Abstract
Purpose
African Americans (AA) exhibit higher rates of prostate cancer (PCa) incidence and mortality compared to European American (EA) men. In addition to socioeconomic influences, biological factors are believed to play a critical role in PCa disparities. We investigated whether population-specific and -enriched miRNA-mRNA interactions might contribute to PCa disparities.
Experimental Design
Integrative genomics was employed, combining miRNA and mRNA profiling, miRNA target prediction, pathway analysis and functional validation, to map miRNA-mRNA interactions associated with PCa disparities.
Results
We identified 22 AA-specific and 18 EA-specific miRNAs in PCa versus patient-matched normal prostate, and 10 ‘AA-enriched/-depleted’ miRNAs in AA PCa versus EA PCa comparisons. Many of these population-specific/-enriched miRNAs could be paired with target mRNAs that exhibited an inverse pattern of differential expression. Pathway analysis revealed epidermal growth factor receptor (EGFR or ERBB) signaling as a critical pathway significantly regulated by AA-specific/-enriched mRNAs and miRNA-mRNA pairings. Novel miRNA-mRNA pairings were validated by qRT-PCR, western blot and/or IHC analyses in PCa specimens. Loss/gain of function assays performed in population-specific PCa cell lines confirmed miR-133a/MCL1, miR-513c/STAT1, miR-96/FOXO3A, miR-145/ITPR2 and miR-34a/PPP2R2A as critical miRNA-mRNA pairings driving oncogenesis. Manipulating the balance of these pairings resulted in decreased proliferation and invasion, and enhanced sensitization to docetaxel-induced cytotoxicity in AA PCa cells.
Conclusion
Our data suggest that AA-specific/-enriched miRNA-mRNA pairings may play a critical role in the activation of oncogenic pathways in AA PCa. Our findings also suggest that miR-133a/MCL1, miR-513c/STAT1 and miR-96/FOXO3A may have clinical significance in the development of novel strategies for treating aggressive PCa.
INTRODUCTION
MiRNAs (miRNAs) are small regulatory RNAs of ~21-25 nucleotides in length that complementarily target mRNAs to inhibit translation and/or promote mRNA degradation. Recently, several reports have suggested that miRNA aberrations may be an important factor in cancer development (1, 2). The potential connection between miRNA regulation and cancer has been made at several levels, suggesting that miRNAs play critical roles in cellular growth and differentiation, which are two cellular processes commonly defective in tumor cells (3). Additional evidence for the involvement of miRNAs in human cancer comes from observations that ~50% of these small regulatory RNAs are transcribed from genomic regions associated with a loss of heterozygosity, minimal amplicons, or breakpoint cluster regions (4). Cancer-related miRNAs have been identified in various cancers (5). In general, oncogenic miRNAs up-regulated in tumors act as oncogenes (repressing tumor suppressor and apoptosis-associated genes), while tumor suppressor miRNAs are down-regulated leading to derepression of oncogenes and proliferation-related genes) (6). Although many miRNAs are differentially expressed in various cancers, the identity of the mRNAs specifically targeted by these miRNAs, functional consequences of miRNA-mRNA pairings and their contributions to cancer pathogenesis remain to be elucidated.
PCa is now the most frequently diagnosed cancer and the second most common cause of cancer deaths in men residing in the United States (7). AAs have among the highest incidence of PCa and mortality attributable to this disease, being 1.6 times more likely to develop PCa, and 2.4 times more likely to die from PCa compared to their EA counterparts (8). Multiple socioeconomic and environmental factors have been postulated to explain the observed PCa health disparities, such as access to care, attitudes toward health care, socioeconomic differences, diet and differences in the type and aggressiveness of treatment (8). However, adjustment for these factors does not preclude the higher mortality and recurrence rate in AA men and suggests intrinsic biological differences exist (9). The application of epidemiology and genomics has revealed biological factors implicated in PCa health disparities between AA and EA, such as differences in the hormonal milieu of the tumor (10), oncogenic activation (11) and tumor immunobiology (12). More recently, our genomic analysis identified multiple signaling pathways converging on the androgen receptor (AR) to activate transcription of AR-target genes promoting PCa progression and aggressiveness in AA patients (13).
Given the importance of miRNAs in cancer, studies have been forthcoming on the association of miRNAs in PCa pathogenesis. Volinia et al. performed large-scale analysis of miRNA expression profiles in 540 samples derived from six types of solid tumors, and demonstrated that 46 miRNAs were differentially expressed when comparing PCa with patient-matched normal prostate (NP), including up-regulated let-7d*, miR-17-5p and miR-21, and down-regulated miR-24, miR-29 and miR-128a (1). A miRNA profiling study by Ozen et al. revealed 76 of the 85 differentially expressed miRNAs were down-regulated (such as let-7c, miR-145, and miR-125b) in the PCa clinical samples compared with normal tissues (14). More recently, Wang et al. identified a set of deregulated miRNAs associated with cell cycle regulation in aggressive PCa by combining miRNA expression profiling and coexpression network analysis (15). While these profiling studies have begun to shed light on the involvement of miRNAs in PCa development, questions on the role of miRNAs in PCa disparities still remain. A recent study evaluated the impact of miRNAs contained in the region of 8q24, a genetic risk locus conferring PCa in AAs. However, no empirical evidence of miRNA transcription was found within the 8q24 PCa risk locus (16). In the present study, we applied a systems biology approach, by combining genome-wide miRNA and mRNA expression profiling in PCa patient specimens, miRNA target predictions, and miRNA-mRNA pairing and pathway analyses, to identify oncogenic signaling pathway most significantly regulated by AA-specific/enriched mRNAs and miRNA-mRNA pairings. The AA-specific/enriched miRNA and mRNA elements were also evaluated in AA and EA PCa cell lines for their functional relevance in cell proliferation, invasion and chemo-sensitivity to cytotoxic agents.
MATERIALS AND METHODS
Acquisition and characteristics of PCa clinical specimens
Tissues were procured from the George Washington University Medical Faculty Associates adhering to IRB approved protocols (IRB#020867), as detailed in Supplemental Methods. High-quality PCa and patient-matched NP biopsy cores from each of 20 AA and 15 EA patients were collected and processed for the microarray analyses. PCa cores were determined by pathologist to have Gleason score of 6-7 (17 AA and 13 EA) or 8-9 (3 AA and 2 EA), while NP cores were negative for cancer. There was no significant difference between the two racial groups with respect to age (average age for AAs was 62.3 ± 8.2, average age for EAs was 63.3 ± 9.2) and Gleason score (Supplementary Table S1A).
PCa Cell Lines
PCa cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA) and passaged less than six months after receipt/resuscitation. Cell lines were tested and authenticated at ATCC by short tandem repeat profiling of multiple unique genetic loci (D5S818, D13S317, D7S820, D16S539, vWA, TH01, Amelogenin, TPOX and CSF1PO).
Gene Expression Microarrays
Total RNA was isolated from PCa and patient-matched NP biopsy cores. For mRNA profiling, total RNA (1 μg) from each biopsy core was purified using the RNeasy micro kit (Qiagen, Valencia, CA) and interrogated with the Affymetrix Human Exon 1.0 ST GeneChip. For miRNA profiling, 250 ng of RNA from each biopsy core was isolated using the miRNeasy kit (Qiagen, Valencia, CA) and interrogated with the Agilent Human miRNA microarray V3 (Agilent Technologies, Santa Clara, CA). High quality RNA samples were confirmed on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Affymetrix exon array data were normalized by quantile normalization with GC-RMA background correction, and data visualization and statistical analysis were performed by Partek Genomics Suite 6.6 software (Partek, St. Louis, MO) as previously described(13). Raw data from Agilent miRNA microarray analysis was quantile normalized and analyzed in GeneSpring GX program version 12.5 (Agilent Technologies, Santa Clara, CA). Identification of statistically significant, differentially expressed/regulated mRNAs and miRNAs was based on ANOVA or paired t-test with a 10% False Discovery Rate (FDR) criterion to correct for multiple testing(13). Microarray data can be assessed at GEO using accession numbers GSE64331 and GSE64318 for Affymetrix exon and Agilent miRNA arrays, respectively.
Principal component analysis (PCA) plots and hierarchical clustering of mRNA and miRNA data was performed using the Partek Genomics Suite 6.6. Two-dimensional hierarchical clustering analysis used average linkage and a Euclidean distance metric.
MiRNA-miRNA pairings and pathway analysis
TargetScanHuman 6.2 was employed to identify mRNAs predicted to be targets of the ANOVA-defined differentially expressed miRNAs. The list of predicted target mRNAs was intersected with the ANOVA-defined differentially expressed mRNAs to generate a catalog of experimental miRNA-mRNA pairings. Pairings were categorized as having reciprocal (e.g. miRNA up and mRNA down, or miRNA down and mRNA up), positive (i.e. miRNA up and mRNA up) or negative correlations (i.e. miRNA down and mRNA down in AA PCa vs. NP or AA PCa vs. EA PCa comparisons). The differentially expressed mRNAs not belonging to any pairings are herein referred to as unpaired mRNAs.
Global test (17) (and Gene Set Enrichment Analysis (GSEA) (18) as a secondary confirmatory approach) was implemented to identify statistically significant canonical signaling pathways containing differentially regulated gene sets that may be associated with AA PCa aggressiveness, based on AA PCa vs. AA NP, AA PCa vs. EA PCa and EA PCa vs. EA NP comparisons (detailed description in Supplementary Materials and Methods). Note that significant genes identified by Global test and ANOVA may be mutually exclusive. Representative genes in different pathways identified by Global test were chosen for validation if these genes were also identified by ANOVA and TargetScan prediction analyses as unpaired mRNAs or mRNAs belonging to miRNA-mRNA pairings. The underlying assumption was that genes fulfilling the above criteria would have a greater likelihood of validation success. Validation of differential gene expression was accomplished by quantitative RT-PCR (qRT-PCR) and immunohistochemistry (IHC) in cohorts of patient specimens separate from those used in microarray analysis (Supplementary Table S1B and S1C). Western analysis and functional assays in PCa cell lines were performed to validate predicted reciprocal miRNA-mRNA pairings.
QRT-PCR validation of mRNAs and miRNAs
QRT-PCR validation was performed as previously described (19, 20). QRT-PCR determinations of mRNAs and miRNAs were performed in duplicate and normalized to levels of house-keeping genes EIF1AX and miR-103, respectively. EIF1AX and miR-103 are constitutively expressed and resistant to expression changes (19, 20). QRT-PCR primer pair sequences for mRNA and miRNA determinations are provided in Supplementary Tables S2 and S3, respectively. Sequences to entire mature miRNA are reported in miRBase database (21).
Tissue processing, IHC and western blot analysis
Serial sections of formalin-fixed, paraffin-embedded (FFPE) PCa specimens from AA and EA patients with Gleason score 6-8 were immunolabeled. Western blot analysis, as previously described (13), was performed on AA and EA PCa cell lines MDA PCa 2b, RC77T/E, VCaP, LNCaP and PC-3. Details for tissue processing, IHC, image capturing/quantification and cell line information can be found in Supplementary Materials and Methods
Antibodies
Antibodies used in IHC assays and western blotting analysis were rabbit monoclonal antibodies for STAT1 and pFOXO3A (Cell Signaling Technology, Danvers, MA), FOXO3A (Millipore, Billerica, MA) and AMACR (Dako, Carpinteria, CA), rabbit polyclonal antibody for MCL-1 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal antibodies for p63 (Biocare Medical, Concord, CA) and β-actin (Santa Cruz, CA).
Functional analysis of PCa cell lines following miRNA mimic or inhibitor transfections
PCa cells were transfected with either miRNA mimics or antagomirs using DharmaFECT4 transfection reagent (Dharmacon), according to the manufacturer's protocol. MiR-133a mimic, miR-513c mimic, miR-96 mimic, miR-34a mimic, miR-145 mimic, miR-133a antagomir, miR-513c antagomir, miR-96 antagomir, and nonsense miRNA mimic and antagomir controls were purchased from Life Technologies (Grand Island, NY).
In vitro functional assays including cell proliferation, apoptosis and invasion assays were conducted following miRNA mimic/antagomir transfections. Cell proliferation and apoptosis assays were performed using BrdU Cell Proliferation Assay kit (Calbiochem, Billerica, MA) and Apo-ONE caspase-3/7 assay kit (Promega, Madison, WI) as described by manufacturers. Detailed experimental design and protocols can be found in Supplementary Methods. Matrigel invasion assays were performed as previously described (19, 20).
RESULTS
Microarray analysis reveals differentially expressed mRNAs and miRNAs in AA and EA PCa patient specimens
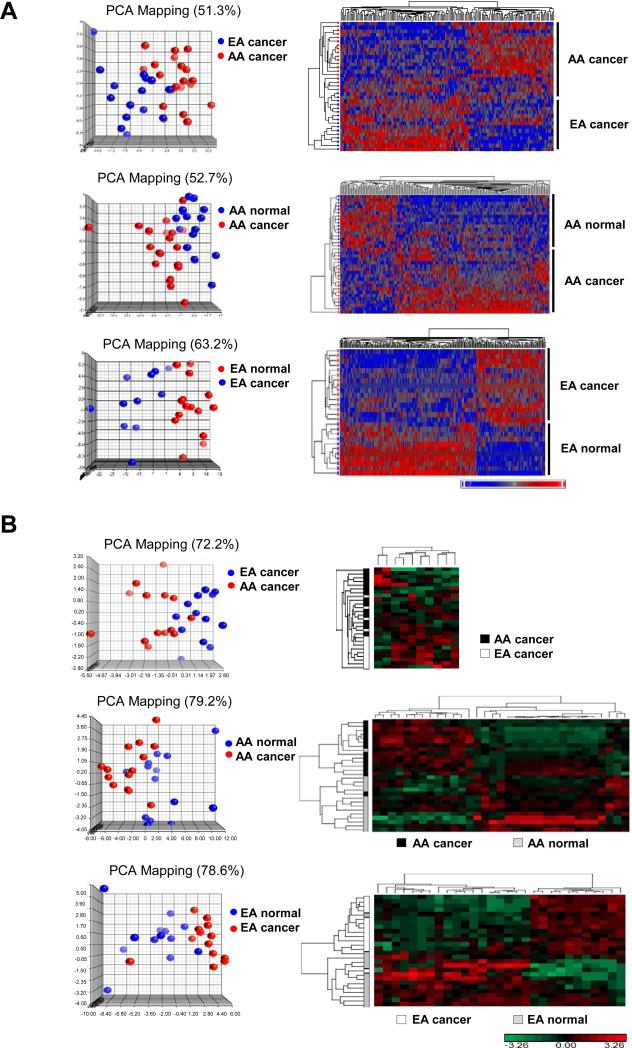
In an earlier study (13), a total of 70 prostate biopsy cores (20 cancerous and 20 patient-matched NP from AA patients; 15 cancerous and 15 patient-matched NP from EA patients) were subjected to mRNA profiling, and a 3-way comparison identified 2908 significant (ANOVA, 10% FDR multiple test correction) differentially expressed unique mRNAs. In the present study, we have classified these mRNAs as follows, 433 mRNAs are ‘AA-enriched’ (significantly over-expressed in AA) and 755 mRNAs are ‘AA-depleted’ (significantly under-expressed in AA) based on the AA PCa vs. EA PCa comparison (Supplementary Table S4). Another 980 mRNAs (up or down) are defined as ‘AA-specific’ based on the AA PCa vs. AA NP comparison (and not significant in EA PCa vs. EA NP), while 740 mRNAs are ‘EA-specific’ based on EA PCa vs. EA NP (and not significant in AA PCa vs. AA NP, Supplementary Table S4). Principal Component Analysis (PCA) and two-dimensional (2D) hierarchical clustering demonstrated clear separation and consistency of gene expression profiles in the three separate comparisons (Figure 1A).
Figure 1.
mRNA and miRNA expression profiling of PCa specimens and patient-matched normal tissues derived from AA and EA patients. (A) PCA plots and hierarchical 2D clustering of mRNA expression in AA PCa versus EA PCa, and PCa versus patient-matched normal tissue. (B) PCA plots and hierarchical clustergrams of miRNA expression in AA PCa versus EA PCa, and PCa versus patient-matched normal tissue. For both (A) and (B), samples are in rows, and mRNAs or miRNAs are in columns. Plots demonstrated clear separation and consistency of mRNA and miRNA expression profiles in group comparisons. For mRNA profiling, n=20, 20, 15, and 15 for AA PCa, AA matched normal, EA PCa and EA matched normal, respectively. For miRNA profiling, n=14, 14, 13, and 13 for AA PCa, AA matched normal, EA PCa and EA matched normal, respectively.
We also sought to investigate the relationship between miRNA and mRNA profiles in the same cohort of patients. Of the original 70 biopsy cores used for mRNA expression analysis, 54 provided sufficient material for miRNA expression profiling (14 cancerous and 14 patient-matched NP from AA patients; 13 cancerous and 13 patient-matched NP from EA patients). MiRNA profiling revealed 10, 33 and 29 miRNAs that were differentially expressed (ANOVA or paired t-test, 10% FDR, fold change≥1.5) between AA PCa vs. EA PCa, AA PCa vs. AA NP and EA PCa versus EA NP, respectively. Eleven of these miRNAs represent race-independent noncoding RNAs (miRNAs found significant in both AA PCa vs. AA NP and EA PCa versus EA NP comparisons), along with 2 AA-enriched, 8 AA-depleted, 22 AA-specific and 18 EA-specific miRNAs (Supplementary Table S5). PCA and 2D hierarchical clustering demonstrated clear separation of miRNA profiles (Figure 1B). In summary, we postulate that AA-enriched, AA-depleted and race-specific miRNAs and mRNAs (but not race-independent mRNAs and miRNAs) may be associated with the biological component of PCa disparities.
Novel reciprocal miRNA-mRNA pairings and dysregulated-unpaired mRNAs in oncogenic signaling pathways promoting PCa disparities
AA-enriched/depleted, AA-specific and EA-specific miRNAs were analyzed by TargetScanHuman 6.2 (implemented in IPA miRNA Target Filter), resulting in the identification of 3,153, 5,244 and 3,812 predicted target mRNAs, respectively. We focused attention on those miRNA-mRNA pairings with the following criteria: i) the predicted target mRNA was also differentially expressed in our microarray analysis (13), and ii) the miRNA exhibited a reciprocal expression relationship with its target mRNA (‘up-down’ or ‘down-up’). Using these criteria, we have compiled 150 reciprocal miRNA/mRNA pairings in AA PCa vs. EA PCa, 103 pairings in AA PCa vs. AA-matched NP and 137 pairings in EA PCa vs. EA-matched NP (Supplementary Table S6).
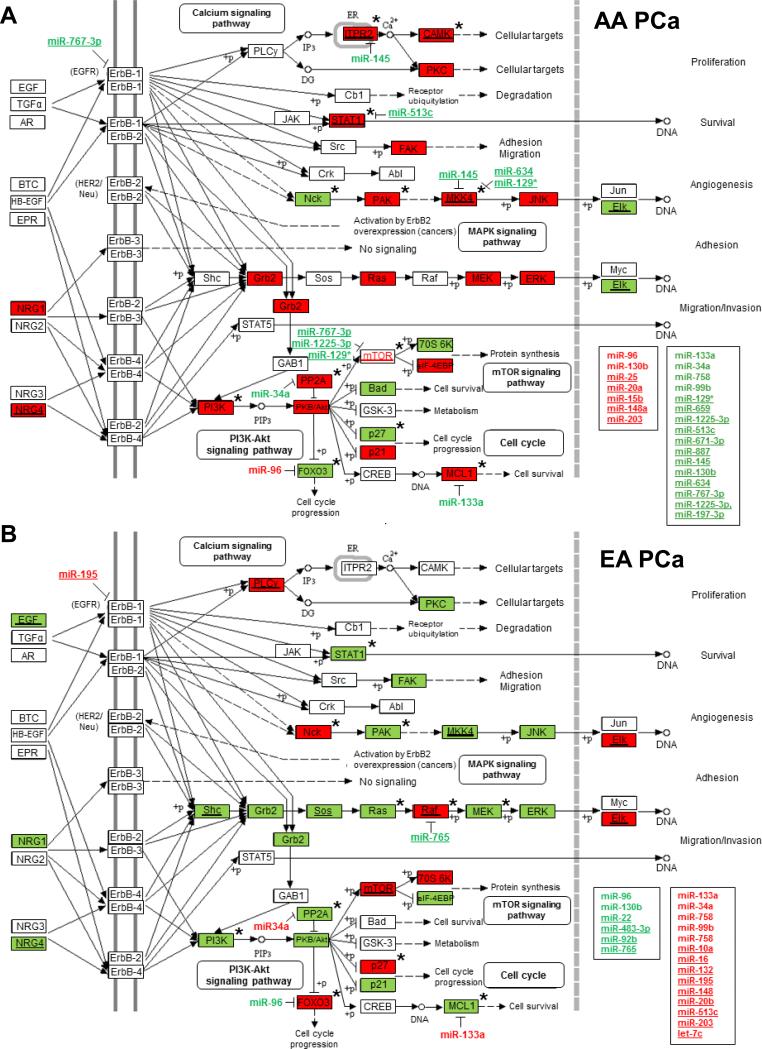
In a separate analysis to identify biological pathways most significantly associated with AA PCa aggressiveness, we applied Global test to our gene expression data from prostate biopsy cores. Global test is a permutation-based approach, coupled with a penalized logistic regression model, to identify gene sets in pathways most significantly associated to clinical phenotypes/outcomes (17). Using this approach, we identified 124, 106 and 137 significant KEGG annotated signaling pathways (FDR< 0.05) in AA PCa vs. EA PCa, AA PCa vs. AA NP and EA PCa vs. EA NP comparisons, respectively (Supplementary Table S7). Among the significant KEGG oncogenic pathways associated with AA PCa were ERBB, MTOR, WNT, JAK-STAT, TGF-β, P53 and VEGF. Noteworthy was the ERBB pathway in AA PCa, where a great majority of pathway genes (mRNAs) identified as significant by Global test were up-regulated in AA PCa vs. EA PCa and AA PCa vs. AA NP comparisons (Figure 2A and Supplementary Table S7). Conversely, the vast majority of significant genes in the ERBB pathway of EA PCa were down-regulated according to Global testing of EA PCa vs. AA PCa and EA PCa vs. EA NP comparisons (Figure 2B and Supplementary Table S7). Similar findings were obtained when analyzing our gene expression data by the GSEA approach (18) (Supplementary Table S8). Collectively, our pathway analysis suggests that differential gene regulation of ERBB signaling components in AA vs. EA PCa may play a critical role towards promoting PCa disparities. A finding that may be particularly relevant given the well-developed targeted therapies for this critical oncogenic pathway (22, 23).
Figure 2.
ERBB signaling pathway is highly activated in AA PCa specimens. Differentially expressed mRNAs (identified by Global test or Global test plus ANOVA (indicated by asterisk) and miRNAs (identified by ANOVA or paired t-test) populating the ERBB signaling pathway in (A) AA PCa and (B) EA PCa. Up- (in red) and down-regulated (in green) miRNAs with underline representing population-specific miRNAs, while miRNAs not underlined represent population-enriched (red) or -depleted (green) miRNAs. The same coloring and underlining scheme is used for differentially expressed mRNAs. ERBB pathway in AA PCa (A) is more highly activated compared to EA PCa (B) as determined by GO-Elite. Eight novel reciprocal miRNA-mRNA pairings are highlighted, including miR-133a/MCL1, miR-96/FOXO3A, miR-513c/STAT1, miR-34a/PPP2R2A, miR-145/ITPR2, miR-145/MKK4, miR-634/MKK4 and miR-129*/MKK4. MiRNAs listed in boxes represent the population-specific (underlined) or -enriched/depleted miRNAs predicted to target genes in the ERBB signaling pathway belonging to positively or negatively correlated pairings or non-differentially expressed targets (see Supplementary Table S6).
Next, we mapped the population-associated miRNAs and miRNA-mRNA pairings (Supplementary Tables S4, S5 and S6) onto the ERBB signaling pathway (Figure 2). Altogether, 17 AA-specific miRNAs (miR-15b, miR-20a, miR-25, miR-148a, miR-203, miR-129*, miR-659, miR-125-3p, miR-513c, miR-671-3p, miR-887, miR-145, miR-130b, miR-634, miR-767-3p, miR-1225-3pand miR-197-3p), 2 AA-enriched miRNAs (miR-96 and miR-130b) and 4 AA-depleted miRNAs (miR-133a, miR-758, miR-34a and miR-99b) were predicted to target 56 of 85 signaling genes of the ERBB pathway in AA PCa (Figure 2A, Supplementary Table S6), leading to a projected overall activation of oncogenic signaling based on GO-Elite analysis (24). Of the reciprocal miRNA-mRNA pairings in the ERBB pathway of AA PCa (Figure 2A), 14 were novel (i.e. predicted miRNA targeting of mRNA not validated in literature), namely miR-133a/MCL1 (down-up), miR-96/PPP2R3A (up-down), miR-133a/PPP2R2D (down-up), miR-767-3p/MTOR (down-up), miR-1225-3p/MTOR (down-up), miR-129*/MTOR (down-up), miR-129*/PIK3AP1 (down-up), miR-96/COL5A1 (up-down), miR-34a/IKBKE (down-up), miR-129*/IKBKB (down-up), mi-933/IKBKB (down-up), miR-145/MKK4 (down-up), miR-634/ MKK4 (down-up) and miR-129*/MKK4 (down-up) (Supplementary Table S6).
In contrast to the projected activation of ERBB signaling in AA PCa, EA PCa was comprised mostly of down-regulated oncogenes and up-regulated EA-specific/enriched miRNAs (predicted to target oncogenes) that were projected by GO-Elite to restrain ERBB pathway activity (Figure 2B). Note that AA- and EA-specific miRNAs do not overlap by definition. Hence, the inverse expression pattern of AA- and EA-specific/enriched/depleted miRNAs targeting different components of the ERBB signaling pathway likely plays a critical role in the differential aggressiveness of PCa progression in the two racial populations.
QRT-PCR validation in AA and EA PCa biopsy specimens
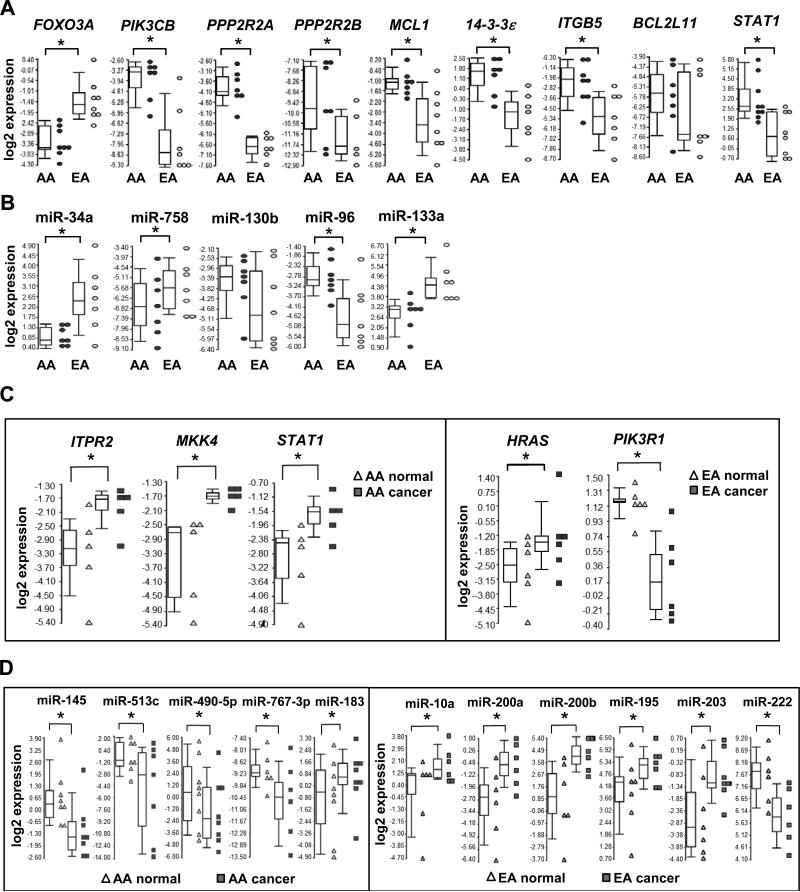
QRT-PCR validation assays were performed in a second cohort of PCa biopsy specimens from patients to validate our microarray analysis (Supplementary Table S1B). We specifically reassessed a combination of 30 differentially expressed miRNAs and mRNAs (identified as significant by both Global test and ANOVA; the exception being BCL2L11 that was identified as significant by ANOVA only) residing in the ERBB signaling pathway, as well as four additional signaling pathways (i.e. non-small cell lung cancer signaling, JAK/STAT pathway, tight junction signaling, phosphatidylinositol signaling). A comparison of the microarray and qRT-PCR results revealed high concordance (28 out of 30) in our expression measurements. Successful validations included AA-enriched and -depleted mRNAs (Figure 3A), AA-enriched and -depleted miRNAs (Figure 3B), population-specific mRNAs (Figure 3C) and population-specific miRNAs (Figure 3D). Encompassed within the validations were the novel reciprocal miRNA-mRNA pairings miR-133a/MCL1 (down-up; target mRNA significant in ERBB pathway by Global test), miR-96/FOXO3A (up-down; non-small cell lung cancer signaling), miR-513c/STAT1 (down-up; JAK/STAT pathway), miR-34a/PPP2R2A (down-up; tight junction signaling), miR-145/ITPR2 (down-up; phosphatidylinositol signaling) and miR-145/MKK4 (down-up; ERBB pathway) (Figures 2A and 3). Interestingly, four of the target mRNAs (FOXO3A, STAT1, PPP2R2A and ITPR2) in these pairings are also known to participate downstream of ERBB signaling and hence included in Figure 2A for illustration (25-29).
Figure 3.
QRT-PCR validation of population-enriched/depleted and -specific mRNAs and miRNAs in AA and EA PCa. (A) QRT-PCR validation of differentially expressed mRNAs in AA PCa versus EA PCa. (B) QRT-PCR validation of differentially expressed miRNAs in AA PCa versus EA PCa. (C) QRT-PCR validation of population-specific mRNAs. (D) QRT-PCR validation of population-specific miRNAs. The expression levels of mRNA or miRNAs from AA and EA patients are presented as Box-and-Whiskers plots (in A-D). Box: upper quantile, median and lower quantile. Whiskers: upper extreme (90 percentile of the dataset) and lower extreme (10 percentile of the dataset). Dot plots represent the relative expression levels of mRNA or miRNA from individual patient samples. * represents p < 0.05 using Student’s t-test (n = 6-9 independent experiments in A and B), or a paired Student's t-test (n= 5-8 independent experiments in C and D).
QRT-PCR assessment of population-specific PCa cell lines
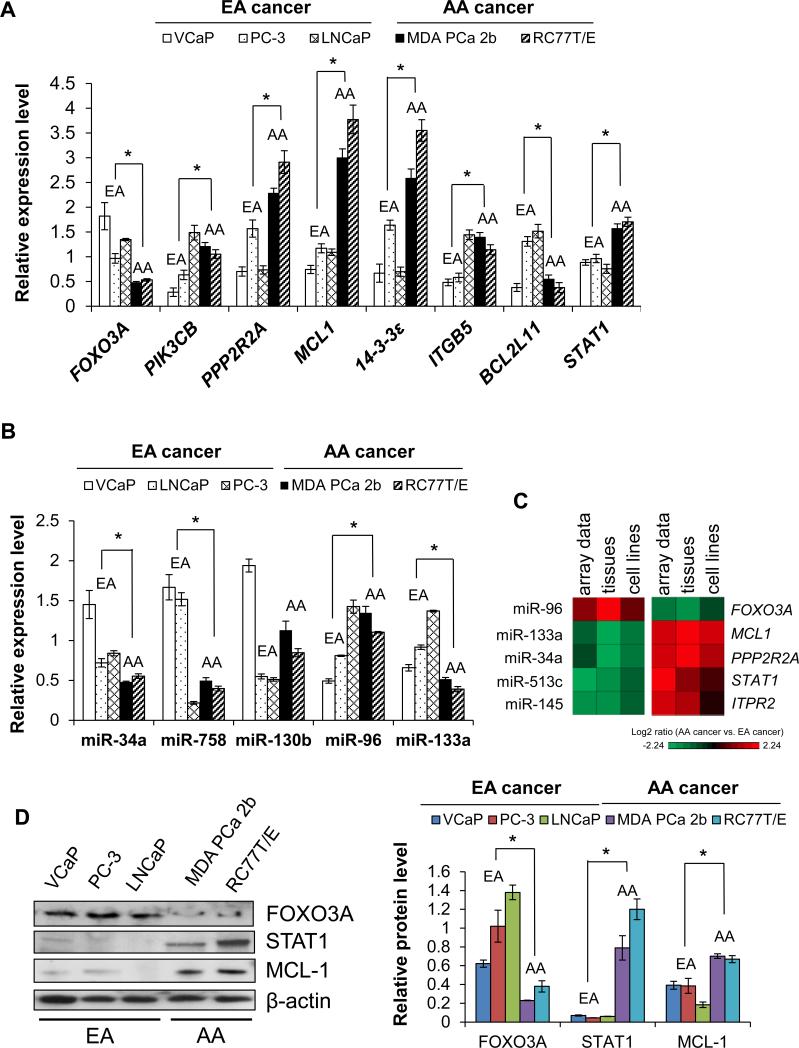
We also assessed the expression of AA-enriched and -depleted miRNAs and mRNAs (depicted in Figure 3) in a panel of PCa cell lines derived from AA (MDA PCa 2b, RC77T/E) and EA patients (VCaP, LNCaP and PC-3) (see Supplementary Methods). There was strong overall agreement between the microarray data from patient specimens and qRT-PCR results of PCa cell lines. Specifically, AA-depleted mRNAs (FOXO3A, BCL2L11) tended to be under-expressed in AA vs. EA PCa cell lines, and AA-enriched mRNAs (PIK3CB, PPP2R2A, MCL1, 14-3-3 ε , ITGB5, STAT1) tended to be over-expressed in AA vs. EA PCa cell lines (Figure 4A). An analogous consistency was observed for the miRNAs (Figure 4B). Again, contained within these validations were the novel reciprocal miRNA-mRNA pairings miR-133a/MCL1 (down-up), miR-96/FOXO3A (up-down), miR-513c/STAT1 (down-up) (Figure 4C). As a final consistency check, miRNA-mRNA pairings were found to be consistent with western blot analysis where FOXO3A was under-expressed while MCL-1 and STAT1 were over-expressed in AA vs. EA PCa cell lines (Figure 4D).
Figure 4.
Population-specific PCa cell lines are in-vitro cell models for PCa disparities. (A) qRT-PCR validation of microarray mRNA data in population-specific PCa cell lines. (B) qRT-PCR validation of microarray miRNA data in population-specific PCa cell lines. (C) Heat maps demonstrating inverse correlation between expression of miRNAs and mRNAs in AA PCa versus EA PCa comparisons. (D) Western blot analysis reveals protein expression correlates with mRNA expression in population-specific cell lines. Relative protein level was normalized to β-actin. Representative blots of 4-6 independent determinations. Data (in A, B and D) are presented as the mean ± SEM, with * p < 0.05 using an unpaired Student's t-test, n = 4-6 independent experiments for each cell line. Means were derived by combining results from AA cell lines versus EA cell lines.
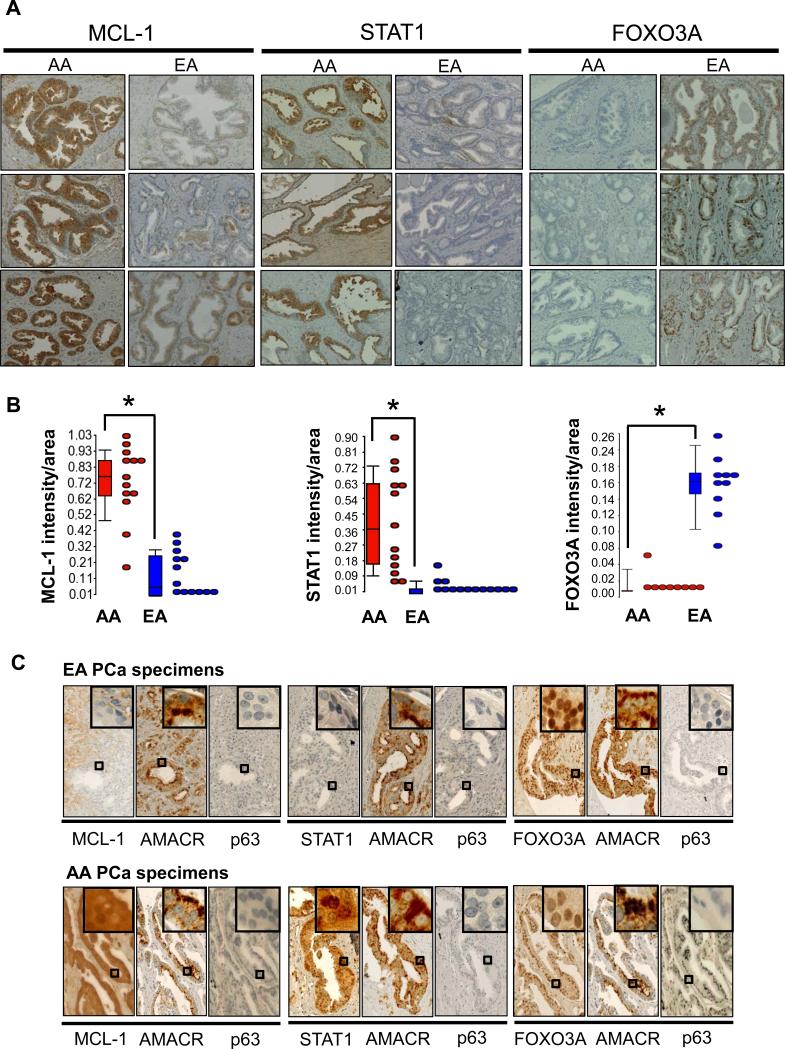
Immunohistochemical assessment of MCL-1, STAT1 and FOXO3A in AA and EA PCa specimens
Next, we examined protein expression of MCL-1, STAT1 and FOXO3A by immunohistochemical examination of archived FFPE PCa specimens from AA and EA patients, representing a third cohort with associated Gleason scores ranging from 6-9 (Figure 5A and 5B; Supplementary Table S1C and Figure S1). To ensure that MCL-1, STAT1 and FOXO3A protein expression was indeed present in cancerous cells, another series of IHC was performed where our proteins of interest were examined along with alpha-methylacyl CoA racemase (AMACR; positive control for cancer cells) and p63 (marker for NP basal cells) in serial sections (30). IHC results demonstrated over-expression of MCL-1 and STAT1 in the cytoplasm of AA vs. EA cancerous cells, and that the equivalent regions in adjacent sections stained strongly for AMACR but negative for p63 (Figure 5B). For FOXO3A, staining was greater in the nuclei of EA vs. AA cancerous cells, and in the equivalent regions of adjacent sections there was strong cytoplasmic staining for AMACR and negative staining for p63 in cancerous cells (Figure 5B). In summary, our IHC findings in patient specimens perfectly match the western results from PCa cell lines.
Figure 5.
Immunohistochemistry reveals differential protein expression in AA PCa versus EA PCa. (A) Paraffin-embedded tissue sections of human PCa specimens show strong MCL-1 and STAT1 expression in the cytoplasm of cancer cells of AA specimens, while FOXO3A immunoreactivity was detected in cancer cell nuclei of EA specimens. Images shown are representative of 13 AA and 13 EA specimens from different patients. (B) The intensities of cytoplasmic MCL-1 and STAT1, and nuclear FOXO3A were quantified by using the ratio of total intensity of immunoreactive MCL-1, STAT1 or FOXO3A over the total area of cells in the images. Data presented as box plots of n = 13 AA or EA samples, with * p < 0.05 using Student's t-test. (C) Serial FFPE sections derived from AA and EA PCa patients were immuno-stained for AMACR (a PCa marker), p63 (a normal basal cell marker) and the protein of interest (MCL-1, STAT1 or FOXO3A). Enlarged pictures (rectangles as indicated) enhance the nuclear or cytoplasmic distribution of these proteins at the cellular level.
Disruption of AA-specific and -enriched reciprocal miRNA-mRNA pairings affect cell proliferation, anti-apoptosis and invasion
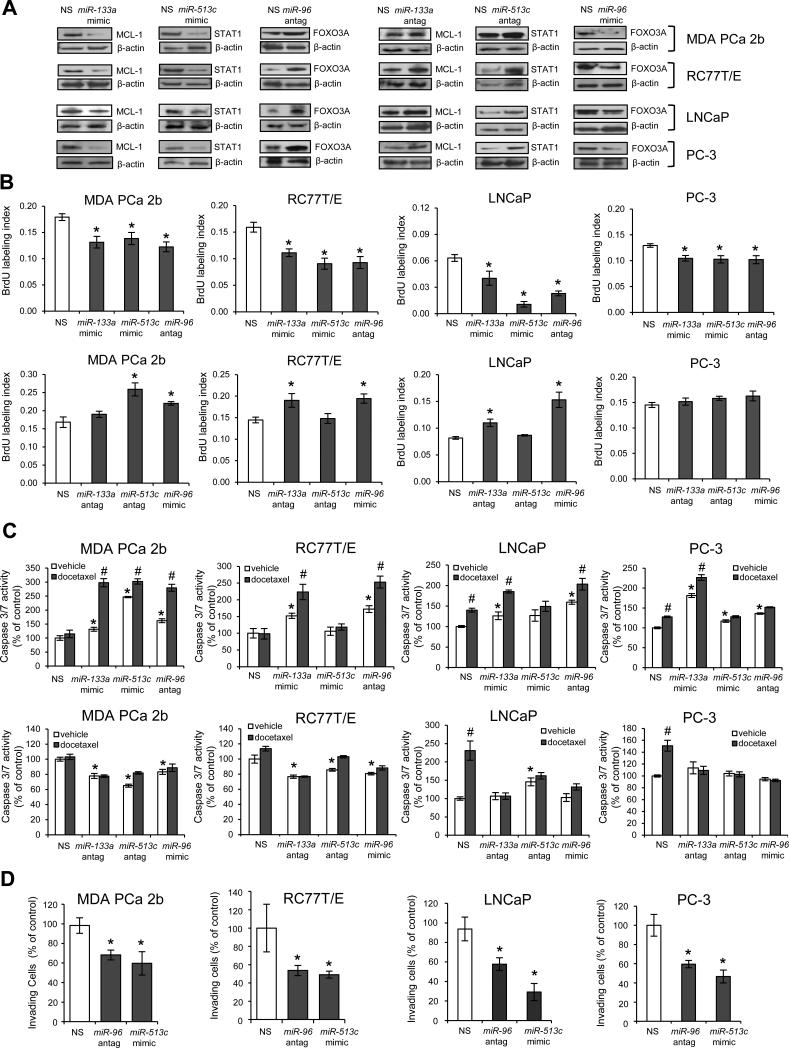
To more firmly establish a causal link among our reciprocal miRNA-mRNA pairings, a series of miRNA mimics and antagomirs were transfected into population-specific PCa cell lines and the protein products of predicted target mRNAs were measured by western blot. Two AA PCa lines (RC77T/E, MDA PCa 2b) and 2 EA PCa lines (LNCaP and PC-3) were chosen for in vitro functional assays on the basis of congruent qRT-PCR, western and immunohistochemical findings (Figures 4 and 5). Transfection of a miR-133a mimetic into AA and EA lines led to a down-regulation of MCL-1 protein compared to cells transfected with nonsense control RNA (Figure 6A, left panel). Conversely, miR-133a antagomir transfection into AA and EA lines led to an up-regulation of MCL-1 protein compared to nonsense control (Figure 6A, right panel). This antagomir-mediated up-regulation in PCa cells was anticipated given the ‘converse’ mimetic-induced down-regulation in PCa cells. Analogous confirmatory findings were also demonstrated for AA-enriched miR-96 (predicted target FOXO3A) and down-regulated AA-specific miR-513c (predicted target STAT1) (Figure 6A). Taken together, our in vitro mimic/antagomir manipulation of population-specific PCa cell lines was consistent with observations in patient specimens (see Figures 3 and 5), providing strong evidence of a causal link between our reciprocal miRNA-mRNA pairings.
Figure 6.
Functional validation of miR-133a/MCL1, miR-513c/STAT1 and miR-96/FOXO3A pairs in PCa aggressiveness. (A) Overexpression of miR-133a mimic, miR-513c mimic or miR-96 antagomir in PCa cell lines resulted in down-regulation of MCL-1 and STAT1, and up-regulation of FOXO3A, respectively. In contrast, inhibition of miR-133a or miR-513c with antagomirs or overexpression of miR-96 mimic resulted in up-regulation of MCL-1 and STAT1, and down-regulation of FOXO3A. AA lines are MDA PCa 2B, RC77T/E, and EA cell lines are LNCaP and PC-3. Representative western blots from 3-6 independent experimets. (B) BrdU-labeled cell proliferation assays of PCa cell lines transfected with miR-133a mimic, miR-513c mimic or miR-96 antagomir, or ‘converse’ antagomir/mimic treatments (miR-133a antagomir, miR-513c antagomir, or miR-96 mimic) were compared to cells treated with NS (nonsense scrambled negative control). Data are presented as mean ± SEM of n = 3-4 independent experiments, with * p < 0.05 using ANOVA and Tukey post-hoc test. (C) Apoptosis assays in population-specific PCa cell lines transfected with mimics or antagomirs. Apoptosis activity was assayed by measuring caspase3/7 activity using Apo-ONE kit (Promega), and the data were normalized to caspase 3/7 level of vehicle-treated NS control. Data are presented as mean ± SEM for n = 3-6 independent experiments, with p < 0.05 using ANOVA and Tukey post-hoc test comparing mimic or antagomir transfection plus vehicle treatment to NS transfection plus vehicle (*), or mimic or antagomir transfection plus vehicle to mimic or antagomir transfection plus docetaxel (#). (D) PCa cells transfected with miR-96 antagomir or miR-513c mimic were significantly less invasive compared to NS control-treated cells. Data are presented as mean ± SEM of n = 4-6 independent experiments, with * p < 0.05 using ANOVA and Holmes post-hoc test. Antag = antagomir.
The oncogenic consequences of disrupting steady-state expression of our prototype reciprocal pairings were assessed in AA lines RC77T/E and MDA PCa 2b, and EA lines LNCaP and PC-3. In the first set of functional assays, PCa lines were transfected with a series of mimics, antagomirs or nonsense control RNA and tested for proliferative activity using a BrdU labeling assay. In each case, the miR-133a mimic, miR-513c mimic and miR-96 antagomir significantly suppressed proliferation of the AA and EA PCa cell lines compared to nonsense control (Figure 6B, upper panels). Conversely, the majority of ‘converse’ antagomir/mimic treatments (miR-133a antagomir, miR-513c antagomir and miR-96 mimic) significantly enhanced proliferation in both AA lines and EA line LNCaP, as anticipated (Figure 6B, bottom panels). Interestingly, EA line PC-3 was completely resistant to the proliferation-inducing effects of all 3 ‘converse’ antagomir/mimic treatments (Figure 6B).
Next, apoptotic sensitivity in the absence and presence of 11nM docetaxel, a cytotoxic agent used in PCa chemotherapy (31), was assessed in PCa cell lines by caspase 3/7 activity assay. In the absence of any antagomir or mimic treatment, AA lines RC77T/E and MDA PCa 2b were chemoresistant to docetaxel-induced apoptosis (see nonsense control transfected cells in Figure 6C, upper panels). In contrast, docetaxel treatment alone significantly induced apoptosis in EA lines LNCaP and PC-3 (see nonsense control transfected cells in Figure 6C, upper panels). In the absence of docetaxel treatment, transfection of AA and EA cell lines with the miR-133a mimic, miR-513c mimic or miR-96 antagomir precipitated a generalized (exception being miR-513c mimic-transfected RC77T/E and LNCaP cells) and significant increase in apoptosis compared to nonsense control transfected cells (Figure 6C, upper panels). Strikingly in AA PCa cells (but not in EA cells), the combination of a mimic or antagomir treatment with docetaxel treatment resulted in apoptotic activity that was greater than either treatment alone, suggesting that disruption of key miRNAs sensitized cells to docetaxel (Figure 6C, upper panels). Interestingly, the ‘converse’ antagomir/mimic treatments (miR-133a antagomir, miR-513c antagomir and miR-96 mimic) in the absence of docetaxel had the effect of rendering AA lines, but not EA lines, more resistant to apoptosis (Figure 6C, bottom panels). Based on the proliferative and apoptotic findings, EA PCa lines compared to AA lines appear to be less susceptible to the oncogenic-promoting effects of the reciprocal pairs miR-133a/MCL1, miR-96/FOXO3A and miR-513c/STAT1.
Finally, the consequences of disrupting steady state expression of our prototype reciprocal pairings on the invasive activity of PCa cell lines were assessed by matrigel assay. Both miR-513c mimic and miR-96 antagomir treatments in AA lines RC77T/E and MDA PCa 2b, and EA lines LNCaP and PC-3 resulted in a significant decrease in invasive activity (Figure 6D), though we cannot discount the possibility that this decrease may be due in part to decreased proliferative activity (Figure 6B, upper panels). In an attempt to identify reciprocal pairings that modulate invasion without affecting proliferation, we tested two additional down-up pairings (miR-145/ITPR2 and miR-34a/PPP2R2A in the EGFR/PI3K/AKT pathway) in the AA PCa lines. Western blot analysis confirmed a causal link for these two reciprocal pairings, as transfection with either the miR-145 mimic or miR-34a mimic in AA lines resulted in a reduction of ITPR2 or PPP2R2A protein levels, respectively (Supplementary Figure S2A). As shown in Supplementary Figure S2B, the miR-145 mimic affected both proliferation and invasion, while the miR-34a mimic was associated with a significant decrease in invasion and had no effect on proliferation in both AA PCa cell lines. Taken together, these findings support the notion that depletion of miR-133a (leading to up-regulation of MCL1), miR-513c (up-regulation of STAT1), miR145 (up-regulation of ITPR2) and miR-34a (up-regulation of PPP2R2A), coupled with enrichment of miR-96 (down-regulation of FOXO3A) collectively drives proliferation, chemoresistance and/or invasion in AA PCa cells.
DISCUSSION
In this study, we performed an integrated analysis of differential miRNA and mRNA expression profiles in PCa and NP specimens derived from AA and EA patients. Our goal was to identify significant oncogenic signaling pathways that are populated with AA-specific/enriched reciprocal miRNA-mRNA pairs. Emphasis was placed on cataloging novel reciprocal pairs (i.e., predicted miRNA targeting of the mRNA has yet to be experimentally validated). The underlying hypothesis being that these novel reciprocal pairs may play a mechanistic role in PCa disparities (i.e. more aggressive nature of AA PCa), which could be assessed by systematically disrupting reciprocal pairs with mimic/antagomir treatment of population-specific PCa cell lines and testing for a loss (or gain) of oncogenic function. To date, the integrated analysis of miRNA-mRNA pairs has been limited to a handful of PCa studies (32, 33) and none have been related to PCa disparities.
There are a number of available miRNA-target mRNA prediction algorithms (34). However, it is estimated that up to 40% of all miRNA-target mRNA predictions are false positives (35), representing a major obstacle in the identification of true miRNA-mRNA interacting partnerships with functional consequences in cancer. An approach exploited by this study was to incorporate both a sequence-based algorithm for miRNA target predictions and focusing on miRNA-mRNA predictions exhibiting reciprocal differential expression profiles (up-down, down-up). Such a strategy has been demonstrated to provide more accurate predictions (35). A total of 390 reciprocal pairings were identified in PCa and NP specimens from AA and EA patients. These pairs (along with unpaired differentially expressed miRNAs and mRNAs) were found populated in 19 and 18 significant cancer signaling pathways from the perspective of AA and EA PCa, respectively.
ERBB signaling pathway in PCa disparities
The ERBB signaling pathway is regarded as a critical oncogenic signaling pathway in cancer, as mutations and/or over-expression of the EGFR and mutations in multiple PI3K isoforms are frequently detected in various types of cancers, including prostate, head and neck, renal, lung, breast, colon, ovarian, glioma, pancreas and bladder cancers (22, 23). In terms of PCa disparities, EGFR over-expression has been shown to be significantly associated with AA patients (11). Our findings suggest that 18 reciprocal miRNA-mRNA pairs populating the EGFR/PI3K/AKT signaling pathway in AA PCa, and likely working in concert with over-expressed EGFR (11), drives AA PCa.
MiR-513c/STAT1 (down-up) represented a novel predicted pairing, and miR-513c has previously been shown to be down-regulated in neuroendocrine lung tumors (36). However, the role of miR-513c in cancer and the identification of its target mRNA(s) have remained undetermined. Our results demonstrate for the first time that STAT1 serves as a target of miR-513c. The STAT1 protein is a transcription factor and its overexpression in PCa cells has been associated with docetaxel-resistance (37). Interestingly, the AA PCa cell lines investigated in this study were resistant to docetaxel-induced apoptosis but became sensitized upon treatment with a miR-513c mimic that down-regulated STAT1. Additional functions of the miR-513c/STAT1 pair in AA PCa cells include proliferation and invasion, as disruption of this pairing with a miR-513c mimic resulted in a loss of proliferative and invasive activities. The role of miR-513c/STAT1 in driving AA PCa was further supported by experiments employing a ‘converse’ targeting approach (i.e. miR-513c antagomir) in EA PCa cell lines, resulting in STAT1 up-regulation and a more aggressive phenotype reminiscent of the AA PCa lines (i.e. increased proliferation and chemoresistance).
Down-regulation of miR-133a has been observed in various cancers (38), acting as a tumor suppressor by targeting multiple oncogenes, such as FSCN1, MMP14, LASP1, EGFR, IGF1R and GSTP1 (39). In our study, MCL1 was identified as a novel target of miR-133a, and overexpression of a miR-133a mimic in PCa cell lines led to a down-regulation of MCL-1 protein and a corresponding decrease in proliferative activity, as well as loss of chemoresistance to docetaxel. MCL-1 has been demonstrated to be overexpressed in PCa and is linked to higher Gleason scores and increased bone metastasis in PCa patients (29). As was the case for miR-513c/STAT1, we demonstrated a role of miR-133a/MCL1 in driving AA PCa by employing a ‘converse’ targeting approach (i.e. miR-133a antagomir) in EA PCa cell lines, resulting in MCL-1 up-regulation and a more aggressive phenotype, again reminiscent of the AA PCa lines.
Up-regulation of miR-96 has been observed in lung, breast, bladder and colorectal cancers (40). MiR-96 promotes cell proliferation by targeting the FOXO1 gene, encoding a transcription factor, in breast and prostate cancer (41, 42); and enhances proliferative, invasive and migratory activity by targeting FOXO1 and RECK in breast cancer, bladder and lung cancers (43, 44). In this study, we further demonstrated that FOXO3A targeted miR-96 in PCa, confirming a previous observation in breast cancer (45). Disruption of miR-96/FOXO3A (up-down) in AA PCa cell lines with a miR-96 antagomir resulted in FOXO3A protein up-regulation and a corresponding decrease in proliferative, invasive and chemoresistant activities. Conversely, introduction of a miR-96 mimic into EA PCa cell lines had the opposite effect by down-regulating FOXO3A protein and promoting proliferation and chemoresistance. In essence, the EA PCa cell lines transformed into a more aggressive AA PCa-like phenotype. Taken together, these findings are consistent with the known tumor suppressor effect of FOXO3A in PCa (46).
Another intriguing miRNA-mRNA pair residing in the ERBB signaling pathway of AA PCa is miR-145/ITPR2 (down-up). Recent genome-wide association studies have implicated the inositol 1,4,5-triphosphate receptor type 2 (ITPR2) gene as a novel risk locus for renal cell carcinoma (47, 48). MiR-145 has been implicated as a tumor suppressive miRNA as it is down-regulated in different cancers and its expression has been associated with an inhibition of PCa cell invasion and migration in vitro (49). Our findings link miR-145 and ITPR2 for the first time as a functional reciprocal pair that promotes invasion and proliferation in AA PCa.
It should also be noted that AA PCa was associated with a large number of up-regulated oncogenes (such as ITGA5, PIK3CB, PIK3AP, ITPR2, STAT1, CSNK2A1, MKK4, 14-3-3ε, MTOR and MCL1) as well as dysregulated unpaired miRNAs that are unique to AA PCa (e.g. AA-specific/depleted miRNAs) and computationally predicted to target EGFR/PI3K/AKT signaling components (such as EGFR, AKT3, GSK3, JAK1, JUN and KRAS) leading to pathway activation. Conversely, our analysis identified an equally large number of dysregulated oncogenes plus unpaired miRNAs that were specific to EA PCa and computationally predicted to target a different set of EGFR/PI3K/AKT signaling components leading to pathway suppression. Also noteworthy, unpaired AA-specific miR-767-3p (down-regulated in AA PCa vs. AA NP) and unpaired EA-specific miR-195 (up-regulated in EA PCa vs. EA NP) were both predicted to target the EGFR mRNA, resulting in an anticipated up- and down-regulation of the EGFR protein, respectively. This finding would be consistent with the observed racial disparity of EGFR over-expression in AA PCa (11). While our analysis has focused on 5 reciprocal miRNA-mRNA pairings, it is important to stress that the miRNAs in these pairings would be expected to coordinately target other mRNAs (i.e., MCL1, FSCN1, MMP14, LASP1, EGFR, IGF1R and GSTP1 by miR-133a, and FOXO3A, FOXO1 and RECK by miR-96), presumably leading to the aggressive phenotypic features found in AA PCa. Lastly, our findings suggest that these deregulated miRNA-mRNA pairs, uniquely found in AA PCa, appear to target the EGFR-PI3K-AKT axis, thus driving PCa aggressiveness in the AA population.
Understanding the origins and etiology of cancer disparities is a complex endeavor and it is imperative that such disparities be addressed at all levels of intervention, both social and biological. Evidence exists indicating that one component of the disparity may be related to biological differences in the molecular etiology of the disease resulting in tumor aggressiveness. We have employed a population-based comparative approach in an attempt to discern potential drivers of PCa aggressiveness and have identified novel pathway alterations in miRNA-mRNA pairs that may contribute to PCa disparities. Given the projected use of miRNA mimics and antagomirs as potential cancer therapeutics (50), our study serves as a first pass catalog of dysregulated miRNA-mRNA pairs residing in key oncogenic signaling pathways in AA PCa.
Supplementary Material
Translational Relevance.
Prostate cancer (PCa) tends to be more aggressive and lethal in African Americans (AA) compared to European Americans (EA). An understanding of the molecular mechanisms associated with PCa disparities can aid in the development of innovative and improved therapeutic options for the AA population. Integrative functional genomics analysis of patient specimens and PCa cell lines has identified novel AA-specific and -enriched miRNA-mRNA pairs, including miR-133a/MCL1, miR-513c/STAT1, miR-96/FOXO3A, miR-145/ITPR2 and miR-34a/PPP2R2A, that reside in key oncogenic signaling pathways. The presence of these miRNA-mRNA pairs is computationally predicted to augment activation of epidermal growth factor receptor (EGFR)/PI3K/AKT signaling in AA compared to EA cancers. Specific manipulation of these pairs reduced cell proliferation/invasion and enhanced docetaxel-induced cytotoxicity in AA PCa cell lines. Converse manipulation resulted in a more aggressive phenotype in EA cell lines. Thus, targeting these novel miRNA-mRNA pairs may provide a potential clinical strategy for reducing AA PCa burden.
Acknowledgments
Grant Support
This work was supported by NCI grant R01 CA120316 (NHL), DOD grant PC121975 (NHL), Affymetrix Collaborations in Cancer Research Award (NHL), NCI supplement grant U01 CA116937 (SRP), and American Cancer Society grant IRG-08-091-01 (BDW).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–7. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi XB, Tepper CG, White RW. MicroRNAs and prostate cancer. J Cell Mol Med. 2008;12:1456–65. doi: 10.1111/j.1582-4934.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 8.Powell IJ. Epidemiology and pathophysiology of prostate cancer in African-American men. J Urol. 2007;177:444–9. doi: 10.1016/j.juro.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 9.Evans S, Metcalfe C, Ibrahim F, Persad R, Ben-Shlomo Y. Investigating Black-White differences in prostate cancer prognosis: A systematic review and meta-analysis. Int J Cancer. 2008;123:430–5. doi: 10.1002/ijc.23500. [DOI] [PubMed] [Google Scholar]
- 10.Devgan SA, Henderson BE, Yu MC, Shi CY, Pike MC, Ross RK, et al. Genetic variation of 3 beta-hydroxysteroid dehydrogenase type II in three racial/ethnic groups: implications for prostate cancer risk. Prostate. 1997;33:9–12. doi: 10.1002/(sici)1097-0045(19970915)33:1<9::aid-pros2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 11.Shuch B, Mikhail M, Satagopan J, Lee P, Yee H, Chang C, et al. Racial disparity of epidermal growth factor receptor expression in prostate cancer. J Clin Oncol. 2004;22:4725–9. doi: 10.1200/JCO.2004.06.134. [DOI] [PubMed] [Google Scholar]
- 12.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 13.Wang BD, Yang Q, Ceniccola K, Bianco F, Andrawis R, Jarrett T, et al. Androgen receptor-target genes in african american prostate cancer disparities. Prostate Cancer. 2013;2013:763569. doi: 10.1155/2013/763569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Tang H, Thayanithy V, Subramanian S, Oberg AL, Cunningham JM, et al. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res. 2009;69:9490–7. doi: 10.1158/0008-5472.CAN-09-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomerantz MM, Beckwith CA, Regan MM, Wyman SK, Petrovics G, Chen Y, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–74. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 18.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A. 2005;102:13544–9. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.House CD, Vaske CJ, Schwartz AM, Obias V, Frank B, Luu T, et al. Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion. Cancer research. 70:6957–67. doi: 10.1158/0008-5472.CAN-10-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang BD, Kline CL, Pastor DM, Olson TL, Frank B, Luu T, et al. Prostate apoptosis response protein 4 sensitizes human colon cancer cells to chemotherapeutic 5-FU through mediation of an NF kappaB and microRNA network. Molecular cancer. 9:98. doi: 10.1186/1476-4598-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seshacharyulu P, Ponnusamy MP, Haridas D, Jain M, Ganti AK, Batra SK. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 28:2209–10. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu G, Fan Z, Ding M, Zhang H, Mu L, Ding Y, et al. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene. doi: 10.1038/onc.2015.45. [DOI] [PubMed] [Google Scholar]
- 26.Han W, Carpenter RL, Cao X, Lo HW. STAT1 gene expression is enhanced by nuclear EGFR and HER2 via cooperation with STAT3. Mol Carcinog. 52:959–69. doi: 10.1002/mc.21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cronshaw DG, Kouroumalis A, Parry R, Webb A, Brown Z, Ward SG. Evidence that phospholipase-C-dependent, calcium-independent mechanisms are required for directional migration of T-lymphocytes in response to the CCR4 ligands CCL17 and CCL22. J Leukoc Biol. 2006;79:1369–80. doi: 10.1189/jlb.0106035. [DOI] [PubMed] [Google Scholar]
- 28.Krol J, Francis RE, Albergaria A, Sunters A, Polychronis A, Coombes RC, et al. The transcription factor FOXO3a is a crucial cellular target of gefitinib (Iressa) in breast cancer cells. Mol Cancer Ther. 2007;6:3169–79. doi: 10.1158/1535-7163.MCT-07-0507. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Zhau HE, Osunkoya AO, Iqbal S, Yang X, Fan S, et al. Vascular endothelial growth factor regulates myeloid cell leukemia-1 expression through neuropilin-1-dependent activation of c-MET signaling in human prostate cancer cells. Mol Cancer. 2010;9:9. doi: 10.1186/1476-4598-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–75. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–12. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 32.Gade S, Porzelius C, Falth M, Brase JC, Wuttig D, Kuner R, et al. Graph based fusion of miRNA and mRNA expression data improves clinical outcome prediction in prostate cancer. BMC Bioinformatics. 2011;12:488. doi: 10.1186/1471-2105-12-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng J, Huang C, Diao X, Fan M, Wang P, Xiao Y, et al. Screening biomarkers of prostate cancer by integrating microRNA and mRNA microarrays. Genet Test Mol Biomarkers. 2013;17:807–13. doi: 10.1089/gtmb.2013.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng H, Fu R, Wang JT, Liu Q, Chen H, Jiang SW. Advances in the Techniques for the Prediction of microRNA Targets. Int J Mol Sci. 2013;14:8179–87. doi: 10.3390/ijms14048179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, et al. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–9. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 36.Mairinger FD, Ting S, Werner R, Walter RF, Hager T, Vollbrecht C, et al. Different micro-RNA expression profiles distinguish subtypes of neuroendocrine tumors of the lung: results of a profiling study. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.74. [DOI] [PubMed] [Google Scholar]
- 37.Patterson SG, Wei S, Chen X, Sallman DA, Gilvary DL, Zhong B, et al. Novel role of Stat1 in the development of docetaxel resistance in prostate tumor cells. Oncogene. 2006;25:6113–22. doi: 10.1038/sj.onc.1209632. [DOI] [PubMed] [Google Scholar]
- 38.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K, et al. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol. 2013;31:115–23. doi: 10.1016/j.urolonc.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Xu XM, Qian JC, Deng ZL, Cai Z, Tang T, Wang P, et al. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett. 2012;4:339–45. doi: 10.3892/ol.2012.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haflidadottir BS, Larne O, Martin M, Persson M, Edsjo A, Bjartell A, et al. Upregulation of miR-96 enhances cellular proliferation of prostate cancer cells through FOXO1. PLoS One. 2013;8:e72400. doi: 10.1371/journal.pone.0072400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H, Li Q, Li W, Zheng T, Zhao S, Liu Z. miR-96 downregulates RECK to promote growth and motility of non-small cell lung cancer cells. Mol Cell Biochem. 2014 doi: 10.1007/s11010-014-1966-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Kong X, Li J, Luo Q, Li X, Shen L, et al. miR-96 promotes tumor proliferation and invasion by targeting RECK in breast cancer. Oncol Rep. 2014;31:1357–63. doi: 10.3892/or.2013.2934. [DOI] [PubMed] [Google Scholar]
- 45.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, et al. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shukla S, Bhaskaran N, Maclennan GT, Gupta S. Deregulation of FoxO3a accelerates prostate cancer progression in TRAMP mice. Prostate. 2013;73:1507–17. doi: 10.1002/pros.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Audenet F, Cancel-Tassin G, Bigot P, Audouin M, Gaffory C, Ondet V, et al. Germline genetic variations at 11q13 and 12p11 locus modulate age at onset for renal cell carcinoma. J Urol. 2014;191:487–92. doi: 10.1016/j.juro.2013.07.064. [DOI] [PubMed] [Google Scholar]
- 48.Wu X, Scelo G, Purdue MP, Rothman N, Johansson M, Ye Y, et al. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 2012;21:456–62. doi: 10.1093/hmg/ddr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59:78–87. doi: 10.1038/jhg.2013.121. [DOI] [PubMed] [Google Scholar]
- 50.Melo SA, Kalluri R. Molecular pathways: microRNAs as cancer therapeutics. Clin Cancer Res. 2012;18:4234–9. doi: 10.1158/1078-0432.CCR-11-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.