ABSTRACT
Linezolid is an oxazolidinone with potent activity against Mycobacterium tuberculosis. Linezolid toxicity in patients correlates with the dose and duration of therapy. These toxicities are attributable to the inhibition of mitochondrial protein synthesis. Clinically relevant linezolid regimens were simulated in the in vitro hollow-fiber infection model (HFIM) system to identify the linezolid therapies that minimize toxicity, maximize antibacterial activity, and prevent drug resistance. Linezolid inhibited mitochondrial proteins in an exposure-dependent manner, with toxicity being driven by trough concentrations. Once-daily linezolid killed M. tuberculosis in an exposure-dependent manner. Further, 300 mg linezolid given every 12 hours generated more bacterial kill but more toxicity than 600 mg linezolid given once daily. None of the regimens prevented linezolid resistance. These findings show that with linezolid monotherapy, a clear tradeoff exists between antibacterial activity and toxicity. By identifying the pharmacokinetic parameters linked with toxicity and antibacterial activity, these data can provide guidance for clinical trials evaluating linezolid in multidrug antituberculosis regimens.
IMPORTANCE
The emergence and spread of multidrug-resistant M. tuberculosis are a major threat to global public health. Linezolid is an oxazolidinone that is licensed for human use and has demonstrated potent activity against multidrug-resistant M. tuberculosis. However, long-term use of linezolid has shown to be toxic in patients, often resulting in thrombocytopenia. We examined therapeutic linezolid regimens in an in vitro model to characterize the exposure-toxicity relationship. The antibacterial activity against M. tuberculosis was also assessed for these regimens, including the amplification or suppression of resistant mutant subpopulations by the chosen regimen. Higher exposures of linezolid resulted in greater antibacterial activity, but with more toxicity and, for some regimens, increased resistant mutant subpopulation amplification, illustrating the trade-off between activity and toxicity. These findings can provide valuable insight for designing optimal dosage regimens for linezolid that are part of the long combination courses used to treat multidrug-resistant M. tuberculosis.
INTRODUCTION
Multiply drug-resistant (MDR) and extensively drug-resistant (XDR) Mycobacterium tuberculosis strains represent major public health challenges and are causes of much morbidity and mortality (1–3). Because chemotherapy is nonoptimal for these pathogens and is poorly tolerated, failure rates are higher, treatment courses are longer, and, consequently, failure and substantial attendant drug-related morbidity are too frequently seen.
Linezolid is an oxazolidinone antibiotic which has only recently entered the clinician’s therapeutic armamentarium for M. tuberculosis. This agent has been employed in the therapy of XDR M. tuberculosis as an add-on drug to “optimized background therapy” for patients not responding to their current drug regimen (4). The results were surprisingly good, with 87% having a therapeutic response and only 4 therapeutic failures. These therapeutic outcomes were driven by an initial dose of 600 mg daily. The responses were also attended by frequent occurrence of possibly or probably drug-related toxic events (82%). A number of toxicities have been associated with linezolid therapy, including gastrointestinal side effects and optic and peripheral neuropathies (4), but the earliest (and a frequent) toxicity is hematological toxicity, resulting in reduced numbers of platelets. This effect has been demonstrated to be related to mitochondrial toxicity (5, 6). However, drug-related toxicities observed in patients receiving 600 mg of linezolid daily were reduced in frequency after a reduction in dose to 300 mg daily (4).
In order to obtain the maximal benefits of this antibiotic, understanding the relationships between drug exposure and M. tuberculosis kill, emergence of isolates resistant to linezolid, and toxicity is required. Our group has previously published work describing the relationships between drug exposure and M. tuberculosis kill and resistance suppression (7–9) in the hollow-fiber infection model (HFIM) system. Recently, our laboratory developed an approach to examining drug exposure and toxicity for oxazolidinones (10). A major objective of this current project was to characterize the exposure-mitochondrial toxicity relationship for linezolid. To that end, we employed a human myeloid leukemia cell line with platelet properties (K562 cells) and exposed them to various concentrations of linezolid, simulating pharmacokinetic profiles that would be achieved in a patient receiving this antibiotic. K562 cells have been used previously to evaluate mitochondrial toxicity related to oxazolidinone therapy (6). K562 cells were then evaluated for mitochondrial toxicity by screening for known toxicity biomarkers, including synthesis of proteins involved with energy metabolism, disruption of ATP production, and altered apoptosis. These different uses of the HFIM give us the ability to relate exposure to both antimicrobial effect and drug concentration-driven mitochondrial toxicity.
The aim of these experiments was to identify the optimal dose and dosing interval of linezolid as monotherapy to obtain optimal cell kill, minimize resistance emergence, and cause the least toxicity for the patient.
RESULTS
Linezolid-associated mitochondrial toxicity of K562 cells in the HFIM system.
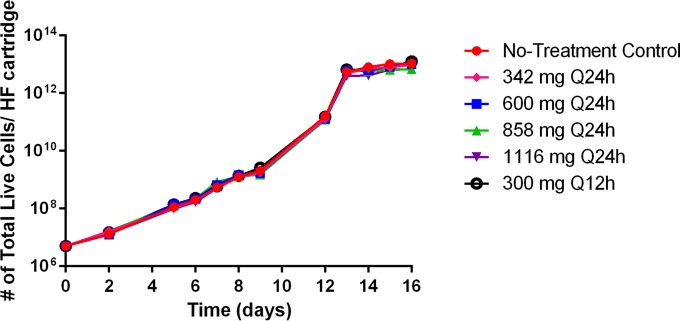
K562 cells grew well in the HFIM system, reaching peak concentrations of approximately 1013 live cells per hollow-fiber cartridge by the end of the 16-day study in the no-treatment control arm (Fig. 1). Cell growth was similar among all treatment arms (Fig. 1), indicating that linezolid exposure had minimal effect on cellular proliferation in the HFIM system.
FIG 1 .
Cell growth of K562 cells in the HFIMS system during linezolid therapy. Linezolid exposure had a minimal effect on K562 cellular proliferation after 16 days in the HFIM system. At various time points, cells were harvested from the HFIM system, and live cells were enumerated using the trypan blue exclusion method.
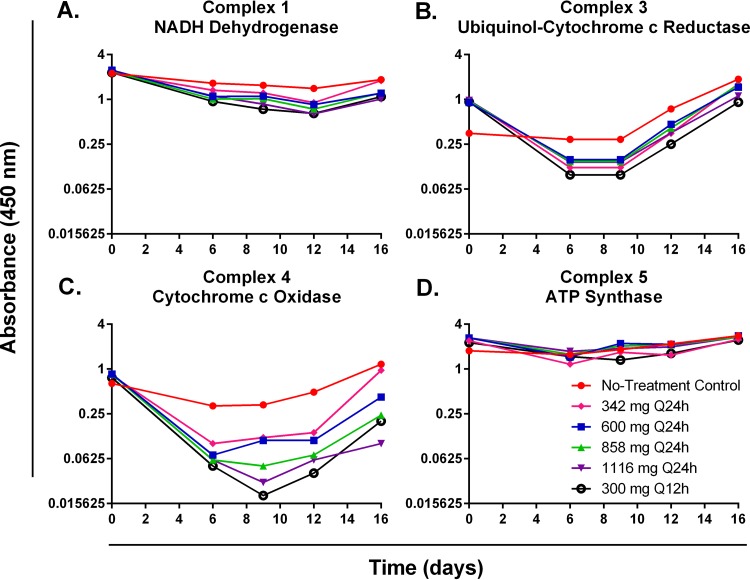
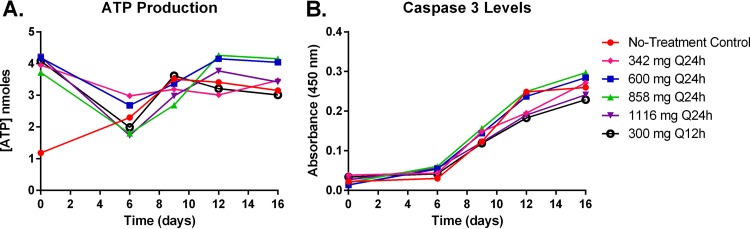
Mitochondrial toxicity of K562 cells was evaluated by assessing two characterized mitochondrial toxicity biomarkers: energy metabolism disruption and altered apoptosis. Disruption of cellular energy metabolism was determined by quantifying oxidative-phosphorylation (OXPHOS) protein complexes 1, 3, 4, and 5 and measuring ATP production. Linezolid therapy had little effect on the protein synthesis of OXPHOS complex 1, 3, or 5 proteins (Fig. 2A, B, and D). In contrast, OXPHOS complex 4 proteins were markedly inhibited in the linezolid treatment arms relative to the no-treatment control, and protein levels were suppressed in a clearly exposure-dependent manner for the once-daily (q24h) regimens (Fig. 2C). At day 9 (when maximal protein inhibition was observed), complex 4 protein levels were 36.36% for the 342-mg q24h regimen, 33.33% for the 600-mg q24h regimen, 15.15% for the 858-mg q24h regimen, and 9.09% for the 1,116-mg q24h regimen relative to the no-treatment control arm (Fig. 2C). Interestingly, the twice-daily regimen (300 mg q12h) resulted in the lowest levels of complex 4 proteins (6.06% relative to the no-treatment control at day 9), indicating that mitochondrial protein synthesis is sensitive to linezolid dosing interval. Cellular ATP production was not influenced by linezolid, as an exposure-response relationship was not observed over time for linezolid-treated K562 cells (Fig. 3A). Finally, mitochondrial toxicity was evaluated by assessing the induction of apoptosis (a mitochondrial toxicity biomarker) in K562 cells treated with linezolid by measuring caspase 3 activation. Caspase 3 activity levels increased in all treatment arms throughout the duration of the 16-day study (Fig. 3B). The degree of apoptosis did not correlate with different exposures of linezolid (Fig. 3B), suggesting that linezolid-induced mitochondrial toxicity is not the result of altered apoptosis in treated K562 cells.
FIG 2 .
OXPHOS mitochondrial protein levels for complexes 1, 3, 4, and 5 from K562 cells treated with linezolid in the HFIM system. The effects of linezolid on the production of OXPHOS protein complexes 1 (A), 3 (B), 4 (C), and 5 (D) in K562 cells harvested from the HFIM system were assessed. Cell pellets were lysed, and OXPHOS mitochondrial protein levels were quantified by ELISA.
FIG 3 .
ATP production and caspase 3 activation in K562 cells treated with linezolid in the HFIM system. The effects of linezolid exposure on the production of ATP (A) and caspase 3 activation (B) in K562 cells harvested from the HFIM system were assessed. ATP levels were quantified using a colorimetric ATP assay, and caspase 3 activation was assessed via a colorimetric caspase 3 assay.
Pharmacokinetic/pharmacodynamic (PK/PD) analysis of linezolid-associated mitochondrial toxicity.
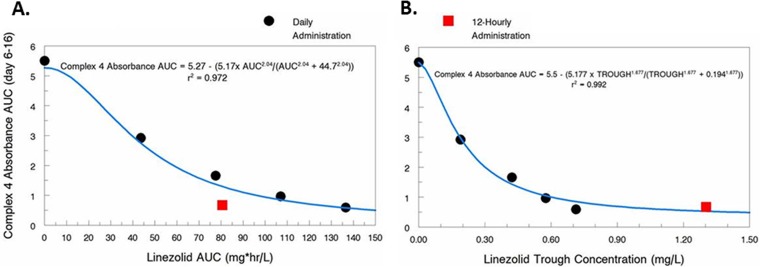
To identify the PD index for linezolid that is most closely linked with toxicity, we graphed the area under the OXPHOS complex 4 protein concentration-time curve (AUCOXPHOS4) from day 6 through day 16 (Fig. 2C) for each hollow-fiber treatment arm against the free-drug linezolid 24-h AUC exposure and trough concentrations that were achieved in the HFIM system. An inhibitory sigmoid-Emax effect model was fit to the data, and the results are shown in Fig. 4. The model fit the data well, resulting in r2 values of 0.972 for linezolid 24-h AUC exposure and 0.992 for linezolid trough concentration. These data suggest that linezolid trough concentration is the PD index that is most closely linked with linezolid-associated mitochondrial toxicity and that 0.19 mg/liter is the trough concentration associated with 50% maximal toxicity.
FIG 4 .
Influence of linezolid exposure (24-h AUC) and trough concentration on mitochondrial toxicity. The area under the OXPHOS protein complex 4-time curve (AUCOXPHOS4) illustrated in Fig. 2C was calculated for all hollow-fiber arms and plotted against the linezolid 24-h AUC exposures and trough concentrations achieved for each regimen in the HFIM system. Black circles correspond to the q24h regimens, and red squares represent the q12h regimens. An inhibitory sigmoid-Emax effect model was fitted to the data (blue line).
Linezolid MICs and mutation frequencies for M. tuberculosis.
The linezolid MIC for the H37Rv M. tuberculosis strain used in these studies was 1 mg/liter. The frequency of mutation to resistance in response to 2.5 mg/liter of linezolid for this organism was −7.4 log CFU.
Antibacterial activity of linezolid against M. tuberculosis in the HFIM system.
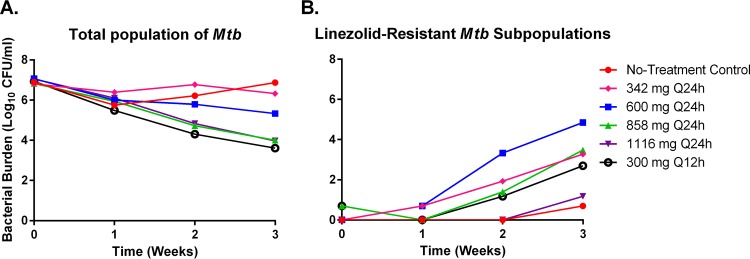
We evaluated the antibacterial activity of the same linezolid dosage regimens described above for the toxicity studies against the H37Rv strain of M. tuberculosis in the HFIM system to identify an optimal regimen that is the most effective and least toxic. The overall microbial kill for each regimen over time is displayed in Fig. 5A. Linezolid killed M. tuberculosis in an exposure-dependent manner, with the two highest exposures associated with 858 mg q24h and 1,116 mg q24h, resulting in a 3-log reduction in bacterial burden at the end of the 3-week study (Fig. 5A). Conversely, the 342-mg q24h and 600-mg q24h regimens yielded an overall bacterial decline of 0.5 and 1.7 logs, respectively. The 300-mg q12h regimen provided the greatest amount of antibacterial activity and provided 3.3 logs of bacterial cell kill at the termination of the study (Fig. 5A).
FIG 5 .
Antibacterial activity of linezolid on M. tuberculosis in the HFIM system. The antibacterial effect of linezolid on the total population of M. tuberculosis (A) and the linezolid-resistant M. tuberculosis subpopulation (B) was determined. Hollow-fiber experiments were performed as two independent studies, and the data are the combined results of both experiments. The data points represent the mean values from both experiments, and the error bars correspond to the standard error of the mean.
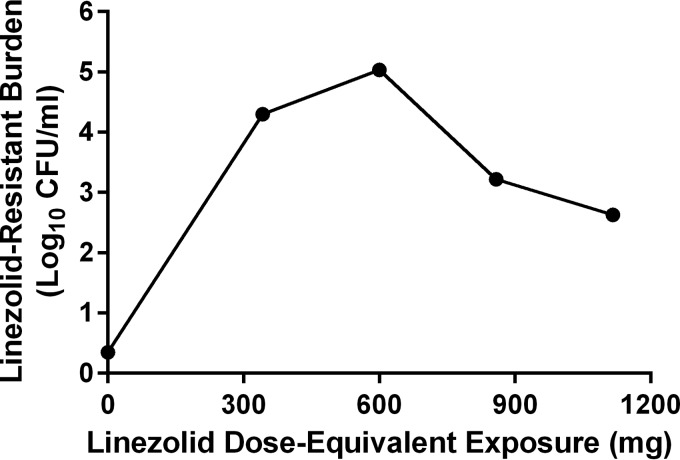
All therapeutic regimens of linezolid allowed the emergence of linezolid-resistant M. tuberculosis subpopulations (Fig. 5B). The number of drug-resistant mutants was heavily influenced by linezolid exposure (Fig. 5B and 6), with the 600-mg q24h regimen resulting in the highest number of mutants and the 1,116-mg q24h regimen resulting in the smallest mutant population after 3 weeks of therapy. The relationship between linezolid exposure and the bacterial burden of the resistant subpopulation resembles an inverted “U” (Fig. 6), a phenomenon that has been previously described for resistance selection for other antibiotics (11). The emergence of resistant subpopulations allowed by the 300-mg q12h dosage regimen was similar to those reported for the 342-mg q24h and 858-mg q24h therapeutic arms.
FIG 6 .
Influence of linezolid exposure on the amplification of linezolid-resistant bacterial mutants after 3 weeks of therapy. Hollow-fiber studies were performed twice as two independent experiments. The data points represent the geometric means from the two experiments for each linezolid exposure.
Mathematical model for antibacterial efficacy in the HFIM system.
A large mathematical model was simultaneously fitted to the linezolid PK, the total bacterial M. tuberculosis burden, and the linezolid-resistant M. tuberculosis subpopulation burden data generated by the studies performed in the HFIM system. The model fit all of the data well, resulting in precise and unbiased curve fits, as demonstrated by the predicted-versus-observed graphs (employing Bayesian posterior parameter values) for linezolid PK and bacterial burden (total population and resistant subpopulation [see Fig. S3 in the supplemental material]) as well as measures of bias and precision (mean weighted error; bias-adjusted mean weighted squared error). The predicted-observed plots for the three system outputs yielded the following: for linezolid concentration, a slope of 1.125, an intercept of 0.598, and an r2 of 0.941; for total bacterial burden, a slope of 1.030, an intercept of −0.130, and an r2 of 0.826; and for the less linezolid-susceptible subpopulation, a slope of 1.056, an intercept of −0.133, and an r2 of 0.949 (see Fig. S3).
The means, medians, and standard deviations of the parameter values are displayed in Table 1. The pharmacokinetic parameter values of volume and clearance (CL) are for protein binding-corrected free linezolid. The maximal growth rate constants for the susceptible and less-susceptible populations demonstrate that the less-susceptible population grows substantially less well. The maximal kill rate constants demonstrate that it is substantially more difficult to kill the less linezolid-susceptible subpopulation, leading to the difficulty in suppressing resistance amplification, even with higher exposures. Finally, the linezolid concentration at which the M. tuberculosis kill rate is 50% of the maximal rate is 5-fold higher for the less-susceptible population. These are in line with expectations for these populations for linezolid.
TABLE 1 .
Parameter values for the full model for estimation of linezolid impact on total MTB burden and the less linezolid-susceptible subpopulationa
| Parameter (units) | Mean | Median | SD |
|---|---|---|---|
| Vol (liters) | 65.2 | 65.2 | 1.21 |
| CL (liters/h) | 12.2 | 13.2 | 1.79 |
| Kg,max-s (h−1) | 0.754 | 0.699 | 0.206 |
| Kg,max-r (h−1) | 0.0208 | 0.0210 | 0.00518 |
| Kk,max-s (h−1) | 3.25 | 3.66 | 0.624 |
| Kk,max-r (h−1) | 1.21 | 0.752 | 0.885 |
| C50,k-s (mg/liter) | 3.68 | 3.74 | 0.496 |
| C50,k-r (mg/liter) | 18.7 | 17.0 | 2.873 |
| Hk-s | 10.9 | 14.3 | 4.39 |
| Hk-r | 13.8 | 14.6 | 10.1 |
| POPMAX (CFU/ml) | 3.72 × 107 | 4.84 × 107 | 1.50 × 107 |
| Initcond_2 (CFU/ml) | 1.81 × 107 | 1.43 × 107 | 9.66 × 106 |
| Initcond_3 (CFU/ml) | 5.90 | 0.601 | 8.35 |
Vol is the apparent volume of the central compartment; CL is the apparent plasma clearance; Kg,max-s and Kg,max-r are maximal growth rate constants for the susceptible and resistant populations; Kk,max-s and Kk,max-r are maximal kill rate constants for the respective populations; C50,k-s and C50,k-r are linezolid concentrations at which the kill rate is half maximal for the respective populations; Hk-s and Hk-r are Hill’s constants for the respective populations; POPMAX is the population burden at which stationary phase is achieved; Initcond_2 is the initial total MTB population burden; Initcond_3 is the initial linezolid-resistant MTB burden. It was assumed that the initial linezolid concentration was 0.0.
DISCUSSION
MDR and XDR TB pose substantial public health problems all around the world. Rates of response to antibiotic intervention are lower than we have come to expect in the chemotherapy of tuberculosis. Treatment courses are long and fraught with toxicities, which leads to decreased patient compliance. These difficulties are directly attributable to the fact that in both instances, our first-line agents isoniazid and rifampin have been rendered ineffective due to the emergence of resistance. There is a need for new agents that have good activity like isoniazid and rifampin. Linezolid and other oxazolidinones have the promise of being one of the new agents to fill this niche for the combination therapy of MDR and XDR M. tuberculosis infection.
In order for linezolid to fulfill this promise, it is important that an optimized dose and dosing interval be chosen for therapy. The most appropriate dosage regimen will have a major impact on the burden of M. tuberculosis, will have a low probability of resistance emergence, and will be relatively nontoxic. In order to identify the optimal dose, understanding the relationships between drug exposure and M. tuberculosis kill, resistance emergence, and the likelihood concentration-driven toxicities is necessary. Understanding that a fixed drug dose will generate a distribution of exposures in a population of patients is also important. Consequently, these considerations must be factored into the design of experiments for both efficacy and toxicity so that not only a population mean exposure is studied, but also exposure equivalents corresponding to +1, +2, and −1 standard deviations from the mean exposure.
In this set of experiments, we employed the hollow-fiber infection model (HFIM) to examine all of these issues. We used this model previously to examine the microbiological endpoints for M. tuberculosis therapy (7–9). In the current project, we developed an approach to link linezolid exposure to an in vitro measure of linezolid-associated toxic effect. One of the most common toxic effects of linezolid and other oxazolidinone therapy is the reduction of the number of hematological cells. In particular, thrombocytopenia and anemia are major problems with long-term oxazolidinone administration (12).
Oxazolidinone toxicity is thought to be mediated by inhibiting mitochondrial protein synthesis (5, 6). Others have shown that OXPHOS complex 4 (cytochrome c oxidase) proteins are consistently and significantly decreased in both in vitro and in vivo systems exposed to oxazolidinone therapy (5, 6, 13). Furthermore, OXPHOS complex 4 levels have been shown to be well below the normal range in patients treated with prolonged courses of linezolid (5, 13). These data identify protein complex 4 as a marker for oxazolidinone-related mitochondrial toxicity. To that end, we employed K562 cells, a human erythromyeloblastoid leukemia cell line, and exposed these cells to linezolid concentration-time profiles similar to that seen in humans using the HFIM system. We sampled the system over time and assayed for OXPHOS protein complexes 1 (NADH dehydrogenase), 3 (cytochrome c reductase), 4 (cytochrome c oxidase), and 5 (ATP synthase). OXPHOS complex 2 proteins (succinate dehydrogenase) have been previously shown to not be altered in the presence of linezolid (5), as these protein subunits are mainly encoded by nuclear DNA (14). Thus, we did not assess OXPHOS complex 2 protein levels as part of this investigation. It was our hypothesis that only the complex 4 proteins would yield a clear exposure-response relationship. In Fig. 2, it is obvious that only complex 4 demonstrates a definite exposure-response with a range of linezolid exposures. We calculated the AUCOXPHOS4 as an integrated measure of the toxic effect of linezolid. The results, seen in Fig. 4, indicate that an inhibitory sigmoid-Emax model describes the data very well. We had employed two measures of exposure as the independent variable, the 24-h AUC of linezolid and the linezolid trough concentration. In the experiment, we examined 5 linezolid exposures plus a no-treatment control. Four of the 5 linezolid exposures were administered daily. For the daily exposures, we evaluated the mean exposure from a 600-mg linezolid dose, as well as +1 standard deviation (mean 24-h AUC exposure equivalent to 858 mg), +2 SD (1,116-mg equivalent), and −1 SD (342-mg equivalent). Additionally, we examined two different dosing intervals for the mean exposure of a 600-mg daily dose, administered either once daily or as 300 mg q12h. As seen in Fig. 4A, when AUC is the independent variable, all the daily exposures are well described by the mathematical model, but the q12h administration results in more toxicity than suggested by the model. When trough concentrations are employed as the independent variable, the model is more predictive of the results. We conclude that the effect of linezolid on OXPHOS complex 4 proteins is most closely linked to trough concentrations. This finding has been seen in clinical trials as well (15, 16).
If toxicity were the only issue, we would choose the lower exposure and we would employ q24h or possibly less frequent dosing to minimize adverse drug effects; however, in order for linezolid therapy to be most effective against M. tuberculosis clinically, it is imperative that a regimen result in major kill of the total M. tuberculosis population burden and also not amplify resistant subpopulations. Linezolid’s mechanism of action for M. tuberculosis kill and for toxicity is inhibition of protein synthesis, bacterial and mitochondrial, respectively. Consequently, as antimicrobial activity increases, an increase in toxicity would be predicted to accompany it.
In Fig. 5, we see the change over time in the total M. tuberculosis burden. The decline in M. tuberculosis total population reaches a maximum at about 2 weeks. Thereafter, the total burden increases due to resistance emergence. The two higher exposures (+1 SD [858-mg equivalents] and +2 SD [1,116-mg equivalents]) produce more overall kill than the two smaller exposures, with +1 SD killing ca. 1.2 log10 CFU/ml more, which rises to 1.36 log10 CFU/ml with the +2 SD exposure, relative to the mean (600 mg). Conversely, the −1 SD (342-mg equivalents) kills substantially less (~1.1 log10 CFU/ml) than the mean exposure (maximum kill of 0.7 log10 CFU/ml for the −1 SD exposure compared to 1.8 log10 CFU/ml for the mean exposure). Clearly, increasing exposure to increase antimicrobial activity has to be balanced for the concomitant increase in toxicity.
We also examined half the mean exposure given every 12 h (q12h) to identify whether dosing interval influences M. tuberculosis cell kill. Somewhat to our surprise, the 300-mg-equivalent exposure given q12h resulted in significantly better M. tuberculosis kill than 600 mg daily, indicating that trough concentrations (relative to the MIC) were driving M. tuberculosis kill. A similar finding was observed by Wallis et al. (17). As described above, however, the price is one of toxicity. This is important for design and interpretation of early bactericidal activity (EBA) trials for oxazolidinones. A 14-day EBA study of linezolid may demonstrate maximal killing activity for a twice-daily regimen of linezolid compared with daily dosing of the same total amount. Such a result must be balanced by the increased toxicity that will result from this dosing interval and the fact that the toxicity will not be evident clinically until after 14 days of treatment. We have noted in other studies evaluating oxazolidinone pharmacodynamics (18) that the improved bacterial kill with twice-daily dosing is seen only with monotherapy and that the advantage is abrogated in the face of adequate combination chemotherapy. Also, for the advantage of better patient adherence to regimen or ease of directly observed therapy, it is likely that a daily regimen will be optimal.
Finally, what of resistance emergence? Figure 5B shows a somewhat surprising finding. The order of resistance emergence does not follow a simple exposure response. The mean exposure actually drives the greatest resistance emergence, followed by the −1 SD exposure. The +1 and +2 SD exposures have considerably less resistant subpopulation amplification at the end of the third week, but both are still considerably above baseline. The reason for this is shown in Fig. 6. The data describe a nonmonotonic function, which our laboratory has described as an inverted “U” (11). At lower exposures, there is some, but not maximal, resistant subpopulation amplification. As greater pressure is exerted, there is more selective pressure, and the resistance amplification increases to a maximum. At yet-higher exposures, some control is exerted over the less-susceptible subpopulation, and its numbers decline from the maximum. In this instance, even the +2-SD exposure did not fully control the less-susceptible subpopulation amplification. Consequently, increasing exposure that would partially control the resistance amplification would result in considerable more toxicity with higher linezolid doses. In addition, linezolid (and other oxazolidinones) will be given as part of combination chemotherapy, where there will be a much greater likelihood of controlling resistance amplification, as long as there is sufficient consideration given to the exposures for the other drug(s) in the combination.
These data and the analysis provide some guidance for clinical trials to study linezolid in clinical trials of M. tuberculosis-infected patients. The exposure-responses for M. tuberculosis kill and toxicity show a clear trade-off with increasing dosage. The magnitude of the M. tuberculosis kill for −1 SD exposure is likely to be suboptimal for many patients. The higher exposures (+1 and +2 SD) induce a rapid increase in the measure of toxicity examined here. Therefore, if linezolid is to be incorporated into a multidrug regimen in a phase II study, doses slightly smaller and slightly larger than 600 mg (e.g., perhaps 450 and 900 mg) should be examined. However, no 450-mg dosing is available, so a 300-mg/day regimen might be considered. Several centers with experience in MDR treatment now initiate therapy with 300 mg/day. In addition, studies of initial dosing with 900 or 600 mg/day should allow dose reductions to be made when exposure-related toxicities are detected. While the amplification of a less susceptible subpopulation is worrisome, if linezolid is included in a robust regimen (i.e., not added as a single agent to a failing regimen), there is a high likelihood of protection from resistance emergence. We are currently conducting studies evaluating q24h dosing intervals of linezolid in combination with moxifloxacin to determine relationships to toxicity and antimicrobial activity.
These in vitro models can provide guidance and decision support for the choice of doses and schedules in phase II clinical trials to ultimately result in regimens that will have substantial M. tuberculosis kill with moderate toxic liability and which will (in combination regimens) suppress emergence of resistance.
MATERIALS AND METHODS
K562 cells and bacterial strain.
For the linezolid toxicity studies, human erythromyeloblastoid leukemia K562 (ATCC CCL-243) cells were used. Cells were maintained in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (HyClone) at 37°C and 5% CO2 and were subcultured twice weekly.
The H37Rv strain of M. tuberculosis was used for the linezolid antibacterial studies. Bacterial stocks were stored at −80°C and grown as previously described (7).
Compound.
Pharmaceutical-grade linezolid was purchased from CuraScript (Orlando, FL) as a solution for injection and stored at room temperature in the dark. Prior to experimental use, linezolid was diluted to the desired concentrations in sterile deionized water.
Linezolid susceptibility.
The susceptibility of the H37Rv strain to linezolid was determined using the absolute serial dilution method as previously described (7, 9). Briefly, 104 CFU of H37Rv in log-phase growth was plated on Middlebrook 7H10 agar (Becton Dickinson Microbiology Systems, Sparks, MD) supplemented with 10% oleic acid, albumin, dextrose, and catalase (OADC; Becton Dickinson Microbiology Systems) containing 2-fold dilutions of linezolid. The cultures were incubated at 37°C and 5% CO2. After 4 weeks of incubation, the MICs were determined by identifying the lowest drug concentration at which there was no bacterial growth on the agar plate.
Mutation frequency determination.
The mutation frequency of the H37Rv strain was evaluated using methods that are described elsewhere (7, 9). Briefly, H37Rv cultures in log-phase growth were inoculated onto plates containing Middlebrook 7H10 agar plus 10% Middlebrook OADC with linezolid at a concentration equivalent to 2.5× the MIC. The mutation frequency was identified after 4 weeks of incubation at 37°C and 5% CO2.
Toxicity studies in the HFIM system.
Drug-related toxicities associated with prolonged linezolid therapy were assessed in the HFIM system. A detailed description of the HFIM system is found elsewhere (7, 9, 10, 19, 20). For these studies, the extracapillary space (ECS) of six polysulfone hollow-fiber cartridges (FiberCellSystems, Frederick, MD) were inoculated with 5 × 106 K562 cells. Linezolid was administered into five cartridges as a 1-h infusion via computer-controlled syringe pumps to simulate the free (non-protein-bound)-drug 24-h area under the concentration-time curve (AUC) that is associated with the mean free-drug plasma AUC of linezolid at steady state after oral dosing at 600 mg daily (q24h), one standard deviation above the mean free 24-h-AUC exposure (858-mg mean equivalent exposure q24h), two standard deviations above the mean free 24-h-AUC exposure (1,116-mg mean equivalent exposure q24h), and one standard deviation below the mean free 24-h-AUC exposure (342-mg mean equivalent exposure q24h). The fifth cartridge received linezolid as the mean free 24-h AUC exposure for 300 mg administered twice daily (q12h). Finally, one cartridge did not receive drug treatment and served as a no-treatment control. Linezolid containing medium was isovolumetrically replaced with drug-free medium to simulate the mean clinical half-life of 4.26 h. Human pharmacokinetic parameters for linezolid were obtained from the Zyvox package insert (21).
Serial drug concentrations were collected from the HFIM system over the course of the study and quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (see below). Intensive PK sampling was conducted for the first 48 h of the study, followed by sparse PK sampling for the duration of the study. The measured linezolid concentrations in the HIFM system were within 10% of the targeted value at all time points, indicating that the desired PK profiles for linezolid were achieved in the HFIM system (see Fig. S1 in the supplemental material).
Mitochondrial toxicity of K562 cells was evaluated at various time points over the course of the 16-day study. K562 cells were harvested from the HFIM system, enumerated, and assessed for viability via the trypan blue exclusion method. Cell pellets were collected on days 0, 6, 9, 12, and 16 after linezolid exposure and were frozen at −80°C until the end of the study. Cell pellets were lysed using the cell extraction buffer provided with the cytochrome c oxidase (complex IV) human profiling enzyme-linked immunosorbent assay (ELISA) kit (Abcam, Cambridge, MA) according to the manufacturer’s instructions, and total cellular protein concentration was determined for each sample using a Thermo Scientific Pierce bincinchoninic acid (BCA) protein assay kit (Life Technologies, Grand Island, NY). Cell lysates were assessed for energy metabolism disruption and altered apoptosis, two biomarkers of mitochondrial toxicity. Energy metabolism disruption was evaluated by quantifying oxidative phosphorylation (OXPHOS) protein complexes using a NADH dehydrogenase (complex 1) human profiling ELISA kit (Abcam), a cytochrome c reductase (complex 3) human profiling ELISA kit (Abcam), a cytochrome c oxidase (complex 4) human profiling ELISA kit (Abcam), and an ATP synthase (complex 5) human profiling ELISA kit (Abcam). Because OXPHOS complex 2 (succinate dehydrogenase) protein subunits are encoded mainly by nuclear DNA (14) and have been shown to not be altered in the presence of linezolid (5), we did not assess these protein levels to conserve cell lysates for additional assays. Energy metabolism disruption of cell lysates was also evaluated by measuring total ATP production using a colorimetric ATP assay kit (Abcam). Finally, the mitochondrial toxicity marker of altered apoptosis was examined by measuring the increase in caspase 3 activation using a colorimetric assay (Abcam) as per the manufacturer’s instructions.
Assessment of antibacterial activity for linezolid against M. tuberculosis in the HFIM system.
The same linezolid dosage regimens and pharmacokinetic profiles described above for the toxicity study were evaluated for antibacterial activity against M. tuberculosis in the HFIM system. The HFIM system for M. tuberculosis has been described elsewhere (9). Briefly, 10 ml of H37Rv M. tuberculosis was inoculated into the HFIM system (FiberCell Systems) at a concentration of 1 × 107 CFU per ml. Linezolid was administered into cartridges as described above for the toxicity studies. Intensive PK sampling was conducted for the first 48 h of the study, followed by sparse PK sampling for the duration of the study (see Fig. S2 in the supplemental material). At various times throughout the 3-week study, bacterial cultures were collected. The samples were washed by centrifugation (1,500 × g for 5 min) to remove linezolid and then plated onto drug-free 7H10 OADC agar and agar supplemented with linezolid at 2.5× the MIC for quantitative culture. After 28 days of incubation, the colonies on each plate were enumerated, and the bacterial burden was calculated. Two independent hollow-fiber studies were performed.
Linezolid LC-MS/MS assay.
Samples collected from the HFIM system experiments were diluted with water and analyzed by high-pressure liquid chromatography-tandem mass spectrometry (LC-MS/MS; AB Sciex, Foster City, CA) for linezolid concentrations. The HPLC parameters were as follows: the column used was a Thermoscientific Hypersil Gold C18 column, 150 by 4.6 mm, with a 5-µm particle diameter, and mobile phases A and B were 5 mM ammonium formate adjusted to pH 3.00 using formic acid in water and acetonitrile, respectively, and ran in isocratic mode at 34% B with a flow rate of 0.750 ml/min. Linezolid concentrations were measured using the MS/MS transition from m/z 338 to m/z 235. The analysis run time was 5 min, and the target concentration range was 0.125 to 20.0 µg/ml (r2 > 0.997). Quality control samples (0.125, 2.50, and 20.0 µg/ml) had coefficients of variation (CVs) ranging from 2.9 to 4.3% and accuracies (percent recovery) between 85 and 103%.
Statistical analysis and mathematical modeling.
The area under the OXPHOS protein complex 4-time curve (AUCOXPHOS4) from days 6 to 16 was calculated for each hollow-fiber arm from the toxicity studies using Prism software version 6.01 (GraphPad Software, Inc., La Jolla, CA). The complex 4 AUCOXPHOS from days 6 to 16 was graphed against the linezolid free-drug 24-h AUC exposure and free-drug trough concentrations achieved for each regimen in the HFIM system. An inhibitory sigmoid-Emax effect model was fitted to the data using ADAPT V software. Parameter estimates were obtained employing maximum-likelihood estimation.
The impact of linezolid on both the total M. tuberculosis burden and the less-susceptible subpopulation burden was analyzed in a population sense employing the program BigNPAG, a nonparametric (for parameter distributions) adaptive grid program written by Leary et al. and Neely et al. (22, 23). Starting weights were as estimates of the inverse of the observation variance for each of the three system outputs. The adaptive γ function in BigNPAG was employed to optimize the weights so as to give the best approximation to the homoscedastic assumption. The three system outputs were (i) measurement of linezolid concentration-time profiles in each HFIM arm, (ii) measurement of total M. tuberculosis burden in each HFIM arm, and (iii) measurement of the less linezolid-susceptible subpopulation in each HFIM arm.
The system of nonhomogeneous differential equations to describe this system was published previously (24).
SUPPLEMENTAL MATERIAL
PK profiles achieved in the HFIM system for the toxicity studies. The solid black lines correspond to the “desired” or targeted linezolid concentrations, and the red squares represent the actual linezolid concentrations measured in the hollow-fiber cartridges at the designated time points. Download
PK profiles achieved in the HFIM system for the antibacterial activity studies. The solid black lines correspond to the “desired” or targeted linezolid concentrations, and the red squares represent the actual linezolid concentrations measured in the hollow-fiber cartridges at the designated time points. Download
Linear regression analysis of the predicted-versus-observed plots determined by the mathematical model. The mathematical model described in Materials and Methods was fitted to the data generated from the antibacterial activity studies for linezolid in the HFIM system with the H37Rv strain of M. tuberculosis. The predicted-versus-observed plots for the linezolid PK (A), the total bacterial burden (B), and the bacterial burden of the linezolid-resistant subpopulation (C) are shown. The model was fitted to the data sets resulting from the two independent hollow-fiber experiments. The first data set is represented by the black circles, the second data set corresponds to the blue circles, and the linear regression is designated by the red line. Download
ACKNOWLEDGMENTS
This work was supported by a 2012 grant from the Lifespan/Tufts/Brown Center for AIDS Research (CFAR), an NIH/NIAID/Miriam Hospital funded program (P30-AI-042853) and also by the NIAID/DAIDS of the National Institutes of Health under contract award No. HHSN266200700043C and HHSN272201400006I.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We have no conflicts of interest to declare.
Footnotes
Citation Brown AN, Drusano GL, Adams JR, Rodriquez JL, Jambunathan K, Baluya DL, Brown DL, Kwara A, Mirsalis JC, Hafner R, Louie A. 2015. Preclinical evaluations to identify optimal linezolid regimens for tuberculosis therapy. mBio 6(6):e01741-15. doi:10.1128/mBio.01741-15.
REFERENCES
- 1.Aziz MA, Wright A, Laszlo A, De MA, Portaels F, Van DA, Wells C, Nunn P, Blanc L, Raviglione M, WHO/International Union Against Tuberculosis And Lung Disease Global Project on Anti-tuberculosis Drug Resistance Surveillance . 2006. Epidemiology of antituberculosis drug resistance (the Global Project on Anti-tuberculosis Drug Resistance Surveillance): an updated analysis. Lancet 368:2142–2154. [DOI] [PubMed] [Google Scholar]
- 2.Shah NS, Wright A, Bai G, Barrera L, Boulahbal F, Martín-Casabona N, Drobniewski F, Gilpin C, Havelková M, Lepe R, Lumb R, Metchock B, Portaels F, Rodrigues MF, Rüsch-Gerdes S, Van Deun A, Vincent V, Laserson K, Wells C, Cegielski JP. 2007. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis 13:380–387. doi: 10.3201/eid1303.061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zignol M, Hosseini M, Wright A, Weezenbeek C, Nunn P, Watt C, Williams B, Dye C. 2006. Global incidence of multidrug-resistant tuberculosis. J Infect Dis 194:479–485. doi: 10.1086/505877. [DOI] [PubMed] [Google Scholar]
- 4.Lee M, Lee J, Carroll MW, Choi H, Min S, Song T, Via LE, Goldfeder LC, Kang E, Jin B, Park H, Kwak H, Kim H, Jeon H, Jeong I, Joh JS, Chen RY, Olivier KN, Shaw PA, Follmann D, Song SD, Lee JK, Lee D, Kim CT, Dartois V, Park SK, Cho SN, Barry CE III. 2012. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med 367:1508–1518. doi: 10.1056/NEJMoa1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Vriese AS, Coster R, Smet J, Seneca S, Lovering A, Van Haute LL, Vanopdenbosch LJ, Martin J-J, Groote C, Vandecasteele S, Boelaert JR. 2006. Linezolid-induced inhibition of mitochondrial protein synthesis. Clin Infect Dis 42:1111–1117. doi: 10.1086/501356. [DOI] [PubMed] [Google Scholar]
- 6.Nagiec EE, Wu L, Swaney SM, Chosay JG, Ross DE, Brieland JK, Leach KL. 2005. Oxazolidinones inhibit cellular proliferation via inhibition of mitochondrial protein synthesis. Antimicrob Agents Chemother 49:3896–3902. doi: 10.1128/AAC.49.9.3896-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drusano GL, Neely M, Van Guilder M, Schumitzky A, Brown D, Fikes S, Peloquin C, Louie A. 2014. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS One 9:e101311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drusano GL, Sgambati N, Eichas A, Brown D, Kulawy R, Louie A. 2011. Effect of administration of moxifloxacin plus rifampin against Mycobacterium tuberculosis for 7 of 7 days versus 5 of 7 days in an in vitro pharmacodynamic system. mBio 2:e00108-11. doi: 10.1371/journal.pone.0101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drusano GL, Sgambati N, Eichas A, Brown DL, Kulawy R, Louie A. 2010. The combination of rifampin plus moxifloxacin is synergistic for suppression of resistance but antagonistic for cell kill of Mycobacterium tuberculosis as determined in a hollow-fiber infection model. mBio 1:e00139-10. doi: 10.1128/mBio.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown AN, Louie A, Adams JR, Jambunathan K, Baluya D, Hafner R, Drusano GL. 2014. Relationship between linezolid (LZD) exposure profiles and toxicity in the hollow fiber infection model (HFIM) system, abstr. A-026b. In Late Breaker Abstr 54th Int Conf Antimicrob Agents Chemother, 5 to 9 September 2014, Washington, DC. [Google Scholar]
- 11.Tam VH, Louie A, Deziel MR, Liu W, Drusano GL. 2007. The relationship between quinolone exposures and resistance amplification is characterized by an inverted U: a new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrob Agents Chemother 51:744–747. doi: 10.1128/AAC.00334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuter DJ, Tillotson GS. 2001. Hematologic effects of antimicrobials: focus on the oxazolidinone linezolid. Pharmacotherapy 21:1010–1013. doi: 10.1592/phco.21.11.1010.34517. [DOI] [PubMed] [Google Scholar]
- 13.Soriano A, Miró O, Mensa J. 2005. Mitochondrial toxicity associated with linezolid. N Engl J Med 353:2305–2306. doi: 10.1056/NEJM200511243532123. [DOI] [PubMed] [Google Scholar]
- 14.Saraste M. 1999. Oxidative phosphorylation at the fin de siècle. Science 283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 15.Pea F, Furlanut M, Cojutti P, Cristini F, Zamparini E, Franceschi L, Viale P. 2010. Therapeutic drug monitoring of linezolid: a retrospective monocentric analysis. Antimicrob Agents Chemother 54:4605–4610. doi: 10.1128/AAC.00177-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pea F, Viale P, Cojutti P, Del Pin B, Zamparini E, Furlanut M. 2012. Therapeutic drug monitoring may improve safety outcomes of long-term treatment with linezolid in adult patients. J Antimicrob Chemother 67:2034–2042. doi: 10.1093/jac/dks153. [DOI] [PubMed] [Google Scholar]
- 17.Wallis R, Jakubiec W, Kumar V, Silvia A, Paige D, Dimitrova D, Li X, Ladutko L, Campbell S, Friedland G, Mitton-Fry M, Miller P. 2010. Pharmacokinetics and whole-blood bactericidal activity against Mycobacterium tuberculosis of single doses of PNU-100480 in healthy volunteers. J Infect Dis 202:745–751. doi: 10.1086/655471. [DOI] [PubMed] [Google Scholar]
- 18.Louie A, Brown D, Files K, Swift M, Fikes S, Drusano GL. 2012. Pharmacodynamics of PNU-10048 (U, sutezolid), a new oxazolidinone, in combination with its active metabolite in the killing of Mycobacterium tuberculosis (Mtb) in an in vitro hollow fiber infection model (HFIM). In Abstr 52nd Int Conf Antimicrob Agents Chemother, 9 to 12 September 2012, San Francisco, CA. [Google Scholar]
- 19.McSharry JJ, Deziel MR, Zager K, Weng Q, Drusano GL. 2009. Pharmacodynamics of cidofovir for vaccinia virus infection in an in vitro hollow-fiber infection model system. Antimicrob Agents Chemother 53:129–135. doi: 10.1128/AAC.00708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McSharry JJ, Weng Q, Brown A, Kulawy R, Drusano GL. 2009. Prediction of the pharmacodynamically linked variable of oseltamivir carboxylate for influenza A virus using an in vitro hollow-fiber infection model system. Antimicrob Agents Chemother 53:2375–2381. doi: 10.1128/AAC.00167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfizer Inc 2000. ZYVOX (linezolid) injection, tablets, and oral suspension: full U.S. prescribing information. Pfizer Inc., New York, NY. [Google Scholar]
- 22.Leary R, Jelliffe R, Schumitzky A, Van Guilder M. 2001. An adaptive grid non-parametric approach to pharmacokinetic and dynamic (PK/PD) models, p 389-394. In Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems. IEEE Computer Society, Bethesda, MD. [Google Scholar]
- 23.Neely MN, van Guilder MG, Yamada WM, Schumitzky A, Jelliffe RW. 2012. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 34:467–476. doi: 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louie A, Heine HS, Kim K, Brown DL, VanScoy B, Liu W, Kinzig-Schippers M, Sorgel F, Drusano GL. 2008. Use of an in vitro pharmacodynamic model to derive a linezolid regimen that optimizes bacterial kill and prevents emergence of resistance in Bacillus anthracis. Antimicrob Agents Chemother 52:2486–2496. doi: 10.1128/AAC.01439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PK profiles achieved in the HFIM system for the toxicity studies. The solid black lines correspond to the “desired” or targeted linezolid concentrations, and the red squares represent the actual linezolid concentrations measured in the hollow-fiber cartridges at the designated time points. Download
PK profiles achieved in the HFIM system for the antibacterial activity studies. The solid black lines correspond to the “desired” or targeted linezolid concentrations, and the red squares represent the actual linezolid concentrations measured in the hollow-fiber cartridges at the designated time points. Download
Linear regression analysis of the predicted-versus-observed plots determined by the mathematical model. The mathematical model described in Materials and Methods was fitted to the data generated from the antibacterial activity studies for linezolid in the HFIM system with the H37Rv strain of M. tuberculosis. The predicted-versus-observed plots for the linezolid PK (A), the total bacterial burden (B), and the bacterial burden of the linezolid-resistant subpopulation (C) are shown. The model was fitted to the data sets resulting from the two independent hollow-fiber experiments. The first data set is represented by the black circles, the second data set corresponds to the blue circles, and the linear regression is designated by the red line. Download