extension (Mardia-Watson-Wheeler, P<1×10−6). Red bar denotes the circular mean and deviation. The distribution of ced-10(CA) angles was not statistically significantly different from random (Rayleigh's R, P=0.86). (D) Medial protrusions are decreased in ced-10(DN) and mig-2 null (mu28) cells. Yellow lines indicate medial areas devoid of protrusive activity. Ectopic, lateral protrusions or blebs (yellow arrows) are observed in ced-10(CA) and, to a lesser extent, in mig-2(gm103gf) cells. Green asterisks indicate medial protrusions. Scale bar: 5 µm.
Conditional expression of CED-10(DN) in dorsal epidermal cells led to blunt medial edges in a subset of cells (29.4%, n=13), which were devoid of F-actin protrusions (Fig. 2D, Movie 1). This phenotype was also seen in the ced-10 null mutants tm597 (Shakir et al., 2006) and n3417 (Lundquist et al., 2001; Soto et al., 2002), but was harder to detect due to concurrent morphological defects (Fig. S2). Expression of CED-10(CA), on the other hand, led to rounded cells that were often unable to complete intercalation (Fig. 2D) and which exhibited excessive, unpolarized protrusions (Fig. 2B,C, Movie 2). These results suggest that CED-10 controls the intercalation of epidermal cells cell-autonomously. However, owing to the partial penetrance of these defects, we hypothesized that another Rho family GTPase functions redundantly with CED-10.
CED-10/Rac and MIG-2/RhoG function redundantly during dorsal intercalation
Embryonic RNAseq revealed no stage-appropriate expression of the other C. elegans Rac ortholog rac-2 (Celniker et al., 2009) (Fig. S3, supplementary materials and methods). Therefore, we focused on mig-2, which can act redundantly with ced-10 in other contexts (Lundquist et al., 2001). Examination of translational reporters of mig-2 and ced-10 confirmed that both proteins were present at the plasma membrane in intercalating cells (Fig. S3). Using an F-actin reporter in the dorsal epidermis, we observed that intercalating dorsal epidermal cells in mig-2(mu28) null mutants had significantly fewer protrusions than in wild type, whereas CA mig-2(gm103gf) mutants (Zipkin et al., 1997) had significantly more, unpolarized protrusions (Fig. 2B-D). These data indicate that, in addition to CED-10, MIG-2 also regulates protrusive activity during dorsal intercalation.
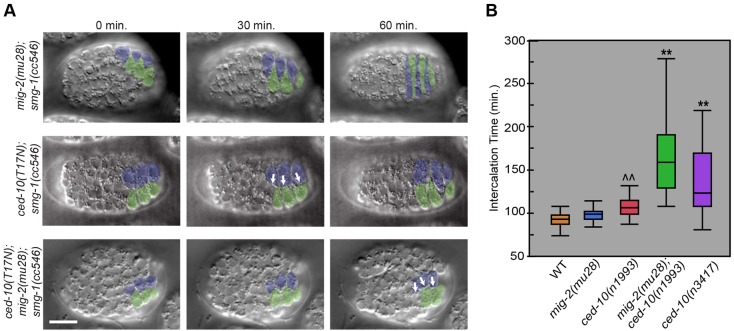
We next assessed whether mig-2 and ced-10 cooperatively regulate dorsal intercalation. Unfortunately, our attempts to generate mig-2; ced-10 loss-of-function animals carrying the F-actin reporter repeatedly failed. However, based on DIC microscopy, mig-2(mu28) mutants closely resembled wild type; however, mig-2(mu28) strongly enhanced intercalation defects in both an epidermal-specific CED-10(DN) (Fig. 3A) or a ced-10(n1993) reduced-function background (Fig. 3, Fig. S4). Moreover, these defects were more severe than in either mig-2 or ced-10 null mutants alone, suggesting that CED-10 and MIG-2 function in parallel to control intercalation. Taken together, our results indicate that the spatiotemporal regulation of F-actin protrusions is essential for proper dorsal intercalation, and that these protrusions are regulated by both CED-10/Rac and MIG-2/RhoG.
Fig. 3.
CED-10 and MIG-2 function redundantly during dorsal intercalation. (A) ced-10(T17N/DN) intercalation defects are enhanced by loss of mig-2 function. Blunt medial edges (white arrows) in ced-10(DN) mutants often resolve, whereas intercalation completely fails in ced-10(DN); mig-2(mu28) embryos. DIC images, dorsal view, 25°C. Scale bar: 10 µm. (B) Box plots showing enhancement of intercalation defects in the moderate ced-10(n1993) mutant by mig-2(mu28). Note that ced-10(n1993); mig-2(mu28) double mutants show significantly more defects (P≤3×10−4, ANOVA) than ced-10(n3417) null mutants. ^^P=0.0325 (ANOVA) versus wild type (WT); **P≤3×10−4 (ANOVA) versus all other groups.
The actin nucleation-promoting factors WVE-1 and WSP-1 function downstream of CED-10 and MIG-2, respectively
We posited that CED-10 and MIG-2 function during intercalation by interacting with effectors known to direct dendritic actin polymerization. To determine the relationship of WVE-1/WASP to CED-10/Rac and MIG-2/RhoG we performed pairwise epistasis tests. The strong wsp-1 loss-of-function allele gm324 enhanced intercalation defects in ced-10(e1993) embryos but not mig-2(mu28) embryos (Fig. S5), suggesting that wsp-1 functions in parallel to ced-10 but in the same pathway as mig-2 during intercalation. Consistent with this interpretation, wsp-1(gm324) was unable to suppress epidermal-specific CED-10(CA) (Fig. 4B), whereas it significantly suppressed intercalation defects of embryos heterozygous for mig-2(gm103gf) (Fig. 4A). During intercalation, mig-2(gm103gf) acts dominantly [heterozygotes are not significantly different from homozygotes but both are significantly different from wild type (P<0.0001, Student's t-test); Fig. 4A].
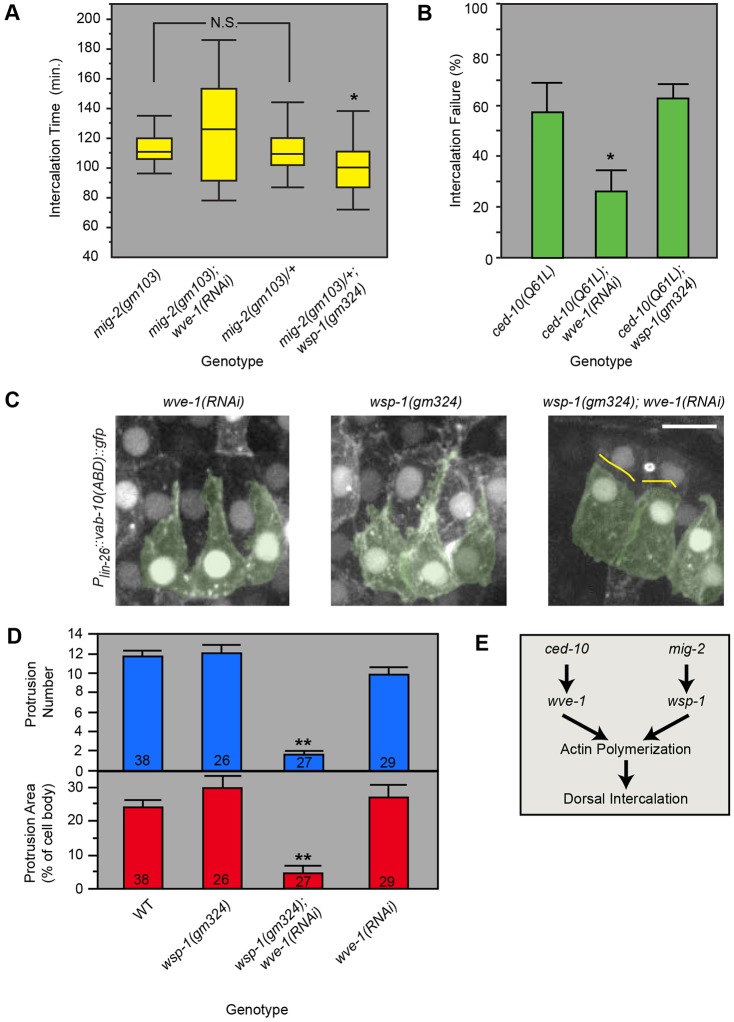
Fig. 4.
The actin nucleation activators WVE-1 and WSP-1 function downstream of CED-10 and MIG-2, respectively. (A) Box plots of intercalation time for embryos that successfully intercalated in various Rac/WAVE/WASP loss-of-function conditions. *P=0.03, Student's t-test; N.S., not significant. (B) Intercalation failure of epidermal ced-10(Q61L/CA) is suppressed by weak wve-1(RNAi) (*P=0.03, Student's t-test) but not wsp-1(gm324). Error bars indicate s.e.m. (C) WSP-1/WASP and WVE-1/WAVE function redundantly to promote F-actin protrusions in intercalating cells. Left dorsal cells are pseudocolored green. Protrusions were absent on the medial edges of wsp-1(gm324); wve-1(RNAi) cells (yellow lines). Scale bar: 5 µm. (D) Protrusion number and protrusion area were significantly lower in wsp-1(gm324); wve-1(RNAi) embryos than in all other groups (**P≤1×10−4, ANOVA). Error bars indicate s.e.m. Sample size (number of cells) is given at the bottom of each bar. WT, wild type. (E) An interpretation of genetic relationships between Rac/RhoG orthologs and actin nucleation activators during intercalation.
WAVE is encoded by wve-1 in C. elegans. There is strong evidence in C. elegans that null mutations in wve-1/WAVE, along with other components of the WAVE complex, namely gex-2/Sra1 and gex-3/Nap1, eliminate epithelial morphogenesis (Patel et al., 2008; Soto et al., 2002). Indeed, we find that dorsal epidermal cells in wve-1(ne350) or gex-2(ok1603) embryos show negligible motility (Fig. S6). To investigate the role of wve-1 and its genetic interactions during intercalation more informatively, we sought to achieve weak loss of wve-1 function. To do so, we performed RNAi, which as described previously yielded partially penetrant wve-1 knockdown (Patel et al., 2008) and intercalation failure (19.3%, n=31). The proportion of embryos that failed to intercalate was enhanced in the mig-2(mu28) background (Fig. S6), suggesting that MIG-2 and WVE-1 are in separate pathways. However, wve-1(RNAi) also enhanced defects in the ced-10(n1993) background. Based on previous studies in C. elegans growth cones (Shakir et al., 2008), we hypothesize that weak perturbation of two components in the same pathway leads to stronger loss of that pathway's function during intercalation. Providing further evidence for this interpretation, we found that wve-1(RNAi) could suppress epidermal-specific, CED-10(CA) (Fig. 4B) but not overactive mig-2(gm103gf) (Fig. 4A, Fig. S7).
In summary, these epistasis tests suggest that, as in neuronal growth cones in C. elegans (Shakir et al., 2008), the two Rho family members CED-10/Rac and MIG-2/RhoG function redundantly and act via different downstream effectors: MIG-2/RhoG acts upstream of WSP-1/WASP and CED-10/Rac acts upstream of WVE-1/WAVE (Fig. 4E). As expected given this ordering of gene function, we found that wsp-1(gm324) enhances both the intercalation failure (Fig. S6) and F-actin defects in wve-1(RNAi) (Fig. 4C,D).
The GEF UNC-73/Trio promotes protrusive activity during dorsal intercalation
To narrow down the candidate list of C. elegans GEFs that activate Rac/RhoG during dorsal intercalation, we mined the literature on GEFs for high-throughput expression data and GTPase exchange specificities (Fig. S8). We predicted that the C. elegans Trio ortholog, UNC-73, was likely to activate Rac/RhoG during dorsal intercalation. UNC-73 is part of a unique family of proteins that contain two separate GEF domains with different GTPase specificities. Each GEF domain is defined by sequential Dbl homology (DH) and Plekstrin homology (PH) domains. The first GEF domain, GEF1, activates CED-10/Rac and MIG-2/RhoG, whereas the second GEF domain, GEF2, activates RHO-1/RhoA (Fig. 5A) (Bellanger et al., 1998; Blangy et al., 2000; Debant et al., 1996; Kubiseski et al., 2003; Steven et al., 1998; Wu et al., 2002).
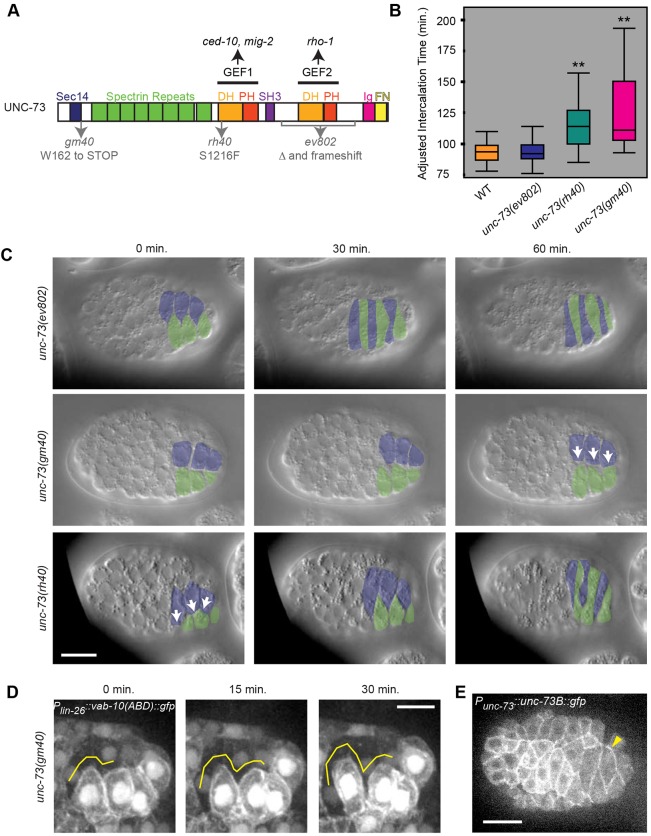
Fig. 5.
UNC-73/Trio activates protrusive activity through CED-10 and MIG-2. (A) The two RhoGEF domains in UNC-73 have different specificities. Lesions used in this study are noted at the bottom; gm40 and rh40 are GEF1 mutations, whereas ev802 is a GEF2 deletion (Δ). Domain annotations: DH, Dbl homology; PH, plekstrin homology; SH3, Src homology 3; Ig, immunoglobulin; FN, fibronectin. (B) Intercalation times of UNC-73 GEF1 domain (rh40, gm40) and UNC-73 GEF2 domain (ev802) mutants. rh40 and gm40 were significantly different from wild type (WT) and ev802 (**P<1×104), but not from one another (P=0.2776, ANOVA). Intercalation time was normalized based on timing of a landmark terminal cell division within the epidermal lineage for all groups due to differences in overall developmental timing. (C) DIC images of unc-73 mutants during intercalation reveal blunt medial edges (white arrows) in GEF1 mutants. The first time point is 1 h after terminal epidermal cell divisions. (D) UNC-73 GEF1 controls protrusive activity in dorsal intercalating cells. Medial edges are devoid of protrusions (yellow lines) in the GEF1 mutant gm40. (E) A GFP fusion of the GEF1 isoform, UNC-73B, is expressed uniformly at the periphery of intercalating cells (yellow arrowhead points to lateral UNC-73B::GFP). Scale bars: 10 µm in C,E; 5 µm in D.
Domain-specific lesions in unc-73 allowed us to take a structure/function approach to ask which GEF domains in UNC-73 are required for intercalation. gm40 is a premature stop codon that results in a protein truncated before both GEF domains; however, alternative promoters within unc-73 allow multiple GEF2 isoforms to be expressed (Steven et al., 2005). rh40 is a missense mutation within the GEF1 domain that decreases its ability to activate CED-10 and MIG-2 (Kubiseski et al., 2003). ev802 is a deletion of the GEF2 domain, which can be rescued by unc-73 DNA with the GEF1 domain deleted, suggesting that it is a GEF2-specific mutant (Steven et al., 2005). As expected, mutations disrupting GEF1 (gm40 and rh40) resulted in significantly longer intercalation times than in wild type due to blunted medial edges, similar to CED-10(DN) (Fig. 5B,C). After normalizing for overall slower development, intercalation time in the GEF2 mutant ev802 was not significantly different than similarly adjusted wild-type controls. As predicted, dorsal cell protrusions were dramatically decreased in unc-73(gm40) mutants (Fig. 5D, Movie 3). The requirement for the GEF1 domain of UNC-73 indicates that the main function of UNC-73 is to act as an upstream regulator of Rac/RhoG-dependent protrusive activity during dorsal intercalation. Previous biochemical studies confirm that the GEF1 domain of UNC-73 can activate both MIG-2 and CED-10, consistent with this conclusion (Kubiseski et al., 2003).
Protrusive activity is inhibited by the UNC-73/Trio regulator CRML-1/CARMIL
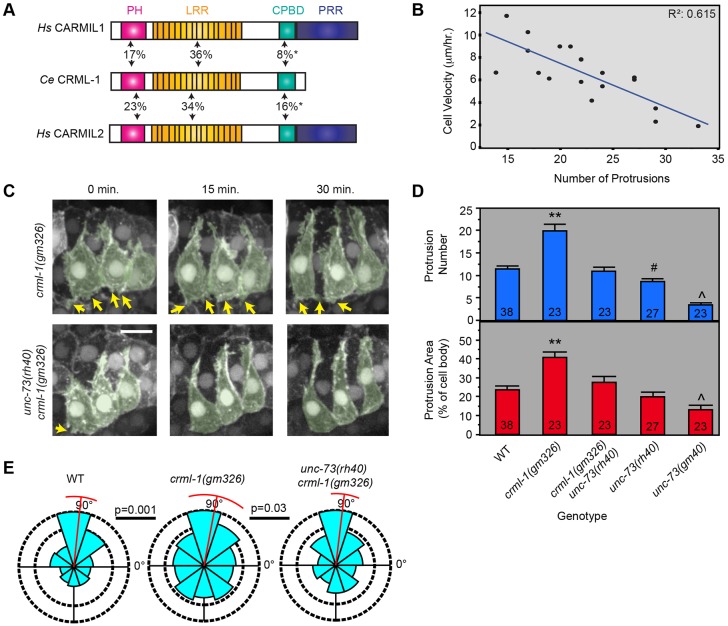
We expected UNC-73B::GFP, a GEF1-specific UNC-73 fusion protein (Steven et al., 2005), to be enriched at protrusively active medial edges of intercalating cells. Instead, we found that during dorsal intercalation UNC-73B::GFP was localized along the entire dorsal cell membrane, even at edges that are not protrusively active (Fig. 5E). Because the protrusions themselves are obviously polarized (Fig. 1D) but the localization of an upstream regulator, UNC-73, is not, we sought to identify proteins that might contribute to the spatial restriction of UNC-73 activity at or near the leading edge. We took a candidate gene approach to determine if known regulators of UNC-73, Trio or the shorter vertebrate isoform Kalirin have intercalation phenotypes, and found that crml-1(n1962) mutants displayed significantly increased intercalation time (Fig. S9A). crml-1 is a member of the conserved CARMIL (capping Arp2/3 myosin I linker) gene family first discovered in Dictyostelium (Jung et al., 2001). CARMILs consist of a non-canonical plekstrin homology (PH) domain, leucine-rich repeats (LRR), a capping protein-binding domain (CPBD) and a proline-rich region (PRR) (Zwolak et al., 2013) (Fig. 6A). In vertebrates, CARMIL proteins can allosterically inhibit the ability of capping protein to bind the barbed ends of F-actin, thereby allowing more actin polymerization and hence protrusive activity (Fujiwara et al., 2014; Kim et al., 2012; Takeda et al., 2010; Yang et al., 2005). However, based on co-immunoprecipitation analysis, human CARMIL1 (LRRC16A) (Liang et al., 2009) and C. elegans CRML-1 (Vanderzalm et al., 2009) are also part of a complex with Trio and UNC-73, respectively. The significance of this interaction is largely uncharacterized. It has been suggested that CRML-1 functions upstream of UNC-73 to inhibit its function, as crml-1 mutations fail to suppress unc-73 mutations in neurons (Vanderzalm et al., 2009). Consistent with the hypothesis that CRML-1/CARMIL inhibits protrusions during intercalation, we find that nonsense mutations in crml-1, gm326 and n1962, or crml-1(RNAi) result in excessive, unpolarized F-actin protrusions in dorsal epidermal cells that are inversely related to intercalating cell velocity (Fig. 6B, Movie 4). Moreover, the extra, nonpolarized protrusions in crml-1(gm326) embryos are suppressed by the unc-73(rh40) mutation (Fig. 6). Together, these data suggest that UNC-73/Trio functions downstream of a negative regulator, CRML-1/CARMIL, to inhibit protrusive activity during dorsal intercalation.
Fig. 6.
UNC-73 function is inhibited by CRML-1. (A) Conservation between human (Hs) and C. elegans (Ce) CARMIL family members. Percent identity is indicated; asterisks mark conserved capping protein-binding sites (Edwards et al., 2014); see main text for domain annotations. (B) Correlation between the number of protrusions and cell velocity in crml-1(gm326) (R2=0.615; P<0.0001, F-test). (C) crml-1(gm326) dorsal cells have excessive protrusive activity, which can be suppressed by GEF1 loss of function in unc-73(rh40) mutants. Left-hand cells are pseudocolored green. Yellow arrows indicate excessive, lateral protrusions. Scale bar: 5 µm. (D) Both the increased number (top) and the increased total area (bottom) of protrusions in crml-1(gm326) are suppressed by unc-73(rh40); P<1×10−4, P=6×10−4, respectively (ANOVA). **, significantly different from all other groups; #, significantly different from wild type and unc-73(gm40); ^, significantly different from wild type (WT) and unc-73(rh40). Error bars indicate s.e.m. Sample size (number of cells) is given at the bottom of each bar. (E) During tip extension, protrusions in crml-1(gm326) are less polarized than in wild type (P=0.001, Mardia-Watson-Wheeler). Protrusions in unc-73(rh40) crml-1(gm326) mutants are significantly more polarized than in crml-1(gm326) alone (P=0.03, Mardia-Watson-Wheeler).
We predicted that CRML-1 might spatially restrict UNC-73 activity at the lateral (rear) edges of dorsal epidermal cells, where protrusions are usually absent, thus confining UNC-73 activity to the medial, extending tip. To examine the spatial localization of CRML-1, we made an epidermal-specific CRML-1::GFP fusion protein (using the lbp-1 promoter; Plenefisch et al., 2000). This construct rescued the excessive F-actin protrusions in dorsal epidermal cells of crml-1(gm326) mutants (Fig. S9), indicating that the construct is functional and specifically required in the epidermis. Moreover, during tip extension we found that CRML-1::GFP consistently accumulated in lateral regions of dorsal epidermal cells (compared with broadly localized PH::mCherry controls) (Fig. 7B), where protrusions are normally absent. Later during intercalation we also observed transient expression of CRML-1::GFP in the advancing tips after cells reached the contralateral side (n=9/10; white arrowheads, Fig. 7A). This late accumulation after protrusions cease further supports the interpretation that CRML-1 inhibits protrusive activity. Together, these data suggest that polarized CRML-1 expression at the rear of intercalating cells spatially inhibits UNC-73 and dampens downstream protrusive activity.
Fig. 7.
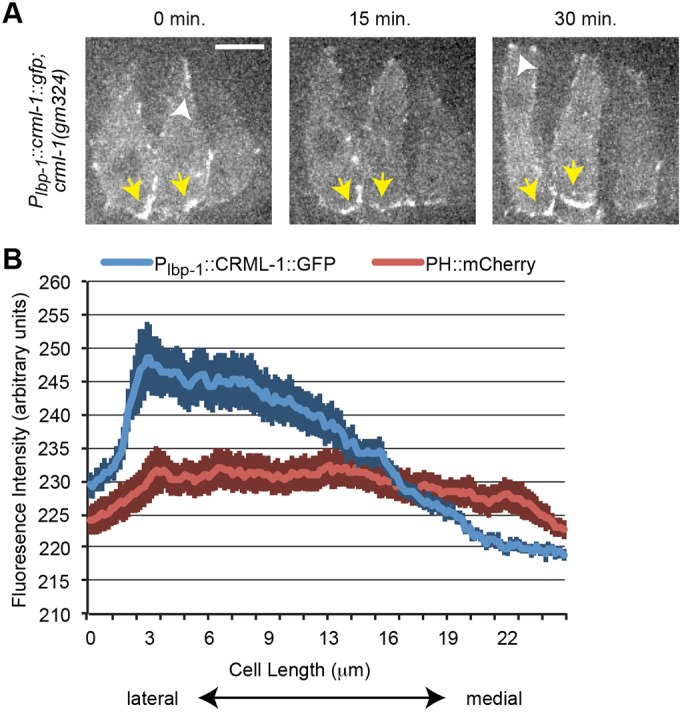
Epidermal CRML-1::GFP is enriched laterally in dorsal cells. (A) A rescuing CRML-1::GFP transgene localizes to the lateral edges of dorsal cells (yellow arrows). White arrowheads point to transient areas of CRML-1::GFP expression at the medial edge, which accumulate after the tip reaches the contralateral edge. Scale bar: 5 µm. (B) CRML-1::GFP is enriched laterally in intercalating dorsal epidermal cells. Average fluorescence intensity along the mediolateral axis for CRML-1::GFP (blue) and PH::mCherry (red). Measurements were averaged from at least seven intercalating dorsal cells during tip extension. Darker vertical bars indicate s.e.m. at each position.
DISCUSSION
Our work defines a new molecular network that promotes epithelial cell intercalation, driven by Rac/RhoG. CED-10/Rac and MIG-2/RhoG function redundantly downstream of activation by the GEF1 domain of UNC-73/Trio to drive dendritic actin polymerization. Our data suggest that CED-10/Rac signals through the WAVE complex, whereas MIG-2/RhoG signals through WSP-1. Moreover, protrusive activity is polarized through the negative UNC-73/Trio regulator CRML-1/CARMIL, which accumulates at the lateral edges of dorsal epidermal cells. Together, these data suggest that cell intercalation in the dorsal epidermis of C. elegans proceeds via protrusions promoted by a Rac/RhoG-dependent pathway that is tightly regulated, both temporally and spatially (Fig. 8). These protrusions provide the major driving force for intercalation. When they are nearly completely abrogated, as in wsp-1(gm324); wve-1(RNAi) or wve-1(ne350) embryos, intercalation fails (Fig. 4C,D, Fig. S6). Moreover, intercalation requires that these protrusions are polarized, as shown in CED-10(CA) (Fig. 2) or crml-1 mutants (Fig. 6), in which unpolarized protrusions result in dorsal intercalation failure (Fig. 4B) or marked delay (Fig. S9A), respectively. Together, these data suggest that polarized protrusions are crucial for the directional translocation of epithelial cells during dorsal intercalation in C. elegans.
Fig. 8.
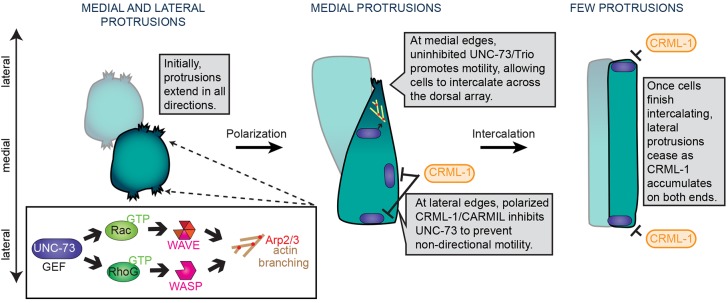
A model for the molecular control of protrusive activity during dorsal intercalation. Early, as intercalating cells begin to adopt a polarized, wedge-shaped morphology, protrusive activity is broadly distributed at the cell periphery. As intercalation proceeds, protrusions focus medially and basolaterally to produce a single, extending tip. As cell bodies translocate, cells migrate past one another and intercalation completes. The GEF UNC-73, which is broadly distributed, activates CED-10 and MIG-2, but only in regions devoid of CRML-1, which inhibits UNC-73 at lateral edges. Accumulation of CRML-1 at the contralateral edges of intercalating cells may promote protrusive downregulation as intercalation completes.
Most of the existing work on epithelial intercalation has focused on the Rho-driven shrinkage of junctions oriented specifically in the axis of tissue shortening (Levayer et al., 2011; Nishimura et al., 2012; Simões et al., 2014; Warrington et al., 2013). In the C. elegans dorsal epidermis, by contrast, zygotic rho-1(ok2418) mutants or epidermal-specific RHO-1(T17N) embryos can still intercalate, even though cytokinesis has failed and the cells are binucleate in the latter (E.W.-S. and J.H., unpublished). This is, to our knowledge, the first study to investigate Rac and RhoG during epithelial intercalation. Previous reports suggest that basolateral protrusions exist in other epithelial tissues as well (Hardin, 1989; Munro and Odell, 2002b; Williams et al., 2014). Therefore, Rac- and/or RhoG-driven basolateral mechanisms of epithelial intercalation might be more widespread than previously appreciated.
Racs are known to act via downstream effectors that stimulate Arp2/3-dependent dendritic actin networks (Heasman and Ridley, 2008). Previously, there had been some ambiguity about whether actin polymerization was required for dorsal intercalation (Sawa, 2003; Soto et al., 2002). Here, we provide several lines of evidence that the WAVE complex and WASP are required for dorsal intercalation downstream of CED-10 and MIG-2, respectively. First, wsp-1(gm324) can both enhance ced-10(n1993) mutants and suppress mig-2(gm103gf) mutants. Second, wve-1(RNAi) can enhance mig-2(mu28) mutants and suppress CED-10(CA) specifically expressed in the epidermis. Third, wsp-1(gm324) can enhance the intercalation failure of wve-1(RNAi). However, given the relatively low penetrance of intercalation defects in ced-10/Rac null mutants compared with wve-1/WAVE null mutants, which display complete intercalation failure, additional upstream activators are likely to regulate the WAVE complex. Recent evidence suggests that within the WAVE complex there is a second, Rac-independent interaction site, which is conserved in C. elegans, that can be activated by a diverse set of receptors (Chen et al., 2014). Future studies focusing on whether this second interaction site is required for the WAVE complex during dorsal intercalation would likely prove informative.
Within intercalating cells, the asymmetric localization of molecules orthogonal to or along the direction of movement is a common strategy to polarize cells during intercalation (reviewed by Bertet and Lecuit, 2009; Gray et al., 2011; Walck-Shannon and Hardin, 2014; Wallingford, 2012; Zallen, 2007). During intercalation of the Drosophila germband, for example, actin, nonmuscle myosin II and its regulators accumulate at shrinking junctions, whereas cadherin is enriched at junctions that persist during intercalation (Blankenship et al., 2006; Levayer et al., 2011; Simões et al., 2010; Zallen and Wieschaus, 2004). Asymmetric localization of proteins in epithelial cells that use basolateral protrusive activity during rearrangement has not previously been examined. We found that a negative regulator of UNC-73/Trio, CRML-1/CARMIL, predominantly localizes to lateral edges, where protrusions are normally absent. We hypothesize that CRML-1 polarization contributes to the directional movement of intercalating cells by preventing non-directional (lateral) protrusions. At a genetic level, crml-1 regulates the activity of unc-73, rather than its expression in dorsal epidermal cells, because UNC-73::GFP localization in the crml-1(gm326) background is indistinguishable from that in wild type (Fig. S10). It remains unclear how CRML-1 becomes spatially restricted in its localization. Repeated attempts to disrupt dorsal intercalation by perturbing the function of conserved homologs of planar cell polarity components have failed to detect defects (King et al., 2009; E.W.-S. and J.H., data not shown); it is therefore likely that additional proteins are involved. CRML-1::GFP also accumulated at the tips of cells as intercalation concluded. This separate pool of CRML-1 might represent highly localized areas of UNC-73 inhibition at the migrating tips as they make contact with contralateral seam cells; alternatively, this pool could have a second, separate function that relates to capping protein at medial tips.
There are multiple isoforms of human CARMIL. Most effort has focused on CARMIL1, which can form a complex with Trio and promotes protrusive activity (Liang et al., 2009). The role of CARMILs in vertebrates has focused almost entirely on their inhibition of capping protein (Edwards et al., 2014). However, C. elegans CRML-1 seems to inhibit protrusive activity (this work; Vanderzalm et al., 2009) rather than promote it. The opposite effect would be expected if CRML-1 acts primarily via anti-capping protein activity. Here we show that CRML-1 has a crucial role in regulating F-actin dynamics through the inhibition of the RacGEF (GEF1) activity of UNC-73. In this regard, crml-1 loss of function more closely mimics CARMIL2 mutant phenotypes in vertebrate cells, which include loss of cell polarity and multiple lamellipodia (Liang et al., 2009). It is possible that C. elegans CRML-1 retains functions of both CARMIL1 (Trio binding/inhibition) and CARMIL2 (regulation of cell polarity/capping protein activity), and that these are each deployed in different contexts in C. elegans. CARMILs might have undergone subfunctionalization in vertebrates for separate, more specialized functions.
Our study unites previous observations (Patel et al., 2008; Williams-Masson et al., 1998) into a concise model for dorsal intercalation in C. elegans, in which Rac/RhoG control of dendritic actin polymerization is the major driver of medial protrusions, and hence cell rearrangement. This observation also raises the intriguing possibility that other epithelia use both mechanisms to varying degrees. Asymmetric Rho-mediated constriction of apical junctions has motivated most study of intercalation within the Drosophila germband, but embryos that carry mutations for eve and Toll-like receptors, which pattern these molecular asymmetries, still have germbands that extend 1.6× their original length, compared with 2× extension in wild type (Irvine and Wieschaus, 1994; Paré et al., 2014). Although oriented cell divisions may contribute to the remaining tissue extension (da Silva and Vincent, 2007), it is also possible that basolateral protrusions contribute to convergent extension in the Drosophila germband but have escaped detection thus far. Further evidence that the Rac/RhoG-dependent mechanism described here may constitute an additional general mechanism for driving intercalation in epithelia is suggested by studies of the mouse neural tube. In this neuroepithelium, the planar cell polarity pathway directs both apical junctional rearrangement and basolateral protrusions to achieve mediolateral intercalation (Williams et al., 2014). These results support the idea that apical and basolateral pathways during intercalation are not mutually exclusive. Instead, different epithelial cell types might utilize each pathway to varying extents. Understanding the extent to which various epithelial cells use one program or the other and the reasons why various epithelia favor one pathway over another is an important question for future research.
MATERIALS AND METHODS
Nematode strains and genetics
C. elegans were maintained on OP50 bacteria as previously described (Brenner, 1974). The wild-type strain was Bristol N2. Experiments were performed at 20°C unless otherwise stated. The following genetic lesions were utilized in this study. LG1: smg-1(cc546ts), unc-73(ev802), unc-73(gm40), unc-73(rh40), crml-1(gm326). LGIV: ced-10(n1993), ced-10(n3417), ced-10(tm597), wsp-1(gm324). LGX: mig-2(gm103gf), mig-2(mu28). The following transgenic arrays were made for, or utilized in, this study: mcEx227[Plin-26::VAB-10(actin binding domain)::GFP, rol-6(su1006)] (Gally et al., 2009), jcEx222[Plin-26::ced-10(T17N/DN)::smg sensitive 3′UTR, sur-5::mCherry], jcEx224[Punc-73::UNC-73::GFP), rol-6(su1006)], reIs6[Plin-26::ced-10(Q61L/CA)::smg sensitive 3′UTR, rol-6(su1006), Pmyo-3::gfp)], ltIs44[Ppie-1::PHPLC1∂1::mCherry, unc-119(+)] (Kachur et al., 2008). Additional strains are described in the supplementary materials and methods.
F-actin imaging
We imaged live embryos mosaically expressing a previously validated F-actin reporter, mcEx227[Plin-26::VAB-10(actin binding domain)::GFP, rol-6(su1006)] (Gally et al., 2009), in the dorsal epidermis with a Nikon Eclipse E600 microscope connected to a Yokogawa CSU10 spinning disk scanhead. Images were gathered with a Hamamatsu ORCA-ER charge-coupled device (CCD) camera and Micromanager software (Edelstein et al., 2010, 2014) with a z-slice spacing of 0.4 μm. ImageJ (Schneider et al., 2012) was used to project 20-25 z-slices, which is shown in dorsal view images. Reslices were also performed in ImageJ. mcEx227 was crossed into all other genetic backgrounds to maintain similar expression levels.
Protrusion quantification
Protrusions were quantified from maximum projections of z-stacks of F-actin reporter expression as described above. Relative protrusion area was calculated as the ratio between a cell's area in maximum projection images (which includes all protrusions) to a cell's lateral area (measured at the depth of the cell nucleus). Protrusion number was obtained by counting aggregations of F-actin reporter signal extending at least 0.5 μm from the cell body. Protrusion position was recorded using the ROI tool in Image J. Angles were obtained using trigonometric functions in Microsoft Excel to compare protrusion position relative to the cell centroid (also calculated in ImageJ). At least 15 cells from at least 10 embryos were analyzed per genotype. JMP software (SAS) was used for statistics and graphing of protrusion area and number; PAST was used for angular statistics and graphs (Hammer et al., 2001). We designated cells as ‘early’ if they were in the process of polarizing into a wedge shape and ‘late' if the cell tip or body had begun to extend. Cell velocity was calculated by measuring medial edge displacement over time. Linear regression and statistical analyses were carried out in JMP.
Dorsal intercalation time measurements
Four-dimensional (4D) DIC movies were gathered on either a Nikon Optiphot-2 connected to a QImaging camera or Olympus BX50 connected to a Scion camera. ImageJ plugins (available at http://worms.zoology.wisc.edu/research/4d/4d.html) were used to analyze movies. For this study, we defined intercalation time as the time between terminal epidermal (Cpaa.a/p) divisions and when contralateral nuclei met at the midline after cell migration was complete. For unc-73 mutants, intercalation time was measured relative to the time between the Cpaa and Cpaa(a/p) divisions. At least 20 embryos from at least 10 mounts were analyzed per genotype. JMP was used to create box plots and for statistical analysis.
NMD-dependent conditional expression system
The epidermal-specific, NMD-sensitive plasmid backbone was made as follows. (1) The hybrid eFGHi lin-26 promoter (including minimal myo-2 promoter) is expressed only in the epidermis and support cells (Landmann et al., 2004): Plin-26(eFGHi) was amplified from pML433 with primers (5′-3′; lowercase letters are restriction sites) TTTTTTgcggccgcGGCGCGCCCGAGGTTAATATCTGAGCTCC and TTTTTTggatccTggccggccACTCATTTTTTCTGAGCTCGGTACCCTCC and cloned into pBluescript cut with NotI/BamHI (pCM1.1). (2) The NMD-sensitive 3′UTR sequence (inverted let-858 coding sequence without a sense polyadenylation sequence) was amplified from pPD118.44 (gift of A. Fire, Stanford University, CA, USA) using primers TTTTatcgatcctaaTCGTCGAGTCGGTCACAATCACC and TTTTgggcccGCTCATGTTTAGATTTGGATTG and cloned into pCM1.1 digested with ClaI/ApaI (pCM1.3). pCM1.1 plasmid for this step was grown in SCS110 bacteria because ClaI is Dam sensitive. The wild-type ced-10 sequence was amplified from pPR37 (Reddien and Horvitz, 2000) using primers AAAAAAAAggccggcctggcATGCAAGCGATCAAATGTGTCGTCG and TTTTTTatcgatTTAGAGCACCGTACACTTGCTCTTTTTGG and cloned into pCM1.3 using FseI/ClaI to generate pCM3.2. To make ced-10(Q61L/CA) (pCM3.3) and ced-10(T17N/DN) (pEWS27) variants, we used PCR site-directed mutagenesis, with pCM3.2 as template and the following primers (forward and reverse; underlining indicates bases mutated relative to wild type): ced-10(Q61L/CA), GGGATACAGCTGGACTGGAAGATTACGATCGAC and GTCGATCGTAATCTTCCAGTCCAGCTGTATCCC; ced-10(T17N/DN), ACTGTCTCCTGATATCCTACAC and TTTTACCGACGGCTCCGTCA.
Gonads of wild-type animals were microinjected with these constructs at 40 ng/μl and a co-injection marker (Mello and Fire, 1995). Resulting lines in the wild-type background were screened for defects using smg-1 feeding RNAi (Kamath et al., 2003). Representative lines were crossed into the smg-1(cc546ts) background, which can be detected by PCR using forward CAGTCGTGAGCTTTGGATGCGTGC and reverse TCGGGGATACGCAGATTCTTTCCC followed by digestion specifically of wild-type product using MslI. At least three lines were analyzed per construct. The resulting extrachromosomal array jcEx222[Plin-26::ced-10(T17N/DN)::smg sensitive 3′UTR, sur-5::mCherry] was used for experiments. ced-10(Q61L/CA) was integrated into the genome using UV irradiation, resulting in reIs6[Plin-26::ced-10(Q61L/CA)::smg sensitive 3′UTR, rol-6(su1006), Pmyo-3::gfp)]. Lines were maintained at 15°C and heat shocked at 25°C for 24 h to induce transgene expression prior to filming (unless otherwise noted). Crosses were performed at 15°C. Filming was at 20°C. For GFP induction using the NMD system, see the supplementary materials and methods.
RNAi
T7 and T3 sites were added to full-length wve-1 cDNA derived from wild-type worms by PCR with primers forward TAATACGATCACTATAGGTCCATCAACAATCCATCGTG and reverse CCCTTTAGTGAGGGTTAATTCCCATTCATCATCATCAGC. From the PCR product, MegaScript kits (Ambion) were used to synthesize double-stranded wve-1 RNA, which was injected (Zipperlen et al., 2001) into the psuedocoelom at 0.5 μg/μl. As described previously for feeding RNAi, injection RNAi elicited partial knockdown of wve-1 (Patel et al., 2008). For ced-10(Q61L/CA) suppression, animals were incubated at a semi-permissive temperature of 18°C for 6 h prior to mounting. For mig-2(gm103/CA) suppression, animals were incubated at 20°C for 16 h. For vav-1 RNA, see the supplementary materials and methods.
UNC-73B::GFP
A rescuing unc-73 GEF1 construct, pNW248 (Punc-73::unc-73B::GFP) (Steven et al., 2005), was microinjected into the gonads of wild-type animals (Mello and Fire, 1995) at 10 ng/μl to create the extrachromosomal array jcEx224[Punc-73::UNC-73::GFP), rol-6(su1006)].
Epidermal-driven CRML-1::GFP
lbp-1 is expressed predominantly in the epidermis at embryonic stages (Plenefisch et al., 2000). crml-1 cDNA was cloned into an existing Plbp-1::GFP vector, pSL500 (Fridolfsson and Starr, 2010), using Gibson assembly (Gibson, 2011). The following primers were used (forward and reverse; uppercase letters indicate regions complementary to template and lowercase letters indicate overhangs complementary to vector): pSL500, ATGAGTAAAGGAGAAGAACTTTTCAC and CATGGTTGGCTAGGTCGATGAATG; crml-1 cDNA, tcgacctagccaaccatgTCCATGTCGAGGAGCACC and agttcttctcctttactcatTTTTTGGAAAATTCTAGCCATATCAGC. The resulting construct (pEWS38) was injected into crml-1(gm326) mutants at 10 ng/μl. crml-1(gm326) can be detected using PJV98/PJV99 (Vanderzalm et al., 2009) and AccI digestion. Three independently derived lines were analyzed. Intensity measurements of CRML-1::GFP were performed using the Plot Profile tool with congruent rectangular selections outlining each dorsal cell in ImageJ. Cells came from at least five different embryos.
Acknowledgements
Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs [P40 OD010440]. Bethany Lucas created the wve-1 cDNA clone used for RNAi. We thank the laboratories of E. Lundquist and G. Garriga for graciously providing many worm strains used in this study and R. Steven for providing pNW248 DNA. The SAGE data were produced at the Michael Smith Genome Sciences Centre with funding from Genome Canada; the embryonic RNA sequencing data were produced by the Waterston laboratory at the University of Washington. We are grateful to members of the J.H. laboratory for helpful input.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
E.W.-S. developed concepts and approach, performed experiments, analyzed data and prepared the manuscript. D.R. developed the nonsense-mediated mRNA decay method for inducible expression of dominant transgenes and edited the manuscript. J.H. developed concepts and approach, developed software tools and analysis procedures and edited the manuscript.
Funding
This work was supported by grants from the National Science Foundation [IOB 1426555] and National Institutes of Health (NIH) [GM058038 to J.H. and GM085309 to D.R.]. E.W.-S. was supported by a Genetics Training Grant from the NIH [GM07133]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.127597/-/DC1
References
- Bellanger J.-M., Lazaro J.-B., Diriong S., Fernandez A., Lamb N. and Debant A. (1998). The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene 16, 147-152. 10.1038/sj.onc.1201532 [DOI] [PubMed] [Google Scholar]
- Bertet C. and Lecuit T. (2009). Planar polarity and short-range polarization in Drosophila embryos. Semin. Cell Dev. Biol. 20, 1006-1013. 10.1016/j.semcdb.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Bertet C., Sulak L. and Lecuit T. (2004). Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667-671. 10.1038/nature02590 [DOI] [PubMed] [Google Scholar]
- Blangy A., Vignal E., Schmidt S., Debant A., Gauthier-Rouvière C. and Fort P. (2000). TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J. Cell Sci. 113, 729-739. [DOI] [PubMed] [Google Scholar]
- Blankenship J. T., Backovic S. T., Sanny J. S. P., Weitz O. and Zallen J. A. (2006). Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 11, 459-470. 10.1016/j.devcel.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A. and McCormick F. (1991). The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117-127. 10.1038/349117a0 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A. L., Gerstein M. B., Gunsalus K. C., Henikoff S., Karpen G. H., Kellis M., Lai E. C., Lieb J. D., MacAlpine D. M. et al. (2009). Unlocking the secrets of the genome. Nature 459, 927-930. 10.1038/459927a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Heszele M. F., Ren D., Reynolds A. B., Chi F. and Chen P. (2012). Regulation of cochlear convergent extension by the vertebrate planar cell polarity pathway is dependent on p120-catenin. Development 139, 968-978. 10.1242/dev.065326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-F., Imam J. S. and Wilkinson M. F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51-74. 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- Chen B., Brinkmann K., Chen Z., Pak C. W., Liao Y., Shi S., Henry L., Grishin N. V., Bogdan S. and Rosen M. K. (2014). The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156, 195-207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Raich W., Agbunag C., Leung B., Hardin J. and Priess J. R. (1998). A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141, 297-308. 10.1083/jcb.141.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. M. and Vincent J.-P. (2007). Oriented cell divisions in the extending germband of Drosophila. Development 134, 3049-3054. 10.1242/dev.004911 [DOI] [PubMed] [Google Scholar]
- Debant A., Serra-Pagès C., Seipel K., O'Brien S., Tang M., Park S. H. and Streuli M. (1996). The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. USA 93, 5466-5471. 10.1073/pnas.93.11.5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeier M. E., Morse D. P., Knight S. W., Portereiko M., Bass B. L. and Mango S. E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289, 1928-1930. 10.1126/science.289.5486.1928 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R. and Stuurman N. (2010). Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14, Unit14.20 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A. D., Tsuchida M. A., Amodaj N., Pinkard H., Vale R. D. and Stuurman N. (2014). Advanced methods of microscope control using μManager software. J. Biol. Methods 1, 10 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M., Zwolak A., Schafer D. A., Sept D., Dominguez R. and Cooper J. A. (2014). Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15, 677-689. 10.1038/nrm3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson H. N. and Starr D. A. (2010). Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191, 115-128. 10.1083/jcb.201004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I., Remmert K., Piszczek G. and Hammer J. A. (2014). Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc. Natl. Acad. Sci. USA 111, E1970-E1979. 10.1073/pnas.1313738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally C., Wissler F., Zahreddine H., Quintin S., Landmann F. and Labouesse M. (2009). Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development 136, 3109-3119. 10.1242/dev.039412 [DOI] [PubMed] [Google Scholar]
- Gibson D. G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349-361. 10.1016/B978-0-12-385120-8.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R. S., Roszko I. and Solnica-Krezel L. (2011). Planar cell polarity: coordinating morphogenetic cell behaviors with embryonic polarity. Dev. Cell 21, 120-133. 10.1016/j.devcel.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T. and Ryan P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9. [Google Scholar]
- Hardin J. (1989). Local shifts in position and polarized motility drive cell rearrangement during sea urchin gastrulation. Dev. Biol. 136, 430-445. 10.1016/0012-1606(89)90268-6 [DOI] [PubMed] [Google Scholar]
- Heasman S. J. and Ridley A. J. (2008). Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690-701. 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Heid P. J., Raich W. B., Smith R., Mohler W. A., Simokat K., Gendreau S. B., Rothman J. H. and Hardin J. (2001). The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev. Biol. 236, 165-180. 10.1006/dbio.2001.0315 [DOI] [PubMed] [Google Scholar]
- Irvine K. D. and Wieschaus E. (1994). Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827-841. [DOI] [PubMed] [Google Scholar]
- Jung G., Remmert K., Wu X., Volosky J. M. and Hammer J. A. (2001). The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153, 1479-1498. 10.1083/jcb.153.7.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur T. M., Audhya A. and Pilgrim D. B. (2008). UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev. Biol. 314, 287-299. 10.1016/j.ydbio.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M. et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Karner C. M., Chirumamilla R., Aoki S., Igarashi P., Wallingford J. B. and Carroll T. J. (2009). Wnt9b signaling regulates planar cell polarity and kidney tubule morphogenesis. Nat. Genet. 41, 793-799. 10.1038/ng.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A. and Rosen M. K. (2000). Autoinhibition and activation mechanisms of the Wiskott–Aldrich syndrome protein. Nature 404, 151-158. 10.1038/35004513 [DOI] [PubMed] [Google Scholar]
- Kim T., Ravilious G. E., Sept D. and Cooper J. A. (2012). Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. J. Biol. Chem. 287, 15251-15262. 10.1074/jbc.M112.345447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. S., Maiden S. L., Hawkins N. C., Kidd A. R., Kimble J., Hardin J. and Walston T. D. (2009). The N- or C-terminal domains of DSH-2 can activate the C. elegans Wnt/beta-catenin asymmetry pathway. Dev. Biol. 328, 234-244. 10.1016/j.ydbio.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiseski T. J., Culotti J. and Pawson T. (2003). Functional analysis of the Caenorhabditis elegans UNC-73B PH domain demonstrates a role in activation of the Rac GTPase in vitro and axon guidance in vivo. Mol. Cell. Biol. 23, 6823-6835. 10.1128/MCB.23.19.6823-6835.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F., Quintin S. and Labouesse M. (2004). Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev. Biol. 265, 478-490. 10.1016/j.ydbio.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Levayer R., Pelissier-Monier A. and Lecuit T. (2011). Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol. 13, 529-540. 10.1038/ncb2224 [DOI] [PubMed] [Google Scholar]
- Liang Y., Niederstrasser H., Edwards M., Jackson C. E. and Cooper J. A. (2009). Distinct roles for CARMIL isoforms in cell migration. Mol. Biol. Cell 20, 5290-5305. 10.1091/mbc.E08-10-1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp S. S., Liu K., Karner C. M., Carroll T. J., Ronneberger O., Wallingford J. B. and Walz G. (2012). Vertebrate kidney tubules elongate using a planar cell polarity–dependent, rosette-based mechanism of convergent extension. Nat. Genet. 44, 1382-1387. 10.1038/ng.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist E. A. (2006). Small GTPases. WormBook 1-18. 10.1895/wormbook.1.67.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist E. A., Reddien P. W., Hartwieg E., Horvitz H. R. and Bargmann C. I. (2001). Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128, 4475-4488. [DOI] [PubMed] [Google Scholar]
- McMahon L., Legouis R., Vonesch J. L. and Labouesse M. (2001). Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J. Cell Sci. 114, 2265-2277. [DOI] [PubMed] [Google Scholar]
- Mello C. and Fire A. (1995). DNA transformation. Methods Cell Biol. 48, 451-482. 10.1016/S0091-679X(08)61399-0 [DOI] [PubMed] [Google Scholar]
- Miki H., Suetsugu S. and Takenawa T. (1998). WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17, 6932-6941. 10.1093/emboj/17.23.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E. M. and Odell G. (2002a). Morphogenetic pattern formation during ascidian notochord formation is regulative and highly robust. Development 129, 1-12. [DOI] [PubMed] [Google Scholar]
- Munro E. M. and Odell G. M. (2002b). Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development 129, 13-24. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Honda H. and Takeichi M. (2012). Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149, 1084-1097. 10.1016/j.cell.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Nobes C. D. and Hall A. (1995). Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53-62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Paré A. C., Vichas A., Fincher C. T., Mirman Z., Farrell D. L., Mainieri A. and Zallen J. A. (2014). A positional Toll receptor code directs convergent extension in Drosophila. Nature 515, 523-527. 10.1038/nature13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel F. B., Bernadskaya Y. Y., Chen E., Jobanputra A., Pooladi Z., Freeman K. L., Gally C., Mohler W. A. and Soto M. C. (2008). The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev. Biol. 324, 297-309. 10.1016/j.ydbio.2008.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenefisch J., Xiao H., Mei B., Geng J., Komuniecki P. R. and Komuniecki R. (2000). Secretion of a novel class of iFABPs in nematodes: coordinate use of the Ascaris/Caenorhabditis model systems. Mol. Biochem. Parasitol. 105, 223-236. 10.1016/S0166-6851(99)00179-6 [DOI] [PubMed] [Google Scholar]
- Reddien P. W. and Horvitz H. R. (2000). CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2, 131-136. 10.1038/35004000 [DOI] [PubMed] [Google Scholar]
- Sawa M. (2003). Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116, 1505-1518. 10.1242/jcs.00362 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir M. A., Gill J. S. and Lundquist E. A. (2006). Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics 172, 893-913. 10.1534/genetics.105.046359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir M. A., Jiang K., Struckhoff E. C., Demarco R. S., Patel F. B., Soto M. C. and Lundquist E. A. (2008). The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in Caenorhabditis elegans axon guidance. Genetics 179, 1957-1971. 10.1534/genetics.108.088963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S. d. M., Blankenship J. T., Weitz O., Farrell D. L., Tamada M., Fernandez-Gonzalez R. and Zallen J. A. (2010). Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell 19, 377-388. 10.1016/j.devcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S. d. M., Mainieri A. and Zallen J. A. (2014). Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J. Cell Biol. 204, 575-589. 10.1083/jcb.201307070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M. C., Qadota H., Kasuya K., Inoue M., Tsuboi D., Mello C. C. and Kaibuchi K. (2002). The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 16, 620-632. 10.1101/gad.955702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven R., Kubiseski T. J., Zheng H., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C. W. V., Pawson T. and Culotti J. (1998). UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92, 785-795. 10.1016/S0092-8674(00)81406-3 [DOI] [PubMed] [Google Scholar]
- Steven R., Zhang L., Culotti J. and Pawson T. (2005). The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans. Genes Dev. 19, 2016-2029. 10.1101/gad.1319905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G. and Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Takeda S., Minakata S., Koike R., Kawahata I., Narita A., Kitazawa M., Ota M., Yamakuni T., Maéda Y. and Nitanai Y. (2010). Two distinct mechanisms for actin capping protein regulation—steric and allosteric inhibition. PLoS Biol. 8, e1000416 10.1371/journal.pbio.1000416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T. and Suetsugu S. (2007). The WASP–WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37-48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Vanderzalm P. J., Pandey A., Hurwitz M. E., Bloom L., Horvitz H. R. and Garriga G. (2009). C. elegans CARMIL negatively regulates UNC-73/Trio function during neuronal development. Development 136, 1201-1210. 10.1242/dev.026666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walck-Shannon E. and Hardin J. (2014). Cell intercalation from top to bottom. Nat. Rev. Mol. Cell Biol. 15, 34-48. 10.1038/nrm3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford J. B. (2012). Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28, 627-653. 10.1146/annurev-cellbio-092910-154208 [DOI] [PubMed] [Google Scholar]
- Warrington S. J., Strutt H. and Strutt D. (2013). The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Development 140, 1045-1054. 10.1242/dev.088724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M., Yen W., Lu X. and Sutherland A. (2014). Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Dev. Cell 29, 34-46. 10.1016/j.devcel.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Masson E. M., Heid P. J., Lavin C. A. and Hardin J. (1998). The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev. Biol. 204, 263-276. 10.1006/dbio.1998.9048 [DOI] [PubMed] [Google Scholar]
- Withee J., Galligan B., Hawkins N. and Garriga G. (2004). Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics 167, 1165-1176. 10.1534/genetics.103.025676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.-C., Cheng T.-W., Lee M.-C. and Weng N.-Y. (2002). Distinct rac activation pathways control Caenorhabditis elegans cell migration and axon outgrowth. Dev. Biol. 250, 145-155. 10.1006/dbio.2002.0785 [DOI] [PubMed] [Google Scholar]
- Yang C., Pring M., Wear M. A., Huang M., Cooper J. A., Svitkina T. M. and Zigmond S. H. (2005). Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 9, 209-221. 10.1016/j.devcel.2005.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A. (2007). Planar polarity and tissue morphogenesis. Cell 129, 1051-1063. 10.1016/j.cell.2007.05.050 [DOI] [PubMed] [Google Scholar]
- Zallen J. A. and Wieschaus E. (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343-355. 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
- Zipkin I. D., Kindt R. M. and Kenyon C. J. (1997). Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90, 883-894. 10.1016/S0092-8674(00)80353-0 [DOI] [PubMed] [Google Scholar]
- Zipperlen P., Fraser A. G., Kamath R. S., Martinez-Campos M. and Ahringer J. (2001). Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20, 3984-3992. 10.1093/emboj/20.15.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwolak A., Yang C., Feeser E. A., Michael Ostap E., Svitkina T. and Dominguez R. (2013). CARMIL leading edge localization depends on a non-canonical PH domain and dimerization. Nat. Commun. 4, 2523 10.1038/ncomms3523 [DOI] [PMC free article] [PubMed] [Google Scholar]