Abstract
Brassinosteroids (BRs) are a class of steroidal plant hormones that play diverse roles in plant growth and developmental processes. Recently, the easy availability of biological resources, and development of new molecular tools and approaches have provided the required impetus for deeper understanding of the processes involved in BRs biosynthesis, transport, signaling and degradation pathways. From recent studies it is also evident that BRs interact with other phytohormones such as auxin, cytokinin, ethylene, gibberellin, jasmonic acid, abscisic acid, salicylic acid and polyamine in regulating wide range of physiological and developmental processes in plants. The inputs from these studies are now being linked to the versatile roles of BRs. The present review highlights the conceptual development with regard to BR homeostasis, signaling and its crosstalk with other phytohormones. This information will assist in developing predictive models to modulate various useful traits in plants and address current challenges in agriculture.
Keywords: plant hormones, brassinosteroids, signaling, crosstalk, development
Introduction
Brassinosteroids (BRs) are plant specific steroidal hormones, characterized by their polyhydroxylated sterol structure and were first isolated from Brassica napus pollen (Grove et al., 1979). BR are regarded as a class of essential plant hormones that plays diverse roles in monitoring broad spectrum of plant growth and developmental processes. They regulate multiple physiological functions including seed germination, cell elongation, cell division, senescence, vascular-differentiation, reproduction, root development, photomorphogenesis, and also respond to various biotic and abiotic stresses (Clouse and Sasse, 1998; Li and Chory, 1999; Sreeramulu et al., 2013; Sharma et al., 2015). Owing to their diverse functions, extensive research has been conducted to promote BR as essential plant growth regulators for modern agriculture (Ikekawa and Zhao, 1991; Divi and Krishna, 2009).
The maintenance and regulation of endogenous level of BR is crucial for various biological functions in plants (Tanaka et al., 2005). BR biosynthesis, transport and degradation are critical components of BR homeostasis and for maintaining the endogenous level of BR in plant. It has been observed that BR-deficient mutants exhibits extreme dwarfism, altered leaf morphology, abnormal vascular development, delayed flowering and senescence, and reduced male fertility (Clouse, 2011). However, excessive application of bioactive BR leads to downregulation of BR-specific biosynthesis genes and an upregulation of BR-inactivation gene, hampering normal development of plants (Bishop and Yokota, 2001; Tanaka et al., 2003, 2005; Zhu et al., 2013a). Moreover, a finely tuned cellular regulation of BR levels is evident from the observation that increase in endogenous BR concentration lead to feedback regulation of the BR metabolic genes, while BR deficient conditions elicit the expression of BR biosynthesis genes to maintain BR homeostasis (Tanaka et al., 2005). Further to understand the BR mediated regulation of several key molecular and physiological functions in plants, extensive research have been conducted over past two decades (Zhu et al., 2013b). BR signaling involves its perception by the cell membrane receptor followed by activation of cascade of phosphorylation events (Figure 1) to relay the signal to the downstream partners resulting in the BR-induced gene expression (Belkhadir and Jaillais, 2015). The use of different biological approaches such as mutant screening, microarray, proteomics, protein–protein interaction studies and bioinformatics played vital role in identification and characterization of various components involved in BR signaling (Divi et al., 2010, 2015). Recent studies demonstrate that BR interacts at various level with the signaling components of other phytohormones and regulate process like plant growth and development and stress responses (Hu and Yu, 2014; Tong et al., 2014; Chaiwanon and Wang, 2015; Divi et al., 2015; Yuan et al., 2015). In the above background, the present review focuses on the recent advances in our understanding of the process of BR biosynthesis, transport, degradation and signaling. This information is useful in getting insights into dynamics of BR homeostasis and its implication in modulating various critical functions in plants. Present update also emphasizes the interaction between the key genes and transcription factors of BR with the signaling components of other phytohormones. This information will facilitate in getting insights into a fairly complex process of BR- mediated plant responses.
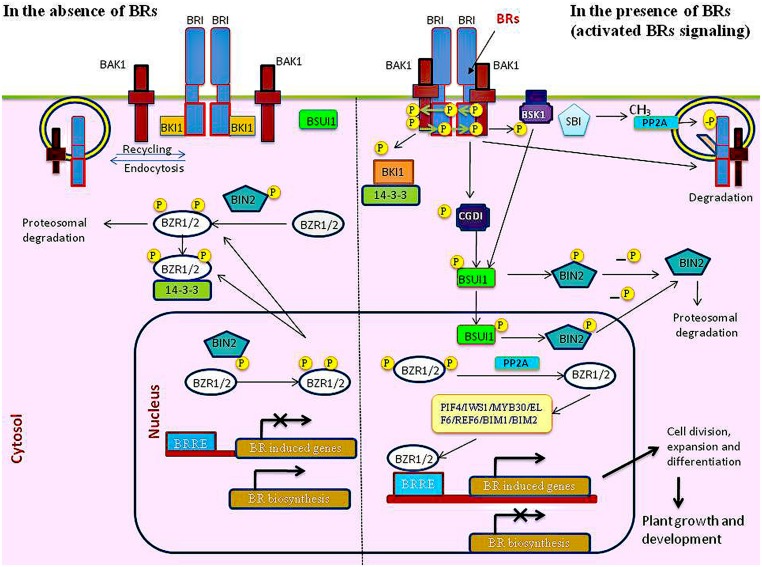
FIGURE 1.
Model for brassinosteroid (BR) signaling pathway in the absence and in the presence of BR.
Brassinosteroid Homeostasis and its Regulation
BR biosynthesis ensues from intricate network pathways and is mostly modulated by transcriptional regulation of BR biosynthetic genes (Chung and Choe, 2013; Vriet et al., 2013). Various genetic and biochemical studies have elucidated BR biosynthetic pathway which commences with campesterol, a precursor for synthesis of the most active form of BR, brassinolide (BL). Firstly campesterol is converted to campestenal which was initially believed to branch into two parallel pathways, namely the early and late C-6 oxidation pathways involving a chain of reductions, hydroxylations, epimerizations and oxidations which eventually converge at castesterone that leads to the formation of BL (Fujioka et al., 1998). Later studies revealed that BR biosynthetic pathway is a triterpenoid pathway (Choe, 2007; Chung and Choe, 2013). Mevalonic acid serves as a precursor of the triterpenoid pathway and is condensed and transformed to 2,3-oxidosqualene which further undergoes modification to form major plant sterols like sitosterols and campesterols. Depending on the availability of substrate and enzymes, campesterol can be modified by two different enzymes: a C-22 hydroxylase dwarf4/CYP90B1 (DWF4) and a C-3 hydrogenase constitutive photomorphogenesis and dwarf/CYP90A1 (CPD). Since DWF4 can act on multiple biosynthetic intermediates including campesterol and campestenol, the pathway branches to a third early C-22 hydroxylation pathway (Choe et al., 2001; Fujioka and Yokota, 2003). LC-MS and various genetic studies have shown that several other branches are formed by CPD, a C-23 hydroxylase which metabolizes campesterol and other intermediates and has recently been found to participate in a C-3 oxidation as well (Ohnishi et al., 2012). The intermediates formed in above mentioned reactions are further modified and later merge into late C-6 oxidation pathway thus, revealing a certain degree of crosstalk between the parallel pathways manifesting complex networking of BR biosynthesis. Lately, several key genes involved in BR biosynthesis such as de-etiolated-2 (DET2), which is a 5α-reductase, CPD, a C-3 oxidase, DWF4, a C-22 hydroxylase, rotundifolia3/CYP90C1 (ROT3) and CYP90D1, C-23 hydroxylases have been characterized to advance the perspective of BR biosynthesis (Wang et al., 2012a; Vriet et al., 2013). Interestingly, monocots like rice and maize, lack the enzyme (CYP85A2) responsible for C-6 oxidation reaction implying that BR synthesis culminates at castesterone in rice (Kim et al., 2008). Recently, it has been shown that in rice, in addition to castesterone, an alternate pathway for biosynthesis of functionally less active C29-BRs exists in order to increase the biological activity of BR in rice. Moreover, C29-BRs even appear to play role in regulation of BR levels by bio-degrading their C-26 demethylated C28-BR analogs to reduce BR activity in planta (Joo et al., 2015).
In contrast to other hormones, BR are not transported to long distances but are rather used in proximity to synthesizing cells. However, they undergo intracellular transport either passively or actively, from their site of synthesis in ER to the plasma membrane where its perception occurs (Symons et al., 2008). Nevertheless, BR are able to exert a long-distance effect by their crosstalk with other hormones like auxins (Symons et al., 2008; Vriet et al., 2013). The short distance transport of BR are suggested to be mediated by some carrier mechanism (BR conjugates formed by binding of BR to fatty acids or glucose) or through specific proteinaceous transporters (Fujioka and Yokota, 2003; Symons et al., 2008). Several proteins belonging to the class of pathogenesis-related (PR) 10 family of proteins, protein family belonging to A or G classes of ABC transporters (ATP binding cassette) and several Sec-14 proteins are potential candidates for mediating BR transport (Markovic-Housley et al., 2003; Deng et al., 2007; Kerr et al., 2011). In the absence of a mode for long distance transport of BR, the spatial and temporal regulation of its homeostasis at the tissue or at the cellular level is extremely crucial for normal growth and development (Symons et al., 2008). BR biosynthesis undergoes two-way regulation mechanism: firstly at the level of endogenous BR by modulating the expression of biosynthetic genes and secondly by inactivating bioactive BR. Most of the BR-specific biosynthesis genes (DET2, DWF4, CPD, BR6ox1, and ROT3) are feedback regulated (Sun et al., 2010; Yu et al., 2011). Moreover, BR signaling mutant brassinosteroid insensitive 1 (bri1) shows considerable accumulation of endogenous BR as their feedback regulation requires intact BR perception and signaling pathway (Sun et al., 2010). Regulation at the level of transcription is mediated by two major BR signaling transcription factors brassinazole-resistant 1 (BZR1) and BRI1-EMS-suppressor1 (BZR2/BES1) as well as by several other novel transcription factors (CESTA, RAVL1, TCP1) that have been lately identified to regulate the expression of key BR biosynthetic genes such as CPD and DWF4 (Je et al., 2010; Sun et al., 2010; Poppenberger et al., 2011; Guo et al., 2013) Some bHLH like transcription factors like CESTA and TCP are involved in positive regulation of BR signaling (Poppenberger et al., 2011; Wang et al., 2012a; Guo et al., 2013). In rice, another set of transcription factors, ABA insensitive 3/vivaparous1 (ABI3/VP1) and related to ABI3/VP1 (RAV1), not only helps in BR homeostasis by positively regulating biosynthetic genes (D2, D11 and BRD1) but also helps in enhancing the expression of OsBRI1 (Je et al., 2010).
Brassinosteroid catabolism/metabolism involving various process like acylation, sulphonation, glycosylation etc. play a crucial role in maintaining the optimum levels of bioactive BR in the cell. Novel genes belonging to the family of BAHD acyltransferases, brassinosteroid inactivator1 (BIA1) and abnormal shoot-1 (abs-1) (Roh et al., 2012; Wang et al., 2012a) have been identified to be involved in BR acylation to inactivate BR. Another BAHD acyltransferases pizza (PIZ), have a redundant role with BIA1 in BR inactivation (Schneider et al., 2012). Similarly, bri1-5 enhanced 1 (BEN1) and Brassica napus sulfotransferase 3 (BNST3) possessing differential specificities to castesterone and BL inactivate active BR by various mechanism involving reduction and sulfonation (Marsolais et al., 2007; Yuan et al., 2007). A set of glycotransferases enzymes UGT73C6 and its close homolog UGT73C5, catalyze the 23 O-glycosylation of CS and BL as part of the inactivation process (Poppenberger et al., 2005). Evidence reveal that these conjugations may serve as temporary storage forms of pool of inactive BR and believed to serve additional functions such as irreversible inactivation, transport, compartmentalization, and protection against cellular removal (Bajguz, 2007; Husar et al., 2011; Piotrowska and Bajguz, 2011). Recent studies have also shown that BR biosynthesis can be regulated by external stimuli like salt and temperature stress (Maharjan and Choe, 2011; Sharma et al., 2013).
Brassinosteroids Signaling
In the recent past, with the use of various biochemical, genetic and proteomic approaches, a great advancement has been made in our understanding of the BR signaling pathway (Zhu et al., 2013b; Fàbregas and Caño-Delgado, 2014; Belkhadir and Jaillais, 2015). At the plasma membrane (Figure 1), BR are perceived by the extracellular domain of the plasma membrane localized leucine rich repeat receptor like kinase (LRR-RLK) BRI1 and its two closely related homologs, BRI1-LIKE 1 (BRL1) and BRI1 LIKE 3 (BRL3), in the nanomolar range (Cano-Delgado et al., 2004). With the recent elucidation of atomic structures of the BRI1 and BRL1 in complex with BL has facilitated in structural understanding of BR perception at the cell surface (Hothorn et al., 2011; She et al., 2011, 2013). BRI1 and BRL1 have similar basic skeleton with 25 and 24 units of LRRs, respectively with a great degree of similarity in overall shape and curvature of the horseshoe-like structure of extracellular domain which is the seat for binding of BR. However, minor structural differences in the BR binding pocket of BRL1 and BRI1 reveals that BR have less affinity to BRI1 as compared to BRL1 as BR binds more strongly to BRL1 as compared to BRI1 (She et al., 2011, 2013). BRL2, which is considered as another BRL1 homolog and responsible for vascular development has also been studied for its ability to bind to BR (Ceserani et al., 2009). It is interesting to note that despite having high sequence identity of BRL2 with BRL1 and possessing conserved Arg588 and Gly690 residues responsible for higher affinity of BRL1 for BL, BRL2 lacks the ability to bind to BR. This could be due to the presence of heavily negatively charged residues located at the inner side of the binding pocket in BRL2 that result in the change in its hydrophobicity to BRs (She et al., 2013).
In the absence of BR (Figure 1), BRI1 homodimer remains in an inactive form by the interaction with inhibitory protein BRI1 kinase inhibitor 1 (BKI1) as well as by autoinhibitory function of its cytoplasmic kinase domain (Wang et al., 2005a; Wang and Chory, 2006; Jaillais et al., 2011). Concurrently, active BR insensitive 2 (BIN2), a GSK3/Shaggy-like kinase, phosphorylates BES1/BZR2 family transcription factors and inhibits their activity by protein degradation, reduced DNA binding, and/or cytoplasmic retention by 14–3–3 proteins (Ye et al., 2011; Hao et al., 2013). However, for fully activation of BR signaling, BRI1 heteroligomerise with its co-receptor BRI1-associated receptor kinase1 (BAK1), also known as somatic embryogenesis receptor-like kinase 3 (SERK3) (Li et al., 2002; Nam and Li, 2002). Binding of BR to the 70-amino acids island domain of BRI1 triggers a change in the receptor either in the form of conformational change in the preformed homodimer or receptor dimerisation (Wang et al., 2005a). It results in the autophosphorylation of BRI1 kinase domain activation loop which induces a partial kinase activity of BRI1 leading to the transphosphorylation at Tyr211 residue of BKI1 and resulting in its dissociation from the membrane and thus allowing BRI1 to interact with BAK1 (Wang et al., 2005b; Wang and Chory, 2006). Thus, BR induces the conformational changes in its receptor that are necessary for BRI1-BAK1 interaction (Santiago et al., 2013; Sun et al., 2013). BKI1 also promotes BRI1 signaling by binding to 14–3–3 protein and repressing their negative functions in BR signaling (Wang et al., 2011). BRI1 then interacts with BAK1 to transphosphorylate each other at multiple residues (Wang et al., 2008). Forster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) approaches have shown that a significant amount of BRI1-BAK1 hetero-oligomers are present at the cell surface even in the absence of BR (Bucherl et al., 2013). It is hypothesized that preassembled BRI1-BAK1 after conjugation with ligand might undergo discrete rearrangements of their respective intracellular kinase domains. This hypothesis derives its significance as similar signaling paradigms is observed in receptor tyrosine kinases in animals (RTKs) which undergo ligand-independent receptor dimerization, followed by dimer reorganization upon ligand binding (Lemmon and Schlessinger, 2010; Bucherl et al., 2013). Activated BRI1 initiates a cascade of phosphorylation events of its downstream plasma membrane bound receptor-like cytoplasmic kinases (RLCKs), BR signaling kinases (BSKs) and constitutive differential growth 1 (CDG1) to transmit the signal from membrane receptors to cytoplasmic regulators of BR signaling (Tang et al., 2008; Kim et al., 2011; Sreeramulu et al., 2013). Mass-spectrometric analysis has shown that BRI1 phosphorylates CGD1 and BSK1 at Ser-234 and Ser-230, respectively and subsequently phosphorylate and activate BRI1-suppressor 1 (BSU1). It has been found that BSU1 can be activated by BRI1 either through BSK1 or CDG1. Furthermore, BSK1 induced activation requires BRI1 kinase activity, whereas CDG1 induced activation does not require BRI1 but is enhanced by BRI1. Thus, either of one set of interacting partners, BSU1-CDG1 or BSUI-BSK1 is the minimum set of components required for transducing the signal from the receptor kinase BRI1 to the GSK3-like kinase BIN2 (Kim et al., 2011). BSU1 then dephosphorylates GSK3 like kinase BR insensitive 2 (BIN2) (Kim and Wang, 2010) to inhibit its function and relieves the inhibitory effect on two master transcription factors of BR signaling, BZR1 and BZR2 also known as BRI1-EMS suppressor1 (BES1) (Wang et al., 2002; Yin et al., 2002, 2005; He et al., 2005). BZR1 and BES1 are then dephosphorylated by protein phosphatase 2A (PP2A) and released from 14–3–3 proteins (Tang et al., 2011), resulting in their nuclear localization to bind to the promoter of their target genes to regulate their gene expression (Sun et al., 2010; Yu et al., 2011; Wang et al., 2012b). Other BRI1 substrates like Arabidopsis TGF-β receptor-interacting protein-1 (TRIP-1) and transthyretin-like protein (TTL) have also been identified with TTL being putatively linked to inhibition of BRI1 signaling as it binds with higher affinity to kinase-active BRI1 while TRIP-1 being an essential subunit of eIF3 protein translation initiation complex, its phosphorylation by BRI1 is thought to modulate its activity and influence protein translation (Ehsan et al., 2005). Though both BRI1 and BAK1 are classified as serine/threonine kinases, the studies done in past decade has shown that BRI1 has structural features reminiscent of both serine/threonine and tyrosine kinases like insulin receptor, thus providing insights into the evolution of dual-specificity kinases in plants (Wang et al., 2005b; Oh et al., 2009; Macho et al., 2015). Infact, BRI1 possess significant tyrosine kinase activity and it can undergo autophosphorylation on tyrosine residues within the kinase and juxtamembrane domains and can lead to transphosphorylation of Tyr211 in BKI1 as well as tyrosines in other proteins (Oh et al., 2009; Jaillais et al., 2011; Wu et al., 2012). Tyr-831 and Tyr-956 are identified as autophosphorylation sites in vitro and in vivo with Tyr-956 in kinase subdomain V being essential for activity, while Tyr-831 in the juxtamembrane domain is not essential for kinase activity but plays an important role in BR signaling in vivo (Wang et al., 2005b; Oh et al., 2009; Macho et al., 2015).
Brassinosteroid Regulated Transcriptional Networks
Brassinazole-resistant 1 and BZR2/BES1 are the two major transcription factors of the BR signaling pathway that mediate BR function by regulating the expression of several thousands of genes amounting to about 20% of the genome in Arabidopsis (Guo et al., 2013). They share a significant 88% sequence identity at the protein level and a 97% identity in their DNA-binding domain (Wang et al., 2012b). Their structure basically comprises of a bHLH DNA binding domain (DBD), a BIN2 phosphorylation domain containing 22 putative BIN2 phosphorylation sites, a PEST motif (Pro-, Glu-, Ser-, and Thr-rich) involved in protein degradation, and a 14–3–3 binding motif interacting with 14–3–3 upon phosphorylation of BES1 and BZR1 (Tang et al., 2011; Hao et al., 2013). The highly conserved C-terminal domain between two proteins is implicated in the interaction of BZR1 and BES1 with BIN2 (Wang et al., 2002). Though BZR1 and BZR2/BES1 render various similar biochemical and genetically redundant functions, yet the studies conducted on the light grown mutants (bzr1-1D and bes1-D) show distinctive phenotypes revealing a variation in the level of two proteins either at the level of expression pattern or interaction with other protein partners (He et al., 2005; Sun et al., 2010). With the help of chromatin immunoprecipitation (ChIP) coupled with Arabidopsis tiling arrays and promoter element analysis, it is evident that both BZR1 and BES1 have similar DNA-binding specificities. It has been found recently that both BES1 and BZR1 can bind to the BRRE and E-boxes with BRRE primarily enriched in BR-repressed genes and E-boxes are mostly enriched in BR-induced genes (Sun et al., 2010; Yu et al., 2011; Zhu et al., 2013b). Genome-wide protein-DNA interaction analyses as well as the expression profiling have identified about 953 genes targeted by BZR1 downstream from the BRI1-mediated signaling pathway, while BES1 controls 250 genes out of which a small set of 120 genes show an overlap with the BZR1 target genes and thus regulate various genes depending on specific target gene promoter and dimerization partner (Sun et al., 2010; Gudesblat and Russinova, 2011; Yu et al., 2011). Upon activation of BR signaling (Figure 1), BES1 and BZR1 bind to their own promoter sequence to induce their expression through a positive feedback loop (Yu et al., 2011). BES1 and BZR1 can regulate or interact with additional transcription factors like AtIWS, BIM and GATA-binding TFs to regulate secondary BR-responsive genes (Sun et al., 2010). BES1 interacts with a bHLH transcription factor BIM1 to synergistically bind to E box sequences present in many BR-induced gene promoters (Yin et al., 2005; Guo et al., 2013). BES1 can also physically interact with interacting-with-spt6 1 (IWS1), which is involved in RNA polymerase II (RNAPII) post-recruitment, transcriptional elongation, RNA export and histone modifications. IWS1 is recruited to target genes by BES1 to enhance gene expression during transcription elongation (Li et al., 2010). BR mediated regulation of gene expression involves epigenetic mechanism also. BES1 directly interacts with a pair of histone demethylases, relative of early flowering6 (REF6) and its homolog ELF6 (early flowering 6) belonging to the class of jumonji domain proteins, to regulate various physiological processes such as flowering time (Noh et al., 2004; Yu et al., 2008). BES1 can also recruit, a histone lysine methyltransferase called set domain group8 (SDG8), which is implicated in histone H3 Lys-36 di- and trimethylation which suggests that epigenetic mechanisms are also involved in regulating the expression of a subset of BR target genes. (Xu et al., 2008; Wang et al., 2014). Topless (TPL), TPL-related proteins (TPRs), and histone deacetylase 19 (HDAC19) were recently found to assemble into a transcriptional repressor module with BES1 (Ryu et al., 2014). Recently, it has been found that BES1 physically interacts to repress transcription factor, brassinosteroids at vascular and organizing center (BRAVO) which acts as a cell-specific repressor of quiescent center divisions in the primary root of Arabidopsis. Thus, BES1 modulates quiescent center divisions at the root stem cell niche, by utilizing BRAVO as a regulatory node to control BR-mediated regulation of stem cell quiescence in plants (Vilarrasa-Blasi et al., 2014). Another transcription repressor, myeloblastosis family transcription factor-like 2 (MYBL2) is known to interact with BES1 to down-regulate the expression of BR-repressed genes which is required for optimal BR response (Ye et al., 2012). BR-activated BZR1 have been found to interact with dark and heat-activated transcription factor phytochrome-interacting factor4 (PIF4). They bind to nearly two thousand common target genes, and synergistically regulate these genes, many of which encode transcription factors and proteins that function in the cell wall and chloroplast (Oh et al., 2012). Promotion of cell elongation by BZR1 and PIFs requires activation of the group of a typical bHLH transcription factors, comprising six proteins paclobutrazol resistant (PREs) (Bai et al., 2012; Oh et al., 2012). BZR1 binds to the promoter of the PRE1 gene to enhance its expression while PREs further interact with and inhibit several other HLH/bHLH factors, including ILI1 binding bHLH Protein 1 (IBH1), a negative regulator BR-dependent gene expression (Zhang et al., 2009b). Another representative of the PRE family PRE3, also named activation-tagged BRI1-suppressor1 (ATBS1), is a 93-amino acids atypical bHLH protein, which acts as a positive regulator of BR-dependent gene expression. Since ATBS1 lacks a DNA binding domain, its positive effect on BR signaling results from heterodimerizing and sequestering other bHLH proteins that were negative regulators of BR signaling. It functions by blocking the DNA-binding activity of bHLH transcription factor ATBS1-interacting factor/ILI1–binding bHLH (AIFs/IBH1) that functions as a negative regulator of the BR pathway (Wang et al., 2009). Phytochrome rapidly regulated1 (PAR1) is another atypical HLH proteins lacking proper DNA binding domain. It functions by forming a heterodimer complex with BR regulated PIF4 (PAR-PIF4) and PRE1 (PAR-PRE1) regulating cell elongation and plant development (Hao et al., 2012).
BR regulate various aspects of photomorphogenesis by interacting with other transcription factors like HY5 and GATA. HY5 is a basic leucine zipper transcription factor that functions as a positive regulator of photomorphogenesis and binds to the promoter of over a thousand genes that are also the direct targets of BZR1. Both BZR1 and HY5 regulate the transcription of these genes in opposite ways thereby supporting an antagonistic interaction between BR and light signals (Sun et al., 2010). A novel B-box zinc finger transcritpion factor, BZR1 suppressor1 (BZS1), which functions downstream of both BR and light signaling pathways has been identified to be repressed by BZR1. BZS1 may also interact with Hy5 transcription factor to control expression of a subset of light-responsive genes. Thus, BZS1 is oppositely regulated by light and BR signals to regulate photomorhogenesis responses (Fan et al., 2012). BZR1 represses expression of another positive regulator of photomorphogenesis, GATA2 that controls a subset of genes co-regulated by light and BR. BZR1 directly binds to the promoter of GATA2 to repress its function, while it is post-transcriptionally activated by light signals (Luo et al., 2010). Another connection between light and BR pathways in regulating morphogenesis is provided by two related transcription factors, golden2-like 1 (GLK1) and GLK2. BES1 directly represses the expression of two related transcription factors, (GLK1) and GLK2, that promote the expression of large number of photosynthetic genes and are required for chloroplast development (Waters et al., 2009; Yu et al., 2011) thus, correlating to BR-mediated inhibition of chloroplast development in the dark.
Interplay of Brassinosteroids with other Phytohormones
Brassinosteroids perform diverse functions due to its interplay with other phytohormones. In response to environmental cues BR interact with different phytohormones such as abscisic acid (ABA), auxin, cytokinin (CK), ethylene, gibberellins (GA), jasmonic acid (JA), polyamines (PA) and salicylic acid (SA), and to regulate myriad aspects of plant growth and developmental processes in plants (Choudhary et al., 2012b; Gruszka, 2013). Recent studies clearly indicate that in response to various intrinsic and extrinsic factors, the signaling components of BR crosstalks with the key genes and transcription factors of other phytohormones and thereby regulates multiple functions in plants. The unraveling of these complicated mechanisms of BR signaling and its collaboration with other molecular networks will be of great importance in improving modern agriculture.
Brassinosteroid-Abscisic Acid Crosstalk
It is well documented that ABA is required to inhibit seed germination and is also mandatory to establish seed dormancy during embryo maturation. On contrary, BR promotes seed germination indicating the antagonistic interaction between both these hormones (Steber and McCourt, 2001; Finkelstein et al., 2008). Genetic, physiological and biochemical studies have revealed that BR and ABA can co-regulate the expression of 100s of genes (Nemhauser et al., 2006; Zhang et al., 2009a). However, the underlying molecular mechanism and the signaling components involved in this crosstalk are largely unknown. BR and ABA signaling mutants have been analyzed to investigate how ABA inhibits BR signaling (Zhang et al., 2009a). It has been observed that in BR biosynthetic and signaling mutants such as det2-1 and bri1, respectively, the effect of ABA on BR signaling does not rely upon BR perception, but depends on BIN2 (Figure 2A), a negative regulator of BR signaling (Zhang et al., 2009a). However, on analyzing ABA signaling mutants, it has been demonstrated that regulatory effect of ABA on BR signaling largely depends upon ABI2 and slightly on ABI1, a PP2C family serine/threonine phosphatases (Zhang et al., 2009a). This study indicates that ABA and BR crosstalks through BR signaling components such as BIN2 and, ABA signaling components such as ABI1 and ABI2. In addition, ABA signaling components act downstream of BRI1 receptor but upstream of BIN2, thus activating BIN2 to negatively regulate BR-mediated responses (Zhang et al., 2009a). Furthermore, BR and ABA have been suggested to play antagonistic roles in regulating seed germination and post-germinative growth processes (Hu and Yu, 2014). ABA inhibits while BR-enhances seed germination and post-germinative growth processes. It has been observed that BIN2, positively regulates ABA responses by physically interacting with ABI5. However, mutations of BIN2 phosphorylation sites on ABI5 made the mutant protein respond to ABA improperly. Hence, the study confirms that BIN2 stabilizes ABI5, by phosphorylating it, thus mediating ABA responses during seed germination. However, BR application inhibits the regulation of ABI5 by BIN2 to antagonize ABA mediated inhibition (Hu and Yu, 2014). Recently, a mutant studies indicate a synergistic correlation between BR and ABA in inducing responses such as, H2O2 production, respiratory burst oxidase homolog1 (RBOH1) gene expression, NADPH oxidase activity and in mediating heat and oxidative stress tolerance (Zhou et al., 2014). In, ABA biosynthetic mutant, not, BR induces transient increase in these responses, however, in BR biosynthetic mutant dˆim, ABA induced strong and prolonged increase in these responses. These results indicate that ABA biosynthesis plays a key role in sustaining stress tolerance in BR-induced pathways in plants (Zhou et al., 2014).
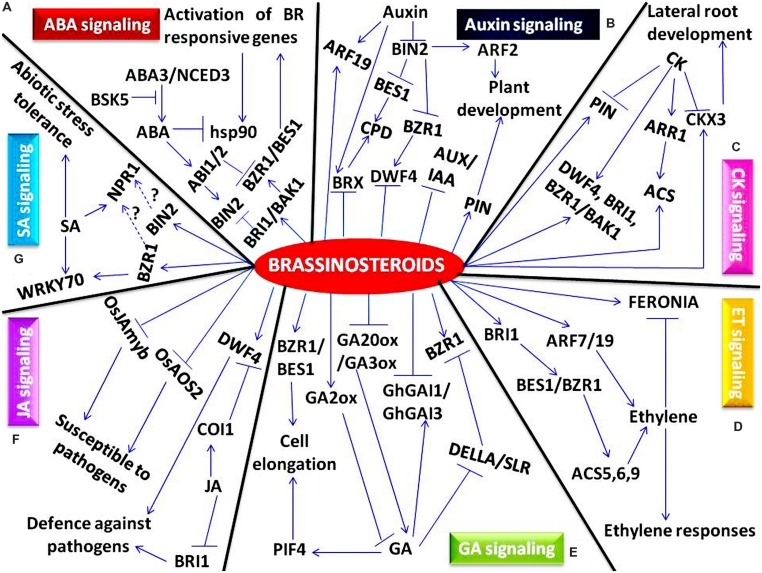
FIGURE 2.
A schematic model showing crosstalk of various pyhtohormones with brassinosteroids. (A) Brassinosteroid-abscisic acid (ABA) crosstalk, (B) Brassinosteroid-auxin crosstalk, (C) Brassinosteroid-cytokinin (CK) crosstalk, (D) Brassinosteroid-ethylene (ET) crosstalk, (E) Brassinosteroid-gibberellin (GA) crosstalk, (F) Brassinosteroid-jasmonic acid (JA) crosstalk, (G) Brassinosteroid-salicylic acid (SA) crosstalk. Involvement of different genes and transcription factors as a key component in transcriptional regulation of plant development has been represented. Positive effects are indicated by arrows, bars indicate repression and unknown interactions are represented by dashed arrows.
It has been demonstrated that high endogenous level of ABA suppresses BR- mediated responses in plants (Divi et al., 2010). In ABA deficient mutant, aba1-1, pronounced effects of BR application were observed under heat stress conditions with respect to survival rate due to higher accumulation of heat shock protein90 (HSP90), it indicates that ABA conceals the effect of BR in plant stress responses (Divi et al., 2010). The antagonistic interaction between BR and ABA, has also been established by studying BSK5 protein (Li et al., 2012b). It has been observed that under abiotic stress (salt and drought stress) and upon exogenous phytohormone application (BR and ABA), the transcript levels of BSK5 gene were enhanced. Moreover, in loss-of-function mutant, bsk5, the transcript levels of ABA biosynthesis genes, ABA3 and 9-cis-epoxycarotenoid dioxygenase (NCED3), were up-regulated leading to higher ABA levels in bsk5 plants (Figure 2A). It indicates that BSK5 mutation alters ABA and salt sensitivity and also modifies ABA biosynthetic pathway in plants (Li et al., 2012b).
Brassinosteroid-Auxin Crosstalk
Crosstalk between BR and auxin regulates myriad aspects of plant growth and developmental processes, (Hao et al., 2013; Saini et al., 2013; Liu et al., 2014; Chaiwanon and Wang, 2015) however, the role of auxin and BR interaction in regulating stress responses has remained elusive (Kissoudis et al., 2014). In Arabidopsis, the maintenance of threshold BR level to permit optimum auxin action required for root growth is mediated by BRAVIS RADIX (BRX). It has been demonstrated that the expression of BRX gene is strongly induced by auxin while repressed mildly by BR (Figure 2B), indicating that BRX acts at the nexus of a feedback loop (Mouchel et al., 2006). Interestingly, BR biosynthesis genes CPD and DWF4 are also positively regulated by BRX (Tanaka et al., 2005) suggesting a connection between BR biosynthesis and auxin signaling (Mouchel et al., 2006). Furthermore, auxin regulates the expression of BR biosynthesis genes, and hence, are linked directly with BR biosynthesis (Chung et al., 2011). In Arabidopsis, the exogenous treatment of auxin dramatically increased the transcript levels of DWF4 gene leading to enhancement in BR biosynthesis probably through induction of BRX protein (Chung et al., 2011). However, when optimum amount of BR is synthesized, there is a feedback inhibition of DWF4 gene by BR (Figure 2B). The study confirms the antagonistic role of BR and auxin in regulating BR biosynthetic gene, DWF4 (Maharjan and Choe, 2011; Maharjan et al., 2011). Further, it has also been observed that the transcription of BR biosynthetic genes such as DWF4 and CPD, which catalyzes the rate-determining step and also acts downstream of auxin signaling pathway, are regulated by BZR1 and BES1 (Figure 2B), a plant-specific family of transcription factors, respectively (Maharjan et al., 2011). Furthermore, a close relationship between BR and auxin in plant growth and development has been established through interaction between BIN2 and auxin response factors (ARF2). BIN2 mediated phosphorylation of ARF2 results in loss of DNA-binding repression activities of ARF2 (Vert et al., 2008). It leads to the up-regulation of BR regulated genes and also promotes the activity of ARF promoters, enhancing auxin signaling, indicating BR-auxin synergistic interaction. Furthermore, auxin/indole-3-acetic acid (AUX/IAA) proteins which are key players in auxin signaling pathway are also involved in BR responses (Figure 2B). Since, AUX/IAA mutants, iaa7/axr2-1 and iaa17/axr3-3, showed aberrant BR sensitivity and aberrant BR induced gene expression due to stabilization of mutants protein caused by failure to interact with SCFTIR1 complex, S-phase kinase-associated protein-1 (skp1), such as Arabidopsis Skp1-like 1, (ASK1), in plants-the ring box protein 1 (rbx1)-cullin (CUL1)-F box protein, transport inhibitor response1 (TIR1). It indicates that BR and auxin interaction is mediated by SCFTIR1 complex and AUX/IAA proteins (Nakamura et al., 2006). Further studies indicate that in rice, auxin treatment enhances the transcript levels of BR receptor gene OsBRI1 suggesting that auxin regulates the level of BR receptors thus enhancing BR perception. Moreover, the promoter studies of OsBRI1 gene also indicates that this gene posseses an upstream auxin-response element (AuxRE) motif which is targeted by ARF transcription factors. Further, mutant studies indicate that upon mutation of AuxRE, the induction of OsBRI1 expression of auxin is abolished and in arf mutant the expression of OsBRI1 gene is down-regulated (Sakamoto et al., 2013). Recently, ChIP and yeast one-hybrid assay demonstrates a link between BR and auxin in controlling lamina inclination which is implicated in plant architecture development and grain yield (Zhang et al., 2014). It has been observed that in OsGH3.5 and OsARF19 overexpressing plants, there is reduced content of free IAA at lamina joint resulting in alteration of lamina inclination. Moreover, OsARF19 also binds to the promoter of OsBRI1 and positively regulates its expression. In turn, OsBRI1 activates the expression of OsBZR1 and its downstream signaling genes to affect lamina inclination (Zhang et al., 2014). The study clearly indicates that OsARF19 links auxin and BR signaling during regulation of lamina inclination in rice (Figure 2B). Furthermore, an antagonistic interaction between BR and auxin has been linked through the transcription factor BZR1 in roots of Arabidopsis, to control the spatiotemporal balance of stem cell dynamics required for optimum root growth (Chaiwanon and Wang, 2015). It has been observed that the optimum amount of BZR1 expression patterns required for root growth is established through local BR catabolism, auxin biosynthesis and BR signaling. BZR1 activates the target genes expressed in the transition-elongation zone, but represses genes in the quiescent center and surrounding stem cells, however, on contrary, auxin has an opposite effect to BR on the spatiotemporal gene expression (Chaiwanon and Wang, 2015). BR are also known to play a key role in regulating auxin-responsive genes involved in polar auxin transport such as pinformed3 (PIN3), PIN4 and influx carriers, auxin-resistant1/like aux1 (AUX1/LAXs) (Figure 2B) (Hacham et al., 2011, 2012) by affecting their cellular localization. Recently, the actin cytoskeleton was reported to play an essential role in integrating BR and auxin responses. Since, it has been demonstrated that BR alter cytoskeletal configuration in a manner similar to that of auxin. Moreover, BR-mediated reconfiguration of actin cytoskeleton causes delocalization of the PIN2 transporters, thus promoting auxin responses (Lanza et al., 2012). Recent study indicates that in rice, out of 12 OsPIN genes, two OsPINs (OsPIN2 and OsPIN5b) were induced by drought, heat and cold stresses, however, the other members in rice were suppressed significantly under abiotic stress (Du et al., 2013). Further, it has also been examined that cold stress inhibits the intracellular-cycling of PIN2 and PIN3 resulting in diminished shootward transport of auxin and reduced root’s capability to form auxin gradient required for root growth and patterning (Friml and Palme, 2002; Harrison and Masson, 2008; Shibasaki et al., 2009; Sukumar et al., 2009). Moreover, the effect of BR on auxin transporters has also been studied and it has been observed that BR also represses the expression of several auxin transport related genes, including PIN3, PIN4, PIN7 and LAX gene (Nemhauser et al., 2004). It indicates toward the possible role of BR and auxin crosstalk in abiotic stress tolerance through auxin transporter genes. Furthermore, rice genome acquires seven YUCCA genes which encode the rate limiting enzymes for auxin biosynthesis (Yamamoto et al., 2007). It has been observed that all six OsYUCCA genes except for OsYUCCA4 were down-regulated under drought stress. However, under cold stress the transcript levels of OsYUCCA2, OsYUCCA3, OsYUCCA6, and OsYUCCA7 were strongly upregulated, whereas, heat stress leads to upto five-fold increase in the transcript levels of OsYUCCA3, OsYUCCA6, and OsYUCCA7 genes (Du et al., 2013). Further, alteration in the transcript levels of the 40% of BR-upregulated genes has been observed in yucca mutants (Nemhauser et al., 2004) indicating BR and auxin crosstalk point. Although the relationship of BR and auxin has been well documented primarily in plant growth and developmental processes, however, further investigations are prerequisite to understand the mechanism of auxin and BR crosstalk involved in abiotic stress tolerance.
Brassinosteroid-Cytokinin Crosstalk
Cytokinin-brassinosteroid indirectly crosstalks through modulation of auxin transport at the molecular level in regulating lateral root development. BR enhances the expression of auxin efflux carriers such as PIN genes (Figure 2C) which probably aids to maintain local auxin maxima required for root primordium development (Bao et al., 2004). On contrary, CK inhibits the establishment of lateral root primordia and disturbs auxin accumulation by down-regulating the expression of PIN genes, indicating an indirect interaction between BR and CK, probably through an unknown mechanism (Benjamins and Scheres, 2008). Furthermore, enhanced root growth due to reduction of CK content was observed in Arabidopsis roots upon overexpression of the cytokinin oxidase/dehydrogenase3 (CKX3) gene driven by root-specific promoter PYK10 (Werner et al., 2010). Exogenous application of BR enhances lateral root and leaf length in P10-CKX3 plants under simultaneous effect of CKX3 overexpression and BRI1 ectopic expression (Figure 2C). These findings suggest that crosstalk between BR and CKs is involved in the regulation of plant growth and development (Vercruyssen et al., 2011). An evidence for the involvement of BR in regulation of CK level in wheat seedlings has also been provided recently (Yuldashev et al., 2012). CK accumulation, particularly, zeatin derivatives were detected in roots and shoots of 4-day-old wheat seedlings on treatment with BR. However, upon hormone removal, revival of CK concentration to the control level in the roots and shoots was observed, which is suggested to be due to inhibition of the gene encoding CKX protein (Yuldashev et al., 2012). In addition, rice plants overexpressing isopentyl transferase, IPT gene, resulted in enhanced CK level just before the onset of senescence leading to increase in tolerance to drought stress. The transgenic plants had shown delayed response to stress and exhibited significantly higher grain yield. The observed increase of CKs corresponds with the upregulation of various BR-related biosynthesis genes (DWF4, DWF5, HYD1) and BR signaling genes (BRI1, BZR1, BAK1, SERK1, BRH1) (Figure 2C). Hence, the crosstalk between BR and CK contribute toward significantly higher grain yield through modification of source–sink relations, thus enhancing drought tolerance (Peleg et al., 2011). Further it has been demonstrated that the CK-mediated inhibition of lateral root initiation does not depend directly on BR level. Since, a report has shown that the brx-2 mutant, in which BR homoeostasis is hindered, is insensitive to CK-mediated inhibition of lateral root development while, the exogenous application of BR could restore this defect. However, in the presence of CK, the post-embryonic BR treatment could not rescue brx-2 mutant phenotype (Li et al., 2009) indicating BR-CK independence in the regulation of lateral root development.
Recent studies indicate that in Chlorella vulgaris, CK stimulates the accumulation of endogenous BR suggesting the synergistic interaction between BR and CK (Bajguz and Piotrowska-Niczyporuk, 2014). Upon exogenous treatment of 10 nM trans-zeatin (tZ) to the C. vulgaris culture, there was considerable increase in the level of all endogenous BR by 27–46%. Moreover, the co-application of both BL and trans-Zeatin (tZ) lead to highest stimulation in the number of C. vulgaris cells and endogenous accumulation of proteins, chlorophylls and monosaccharides, whereas, the lowest was observed upon treatments with 28-homocastasterone (28-homoCS) and 1,3-diphenylurea (DPU) (Bajguz and Piotrowska-Niczyporuk, 2014) indicating BR-CK crosstalk point. Furthermore, BR and CK acts post-transcriptionally to continuously adjust ethylene biosynthesis in response to various environmental factors (Hansen et al., 2009). Crosstalk between CK and BR enhances stability and transcript levels of ethylene biosynthesis gene, ACS. In Arabidopsis, CK is perceived by a family of Arabidopsis histidine kinase receptors (AHK), which upon autophosphorylation transmit phosphoryl groups to Arabidopsis histidine phosphotransfer proteins (AHP). The AHP further activates typeB Arabidopsis response regulator (ARR), ARR1, which prevents ACS degradation and activates ethylene biosynthesis (Hansen et al., 2009). Since, it has been observed that ARR single mutants, arr1, arr2, arr10, arr12, and quadruple mutant arr 1,2,10,12 showed marked decrease in elevation of ethylene biosynthesis. Hence, the study clearly indicates that CK induction of ethylene biosynthesis is mediated via CK signaling component ARR1. Moreover, BR and CK acts through distinct independent targets but their effect is additive in stabilization of ACS proteins, suggesting BR-CK crosstalk (Hansen et al., 2009). A recent study has ellucidated that BR enhances CK-induced anthocyanin biosynthesis in Arabidopsis seedlings (Yuan et al., 2015). Since, it has been observed that mutants defective in BR biosynthesis, dwf4 and those defective in BR signaling brassinosteroid insensitive 1-4 (bri1-4) exhibited delayed expression of the anthocyanin biosynthesis genes and reduced anthocyanin accumulation, as compared to the wild type treated with BR, indicating a positive interaction between BR and CK (Yuan et al., 2015).
Brassinosteroid–Ethylene Crosstalk
Brassinosteroid and ethylene crosstalk regulate different aspects of plant growth and developmental processes. BR have been identified as a negative regulator of shoot gravitropism, whereas ethylene has been shown to promote gravitropic reorientation in light-grown seedlings (Vandenbussche et al., 2013). It has been suggested that BR and ethylene interact indirectly in regulating shoot gravitropic responses through involving auxin signaling genes (Guo et al., 2008). BR activates AUX/IAA (negative regulator of auxin signaling) and inactivates ARF7 and ARF19 (positive regulator of auxin signaling), thus inhibiting shoot gravitropic responses. On contrary, ethylene downregulate AUX/IAA and enhances ARF7 and ARF19 genes to positively regulate shoot gravitropic responses (Vandenbussche et al., 2013). Therefore, ethylene and BR have been found to have an opposite effects on the upward growth of etiolated shoots. Furthermore, ethylene-BR antagonism has also been observed in the case of roots. Ethylene reduces root gravitropic responses (Buer et al., 2006), while BR enhances root gravitropic bending probably by modulating auxin transport (Kim et al., 2007; Vandenbussche et al., 2013). In BR-insensitive mutants, bri1-301 and bak1, delayed root growth and reduced response to the gravitropic stimulus was revealed (Kim et al., 2007). However, in ethylene insensitive mutants, ein2-5 and etr1-3 reduced inhibition toward root gravitropic responses was observed (Buer et al., 2006), indicating antagonistic interaction between BR and ethylene in regulating gravitropic responses in plants.
The interaction between BR and ethylene in regulating cell expansion has been consolidated by studying FERONIA, which encodes a receptor-like kinase and has a key role in pollen tube reception (Huck et al., 2003; Escobar-Restrepo et al., 2007). Although, FERONIA is clearly required for proper fertilization, the knowledge of its role in plant growth and development is scarce. Therefore, mutant studies have been conducted to know the role of FERONIA in vegetative plant growth. Further, in FERONIA knockdown mutants with reduced FERONIA expression, a limited capacity for cell expansion has been observed (Guo et al., 2009). In case of etiolated seedlings, FERONIA-dependent BR response subsequently decreases the effect of ethylene on hypocotyl growth. However, loss-of-function of fer mutations lead to enhanced ethylene response and reduced BR functions due to change of balance between the positive effects of endogenous BR concentration and inhibitory effects of ethylene on hypocotyl growth. Therefore, the study clearly indicates that FERONIA is mandatory for the promotive effects of BR responses in etiolated seedlings (Figure 2D) (Deslauriers and Larsen, 2010).
It has been observed that the exogenous application of BR enhanced ethylene biosynthesis in Arabidopsis seedlings (Hansen et al., 2009). BR up-regulates the expression of 1-aminocyclopropane-1-carboxylate synthase (ACS), the key gene required for ethylene production (Muday et al., 2012). Further, BR acts post-transcriptionally and also increases the stability of ACS proteins such as ACS5, ACS6 and ASC9 (Figure 2D) by preventing its ubiquitination mediated by 26S proteasome. Therefore, in response to various endogenous and exogenous signals, ACS is regulated by BR to continuously adjust ethylene biosynthesis in various tissues (Hansen et al., 2009).
Furthermore, crosstalk between BR and ethylene through ethylene-inducing xylanase (Eix) plays an important role in plant response to biotic stresses. It has been observed that in specific cultivars of tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum) Eix acts as a potential elicitor of plant defense responses. LeEix1 and LeEix2, are the two Eix receptors that belongs to a superclade of leucine-rich repeat receptor-like proteins (RLP). These receptors are able to bind Eix, however, only LeEix2 initiates defense responses. It has been demonstrated that upon application of the Eix elicitor, LeEix1 heterodimerizes with LeEix2 and further, LeEix1 is able to attenuate Eix-induced internalization and signaling of the LeEix2 receptor. Moreover, the activation of attenuating function of LeEix1 requires the binding of LeEix1 to BAK1 and not LeEix2, indicating BR- ethylene crosstalk (Bar et al., 2010). Furthermore, it is also known that BR crosstalks with ethylene to confer abiotic stress tolerance. Since, in another study it has been suggested that ethylene response factor protein (JERF3) activates the expression of oxidative genes, leading to decreased accumulation of ROS and enhanced abiotic stress tolerance (Wu et al., 2008) indicating that ethylene and BR may interact and lead to sequestration of ROS during stress conditions.
The synergistic interaction between ethylene and BR in regulating hyponastic growth has also been demonstrated (Polko et al., 2012). Ethylene is a key regulator of hyponastic growth, which is employed by plants to cope with biotic and abiotic stresses. ROT3/CYP90C1 encodes an enzyme which mediates C-23 hydroxylation of BR. A mutation in ROT3 reduces hyponastic growth leading to impairment of local cell expansion and inhibition of BR biosynthesis, indicating that hyponastic growth induced by ethylene is mainly regulated by BR (Polko et al., 2012).
Brassinosteroid-Gibberellin Crosstalk
Recently, BR are also found to interact with GA to coordinate different physiological processes (Sun et al., 2010; Li et al., 2012a). However, several evidences indicate BR-GA antagonistic interaction in defense related processes against root oomycete Pythium graminicola. It has been demonstrated that in several GA-deficient and/or –insensitive mutants, the disease development was more severe. It implies a positive role of GA in providing resistance against P. graminicola. Further, it has been demonstrated that susceptibility similar to those observed in BR treated plants was detected when endogenous GA level was disrupted using GA biosynthesis inhibitor, uniconazole (De Vleesschauwer et al., 2012). However, co-application of BR and uniconazole did not cause any additive effect. But treatment of uniconazole along with brassinazole, a BR inhibitor, negated the resistance-inducing effect of brassinazole. It indicates that BR dampens the effective immune response led by GA. Further, it has been demonstrated that the abundance of GA repressors, DELLA and SLR1 is positively regulated by BR. This phenomenon leads to BR mediated suppression of the GA biosynthetic genes such as GA20ox and GA3ox3 induce GA2ox expression (Figure 2E) which is involved in suppression of GA signaling and its deactivation (De Vleesschauwer et al., 2012). Recently, the crosstalk between BR and GA has been established in regulating plant cell elongation in rice (Tong et al., 2014). It has been suggested that BR promotes GA accumulation by inducing the expression of D18/GA3ox-2, one of the GA biosynthetic genes. However, application of exogenous high concentration of BR leads to the activation of GA2ox-3, a GA inactivation gene, resulting in inhibition of cell elongation. Moreover, GA inhibits BR signaling as well as its biosynthesis in a feedback inhibiting loop but facilitate cell elongation through activating primary BR signaling pathway upon applying exogenous high GA concentration, indicating BR-GA crosstalk in regulating cell elongation (Tong et al., 2014). The interaction between BR, IAA, and GA on cotton fiber development has been studied in Gossypium hirsutum (Hu et al., 2011). A class of DELLA proteins GhGAII was down-regulated by BR and auxin treatment during cotton fiber initiation and elongation, suggesting its importance in cotton fiber improvement through genetic modulation of phytohormone strategy. However, the expression of GhGAI1 and GhGAI3 were up-regulated by GA during cotton fiber initiation which is an unessential trait for fiber initiation (Hu et al., 2011) indicating BR-GA crosstalk in regulating cotton fiber development. The synergistic interaction between BR and GA leading to cell expansion during photomorphogenesis has been demonstrated through concurrence of BR-activated BZR1 and GA-inactivated DELLA transcription regulators (Gallego-Bartolomé et al., 2012). It has been suggested that GA promotion of cell elongation requires BR signaling, whereas BR or active BZR1 can suppress the GA-deficient dwarf phenotype. DELLA directly interacts with BZR1 and inhibits BZR1-DNA binding both in vitro and in vivo. This leads to the inhibition in the perception of environmental signals required for regulating cell elongation and seedling etiolation (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li and He, 2013). Another study also establishes a link between BR and GA, through DELLAs, that acts as a mediator in regulating cell elongation and plant growth. It has been reported that BZR1 interacts in vitro and in vivo with repressor of ga1-3 (RGA), a member of the DELLA protein family. Mutation studies indicate that in gain-of-function mutant bzr1-1d, RGA suppresses root and hypocotyl elongation. The study suggests that ectopic expression of DELLA proteins reduces the abundance and transcriptional activity of BZR1 indicating that BZR1 and RGA antagonizes each other’s transcriptional activity and also acts as a positive and negative regulators of BR and GA signaling, respectively (Li et al., 2012a).
Brassinosteroid-Jasmonic Acid Crosstalk
The interaction of BR and JA plays crucial roles in plant development, in both biotic and abiotic responses. The F-box protein, coronatine insensitive1 (COI1) is required for JA responses in Arabidopsis and plays a key role in JA signaling. The partially suppressing coi1 (psc1) is a leaky mutation of DWF4, that encodes a key enzyme in BR biosynthesis leading to inhibition of root growth. Further studies indicate that upon exogenous BR treatment, the partial restoration of JA sensitivity by psc1 in coi1-2 background and the JA hypersensitivity of psc1 in wild-type COI1 background, was eliminated (Ren et al., 2009). In the wild type plants attenuation of JA inhibition of root growth was observed on exogenous BR application. Further, the inhibition of the expression of DWF4 by JA was found to be dependent on COI1. These results indicate that DWF4 is located downstream of COI1 and is down-regulated by JA (Figure 2F). Therefore, BR antagonizes JA-signaling pathway in root growth and development (Ren et al., 2009). Furthermore, BAK1 has also been shown to be involved in providing resistance to Nicotiana attenuata, against its specialist herbivore, Manduca sexta (Yang et al., 2010). The wounding- and herbivory-induced responses were examined on empty vector (EV) and NaBAK1-silenced plants generated by virus-induced gene-silencing system. After wounding, NaBAK1- silenced plants showed attenuated JA and JA-isoleucine bursts, required for mediating plant defenses against herbivores indicating the critical role of BAK1 in providing biotic stress resistance against M. sexta. EV levels of defensive secondary metabolites, namely, trypsin proteinase inhibitors (TPIs) were observed on simulating herbivory in NaBAK1-silenced plants having similar levels of resistance to M. sexta larvae. Furthermore, in NaBAK1-silenced plants, JA application elicited higher levels of TPI activity than in EV plants. It suggests that given level of JA enhances the accumulation of TPIs on silencing NaBAK1 (Yang et al., 2010).
It has also been demonstrated that under stress conditions, BR enhances JA level in rice (Kitanaga et al., 2006), which strongly promotes the expression of thionin genes encoding antimicrobial peptides indicating a potential crosstalk point between these two phytohormones. The crosstalk between BR and JA was further studied to understand how these phytohormones interact in the formation of natural defense in tomatoes against insect herbivory. It has been observed that, BR and JA directly affected trichome density and allelochemical content but in a reverse manner (Campos et al., 2009). The defective mutant studies confirmed that JA promotes the traits required for anti-herbivory whereas BR prevented it. Since, the BR-deficient mutant dumpy (dpy) showed enhanced pubescence, zingiberene biosynthesis, and proteinase inhibitor expression, on contrary, opposite effects were seen in JA-insensitive jai1-1 mutant, leading to an increased production of defensive traits. Therefore, results from the defective mutants confirmed that JA promotes the traits required for anti-herbivory whereas, BR prevented it. Moreover, it has also been demonstrated that BR acts upstream of the JA signaling pathway, since, dpy3jai1-1 double mutant showed that jai1-1 is epistatic to dpy. Furthermore, trichome number in jai1-1 mutants was severely reduced as compared to dpy, which presents a high trichome density emphasizing the importance of JA in trichome formation. The study provides BR-JA antagonistic interactions for the defense against herbivory (Campos et al., 2009). Recently, the crosstalk between BR and JA in regulating rice (Oryza sativa) innate immunity during infection with the root-knot nematode Meloidogyne graminicola has been studied (Nahar et al., 2013). The exogenous application of BR at low concentration (0.1 and 1 μM) induced susceptibility, however, at high BR concentrations (5 and 10 μM), systemic defense against this nematode was observed in the roots (Nahar et al., 2013). It has also been demonstrated that transcript levels of JA biosynthesis gene, allene oxidase synthase2 (OsAOS2) (Mei et al., 2006) and JA-induced signaling gene, OsJAmyb (Lee et al., 2001) were strongly down-regulated when the BR concentration was low. However, upon high BR concentration, the transcript levels of JA biosynthesis and signaling genes were up-regulated. These results were further validated through exogenous foliar spraying with JA which leads to strong down-regulation of BR biosynthesis and signaling genes, OsDWF4 and OsBRI1 (Figure 2F), respectively (Nahar et al., 2013), indicating antagonistic interaction between BR and JA in the rice roots. Furthermore, it has been observed that mutants in the BR biosynthesis or signaling pathway accumulate slightly higher amount of the immediate JA-precursor, 12-oxo-phytodienoic acid, and hence consolidating BR and JA pathway antagonism (Nahar et al., 2013).
Brassinosteroid-Polyamine Crosstalk
Despite the significant progress made over recent years, we are just at the beginning to explore the BR and PA crosstalk. However, the preliminary studies indicate that BR and PA crosstalk is involved in enhancing the ability of stress tolerance potential of plants (Liu and Moriguchi, 2007). Under copper (Cu) stress treatment, the effect of BR on PA tissue concentrations and antioxidant potential of 7-day-old Raphanus sativus L. cv. ‘Pusa chetki’ seedlings had been studied (Choudhary et al., 2010). It has been suggested that abiotic and biotic stresses mediate alteration in free PA content (Liu and Moriguchi, 2007). The change in PA level upon BR application might be involved in mitigating the Cu-generated oxidative stress (Choudhary et al., 2010). Furthermore, BR treatment maintains the optimum amount of spermidine concentration required for normal plant growth and specifically increases the production of putrescine required for stress tolerance but decreases the concentration of cadaverine which generates oxidative burst to counteract heavy metal stress (Takahashi and Kakehi, 2010). Further, the co-application of Cu and BR also decreases cadaverine content enhancing superoxide dismutase (SOD) activity required for stress tolerance (Kuznetsov et al., 2009). It indicates the key role of BR-PA interaction in providing abiotic stress tolerance. A recent study provides the effect of exogenously applied BR and spermidine on R. sativus plants exposed to lethal concentrations of Cu (Choudhary et al., 2012a). Their interaction affects the expression of genes involved in Cu homeostasis and leads to enhanced tolerance to Cu stress in R. sativus (Choudhary et al., 2012a). It has been further suggested that upon co-application of BR and spermidine, the enhanced Cu stress tolerance is met either through the modulation of the expression of genes encoding PA enzymes or the genes that mediates the homeostasis of IAA and ABA (Choudhary et al., 2012a). Therefore, the use of these compounds is prerequisite for sustaining modern agriculture and to unravel the genes and transcription factors involved in BR and PA signaling pathway.
Brassinosteroid-Salicylic Acid Crosstalk
The potential crosstalk between BR and SA is mediated via non-expressor of pathogenesis-related genes 1 (NPR1) and WRKY70 (Figure 2G), encoding a transcription factor working downstream of NPR1 (Divi et al., 2010). In mutant studies, it has been observed that npr1-1 genotype was thermosensitive, and also showed defects in the expression of PR genes in response to SA (Clarke et al., 2004, 2009; Larkindale et al., 2005). However, in response to epibrassinolide, mere 2.4 fold increase in percent survival of the npr1-1 seedlings as compared to ninefold increase in wild type was observed (Divi et al., 2010). The study indicates that stress tolerance is mediated by functional NPR1 for manifestation of BR effect, via controlling BR signaling components such as BIN2 and BZR1 (Divi et al., 2010). Further, the existence of the crosstalk between BR and SA plays a pivotal role in response of plants to biotic as well as abiotic stresses. It has been demonstrated that in tobacco as well as in rice, BR acts as an inducer of a broad range of disease resistance. Upon BR treatment, increase in the resistance to the viral pathogen tobacco mosaic virus, the bacterial pathogen Pseudomonas syringae pv. tabaci, and the fungal pathogen Oidium sp. has been observed in tobacco (Nakashita et al., 2003). However, in rice plants, it has been observed that BR enhances resistance to the fungal pathogen Magnaporthe grisea and the bacterial pathogen Xanthomonas oryzae. Moreover, it has been suggested that in tobacco, enhancement in the BR mediated resistance does not require SA which has further been consolidated by measuring SA accumulation and its analysis using NahG transgenic tobacco (Nakashita et al., 2003). It indicates that BR and SA act independently in providing resistance against pathogens. However, the combined effect of BR and systemic acquired resistance (SAR) inducers provides additive resistance against pathogens. Although, crosstalk may exist between BR and SA signaling pathways for inducing resistance but it is distinct and act independently (Nakashita et al., 2003).
In contrast to the previous view that BR positively regulate plant innate immunity, recent study provides evidence that Pythium graminicola exploits BR as virulence factors and takes the hostage of rice BR machinery to cause disease (De Vleesschauwer et al., 2012). Further, it has been suggested that the negative crosstalk between SA and BR pathways leads to immune suppressive effect of BR. Moreover, upon brassinazole treatment, a reduced susceptibility toward P. graminicola had been observed in rice plants due to derepression of the master defense regulators of SA pathway such as NPR1 and OsWRKY45. The study indicates that BR-mediated suppression of SA defense responses occur upstream of NPR1 and OsWRKY45 but downstream of SA biosynthesis (De Vleesschauwer et al., 2012).
Conclusion
Brassinosteroids have emerged as a potent phytohormone due to its versatile functions. The wide range of functions is attributed to its multiple targets and complex regulatory mechanisms. Serious and rigorous global efforts are being carried out in understanding the complexity of the mechanism of BR action. Understanding the dynamics of BR homeostasis and unraveling its interactions with other phytohormones will add new dimension to BR research. With the availability of biological resources and introduction of new experimental tools, it is expected that in coming years there will be a significant addition of knowledge in mode of BR action and this could ultimately culminate in ushering a new era in plant developmental and stress biology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SS acknowledges the financial support received under Innovation in Science Pursuit for Inspired Research (INSPIRE) Programme, Department of Science and Technology, Government of India [Grant no. DST/INSPIRE Fellowship/2010 [IF10123]. The support from University Grants Commision (UGC), Government of India is acknowledged.
References
- Bai M. Y., Shang J. X., Oh E., Fan M., Bai Y., Zentella R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14 810–817. 10.1038/ncb2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz A. (2007). Metabolism of brassinosteroids in plants. Plant Physiol. Biochem. 45 95–107. 10.1016/j.plaphy.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Bajguz A., Piotrowska-Niczyporuk A. (2014). Interactive effect of brassinosteroids and cytokinins on growth, chlorophyll, monosaccharide and protein content in the green alga Chlorella vulgaris (Trebouxiophyceae). Plant Physiol. Biochem. 80 176–183. 10.1016/j.plaphy.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Bao F., Shen J., Brady S. R., Muday G. K., Asami T., Yang Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134 1624–1631. 10.1104/pp.103.036897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Sharfman M., Ron M., Avni A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63 791–800. 10.1111/j.1365-313X.2010.04282.x [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y. (2015). The molecular circuitry of brassinosteroid signaling. New Phytol. 206 522–540. 10.1111/nph.13269 [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59 443–465. 10.1146/annurev.arplant.58.032806.103805 [DOI] [PubMed] [Google Scholar]
- Bishop G. J., Yokota T. (2001). Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 42 114–120. 10.1093/pcp/pce018 [DOI] [PubMed] [Google Scholar]
- Bucherl C. A., Van Esse G. W., Kruis A., Luchtenberg J., Westphal A. H., Aker J., et al. (2013). Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol. 162 1911–1925. 10.1104/pp.113.220152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer C. S., Sukumar P., Muday G. K. (2006). Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 140 1384–1396. 10.1104/pp.105.075671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M. L., de Almeida M., Rossi M. L., Martinelli A. P., Junior C. G. L., Figueira A., et al. (2009). Brassinosteroids interact negatively with jasmonates in the formation of anti-herbivory traits in tomato. J. Exp. Bot. 60 4347–4361. 10.1093/jxb/erp270 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A., Yin Y., Yu C., Vafeados D., Mora-Garcia S., Cheng J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131 5341–5351. 10.1242/dev.01403 [DOI] [PubMed] [Google Scholar]
- Ceserani T., Trofka A., Gandotra N., Nelson T. (2009). VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 57 1000–1014. 10.1111/j.1365-313X.2008.03742.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J., Wang Z. Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25 1031–1042. 10.1016/j.cub.2015.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S. (2007). “Brassinosteroid biosynthesis and metabolism,” in Plant Hormones: Biosynthesis, Signal Transduction, Action, ed. Davies P. J. (Dordrecht: Kluwer Academic Publishers; ), 156–178. [Google Scholar]
- Choe S., Fujioka S., Noguchi T., Takatsuto S., Yoshida S., Feldmann K. A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26 573–582. 10.1046/j.1365-313x.2001.01055.x [DOI] [PubMed] [Google Scholar]
- Choudhary S. P., Bhardwaj R., Gupta B. D., Dutt P., Gupta R. K., Biondid S., et al. (2010). Epibrassinolide induces changes in indole-3-acetic acid, abscisic acid and polyamine concentrations and enhances antioxidant potential of radish seedlings under copper stress. Physiol. Plant. 140 280–296. 10.1111/j.1399-3054.2010.01403.x [DOI] [PubMed] [Google Scholar]
- Choudhary S. P., Oral H. V., Bhardwaj R., Yu J., Tran L. P. (2012a). Interaction of brassinosteroids and polyamine enhances copper stress tolerance in Raphanus sativus. J. Exp. Bot. 63 5659–5670. 10.1093/jxb/ers219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S. P., Yu J. Q., Yamaguchi-Shinozaki K., Shinozaki K., Tran L. S. (2012b). Benefits of brassinosteroid crosstalk. Trends Plant Sci. 17 594–605. 10.1016/j.tplants.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Chung Y., Choe S. (2013). The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit. Rev. Plant Sci. 32 396–410. 10.1080/07352689.2013.797856 [DOI] [Google Scholar]
- Chung Y., Maharjan P. M., Lee O., Fujioka S., Jang S., Kim B., et al. (2011). Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 66 564–578. 10.1111/j.1365-313X.2011.04513.x [DOI] [PubMed] [Google Scholar]
- Clarke S. M., Cristescu S. M., Miersch O., Harren F. J., Wasternack C., Mur L. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182 175–187. 10.1111/j.1469-8137.2008.02735.x [DOI] [PubMed] [Google Scholar]
- Clarke S. M., Mur L. A., Wood J. E., Scott I. M. (2004). Salicylic acid dependent signalling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J. 38 432–447. 10.1111/j.1365-313X.2004.02054.x [DOI] [PubMed] [Google Scholar]
- Clouse S. D. (2011). Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23 1219–1230. 10.1105/tpc.111.084475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D., Sasse J. M. (1998). Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Mol. Biol. 49 427–451. 10.1146/annurev.arplant.49.1.427 [DOI] [PubMed] [Google Scholar]
- Deng Z., Zhang X., Tang W., Oses-Prieto J. A., Suzuki N., Gendron J. M., et al. (2007). A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell. Proteomics 6 2058–2071. 10.1074/mcp.M700123-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslauriers S. D., Larsen P. B. (2010). FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol. Plant 3 626–640. 10.1093/mp/ssq015 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I. R., et al. (2012). Brassinosteroids antagonize gibberellin– and salicylate-mediated root immunity in rice. Plant Physiol. 158 1833–1846. 10.1104/pp.112.193672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi U. K., Krishna P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. N. Biotechnol. 26 131–136. 10.1016/j.nbt.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Divi U. K., Rahman T., Krishna P. (2010). Research article Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10:151 10.1186/1471-2229-10-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi U. K., Rahman T., Krishna P. (2015). Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol. J. 10.1111/pbi.12396 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Liu H., Xiong L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4:397 10.3389/fpls.2013.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsan H., Ray W. K., Phinney B., Wang X., Huber S. C., Clouse S. D. (2005). Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J. 43 251–261. 10.1111/j.1365-313X.2005.02448.x [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo J. M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W. C., et al. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317 656–660. 10.1126/science.1143562 [DOI] [PubMed] [Google Scholar]
- Fàbregas N., Caño-Delgado A. I. (2014). Turning on the microscope turret: a new view for the study of brassinosteroid signaling in plant development. Physiol. Plant. 151 172–183. 10.1111/ppl.12130 [DOI] [PubMed] [Google Scholar]
- Fan X. Y., Sun Y., Cao D. M., Bai M. Y., Luo X. M., Yang H. J., et al. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5 591–600. 10.1093/mp/sss041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59 387–415. 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Friml J., Palme K. (2002). Polar auxin transport–old questions and new concepts? Plant Mol. Biol. 49 273–284. 10.1023/A:1015248926412 [DOI] [PubMed] [Google Scholar]
- Fujioka S., Noguchi T., Yokota T., Takatsuto S., Yoshida S. (1998). Brassinosteroids in Arabidopsis thaliana. Phytochemistry 48 595–599. 10.1016/S0031-9422(98)00065-X [DOI] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54 137–164. 10.1146/annurev.arplant.54.031902.134921 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Mingueta E. G., Grau-Enguixa F., Abbasa M., Locascioa A., Thomas S. G., et al. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 13446–13451. 10.1073/pnas.1119992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove M. D., Spencer G. F., Rohwedder W. K., Mandava N., Worley J. F., Warthen J. D. J., et al. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281 216–217. 10.1038/281216a0 [DOI] [Google Scholar]
- Gruszka D. (2013). The brassinosteroid signaling pathway-new key players and interconnections with other signaling networks crucial for plant development and stress tolerance. Int. J. Mol. Sci. 14 8740–8774. 10.3390/ijms14058740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat G. E., Russinova E. (2011). Plants grow on brassinosteroids. Curr. Opin. Plant Biol. 14 530–537. 10.1016/j.pbi.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Guo D., Gao X., Li H., Zhang T., Chen G., Huang P., et al. (2008). EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Mol. Biol. 66 345–360. 10.1007/s11103-007-9273-5 [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Aluru M., Aluru S., Yin Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16 545–553. 10.1016/j.pbi.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. (2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 106 7648–7653. 10.1073/pnas.0812346106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M. J., Chory J., et al. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138 839–848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Sela A., Friedlander L., Savaldi-Goldstein S. (2012). BRI1 activity in the root meristem involves post-transcriptional regulation of PIN auxin efflux carriers. Plant Signal. Behav. 7 68–70. 10.4161/psb.7.1.18657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Chae H. S., Kieber J. J. (2009). Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 57 606–614. 10.1111/j.1365-313X.2008.03711.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Yin Y., Fei S. (2013). Brassinosteroid signaling network: implications on yield and stress tolerance. Plant Cell Rep. 32 1017–1030. 10.1007/s00299-013-1438-x [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choic G., Lianga Z., Wang Z. Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5 688–697. 10.1093/mp/sss011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B. R., Masson P. H. (2008). ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 53 380–392. 10.1111/j.1365-313X.2007.03351.x [DOI] [PubMed] [Google Scholar]
- He J. X., Gendron J. M., Sun Y., Gampala S. S., Gendron N., Sun C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307 1634–1638. 10.1126/science.1107580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J. P., Wilson I. A., et al. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474 467–472. 10.1038/nature10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Luo M., Xiao Y., Li X., Tan K., Hou L., et al. (2011). Brassinosteroids and auxin down-regulate DELLA genes in fiber initiation and elongation of cotton. Agric. Sci. China 10 1168–1176. 10.1016/S1671-2927(11)60107-7 [DOI] [Google Scholar]
- Hu Y., Yu D. (2014). BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26 4394–4408. 10.1105/tpc.114.130849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N., Moore J. M., Federer M., Grossniklaus U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130 2149–2159. 10.1242/dev.00458 [DOI] [PubMed] [Google Scholar]
- Husar S., Berthiller F., Fujioka S., Rozhon W., Khan M., Kalaivanan F., et al. (2011). Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 11:51 10.1186/1471-2229-11-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikekawa N., Zhao Y. J. (1991). “Application of 24-epibrassinolide in agriculture,” in Brassinosteroids: Chemistry, Bioactivity, and Applications, eds Cutler H. G., Yokota T., Sadam G. (Washington, DC: In ACS Symposium Series, American Chemical Society; ), 280–291. [Google Scholar]
- Jaillais Y., Hothorn M., Belkhadir Y., Dabi T., Nimchuk Z. L., Meyerowitz E. M., et al. (2011). Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25 232–237. 10.1101/gad.2001911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je B. I., Piao H. L., Park S. J., Park S. H., Kim C. M., Xuan Y. H., et al. (2010). RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 221 1777–1791. 10.1105/tpc.109.069575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S. H., Jang M. S., Kim M. K., Lee J. E., Kim S. K. (2015). Biosynthetic relationship between C28-brassinosteroids and C29-brassinosteroids in rice (Oryza sativa) seedlings. Phytochemistry 111 84–90. 10.1016/j.phytochem.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Kerr I., Carrier D., Twycross J. (2011). “Hormone transport,” in The Plant Plasma Membrane, eds Murphy A. S., Schulz B., Peer W. (Berlin: Springer; ), 379–397. [Google Scholar]
- Kim B. K., Fujioka S., Takatsuto S., Tsujimoto M., Choe S. (2008). Castasterone is a likely end product of brassinosteroid biosynthetic pathway in rice. Biochem. Biophys. Res. Commun. 374 614–619. 10.1016/j.bbrc.2008.07.073 [DOI] [PubMed] [Google Scholar]
- Kim T. W., Guan S., Burlingame A. L., Wang Z. Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43 561–571. 10.1016/j.molcel.2011.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W., Lee S. M., Joo S. H., Yun H. S., Lee Y., Kaufman P. B., et al. (2007). Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ. 30 679–689. 10.1111/j.1365-3040.2007.01659.x [DOI] [PubMed] [Google Scholar]
- Kim T. W., Wang Z. Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61 681–704. 10.1146/annurev.arplant.043008.092057 [DOI] [PubMed] [Google Scholar]
- Kissoudis C., Wiel C., Visser R. G. F., Linden G. (2014). Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 5:207 10.3389/fpls.2014.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanaga Y., Jian C., Hasegawa M., Yazaki J., Kishimoto N., Kikuchi S., et al. (2006). Sequential regulation of gibberellin, brassinosteroid, and jasmonic acid biosynthesis occurs in rice coleoptiles to control the transcript levels of anti-microbial thionin genes. Biosci. Biotechnol. Biochem. 70 2410–2419. 10.1271/bbb.60145 [DOI] [PubMed] [Google Scholar]
- Kuznetsov V. V., Stetsenko L. A., Shevyakova N. I. (2009). Exogenous cadaverine induces oxidative burst and reduces cadaverine conjugate content in the common ice plant. J. Plant Physiol. 166 40–51. 10.1016/j.jplph.2008.01.010 [DOI] [PubMed] [Google Scholar]
- Lanza M., Garcia-Ponce B., Castrillo G., Catarecha P., Sauer M., Rodriguez-Serrano M., et al. (2012). Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 22 1275–1285. 10.1016/j.devcel.2012.04.008 [DOI] [PubMed] [Google Scholar]
- Larkindale J., Hall J. D., Knight M. R., Vierling E. (2005). Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 138 882–897. 10.1104/pp.105.062257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. W., Qi M., Yang Y. O. (2001). A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol. Plant Microbe Interact. 14 527–535. 10.1094/MPMI.2001.14.4.527 [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Schlessinger J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141 1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1999). Brassinosteroid actions in plants. J. Exp. Bot. 50 275–282. 10.1093/jxb/50.332.275 [DOI] [Google Scholar]
- Li J., Mo X., Wang J., Chen N., Fan H., Dai C., et al. (2009). BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis. Planta 229 593–603. 10.1007/s00425-008-0854-6 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., Walke J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222. 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Li L., Ye H., Guo H., Yin Y. (2010). Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. U.S.A. 107 3918–3923. 10.1073/pnas.0909198107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. F., He J. X. (2013). Mechanisms of signaling crosstalk between brassinosteroids and gibberellins. Plant Signal. Behav. 8 e24686 10.4161/psb.24686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. F., Wang C., Jiang L., Li S., Sun S. S., He J. X. (2012a). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5 ra72 10.1126/scisignal.2002908 [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Xu Z. S., He G. Y., Yang G. X., Chen M., Li L. C., et al. (2012b). A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 4 522–527. 10.1016/j.bbrc.2012.08.118 [DOI] [PubMed] [Google Scholar]
- Liu J., Rowe J., Lindsey K. (2014). Hormonal crosstalk for root development: a combined experimental and modeling perspective. Front. Plant Sci. 5:116 10.3389/fpls.2014.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Moriguchi T. (2007). Changes in free polyamine titers and expression of polyamine biosynthetic genes during growth of peach in vitro callus. Plant Cell Rep. 26 125–131. 10.1007/s00299-006-0223-5 [DOI] [PubMed] [Google Scholar]
- Luo X. M., Lin W. H., Zhu S., Zhu J. Y., Sun Y., Fan X. Y., et al. (2010). Integration of light and brassinosteroid signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19 872–883. 10.1016/j.devcel.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A. P., Lozano-Durán R., Zipfel C. (2015). Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci. 20 269–272. 10.1016/j.tplants.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Maharjan P. M., Choe S. (2011). High temperature stimulates DWARF4 (DWF4) expression to increase hypocotyle elongation in Arabidopsis. J. Plant Biol. 54 425–429. 10.1007/s12374-011-9183-6 [DOI] [Google Scholar]
- Maharjan P. M., Schulz B., Choe S. (2011). BIN2/DWF12 antagonistically transduces brassinosteroid and auxin signals in the roots of Arabidopsis. J. Plant Biol. 54 126–134. 10.1007/s12374-010-9138-3 [DOI] [Google Scholar]
- Markovic-Housley Z., Degano M., Lamba D., von Roepenack-Lahaye E., Clemens S., Susani M., et al. (2003). Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J. Mol. Biol. 325 123–133. [DOI] [PubMed] [Google Scholar]
- Marsolais F., Boyd J., Paredes Y., Schinas A. M., Garcia M., Elzein S., et al. (2007). Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 225 1233–1244. 10.1007/s00425-006-0413-y [DOI] [PubMed] [Google Scholar]
- Mei C., Qi M., Sheng G., Yang Y. (2006). Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19 1127–1137. 10.1094/MPMI-19-1127 [DOI] [PubMed] [Google Scholar]
- Mouchel C. F., Osmont K. S., Hardtke C. S. (2006). BRX mediates feedback between brassinosteroid and auxin signalling in root growth. Nature 443 458–461. 10.1038/nature05130 [DOI] [PubMed] [Google Scholar]
- Muday G. K., Rahman A., Binder B. M. (2012). Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17 181–195. 10.1016/j.tplants.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Nahar K., Kyndt T., Hause B., Hofte M., Gheysen G. (2013). Brassinosteroids suppress rice defense against root-knot nematodes through antagonism with the jasmonate pathway. Mol. Plant Microbe Interact. 26 106–115. 10.1094/MPMI-05-12-0108-FI [DOI] [PubMed] [Google Scholar]
- Nakamura A., Nakajima N., Goda H., Shimada Y., Hayashi K., Nozaki H., et al. (2006). Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 45 193–205. 10.1111/j.1365-313X.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Nakashita H., Yasuda M., Nitta T., Asami T., Fujioka S., Arai Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33 887–898. 10.1046/j.1365-313X.2003.01675.x [DOI] [PubMed] [Google Scholar]
- Nam K. H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212. 10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. 10.1016/j.cell.2006.05.050 [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Mockler T. C., Chory J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2:e258 10.1371/journal.pbio.0020258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B., Lee S. H., Kim H. J., Yi G., Shin E. A., Lee M., et al. (2004). Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16 2601–2613. 10.1105/tpc.104.025353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J. Y., Wang Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14 802–809. 10.1038/ncb2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M. H., Wang X., Kota U., Goshe M. B., Clouse S. D., Huber S. C. (2009). Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 658–663. 10.1073/pnas.0810249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S. T., Godza B., Watanabe B., Fujioka S., Hategan L., Ide K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287 31551–31560. 10.1074/jbc.M112.392720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z., Reguera M., Tumimbang E., Walia H., Blumwald E. (2011). Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 9 747–758. 10.1111/j.1467-7652.2010.00584.x [DOI] [PubMed] [Google Scholar]
- Piotrowska A., Bajguz A. (2011). Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72 2097–2112. 10.1016/j.phytochem.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Polko J. K., Pierik R. F., Zanten M. V., Tarkowská D., Strnad M., Voesenek L. A. C. J., et al. (2012). Ethylene promotes hyponastic growth through interaction with ROTUNDIFOLIA3/CYP90C1 in Arabidopsis. J. Exp. Bot. 64 613–624. 10.1093/jxb/ers356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G. L., Vaistij F. E., Hiranuma S., et al. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. U.S.A. 102 15253–15258. 10.1073/pnas.0504279102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Rozhon W., Khan M., Husar S., Adam G., Luschnig C., et al. (2011). CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30 1149–1161. 10.1038/emboj.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Han C., Peng W., Huang Y., Peng Z., Xiong X., et al. (2009). A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiol. 151 1412–1420. 10.1104/pp.109.140202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh H., Jeong C. W., Fujioka S., Kim Y. K., Lee S., Ahn J. H., et al. (2012). Genetic evidence for the reduction of brassinosteroid levels by a BAHD acyltransferase-like protein in Arabidopsis. Plant Physiol. 159 696–709. 10.1104/pp.112.197202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Cho H., Bae W., Hwang I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 5 4138 10.1038/ncomms5138 [DOI] [PubMed] [Google Scholar]
- Saini S., Sharma I., Kaur N., Pati P. K. (2013). Auxin: a master regulator in plant root development. Plant Cell Rep. 32 741–757. 10.1007/s00299-013-1430-5 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Morinaka Y., Inukai Y., Kitano H., Fujioka S. (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 73 676–688. 10.1111/tpj.12071 [DOI] [PubMed] [Google Scholar]
- Santiago J., Henzler C., Hothorn M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341 889–892. 10.1126/science.1242468 [DOI] [PubMed] [Google Scholar]
- Schneider K., Breuer C., Kawamura A., Jikumaru Y., Hanada A., Fujioka S., et al. (2012). Arabidopsis PIZZA has the capacity to acylate brassinosteroids. PLoS ONE 7:e46805 10.1371/journal.pone.0046805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma I., Bhardwaj R., Pati P. K. (2015). Exogenous application of 28-Homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J. Plant Growth Regul. 34 509–518. 10.1007/s00344-015-9486-9 [DOI] [Google Scholar]
- Sharma I., Ching E., Saini S., Bhardwaj R., Pati P. K. (2013). Exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety Pusa Basmati-1. Plant Physiol. Biochem. 69 17–26. 10.1016/j.plaphy.2013.04.013 [DOI] [PubMed] [Google Scholar]
- She J., Han Z., Kim T. W., Wang J., Cheng W., Chang J., et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474 472–476. 10.1038/nature10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J., Han Z., Zhou B., Chai J. (2013). Structural basis for differential recognition of brassinolide by its receptors. Protein Cell 4 475–482. 10.1007/s13238-013-3027-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki K., Uemura M., Tsurumi S., Rahman A. (2009). Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21 3823–3838. 10.1105/tpc.109.069906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreeramulu S., Mostizky Y., Sunitha S., Shani E., Nahum H., Salomon D., et al. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74 905–919. 10.1111/tpj.12175 [DOI] [PubMed] [Google Scholar]
- Steber C. M., McCourt P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125 763–769. 10.1104/pp.125.2.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar P., Edwards K. S., Rahman A., DeLong A., Gloria K., Muday G. K. (2009). PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 150 722–735. 10.1104/pp.108.131607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X. Y., Cao D. M., Tang W., He K., Zhu J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19 765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B., et al. (2013). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23 1326–1329. 10.1038/cr.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G. M., Ross J. J., Jager C. E., Reid J. B. (2008). Brassinosteroid transport. J. Exp. Bot. 59 17–24. 10.1093/jxb/erm098 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Kakehi J. I. (2010). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 105 1–6. 10.1093/aob/mcp259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Asami T., Yoshida S., Nakamura Y., Matsuo T., Okamoto S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138 1117–1125. 10.1104/pp.104.058040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakamura Y., Asami T., Yoshida S., Matsuo T., Okamoto S. (2003). Physiological roles of brassinosteroids in early growth of Arabidopsis: brassinosteroids have a synergistic relationship with gibberellin as well as auxin in light-grown hypocotyl elongation. J. Plant Growth Regul. 22 259–271. 10.1007/s00344-003-0119-3 [DOI] [Google Scholar]
- Tang W., Kim T. W., Oses-Prieto J. A., Sun Y., Deng Z., Zhu S., et al. (2008). Brassinosteroid-Signaling Kinases (BSKs) mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321 557–560. 10.1126/science.1156973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Yuan M., Wang R., Yang Y., Wang C., Oses-Prieto J. A., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13 124–131. 10.1038/ncb2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Xiao Y., Liu D., Gao S., Liu L., Yin Y., et al. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26 4376–4393. 10.1105/tpc.114.132092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Callebert P., Zadnikova P., Benkova E., Van Der Straeten D. (2013). Brassinosteroid control of shoot gravitropism interacts with ethylene and depends on auxin signaling components. Am. J. Bot. 100 215–225. 10.3732/ajb.1200264 [DOI] [PubMed] [Google Scholar]
- Vercruyssen L., Gonzalez N., Werner T., Schmülling T., Inzé D. (2011). Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in Arabidopsis. Plant Physiol. 155 1339–1352. 10.1104/pp.110.167049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Walcher C. L., Chory J., Nemhauser J. L. (2008). Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. U.S.A. 105 9829–9834. 10.1073/pnas.0803996105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J., González-García M. P., Frigola D., Fàbregas N., Alexiou K. G., López-Bigas N., et al. (2014). Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev. Cell 14 36–47. 10.1016/j.devcel.2014.05.020 [DOI] [PubMed] [Google Scholar]
- Vriet C., Russinova E., Reuzeau C. (2013). From squalene to brassinolide: the steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 6 1738–1757. 10.1093/mp/sst096 [DOI] [PubMed] [Google Scholar]
- Wang H., Nagegowda D. A., Rawat R., Bouvier-Nave P., Guo D., Bach T. J., et al. (2012a). Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis upregulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol. J. 10 31–42. 10.1111/j.1467-7652.2011.00631.x [DOI] [PubMed] [Google Scholar]
- Wang Z. Y., Bai M. Y., Oh E., Zhu J. Y. (2012b). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46 701–724. 10.1146/annurev-genet-102209-163450 [DOI] [PubMed] [Google Scholar]
- Wang H., Yang C., Zhang C., Wang N., Lu D., Wang J., et al. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21 825–834. 10.1016/j.devcel.2011.08.018 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix–loop–helix proteins. Plant Cell 21 3781–3791. 10.1105/tpc.109.072504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J., Xie Z., Liu S., Nolan T., Ye H., et al. (2014). Histone lysine methyltransferase SDG8 Is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol. Plant 7 1303–1315. 10.1093/mp/ssu056 [DOI] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313 1118–1122. 10.1126/science.1127593 [DOI] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M. B., et al. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15 220–235. 10.1016/j.devcel.2008.06.011 [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., et al. (2005a). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8 855–865. 10.1016/j.devcel.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M. B., Soderblom E. J., Phinney B. S., Kuchar J. A., Li J., et al. (2005b). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17 1685–1703. 10.1105/tpc.105.031393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2 505–513. 10.1016/S1534-5807(02)00153-3 [DOI] [PubMed] [Google Scholar]
- Waters M. T., Wang P., Korkaric M., Capper R. G., Saunders N. J., Langdale J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21 1109–1128. 10.1105/tpc.108.065250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Nehnevajova E., Köllmer I., Novák O., Strnad M., Krämer U., et al. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22 3905–3920. 10.1105/tpc.109.072694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhang Z., Zhang H., Wang X. C., Huang R. (2008). Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 148 1953–1963. 10.1104/pp.108.126813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Oh M.-H., Kim H. S., Schwartz D., Imai B. S., Yau P. M., et al. (2012). Transphosphorylation of E. coli proteins during production of recombinant protein kinases provides a robust system to characterize kinase specificity. Front. Plant Sci. 3:262 10.3389/fpls.2012.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhao Z., Dong A., Soubigou-Taconnat L., Renou J. P., Steinmetz A., et al. (2008). Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28 1348–1360. 10.1128/MCB.01607-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143 1362–1371. 10.1104/pp.106.091561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. H., Hettenhausen C., Baldwin I. T., Wu J. (2010). BAK1 regulates the accumulation of jasmonic acid and the levels of trypsin proteinase inhibitors in Nicotiana attenuata’s responses to herbivory. J. Exp. Bot. 62 641–652. 10.1093/jxb/erq298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 4 20142–20147. 10.1073/pnas.1205232109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Li L., Yin Y. (2011). Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J. Int. Plant Biol. 53 455–468. 10.1111/j.1744-7909.2011.01046.x [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yokoda T., Asami T., Chory J. (2005). A new class of transcription factors mediate brassinosteroid-regulated gene expression in Arabidopsis. Cell 120 249–259. 10.1016/j.cell.2004.11.044 [DOI] [PubMed] [Google Scholar]
- Yin Y. H., Wang Z. Y., Mora-Garcia S., Li J. M., Yoshida S., Asami T., et al. (2002). BES1 accumulates in the nucleus in response to brassionsteroids to regulate gene expression and promote stem elongation. Cell 109 181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroidregulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105 7618–7623. 10.1073/pnas.0802254105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65 634–646. 10.1111/j.1365-313X.2010.04449.x [DOI] [PubMed] [Google Scholar]
- Yuan L. B., Peng Z. H., Zhi T. T., Zho Z., Liu Y., Zhu Q., et al. (2015). Brassinosteroid enhances cytokinin-induced anthocyanin biosynthesis in Arabidopsis seedlings. Biol. Plant. 59 99–105. 10.1111/j.1744-7909.2011.01042.x [DOI] [Google Scholar]
- Yuan T., Fujioka S., Takatsuto S., Matsumoto S., Gou X., He K., et al. (2007). BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J. 51 220–233. 10.1111/j.1365-313X.2007.03129.x [DOI] [PubMed] [Google Scholar]
- Yuldashev R., Avalbaev A., Bezrukova M., Vysotskaya L., Khripach V., Shakirova F. (2012). Cytokinin oxidase is involved in the regulation of cytokinin content by 24-epibrassinolide in wheat seedlings. Plant Physiol. Biochem. 55 1–6. 10.1016/j.plaphy.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Zhang S., Cai Z., Wang X. (2009a). The primary signaling outputs of brassinosteroids are regulated by abscisic acid signalling. Proc. Natl. Acad. Sci. U.S.A. 106 4543–4548. 10.1073/pnas.0900349106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y., Bai M. Y., Wu J., Zhu J. Y., Wang H., Zhang Z., et al. (2009b). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21 3767–3780. 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang S., Xu Y., Yu C., Shen C., Qian Q., et al. (2014). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38 638–654. 10.1111/pce.12397 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang J., Li X., Xia X. J., Zhou Y. H., Shi K., et al. (2014). H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot. 65 4371–4383. 10.1093/jxb/eru217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Wang H., Fujioka S., Zhou T., Tian H., Tian W., et al. (2013a). Homeostasis of brassinosteroids regulated by DRL1, a putative acyltransferase in Arabidopsis. Mol. Plant 6 546–558. 10.1093/mp/sss144 [DOI] [PubMed] [Google Scholar]
- Zhu J. Y., Sae-Seaw J., Wang Z. Y. (2013b). Brassinosteroid signalling. Development 140 1615–1620. 10.1242/dev.060590 [DOI] [PMC free article] [PubMed] [Google Scholar]