Abstract
Several underutilized grasses have excellent potential for use as bioenergy feedstock due to their lignocellulosic biomass. Genomic tools have enabled identification of lignocellulose biosynthesis genes in several sequenced plants. However, the non-availability of whole genome sequence of bioenergy grasses hinders the study on bioenergy genomics and their genomics-assisted crop improvement. Foxtail millet (Setaria italica L.; Si) is a model crop for studying systems biology of bioenergy grasses. In the present study, a systematic approach has been used for identification of gene families involved in cellulose (CesA/Csl), callose (Gsl) and monolignol biosynthesis (PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, COMT, CCR, CAD) and construction of physical map of foxtail millet. Sequence alignment and phylogenetic analysis of identified proteins showed that monolignol biosynthesis proteins were highly diverse, whereas CesA/Csl and Gsl proteins were homologous to rice and Arabidopsis. Comparative mapping of foxtail millet lignocellulose biosynthesis genes with other C4 panicoid genomes revealed maximum homology with switchgrass, followed by sorghum and maize. Expression profiling of candidate lignocellulose genes in response to different abiotic stresses and hormone treatments showed their differential expression pattern, with significant higher expression of SiGsl12, SiPAL2, SiHCT1, SiF5H2, and SiCAD6 genes. Further, due to the evolutionary conservation of grass genomes, the insights gained from the present study could be extrapolated for identifying genes involved in lignocellulose biosynthesis in other biofuel species for further characterization.
Keywords: foxtail millet (Setaria italica L.), secondary cell wall biosynthesis, lignocellulose, bioenergy grasses, genomics, comparative mapping
Introduction
Cell wall polymers of living plants constitute a predominant proportion of their biomass, which is formed by fermentable linked sugars. These polymers form a major structural component of plant cell wall and particularly, secondary cell walls provide mechanical strength and rigidity to vascular plants (Wang et al., 2013; Zhong and Ye, 2015). Secondary cell walls are present in tracheary elements, xylem, phloem, extraxylary and interfascicular fibers, sclereids and seed coats, and are made of cellulose, hemicelluloses and lignin. Cellulose, the primary unit, cross-links with hemicelluloses including xylan and glucomannan, and impregnated with phenolic polymer lignin, and altogether, this complex polymeric network forms secondary cell wall. The proportion of cellulose, hemicelluloses, and lignin varies among different plant species and of note, the composition may also vary in response to diverse developmental and environmental conditions (Zhong and Ye, 2015). Being the prime constituents of wood and fiber, secondary cell walls have been extensively studied to understand and exploit their biofuel prospects. Biochemical and genomic methods have identified the genes encoding for enzymes which participate in the biosynthesis of secondary cell wall components.
Pear et al. (1996) was the first to identify cellulose synthase (CesA) genes in cotton and following this, CesA genes in other plants have been identified and their numbers were shown to vary between plant species. In Arabidopsis, 10 CesA genes have been identified (Richmond and Somerville, 2000), whereas 12 in maize (Appenzeller et al., 2004), 16 in barley (Burton et al., 2004), 18 in poplar (Djerbi et al., 2005) have been reported. The CesA enzymes belong to glycosyltransferase-2 (GT-2) superfamily, which is defined by an eight-transmembrane topology and conserved cytosolic substrate binding and catalytic residues (McFarlane et al., 2014). In addition to CesA, plants also have cellulose synthase-like (Csl) genes, which can be involved in biosynthesis of hemicellulose and other glucans (Lerouxel et al., 2006). Csl genes can synthesize other polysaccharides that are not components of the hemicellulose matrix (Lerouxel et al., 2006). So far, several types of Csl genes have been identified, denoted as CslA to CslK. CslA encodes for (1,4)-β-D-mannan synthases (Dhugga et al., 2004; Liepman et al., 2005), CslF and CslH encode the mixed linkage glucan synthases for (1,3;1,4)-β-glucan biosynthesis (Burton et al., 2006; Doblin et al., 2009), CslC genes are involved in xyloglucan biosynthesis (Cocuron et al., 2007), and CslD in xylan and homogalacturonan synthesis (Hamann et al., 2004; Bernal et al., 2008a,b; Li et al., 2009), whereas the functional roles of other Csl genes remain elusive (Yin et al., 2009). Noteworthy, CslB and CslG are specific to dicots whereas CslF and CslH are found only in monocots (Fincher, 2009; Doblin et al., 2010), but recently two CslG genes were identified in Panicum virgatum (Pavirv00027268m and Pavirv00027269m; Yin et al., 2014).
Callose is a (1,3)-β-D-glucan, which is not present in cell walls but deposited in the walls of specialized tissues such as pollen mother cell walls, plasmodesmatal canals, and sieve plates in dormant phloem during normal growth and development (Stone and Clarke, 1992). In addition, callose is also deposited in response to environmental stimuli including abiotic stress, wounding, and pathogen challenge (Stone and Clarke, 1992; Muthamilarasan and Prasad, 2013). Callose is synthesized by callose synthases, which are encoded by glucan synthase-like (Gsl) genes (Saxena and Brown, 2000; Cui et al., 2001). To date, 12 Gsl genes have been identified in Arabidopsis, 13 in rice, 9 in poplar, and 8 in barley (Farrokhi et al., 2006).
In the case of lignin biosynthesis, phenylalanine is metabolized through the phenylpropanoid pathway to produce hydroxycinnamoyl-CoA esters, which enter the lignin branch of this pathway and are converted to monolignols. The process requires the involvement of phenylalanine ammonia lyase (PAL), trans-cinnamate 4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumaroyl shikimate 3′-hydroxylase (C3H), caffeoyl CoA 3-O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), cinnamoyl CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) (Bonawitz and Chapple, 2010; Zhong and Ye, 2015). Of these enzymes, PAL is the first enzyme of phenylpropanoid pathway which catalyzes the deamination of phenylalanine to generate cinnamic acid and C4H hydroxylates cinnamic acid to generate p-coumaric acid (Harakava, 2005). 4CL performs CoA esterification of p-coumaric acid and caffeic acid, whereas HCT catalyzes the conversion of p-coumaroyl-CoA and caffeoyl-CoA into corresponding shikimate or quinate esters and C3H converts these esters to corresponding caffeoyl esters. Following this, CCoAOMT catalyzes methylation of caffeoyl CoA to produce feruloyl CoA, whereas CCR converts hydroxycinnamoyl CoA esters to their corresponding aldehydes (Harakava, 2005). F5H has been assumed to catalyze the conversion of ferulic acid to 5-hydroxyferulic acid but recombinant DNA studies in Arabidopsis and Liquidambar styraciflua revealed that F5H converts coniferaldehyde and coniferyl alcohol to synapaldehyde and sinapyl alcohol, respectively (Humphreys et al., 1999; Osakabe et al., 1999). COMT is involved in the conversion of 5-hydroxyconiferaldehyde and/or 5-hydroxyconiferyl alcohol to sinapaldehyde and/or sinapyl alcohol, respectively (Osakabe et al., 1999; Parvathi et al., 2001), while CAD catalyzes the conversion of cinnamyl aldehydes into their corresponding alcohols (Harakava, 2005). The genes encoding these enzymes have recently been identified and characterized in several plant species (Raes et al., 2003; Vanholme et al., 2012; Shen et al., 2013; Carocha et al., 2015; van Parijs et al., 2015).
With the raise in the impacts of global climate change, reduction of greenhouse gases is essential, which could be facilitated through generating biorenewables. Importantly, production of lignocellulosic biofuels from secondary cell wall biomass has become a strategic research area, as it holds the potential to enhance energy security. C4 grasses, namely switchgrass (P. virgatum), napier grass (Pennisetum purpureum), pearl millet (P. glaucum), and foxtail millet (Setaria italica) have recently gained momentum in lignocellulosic biofuel research due to their high-efficiency CO2 fixation and efficient conversion of solar energy to biomass through C4 photosynthesis and photorespiration-suppressing modifications, respectively (Schmer et al., 2008; Byrt et al., 2011; van der Weijde et al., 2013). In addition, these grasses also possess better water use efficiency (WUE), higher nitrogen use efficiency (NUE), capacity to grow in arid and semi-arid regions and relatively high tolerance to environmental constraints including heat, drought, salinity and water-logging. For these reasons, C4 photosynthesis is an important trait for lignocellulosic biofuel crops (Byrt et al., 2011; van der Weijde et al., 2013).
Recently, foxtail millet (S. italica) and its wild progenitor, green foxtail (S. viridis) have been recognized as the suitable experimental models for biofuel research owing to their genetic relatedness to several biofuel grasses (Li and Brutnell, 2011; Zhang et al., 2012; Lata et al., 2013; Petti et al., 2013; Diao et al., 2014; Warnasooriya and Brutnell, 2014; Muthamilarasan and Prasad, 2015). The genomes of both foxtail millet and green foxtail have been sequenced (Bennetzen et al., 2012; Zhang et al., 2012), and the availability of foxtail millet draft genome sequence in public domains has facilitated various genetic and genomic studies in this model crop pertaining to stress response and crop improvement (Diao et al., 2014; Muthamilarasan and Prasad, 2015; Muthamilarasan et al., 2015) though no comprehensive genome-wide study on biofuel traits has been performed. Recently, Petti et al. (2013) has compared the lignocellulosic feedstock composition, cellulose biosynthesis inhibitor response, saccharification dynamics and CesA gene family of green foxtail with sorghum, maize and switchgrass. The study identified eight potential CesA gene family members for functional genomic characterization (Petti et al., 2013).
The present study has been performed to identify the gene families participating in lignocellulose biosynthesis using computational approaches. Further, qRT-PCR analysis of few genes has been performed to understand their expression patterns in response to different abiotic stress treatments.
Materials and methods
Identification of lignocellulose biosynthesis gene families
Protein sequences of enzymes involved in cellulose biosynthesis, namely CesA, Csl, and Gsl of rice and Arabidopsis were retrieved from cell wall genomics webserver (https://cellwall.genomics.purdue.edu/intro/index.html). The sequences for PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, COMT, CCR, and CAD reported in other crops (Appenzeller et al., 2004; Burton et al., 2004; Carocha et al., 2015; Zhong and Ye, 2015) were retrieved from respective literatures and HMM profile has been generated for individual families. Precisely, the sequences of respective families were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and HMM profiles were built using hmmbuilt command (http://hmmer.janelia.org/). HMMER tool was used to identify respective homologous proteins in foxtail millet protein dataset retrieved from Phytozome v10.2 (http://phytozome.jgi.doe.gov/) under default parameters (Muthamilarasan et al., 2014a). The protein sequences were confirmed using HMMSCAN (http://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) analysis, and respective genomic, transcript, and CDS sequences were downloaded from Phytozome by BLAST searching the retrieved protein sequences against S. italica database under default parameters (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Sitalica).
Protein properties and phylogenetic analysis
The properties of identified cell wall-related proteins including molecular weight, pI, and instability index were identified using ExPASy ProtPram tool (http://web.expasy.org/protparam/). The amino acid sequences of respective families were imported into MEGA v6 (Tamura et al., 2013) for multiple sequence alignment and phylogenetic tree construction using neighbor-joining method after bootstrap analysis for 1000 replicates (Muthamilarasan et al., 2014b). Sequence alignment and analysis was performed using BioEdit v7.2.5 (http://www.mbio.ncsu.edu/bioedit/bioedit.html).
Physical mapping and gene structure analysis
The chromosomal location of cell wall biosynthesis genes including chromosome number, position of gene start and end, gene length and orientation were obtained from Phytozome and a physical map was constructed using MapChart (Voorrips, 2002). Gene duplications, namely tandem and segmental were identified by performing MCScanx (Wang et al., 2012) according to the protocol of Plant Genome Duplication Database (Lee et al., 2012). Gene structure was predicted using Gene Structure Display Server v2.0 (http://gsds.cbi.pku.edu.cn/).
Promoter analysis, targeting miRNA, and marker prediction
The upstream genomic sequence (~2 kb) of lignocellulose pathway genes of foxtail millet were retrieved from Phytozome and the presence of cis-regulatory elements were identified by Signal Scan Search using New PLACE web server (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?page=analysis&lang=en). Mature miRNA sequences of foxtail millet were downloaded from miRBase v21 (Kozomara and Griffiths-Jones, 2014) and FmMiRNADb (Khan et al., 2014). This information along with the miRNA data of a dehydration stress library (Yadav et al., unpublished data) were used to identify the miRNAs targeting the transcripts of lignocellulose pathway genes using psRNAtarget server (Dai and Zhao, 2011) under default parameters. The large-scale genome-wide molecular markers namely simple sequence repeats (SSR; Pandey et al., 2013), expressed sequence tag (EST)-SSR (eSSR; Kumari et al., 2013), and intron-length polymorphic markers (Muthamilarasan et al., 2014c) were retrieved from the Foxtail millet Marker Database (http://www.nipgr.res.in/foxtail.html; Suresh et al., 2013) and searched for their presence in the genic and promoter regions of lignocellulose biosynthesis genes using in-house perl script.
Comparative genome mapping and evolutionary analysis
Protein sequences of lignocellulose pathway genes of foxtail millet were BLASTP searched against the protein sequences of switchgrass (Panicum virgatum), rice (Oryza sativa), and poplar (Populus trichocarpa), and hits with more than 80% identity were selected. The genomic and CDS sequences along with chromosomal locations for these proteins were retrieved by performing BLAST searches against the corresponding genomes retrieved from Gramene (http://www.gramene.org/) under default parameters and comparative maps were visualized using Circos (Krzywinski et al., 2009). Reciprocal BLAST was also performed to ensure the unique relationship between the homologous genes (Mishra et al., 2013). Estimation of nonsynonymous substitutions per non-synonymous site (Ka) and synonymous substitutions per synonymous site (Ks) for paralogous (tandem and segmentally duplicated genes) as well as homologous (comparative mapping data) gene pairs were calculated by codeml program in PAML using PAL2NAL (Suyama et al., 2006). The Ka/Ks ratios along with estimation of duplication and divergence (as T = Ks/2λ, where, λ = 6.5 × 10−9) were performed according to Puranik et al. (2013).
In silico expression profiling in tissues and drought stress
The transcriptome data of different tissues, namely root (SRX128223), stem (SRX128225), leaf (SRX128224), spica (SRX128226), and a drought stress library (SRR629694) as well as its control (SRR629695) were retrieved from European Nucleotide Archive (http://www.ebi.ac.uk/ena) (Zhang et al., 2012; Qi et al., 2013). The reads were filtered using NGS Toolkit (http://www.nipgr.res.in/ngsqctoolkit.html), mapped on foxtail millet genome using CLC Genomics Workbench v4.7.1, normalized by RPKM method and a heat map was generated using MultiExperiment Viewer (MeV) v4.9 (Saeed et al., 2003).
Plant materials, stress and hormone treatments and quantitative real-time PCR analysis
Seeds of foxtail millet cv. “IC-403579” (dehydration and salinity tolerant) were grown under optimum conditions following Lata et al. (2014). Twenty one day-old seedlings were exposed to 250 mM NaCl (salinity), 20% PEG6000 (dehydration), 4°C (cold), 100 mM abscisic acid (ABA), 100 mM methyl jasmonate (MeJA), and 100 mM salicylic acid (SA) treatments (Mishra et al., 2013; Puranik et al., 2013; Kumar et al., 2015) and whole seedlings were collected at 0 h (h) (control), 1 h (early), and 24 h (late) (Yadav et al., 2015). The samples were frozen immediately in liquid nitrogen and stored at −80°C. RNA isolation, cDNA synthesis and RT-PCR analysis were performed according to Puranik et al. (2013) in three technical replicates for each biological triplicate using the primers mentioned in Supplementary Table S1. All qRT-PCR data were the means of at least three independent experiments and the results were presented as the mean values ± SE. The significance of differences between mean values of control and each stressed samples were statistically performed using One-Way analysis of variance (ANOVA) and comparison among means was carried out through Tukey-Kramer multiple comparisons test using GRAPHPAD INSTAT software v3.10 (http://www.graphpad.com). The differences in the effects of stress treatments on various parameters in 16 foxtail millet genes under study were considered statistically significant at *P < 0.05, **P < 0.01, ***P < 0.001.
Results
CesA/Csl and Gsl superfamily of foxtail millet
HMM searches identified the presence of 14 CesA (SiCesA) and 39 Csl (SiCsl) proteins in foxtail millet (Supplementary Table S2). Among the 14 SiCesA proteins, one was found to be an alternate transcript (Si028766m), whereas in SiCsl, three alternate transcripts (Si029554m, Si035399m, and Si035101m) were identified. Domain analysis of SiCesA proteins revealed the presence of both the cellulose synthase domain (CS; PF03552) and the zinc finger structure (ZF; PF14569) in all the proteins except SiCesA8 and SiCesA10, which have only the CS domain (Supplementary Table S3). In addition, all the SiCesA proteins except SiCesA8 had Glycosyl transferase 2 (GT2; PF13632) domain. In the case of SiCsl proteins, 36 proteins (primary transcripts) were identified, of which 10 belonged to SiCslA, 6 to SiCslC, 5 to SiCslD, 4 to SiCslE, 7 to SiCslF, 2 each to SiCslH and SiCslJ families (Supplementary Table S2). Interestingly, two members of CslJ have been identified in foxtail millet, which was previously considered to be a cereal-specific gene family (Doblin et al., 2010). Domain analysis showed that all the SiCslA and SiCslC proteins possess GT2 domain (PF13641, PF13632, PF00535, and PF13506) (Supplementary Table S3).
All 5 SiCslD proteins possess CS (PF03552) and GT2 (PF13632) domain, and interestingly, SiCslD2, SiCslD4, and SiCslD5 were evidenced to have an additional RING/Ubox like zinc-binding domain (PF14570), whereas SiCslD3 has two CS domains (Supplementary Table S4). All the SiCslE proteins except SiCslE2 have more than one CS domain and SiCslE3 has an additional GT2 domain (PF13641). In the case of SiCslF proteins, all of the members except SiCslF6 have two CS domains and in addition, SiCslF1, SiCslF3, and SiCslF7 possess GT2 domain (PF13632). Two members each belonging to CslH and CslJ family proteins were identified and both the group members have two CS domains (Supplementary Table S4).
A total of 12 Gsl (SiGsl) proteins were identified in foxtail millet and all possessed glucan synthesis (GS) domain (1,3-beta-glucan synthase component; PF02364) (Supplementary Table S5). The number of GS domain within these proteins also varied as SiGsl1, SiGsl6, and SiGsl12 have two GS domains, whereas SiGsl11 had three domains. In addition, SiGsl2, SiGsl3, SiGsl5, SiGsl7, SiGsl8, SiGsl10, and SiGsl11 have a 1,3-beta-glucan synthase subunit FKS1, domain-1 (PF14288). Furthermore, SiGsl08, SiGsl10, and SiGsl11 have an additional Vta1 (VPS20-associated protein 1) like domain (PF04652) (Supplementary Table S5).
Monolignol pathway proteins of foxtail millet
HMM profiling of PAL (SiPAL), C4H (SiC4H), 4CL (Si4CL), HCT (SiHCT), C3H (SiC3H), CCoAOMT (SiCCoAOMT), F5H (SiF5H), COMT (SiCOMT), CCR (SiCCR), and CAD (SiCAD) proteins in foxtail millet identified 10, 3, 20, 2, 2, 6, 2, 4, 33, and 13 members, respectively (Supplementary Table S6). Splice variants were evidenced among these members, including three each in SiCL16 and SiCCR14, two in SiCCR11 and one each in Si4CL5, SiCCoAOMT1, SiCOMT, SiCCR11, and SiCCR17. HMMSCAN revealed a diverse domain organization of these proteins (Supplementary Table S7). All of the SiPAL proteins possess aromatic amino acid lyase (PF00221) domain, whereas Cytochrome P450 (PF00067) was present in all SiC4H, SiC3H, and SiF5H proteins. AMP-binding enzyme (PF00501) and AMP-binding enzyme C-terminal (PF13193) domains were present in all the Si4CL proteins except Si4CL13, which has only an AMP-binding enzyme domain. Both SiHCT1 and SiHCT2 have transferase family (PF02458) domains, and SiCCoAOMT proteins were evidenced to possess O-methyltransferase (PF01596) and methyltransferase (PF13578) domains with an exception of SiCCoAOMT, which has two O-methyltransferase domains (Supplementary Table S7). O-methyltransferase domain was also found to be present in SiCOMT proteins, whereas SiCOMT2 has an additional dimerisation domain (PF13578). A diverse domain composition was observed among SiCCR proteins in addition to the presence of signature NAD-dependent epimerase/dehydratase family (PF01370) and 3-beta hydroxysteroid dehydrogenase/isomerase family (PF01073) domains. Almost all the SiCCR proteins possess additional domains including GDP-mannose-4,6-dehydratase (PF16363), Male sterility protein (PF07993), NmrA-like family (PF05368), NAD(P)H-binding (PF13460), Polysaccharide biosynthesis protein (PF02719), and KR domains (PF08659). Of note, SiCCR7 was devoid of any of these domains except the NAD-dependent epimerase/dehydratase family domain, and SiCCR3 has an additional Alcohol dehydrogenase GroES-like domain (PF08240) (Supplementary Table S7). The presence of Alcohol dehydrogenase GroES-like and Zinc-binding dehydrogenase (PF00107) domains is the characteristic feature of SiCAD proteins and in addition to these, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding domain (PF02826) was present in SiCAD4, SiCAD9, and SiCAD12. Moreover, an alanine dehydrogenase/PNT, C-terminal domain (PF01262) was found to be present in SiCAD12 and SiCAD13 (Supplementary Table S7).
Properties of lignocellulose pathway proteins
Among the SiCesA proteins, SiCesA4 was the largest protein with 1095 amino acids (aa), followed by SiCesA2 (1092 aa), SiCesA11 (1090 aa) and SiCesA3 (1088 aa), and the smallest was SiCesA8 (884 aa) (Supplementary Table S2). The molecular weight of these proteins also varied accordingly, ranging from SiCesA8 (95.5 kDa) to SiCesA11 (123.2 kDa), with an isoelectric pH (pI) of 6.03 (SiCesA10) to 8.15 (SiCesA1). The protein instability index was between 36.07 (SiCesA11) to 50.62 (SiCesA8), which signified that all the SiCesA proteins except SiCesA2, SiCesA8, and SiCesA10 were stable. In the case of SiCsl proteins, the smallest protein was SiCslE2 with 144 aa and the largest was SiCslD1 (1217 aa), and their respective molecular weights ranged from 16.4 kDa (SiCslE2) to 132.2 kDa (SiCslD1). The pI of SiCsl proteins ranged from 4.61 (SiCslE2) to 9.32 (SiCslF7), and their instability index range (31.44–67.71) revealed that a maximum of SiCsl proteins (~33%) were stable. The size and molecular weights of SiGsl proteins ranged from 418 aa (47.8 kDa in SiGsl9) to 1956 aa (225.2 kDa in SiGsl8). Similarly, pI range of these proteins was between 8.61 (SiGsl12) and 9.69 (SiGsl9). The instability index range between 28.89 and 52.08 indicated that ~46% of SiGsl proteins were stable and the rest are unstable (Supplementary Table S2).
The SiPAL class of monolignol pathway proteins showed a narrow range of protein properties, as their sizes varied from 699 (SiPAL1 and SiPAL2) to 851 aa (SiPAL10), with molecular weights from 74.9 kDa (SiPAL2) to 91.1 kDa (SiPAL10) (Supplementary Table S6). The pI range of SiPAL was between 5.82 and 6.52, and their instability index range (28.82–39.84) showed that all the proteins except SiPAL5 were stable. The three members of SiC4H, namely SiC4H1, SiC4H2, and SiC4H3 had molecular sizes of 530 aa (59.7 kDa), 430 aa (49.3 kDa), and 506 aa (57.9 kDa), respectively. Their respective pI were 9.26, 7.72, and 9.33, and their instability index (46.46, 49.84, and 48.61) revealed that SiC4H proteins were stable. Among the Si4CL proteins, Si4CL4 and Si4CL10 were the smallest proteins with 198 aa (21.8 and 21.7 kDa in size, respectively) and the largest was Si4CL9 (642 aa; 68.5 kDa). Their pI range was between 5.14 and 8.98. The protein instability index ranged from 24.76 (Si4CL3) to 47.96 (Si4CL6) hinting that all the Si4CL proteins except Si4CL3 were stable. SiHCT, SiC3H, and SiF5H proteins have two members each, with a narrow range of protein properties, and all these proteins were found to be stable as indicated by their stability index. A significant difference was observed with the sizes of SiF5H members since SiF5H1 was 158 aa (16.7 kDa) and SiF5H2 was 524 aa (57.7 kDa) (Supplementary Table S6). Among SiCCoAOMT proteins, the smallest protein was SiCCoAOMT1 with 243 aa (25.7 kDa) and the largest was SiCCoAOMT5 with 307 aa (33.4 kDa). The pI range was between 5.04 and 8.94, and the protein instability index range (27.69–51.49) showed that except SiCCoAOMT4, all others were stable. The three-member SiCOMT class proteins have molecular sizes of 247 aa (25.8 kDa; SiCOMT1), 402 aa (43.53 kDa; SiCOMT2), and 153 aa (16.71 kDa; SiCOMT3). The pI values were 5.09, 5.97, and 9 for SiCOMT1, SiCOMT2, and SiCOMT3, respectively. The instability index range (42.24–52.75) hinted that all SiCOMT proteins are stable. Among the monolignol pathway proteins, SiCCR class has the highest number (26 members) and their sizes ranged from 27.2 kDa (251 aa; SiCCR26) to 69.13 kDa (625 aa; SiCCR9), with a pI range of 4.72 (SiCCR23) to 9.32 (SiCCR19). The protein instability index ranged from 24.86 (SiCCR18) to 54.11 (SiCCR13), which points out that ~77% of SiCCR proteins were stable. In the case of SiCAD proteins, SiCAD9 and SiCAD13 were the smallest proteins with 336 aa (35.6 and 36.4 kDa in size, respectively) and SiCAD8 was the largest with 495 aa (52.7 kDa). The pI ranged from 5.05 to 9.24, and the instability index (19.35–39.79) showed that ~50% of SiCAD proteins are unstable (Supplementary Table S6).
Sequence alignment and phylogenetic analysis of CesA/Csl and Gsl proteins
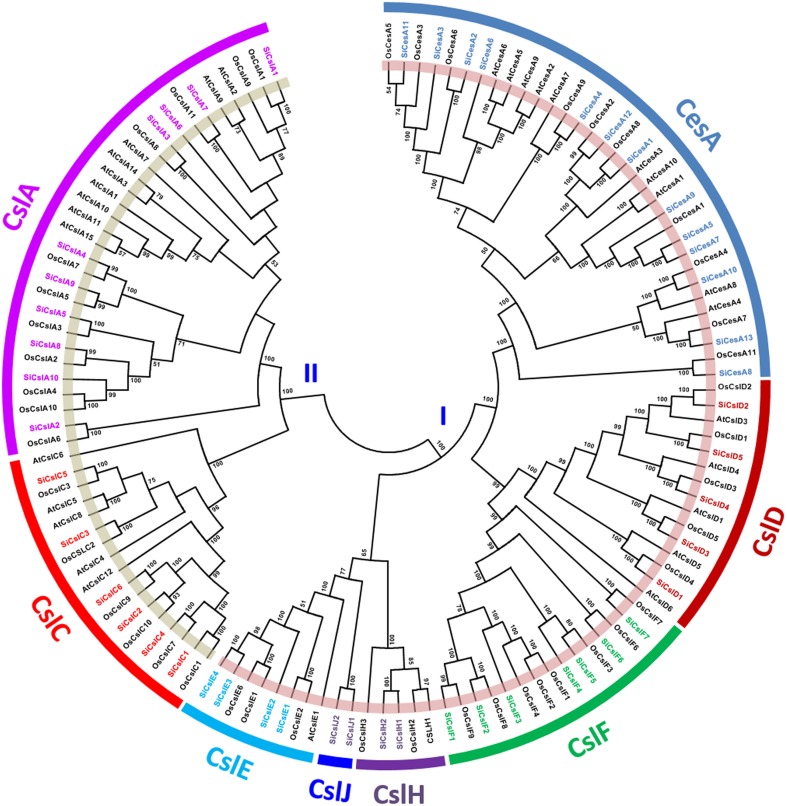
SiCesA and SiCsl proteins were aligned individually, and the alignment revealed the presence of conserved “DXD, D, QXXRW” motif in both the superfamilies. All the SiCesA proteins except SiCesA8 have a “DCD, D, QVLRW” consensus sequence, whereas SiCesA8 had a unique “DYD, D” sequence and the motif “QXXRW” was absent (Supplementary Figure S1). Noteworthy, SiCesA8 protein has only the CS domain, while the other SiCesA proteins possess CS, ZF, and GT2 domains (Supplementary Table S3). In the case of SiCsl proteins, the “DXD” motif is absent in all the members of SiCslA, SiCslC and SiCslE2 (Supplementary Figure S2). This motif was predominantly “DCD,” except in SiCslF1 and SiCslF2, which have “DGD.” The second consensus “D” amino acid is present in all the SiCsl members (as “ED”), except SiCslA6, SiCslE2, and SiCslF4 (Supplementary Figure S2). In addition, SiCslA6 and SiCslE2 did not possess the “QXXRW” motif also, whereas a subgroup-wise conservation was evidenced in this motif in rest of the members. The majority of SiCslA (7) and all the SiCslC members have “QQHRW” motif, whereas SiCslE proteins have “QHKRW,” SiCslH and SiCslJ proteins have “QYKRW” and “QNKRW” motifs, respectively (Supplementary Figure S2). The unrooted phylogenetic tree constructed using the amino acid sequences of SiCesA/Csl proteins along with CesA/Csl proteins of rice and Arabidopsis (https://cellwall.genomics.purdue.edu/intro/index.html) showed 2 distinct clusters, namely I and II (Figure 1). Cluster I was resolved into six branches including CesA, CslD, CslE, CslF, CslH, and CslJ, whereas cluster II had two branches, CslA and CslC.
Figure 1.
Unrooted protein phylogenetic tree constructed with CesA/Csl proteins of Setaria italica (Si), Oryza sativa (Os), and Arabidopsis thaliana (At).
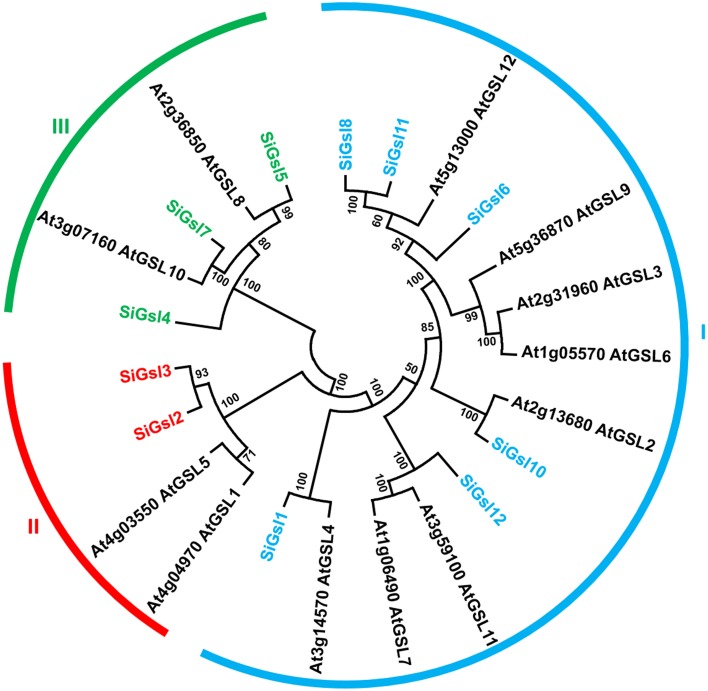
Sequence alignment of SiGsl proteins showed that the N-terminal region of all these proteins was diverse, whereas the C-terminal region was conserved (Supplementary Figure S3). Prediction of transmembrane (TM) helices in these proteins using TMHMM Server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) showed the presence of 7–16 TM helices in SiGsl proteins (Supplementary Figure S4). Phylogeny of foxtail millet, rice and Arabidopsis Gsl proteins showed three clusters (Figure 2). Cluster I included SiGsl4, SiGsl5, and SiGsl7, whereas cluster II comprised SiGsl2 and SiGsl3. SiGsl1, SiGsl6, SiGsl8, SiGsl10, SiGsl11, and SiGsl12 were included in cluster III.
Figure 2.
Unrooted protein phylogenetic tree constructed with Gsl proteins of Setaria italica (Si) and Arabidopsis thaliana (At).
Sequence alignment and phylogenetic analysis of monolignol biosynthesis pathway proteins
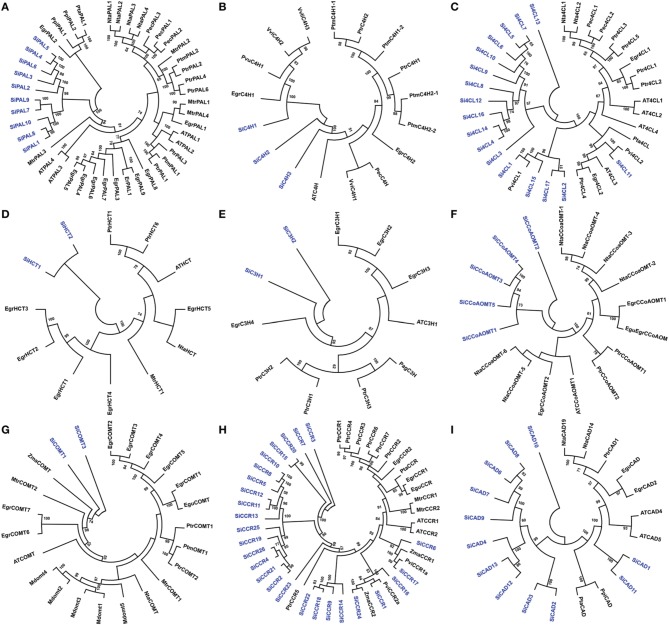
Sequence alignment and analysis of SiPAL proteins showed that all the members are almost completely conserved (Supplementary Figure S5). SiPAL2 was found to possess an extended N-terminal sequence of about 135 amino acids, which is unique to this class of protein. A phylogenetic tree constructed with PAL sequences of foxtail millet, eucalyptus, poplar, tobacco, medicago and Arabidopsis showed that the SiPAL proteins are phylogenetically divergent from the rest (Figure 3A). Sequence alignment of SiC4H showed that all the members share the conserved P450 superfamily domain and P450-featured motifs, namely, haem-iron binding motif (PFGVGRRSCPG), the T-containing binding pocket motif (AAIETT, the E-R-R-E-R-E-R), for optimal orientation of the enzyme (Supplementary Figure S5). Further, presence of conserved substrate recognition sites (SRSs) of C4H/CYP73A5 enzymes, including SRS1 (SRTRNVVFDIFTGKGQDMVFTVY), SRS2 (LSQSFEYNY), SRS4 (IVENINVAAIETTLWS), and SRS5 (RMAIPLLVPH) was also evidenced (Supplementary Figure S5). Phylogeny of SiC4H along with C4H protein sequences of other organisms showed the grouping of SiC4H1 with C4H1 proteins of eucalyptus and Phaseolus vulgaris, whereas SiC4H2 and SiC4H3 were found to be more divergent (Figure 3B).
Figure 3.
Unrooted protein phylogenetic trees constructed with (A) PAL, (B) C4H, (C) 4CL, (D) HCT, (E) C3H, (F) CCoAOMT, (G) COMT, (H) CCR, and (I) CAD proteins of Setaria italica (Si), Eucalyptus gunnii (Egu), E. grandis (Egr), Nicotiana tabacum (Nta), Populus trichocarpa (Ptr), Pinus pinaster (Ppi), Pinus taeda (Pta), Medicago truncatula (Mtr), Panicum virgatum (Pvi), Zea mays (Zma), Malus domestica (Mdom), Vitis vinifera (Vvi), Eucalyptus globulus (Egl), Populus alba x Populus grandidentata (Pag), Petroselinum crispum (Pec), Populus tremuloides (Ptm), Phaseolus vulgaris (Pvu), and Eucalyptus robusta (Er).
Si4CL protein sequence alignment showed the presence of 2 highly conserved peptide motifs “box I” (LPYSSGTTGLPKGV; AMP binding signature) and “box II” (GEICIRG), in addition to other conserved regions (Supplementary Figure S5). Phylogeny of 4CL proteins showed grouping of Si4CL1, Si4CL2, Si4CL15, and Si4CL16 with switchgrass (Pvi4CL1), demonstrating their close proximity and similarly, Si4CL11 was found to be grouped with Pvi4CL2, whereas other Si4CL proteins formed their own distinct cluster (Figure 3C). Alignment of SiHCT sequences showed that all the proteins have the conserved motifs for the acyl transferase family, namely “HXXXDG” and “DFGWG” (Supplementary Figure S5). Multiple sequence alignment of SiC3H proteins showed the presence of Cytochrome P450 cysteine heme-iron ligand signature [FW]-[SGNH]-x-[GD]-{F}-[RKHPT]-{P}-C-[LIVMFAP]-[GAD] (Supplementary Figure S5). The conserved motifs including three putative S-adenosyl-L-methionine binding motifs (A, B, and C) and CCoAOMT signature motifs (D, E, F, G, and H) were identified through multiple sequence alignment of SiCCoAOMT proteins (Supplementary Figure S5). Phylogenetic analysis of SiHCT, SiC3H, and SiCCoAOMT proteins with their respective family members of other organisms revealed the dissimilarity of foxtail millet proteins compared to their homologs (Figures 3D–F). In the case of CCoAOMT, SiCCoAOMT2 formed a distinct clade, whereas other SiCCoAOMT members were grouped together in one clade (Figure 3F).
Being truncated proteins, alignment of SiF5H1 with SiF5H2, and SiCOMT2 with SiCOMT1 and SiCOMT3 were not performed (Supplementary Figure S5). Protein sequence alignment between SiCOMT1 and SiCOMT3 did not highlight any consensus motif and their phylogenetic analysis with COMT proteins of other plants showed grouping of SiCOMT with ZmaCOMT of maize (Figure 3G). Sequence alignment of SiCCR proteins revealed that the conserved “KNWYCYGK” motif, catalytic site or the binding site for the cofactor NADPH (Larsen, 2004) has been diversified in foxtail millet (Supplementary Figure S5). Except SiCCR1 and SiCCR24, other SiCCR proteins have at least one amino acid change in this motif, which could be attributed to the substrate affinity of CCR proteins (Pichon et al., 1998). Phylogenetic analysis of SiCCR proteins showed that a maximum of these proteins were clustered in a separate group, whereas few proteins were grouped with CCR proteins of maize, switchgrass and poplar (Figure 3H). Alignment results of SiCAD highlighted a high degree of similarity in conserved domains and binding residues, including Zn-1 binding domain motif GHE(X)2G(X)5G(X)2V, NADP(H) co-substrate-binding motif GXG(X)2G (glycine-rich repeat) and Zn-2 metal ion binding motif GD(X)9, 10C(X)2C(X)2C(X)7C (Supplementary Figure S5). Phylogenetic tree of SiCAD with CAD proteins of other plant species showed clustering of a maximum of SiCAD proteins in one clade with complete out-grouping of SiCAD10. SiCAD1 and SiCAD11 were found to cluster with poplar CAD proteins (Figure 3I).
Gene structure of lignocellulose pathway genes
The sequence data of genomic DNA, transcript and CDS along with chromosomal locations of confirmed protein sequences of identified lignocellulose biosynthesis pathway enzymes were retrieved and analyzed for gene size, intron-exon and physical position (Supplementary Tables S2, S6). The size of SiCesA genes ranged from 3.1 (SiCesA8) to 6.9 kb (SiCesA9) and few genes including SiCesA3, SiCesA7, SiCesA5, and SiCesA9 have a maximum of 13 introns, whereas SiCesA12 was intronless (Supplementary Figure S6). The gene sizes of SiCsl ranged from 1.7 (SiCslA6 and SiCslE2) to 6.6 kb (SiCslA1 and SiCslF6), and their gene structure analysis revealed that SiCsl genes have up to eight introns (Supplementary Figure S7). The only intronless gene of SiCsl superfamily was SiCslE2. Among the SiGsl gene family members, SiGsl3 was the smallest gene (3.2 kb), whereas the largest one was SiGsl4 (17 kb). Interestingly, SiGsl genes were evidenced to contain numerous introns. SiGsl7 has a maximum of 49 introns, whereas SiGsl2 and SiGsl3 were intronless (Supplementary Figure S8).
SiPAL gene sizes ranged from 2.1 (SiPAL4) to 4.6 kb (SiPAL3), of which SiPAL4, SiPAL5, and SiPAL6 were intronless, SiPAL2 has two introns and other SiPAL genes have 2 introns each (Supplementary Figure S9). Among the Si4CL genes, Si4CL3 was the smallest gene (2 kb), whereas Si4CL15 was the largest (6.7 kb). A total of 10 Si4CL genes have 5 introns each, while maximum number of introns was found in Si4CL5 (6 introns). Si4CL3 has the least number of one intron in its gene (Supplementary Figure S9). The size of SiCCoAOMT genes ranged from 0.8 (SiCCoAOMT4) to 3 kb (SiCCoAOMT2) with a maximum number of introns (7) in SiCCoAOMT2. SiCCoAOMT3 and SiCCoAOMT4 have one intron each (Supplementary Figure S9). Among the SiCCR genes, SiCCR3 was 1.3 kb in size and though it is the smallest gene of this class, it has eight introns. SiCCR9 and SiCCR22 are the largest genes with a size of 5.8 kb and both the genes have 4 introns each. SiCCR2 has a maximum of 10 introns, while SiCCR7 is the only intronless gene in this group. The size of SiCAD genes ranged from 1.4 (SiCAD9) to 4.2 kb (SiCAD1 and SiCAD8), with SiCAD7, SiCAD8, and SiCAD9 having a minimum of 2 introns each whereas SiCAD5 has a maximum of 6 introns (Supplementary Figure S9).
Chromosomal location and gene duplication of lignocellulose pathway genes
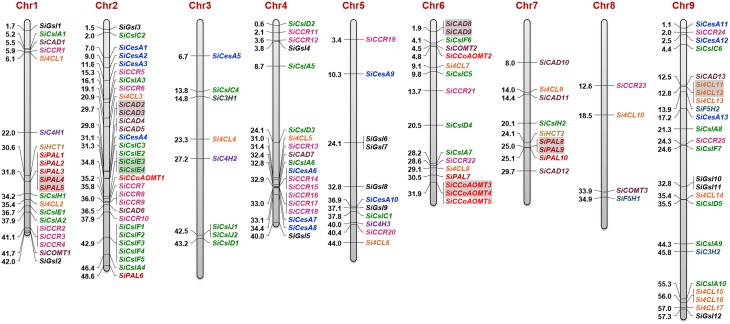
The identified secondary cell wall biosynthesis genes were plotted onto the nine chromosomes of foxtail millet to generate the physical map (Figure 4), which showed that the majority of lignocellulose biosynthesis pathway genes (31; ~22%) were present in chromosome 2, followed by chromosome 9 (24 genes; ~17%) and chromosome 1 (21 genes; ~15%), and a minimum of 4 genes (~3%) were mapped on chromosome 8. Expansion of respective gene families within the genome were analyzed by investigating tandem and segmental duplication, which showed that 7 genes underwent tandem duplication, whereas segmental duplication did not occur among the lignocellulose pathway genes (Figure 4). SiCesA members were distributed on chromosomes 2 (4 genes), 4 (1), 5 (2), and 9 (3) and none of the genes were evidenced to undergo tandem or segmental duplication. SiCsl genes were found to be present in all the chromosomes except chromosome 8, and duplication analysis revealed that SiCslE3 and SiCslE4 were tandemly duplicated gene pairs on chromosome 2. SiGsl members were distributed on chromosomes 1 (2 genes), 2 (1), 4 (2), 5 (4), and 9 (3) and no duplication pattern in this gene family was observed.
Figure 4.
Physical map showing the chromosomal locations of lignocellulose biosynthesis genes. Bars represent chromosomes and the numbers at the left corresponds to location (in Mb). Gene IDs are provided in the right. Tandemly duplicated gene pairs are highlighted with gray shade.
Among the monolignol biosynthesis genes, the majority of SiPAL genes were present in chromosome 1 (5) and 7 (3), and interestingly, SiPAL4 and SiPAL5 as well as SiPAL8 and SiPAL9 were identified to be tandem duplicates. Each of the three SiC4H genes were found in chromosome 1, 3, and 5 (Figure 4). A higher number of Si4CL genes were present in chromosome 9 (7 genes), of which Si4CL11 and Si4CL12 were tandemly duplicated gene pairs. Chromosome 1 and 6 have two Si4CL members each and one member each in chromosome 2, 3, 4, 5, 7, and 8. Two members of SiHCT, SiC3H, and SiF5H as well as three genes of SiCOMT were present in chromosome 1, 3, 6, 7, 8, and 9 (Figure 4). Four out of five SiCCoAOMT genes were present in chromosome 6 and SiCCoAOMT1 was mapped on chromosome 2, and duplication analysis revealed that SiCCoAOMT3 and SiCCoAOMT4 were tandemly duplicated gene-pairs. Among the SiCCR genes, SiCCR26 could not be mapped due to non-availability of its co-ordinates in Phytozome database. Of the 25 SiCCR genes mapped, a maximum of 8 genes were found to be present in chromosome 4 (8), followed by chromosome 2 (6) and 1 (4). Of the 13 SiCAD genes, maximum was in chromosome 2 (5) and a minimum of one each in chromosomes 1, 4, and 9. SiCAD2 and SiCAD3 on chromosome 2 as well as SiCAD8 and SiCAD9 on chromosome 6 were found to be tandemly duplicated gene-pairs (Figure 4).
Promoter analysis on lignocellulose pathway genes
In silico analysis for predicting putative cis-regulatory elements showed the presence of universal as well as gene-specific promoter sequences in the upstream of lignocellulose pathway genes (Supplementary Tables S8, S9). A total of 271 cis-elements were found in CesA/Csl and Gsl genes, of which 15 (5.5%) elements, namely ACGTATERD1, ARR1AT, CAATBOX1, CACTFTPPCA1, DOFCOREZM, EBOXBNNAPA, GATABOX, GT1CONSENSUS, GTGANTG10, MYCCONSENSUSAT, NODCON2GM, OSE2ROOTNODULE, POLLEN1LELAT52, WBOXNTERF3, and WRKY71OS were present in all these genes (Supplementary Table S8). Thirty-nine unique cis-elements (~14%) which were present in any one gene of CesA/Csl and Gsl superfamilies were also found, such as ABADESI1 (SiCslF6), CEREGLUBOX3PSLEGA (SiCesA2), GBOXLERBCS (SiCslA8), ZDNAFORMINGATCAB1 (SiCslA6), TATCCACHVAL21 (SiGsl3), etc. In addition, few promoter sequences were found to be present in all the genes except one or two genes and this includes BIHD1OS (SiCslC4), CCAATBOX1 (SiCslA1, SiCslF4), CURECORECR (SiCslC3, SiGsl1), DPBFCOREDCDC3 (SiCslC2, SiCslC4), EECCRCAH1 (SiGsl5), MYBCORE (SiCesA3, SiCslD4), RAV1AAT (SiCslD1), and SORLIP1AT (SiCesA4, SiGsl8). Of note, no superfamily specific regulatory elements were identified (Supplementary Table S8).
A total of 293 cis-elements were detected in the upstream region of monolignol pathway genes, of which 10 (3.4%) were present in all the genes and 37 (~13%) were unique to any one gene (Supplementary Table S9). The elements which were present in all the genes include ARR1AT, CAATBOX1, CACTFTPPCA1, DOFCOREZM, EBOXBNNAPA, GATABOX, GT1CONSENSUS, GTGANTG10, WBOXNTERF3, and WRKY71OS. Few cis-regulatory elements were found to be present in all except one or two genes and it includes ACGTATERD1 (SiPAL2), CURECORECR (SiPAL2, SiPAL10), and MYBCORE (SiPAL7, SiCCR16). Similar to CesA/Csl and Gsl, no monolignol genes have superfamily specific regulatory elements (Supplementary Table S9).
MicroRNAs and molecular markers of lignocellulose pathway genes
In silico scanning of lignocellulose pathway gene transcripts to identify their targeting miRNAs showed that the transcripts of SiCslC2, SiGsl10, and SiF5H2 could be targeted by the miRNAs sit-miRn29, sit-miR114-npr and sit-miR395b, respectively (Supplementary Table S10). SiGsl3 was predicted to be targeted by two foxtail millet miRNAs, namely sit-miR156d-1 and sit-miR156d-2. These miRNAs would have a putative role in post-transcriptional gene silencing for regulation of lignocellulose pathway gene expression. Identification of previously reported molecular markers in the genic and regulatory regions of lignocellulose pathway genes revealed the presence of SSR and ILP markers in 34 genes (Supplementary Table S11). Of these, three genes have two and three markers each, and the remaining 28 genes possess single markers. Among the markers, SSRs were found to be predominant (~81%) and the rest are ILPs (~19%).
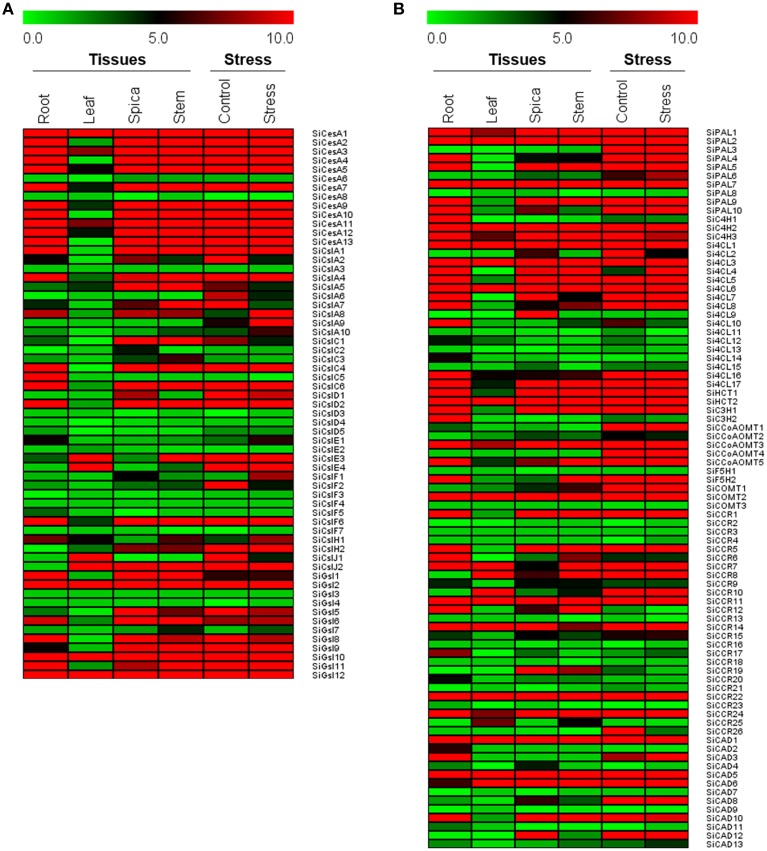
Expression profile of lignocellulose pathway genes in tissues and dehydration stress
Expression of all the genes in four tissues and dehydration stress was calculated using RPKM values derived from RNA-seq data. Tissue-specific expression profile showed differential expression pattern of all the genes with relatively lower expression in leaf (Figure 5). In the case of CesA/Csl and Gsl superfamilies, higher expression of SiCesA1, SiGsl2, SiGsl10, and SiGsl12 was evidenced in all the four tissues when compared to the other members of the same gene family. Tissue-specific higher expression of SiCslD1 in spica, and SiCslE4 and SiCslJ2 in leaf was also observed. Many genes including SiCesA6, SiCesA8, SiCslA3, SiCslC3, SiGsl3, and SiGsl7 were not expressed in these tissues (Figure 5A). Tissue-specific expression profiling of monolignol genes showed higher expression of SiPAL1, SiPAL2, SiPAL7, SiC4H2, Si4CL1, Si4CL3, Si4CL6, SiHCT2, SiCOMT2, SiCCR11, SiCAD1, and SiCAD5 in all the four studied tissues. Tissue-specific higher expression was evidenced with SIPAL4, Si4CL10, and SiCAD3 in root, and Si4CL9 and SiCAD12 in spica. Similar to CesA/Csl and Gsl, monolignol genes also showed a relatively lower expression in leaf tissue (Figure 5B). Expression profiling of all the genes in response to dehydration stress showed almost a uniform expression in both control and stress samples (Figure 5). Comparison of expression patterns between tissues and stress library revealed that the expression of predominant lignocellulose pathway genes was unaltered. Only three genes, namely SiCslA8, SiCslA9, and Si4CL4 showed a higher expression in dehydration stress library compared to control, of which SiCslA8 and SiCslA9 were expressed only during stress and not in any of the tissue-specific RNA-Seq libraries. Few genes which were highly expressed in control were observed to be down regulated during stress and this includes SiCslA5, SiCslA6, SiCslA7, SiCslF2, and SiCCR26 (Figure 5).
Figure 5.
Heat map showing the expression of (A) cellulose biosynthesis genes, and (B) monolignol biosynthesis genes in four different tissues and dehydration stress library. The Illumina RNA-seq data were re-analyzed and the heat map was generated. Bar at the top with the values 0.0, 5.0, and 10.0 represent low, intermediate and high expression, respectively.
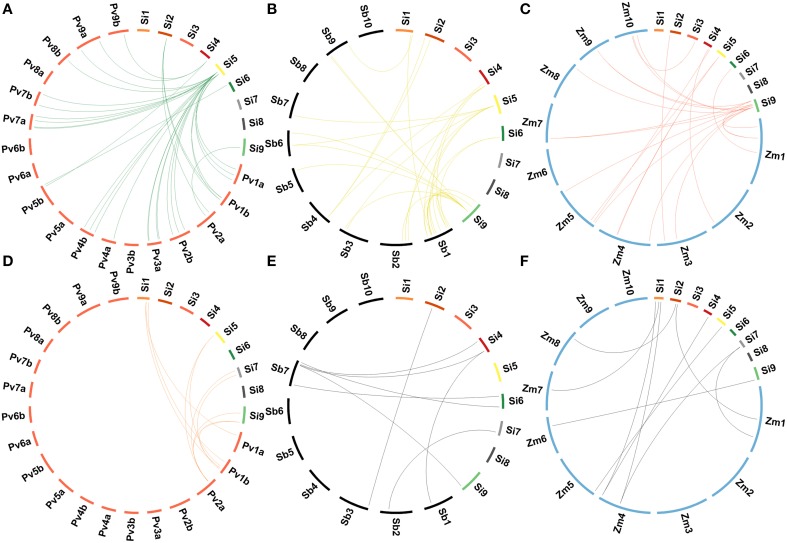
Homologous relationships of lignocellulose pathway genes with other grasses
Homologs of foxtail millet lignocellulose pathway genes in sequenced C4 panicoid genomes, namely switchgrass (Panicum virgatum), sorghum (Sorghum bicolor), and maize (Zea mays) were derived (Figure 6). A maximum lignocellulose pathway gene-based homology was observed between foxtail millet and switchgrass as 19 genes of foxtail millet showed homology with 60 genes of switchgrass (Supplementary Table S12). Of the 19 foxtail millet genes, six belonged to SiGsl, four to SiCCR, three each to SiCsl and SiPAL, and one each to SiHCT, Si4CL and SiCAD. Eighteen foxtail millet genes showed orthologous relationship with 41 sorghum genes, of which SiGsl11 had a maximum of 11 homologs, followed by SiGsl7 (7 homologs) and SiGsl5 and SiCCR17 (3 homologs each) (Supplementary Table S13). In the case of foxtail millet-maize homology, 26 foxtail millet genes showed homologous relationship with 38 maize genes (Supplementary Table S14). Among the foxtail millet genes, SiGsl had a maximum of 7 homologs in maize, followed by SiGsl7 (3 homologs).
Figure 6.
Comparative genome map showing homologous relationships between CesA/Csl and Gsl superfamilies of Setaria italica and (A) Panicum virgatum, (B) Sorghum bicolor, (C) Zea mays, and between monolignol biosynthesis genes of Setaria italica and (D) Panicum virgatum, (E) Sorghum bicolor, (F) Zea mays.
Among the lignocellulose pathway proteins, CADs and COMTs were well characterized as they play key role in secondary cell wall lignification (Saballos et al., 2009; Saathoff et al., 2011a,b, 2012; Sattler et al., 2012; Trabucco et al., 2013). Sequence analysis of these proteins in several grasses identified the presence of conserved motifs in few members, which distinguish them as lignifying proteins from the rest of non-lignifying proteins. Lignifying CADs possess additional 12 amino acids T49, Q53, L58, M60, C95, W119, V276, P286, M289, L290, F299, and I300, which are involved in substrate recognition and binding (Youn et al., 2006). Of the 13 SiCAD proteins, SiCAD11 contains 11 of 12 conserved amino acid residues. Of note, the active substrate-binding residues, W119 and F298, which determine specificity for aromatic alcohols and, the NADP(H) binding site, S212, were present in SiCAD11. Sequence-based homology analysis showed higher percentage of identity between SiCAD11 and lignifying CADs of other grasses namely switchgrass (Pavir.J34526; 91%), sorghum (Sobic.006G211900; 89%) and maize (GRMZM5G844562; 85%). Similarly, the conserved amino acids M130, N131, L136, A162, H166, F176, M180, H183, I319, M320, and N324, which function in substrate-binding and positioning in COMTs (Sattler et al., 2012; Trabucco et al., 2013) are found to be present in SiCOMT02 of foxtail millet. Sequence-based homology with SiCOMT02 showed high percent identity to sorghum (Sobic.007G047300; 94%), switchgrass (Pavir.Fa01907; 85%), and maize (AC196475.3; 89%).
Duplication and divergence of lignocellulose pathway genes
The number of non-synonymous substitutions per non-synonymous site (Ka) and synonymous substitutions per synonymous site (Ks) was calculated for paralogous as well as homologous gene pairs and Ka/Ks ratio along with time of divergence (in million years ago; mya) were derived. The ratio of Ka to Ks for tandemly duplicated gene-pairs ranged from 0.09 to 0.18 with an average value of 0.13, which suggested that these genes were under strong positive purifying selection (Ka/Ks > 1) and the duplication event was predicted to occur around 25 mya (Supplementary Table S15). In the case of Ka/Ks ratio of homologous gene-pairs, it was maximum between foxtail millet-switchgrass (average Ka/Ks = 0.91; Supplementary Table S12), whereas foxtail millet-sorghum and foxtail millet-maize homologs showed an average ratio of 0.19 (Supplementary Tables S13, S14). Since these values were less than 1, it signifies the intense positive selective pressure acted on respective protein-coding genes. The time of divergence between foxtail millet and switchgrass was predicted to occur around 4.7 mya, whereas the divergence of foxtail millet-sorghum and foxtail millet-maize occurred around 27 mya. This demonstrates that duplication and divergence have played a key role in shaping the lignocellulose pathway multigene families in foxtail millet and other C4 grass genomes.
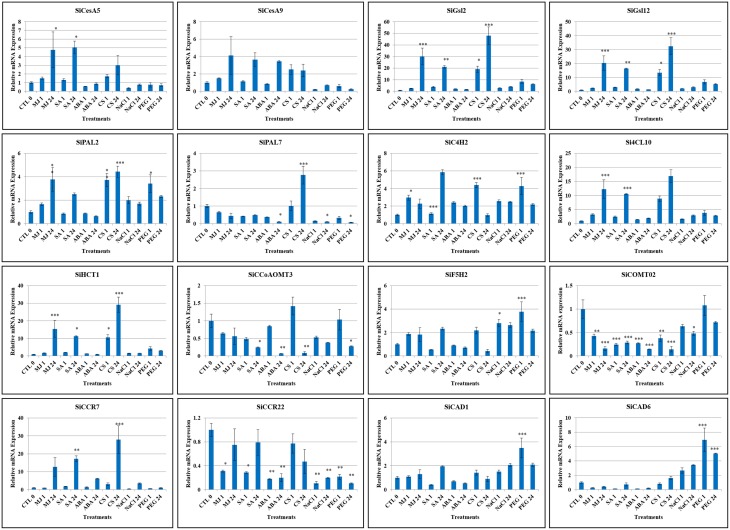
Expression profile of candidate genes during stress and hormone treatments
Expression patterns of sixteen candidate lignocellulose biosynthesis genes, namely SiCesA5, SiCesA9, SiGsl2, SiGsl12, Si4CL10, SiPAL2, SiPAL7, SiC4H2, SiHCT1, SiCCoAOMT3, SiF5H2, SiCOMT2, SiCCR7, SiCCR22, SiCAD1, and SiCAD6 in response to stress (dehydration, salinity, cold) and hormone (abscisic acid, salicylic acid, methyl jasmonate) treatments was performed at two time points (1 h, early; 24 h, late). These candidates were chosen based on; (i) expression profiles deduced in silico using RNA-seq data, (ii) representing the nine chromosomes of foxtail millet, and (iii) their function in secondary cell wall formation such as SiCOMT2 in lignification Overall, the study demonstrated differential expression pattern of these genes during stress and hormone treatments except SiCCR22 which was found to be down-regulated under all conditions (Figure 7). SiGsl2 and SiGsl12 were found to be highly expressed during all the three stress conditions, whereas SiCAD6 was up-regulated during both salinity and dehydration stress. Dehydration stress has been observed to induce the expression of all the genes except SiCCoAOMT3, SiCOMT2, SiCesA5, SiCCR22, SiPAL7, SiCCR7, and SiCesA9, though the degree of expression varied between the genes. Salinity stress showed an induction in expression of SiC4H2, SiCAD6, SiF5H2, SiGsl12, and SiGsl2, while SiPAL2 was induced during early salt stress and SiCAD01, Si4CL10, and SiCCR7 were found to be up-regulated in late phase salinity stress, thus suggesting a significant higher expression among the members of SiGsl and SiCAD family. Significant up-regulation of SiGsl2, SiGsl12, Si4CL10, SiHCT1, and SiCCR7 during cold stress suggests the putative involvement of these genes in strengthening the cell wall for tolerance to low temperature. Higher expression of these genes was also found during both early and late phases of treatment with salicylic acid and methyl jasmonate. Differential expression of candidate genes was observed during the treatment of all the hormones except abscisic acid, which showed no effect on the expression of majority of candidate genes except SiGsl2, which was induced at early phase of ABA treatment, SiCCR7 and SiCes9, which were induced at late phase of ABA treatment, and SiC4H2, which was induced at both the phases of ABA treatment. In addition, expression of SiCCoAOMT3, SiCOMT2, SiCCR22, SiPAL7, SiCAD1, and SiCAD6 was found to be down-regulated during hormone treatments, while SiF5H2 was up-regulated only under late phase of salicylic acid treatment.
Figure 7.
Relative expression of candidate lignocellulose biosynthesis genes analyzed using qRT-PCR under dehydration (PEG), salinity (NaCl) and cold stress (CS) as well as abscisic acid (ABA), salicylic acid (SA) and methyl jasmonate (MJ) treatments for 0 (Control: CTL), 1 and 24 h. Act2 was used as an internal control to normalize the data. The error bars representing standard deviation were calculated based on three technical replicates for biological triplicates. Statistical analysis between treatment and control using Tukey-Kramer multiple comparisons test has been performed and the differences in the effects of stress treatments in all the genes were considered statistically significant at *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Cellulose, hemicelluloses and lignin constitute the complex polymeric structure of secondary cell wall and the lignocellulose biosynthesis pathway involves the action of cellulose synthase (CesA), cellulose synthase-like (Csl), glucan synthase-like (Gsl), phenylalanine ammonia lyase (PAL), trans-cinnamate 4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumaroyl shikimate 3′-hydroxylase (C3H), caffeoyl CoA 3-O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), cinnamoyl CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) genes, which are well studied in several crop plants as well as trees for understanding and improving biofuel traits (Zhong and Ye, 2015). In the present study, all these gene families in foxtail millet were systematically identified and characterized using in silico approaches, and expression profiling of chosen genes was performed in response to several stress as well as hormonal treatments for identifying target genes for functional characterization.
A total of 13 CesA and 36 Csl genes were identified in foxtail millet, and all the SiCesA proteins were found to possess the characteristic cellulose synthase (CS) domain and 12 SiCesA had an additional zinc finger (ZF) structure. Similarly, 11 CesA proteins have been reported in rice, of which 9 contained both CS and ZF domain, and 2 lacked ZF domain (Wang et al., 2010). Role of CesA proteins in cellulose biosynthesis in both primary and secondary cell walls has been well dissected in Arabidopsis. In this plant, 10 CesA genes have been identified (Richmond and Somerville, 2000), of which AtCesA1, AtCesA3, and AtCesA6 were reported to be involved in primary cell wall cellulose synthesis (Persson et al., 2007), AtCesA4, AtCesA7, and AtCesA8 in secondary cell wall development, and AtCesA2, AtCesA5, AtCesA9, and AtCesA10 in tissue-specific cellulose biosynthesis processes (Gardiner et al., 2003; Taylor et al., 2003). Recent functional characterization of AtCesA proteins led to the identification of unidirectional movement of these protein complexes in seed coat epidermal cells, which deposit cellulose that are involved in mucilage extrusion, adherence and ray formation (Griffiths et al., 2015). In flax (Linum usitatissimum), 14 distinct CesA genes were identified and were targeted for silencing using virus-induced gene silencing (VIGS) approach, which showed impacts on outer-stem tissue organization and secondary cell wall formation (Chantreau et al., 2015). A genome-wide association study of single nucleotide polymorphisms (SNPs) developed through re-sequencing of diverse chickpea accessions revealed a superior haplotype and favorable natural allelic variants in the upstream regulatory region of a CesA gene, denoted as Ca_Kabuli_CesA3 (Kujur et al., 2015). Interestingly, up-regulation of this superior gene haplotype resulted in higher transcript expression of Ca_Kabuli_CesA3 gene in pollen and pod of high pod/seed number chickpea accession, thus resulting in enhanced accumulation of cellulose (Kujur et al., 2015). The specific allelic variant caused cellulose changes specifically in pollen tubes of chickpea and therefore, investigating the homologous gene of foxtail millet identified in the present study will provide novel clues on its role, which could be manipulated for achieving greater biomass yield and bioconversion efficiency.
Physical map of SiCesA genes showed their distribution in chromosomes 2, 3, 4, 5, and 9, with a maximum of 4 genes in chromosome 4 and minimum of one gene in chromosome 3 (Figure 4). Extension of gene families is attributed to the occurrence of three major duplication mechanisms, namely segmental, tandem and retroposition (Cannon et al., 2004). However, none of these duplications were found to be involved in the expansion of SiCesA genes as revealed through MCScanX analysis though both tandem and segmental duplication events were reported in OsCesA family (Wang et al., 2010). Being a member of glycosyltransferase 2 (GT2) family, CesA proteins have the conserved “DXD, D, QXXRW” motif (Somerville et al., 2004) and conforming to this, all the SiCesA proteins except SiCesA8 have a “DCD, D, QVLRW” consensus sequence, whereas SiCesA8 had a unique “DYD, D” sequence and the motif “QXXRW” was absent. Similar sequence variations have also been reported by Wang et al. (2010) in rice. Studies on CesA gene family in crop plants have revealed the presence of a large family of cellulose synthase-like (Csl) genes with sequence similarity to CesA (Richmond and Somerville, 2000), and these genes are shown to be involved in biosynthesis of hemicelluloses (Yin et al., 2009). Similar to CesA, Csl proteins also belong to GT2 family and possess the conserved “DXD, D, QXXRW” motif (Somerville et al., 2004). In foxtail millet, 36 Csl genes were identified and categorized as CslA, CslC, CslD, CslE, CslF, CslH, and CslJ in accordance to the classification followed by Wang et al. (2010) in rice. Interestingly, 2 CslJ genes were identified in foxtail millet, which were reported to be specific to cereals though they are not present in rice and Brachypodium (Fincher, 2009). Domain analysis has shown the presence of GT2 domains in all SiCslA and SiCslC proteins, whereas other SiCsl possess CS domain. Similar reports in Arabidopsis and rice have shown the presence of characteristic GT2 domain in CslA and CslC proteins (Yin et al., 2009; Wang et al., 2010). Studies have shown that CslA and CslC subgroups are the most divergent proteins, which have evolved through duplication and divergence from a common ancestral gene (Yin et al., 2009; Del Bem and Vincentz, 2010), and therefore share similar structural and physicochemical properties (Youngs et al., 2007). Nevertheless, membrane topology and enzymatic function of these proteins are contrastingly different (Davis et al., 2010; Liepman and Cavalier, 2012). In addition, predominant SiCslD family proteins have an additional RING/Ubox like zinc-binding domain, which contains a C3HC4 motif capable of binding to zinc cations.
Molecular processes and biological functions of Csl genes have been less explored when compared to CesA genes though Csl proteins are equally important in cell structuring. Numerous reports have supported the involvement of CslA protein in the synthesis of 1,4-β-mannan and glucomannan backbones (Dhugga et al., 2004; Liepman et al., 2005; Suzuki et al., 2006; Goubet et al., 2009; Gille et al., 2011) and heterologous expression of CslA genes has shown the activity of single enzyme in integrating mannose and glucose into glcomannan chains (Suzuki et al., 2006; Liepman et al., 2007; Gille et al., 2011). Similarly, CslC genes encode for xyloglucan glucan synthase, which are involved in xyloglucan biosynthesis (Cocuron et al., 2007). Heterologous expression of AtCslC4 in Pichia pastoris produced soluble 1,4-β-glucans with a low degree of polymerization, whereas expression of AtCslC4 along with AtXXT1 (xyloglucan xylosyltransferase) produced insoluble 1,4-β-glucans with a higher degree of polymerization suggesting the cooperative action of both the enzymes in xyloglucan biosynthesis (Liepman and Cavalier, 2012). Though CslD proteins were speculated to be involved in xylan and homogalacturonan synthesis (Hamann et al., 2004; Bernal et al., 2008a,b; Li et al., 2009), Arabidopsis csld mutants have been shown to possess severe phenotypic defects including deformed root hairs (csld2; Bernal et al., 2008b), root hairs burst (csld3; Bernal et al., 2008b), defective growth of pollen tube (csld1 and csld4; Bernal et al., 2008b; Wang et al., 2011) and reduced plant growth (csld5; Bernal et al., 2008a). These reports suggest the role of CslD in normal growth and development of plants beyond their function in xylan and homogalacturonan synthesis. The present study identified 4 SiCslE genes, whose characterization has not been performed yet in any crop species. One CslE gene in Arabidopsis and two in rice were reported to date. CslF family of genes were considered to be present among grass species and they regulate the synthesis of mixed-linkage glucan (β-1,3; 1,4, glucan) (Hazen et al., 2002; Burton et al., 2006). Mutation of barley CslF6 gene resulted in reduction of (1,3;1,4)-β-D-Glucan and had an impact on chemical composition of barley grains (Hu et al., 2014), whereas overexpression of this gene in Nicotiana benthamiana led to accumulation of (1,3;1,4)-β-D-Glucan (Wong et al., 2015). Recently, Jin et al. (2015) has demonstrated the role of OsCslF6 in affecting phosphate accumulation altering the level of carbon metabolism in rice. Similar to CslF, CslH and CslJ are also grass-specific gene family involved in deposition of (1,3;1,4)-β-D-Glucan (Doblin et al., 2009; Yin et al., 2009, 2014). In the present study, two genes each belonging to CslH and CslJ family were identified.
Similar to CesA/Csl, glucan synthase-like protein (Gsl) family are also involved in polysaccharide biosynthesis, particularly in synthesis 1,3-β-D-glucan callose (Li et al., 1999). Calloses are deposited in developing cell walls of fiber cells, seed hairs and plasmodesmatal canals. Moreover, deposition of callose is also reported in response to pathogen invasion (Muthamilarasan and Prasad, 2013) and abiotic stress including desiccation, wounding and metal toxicity (Stone and Clarke, 1992). In spite of the importance of Gsl genes, limited studies have been performed on elucidating the molecular role of these genes and their respective proteins. In Arabidopsis, 12 Gsl genes have been identified (https://cellwall.genomics.purdue.edu/intro/index.html) and mutating AtGSL5 has been found to confer resistance to powdery mildew infection (Nishimura et al., 2003). A similar report by Jacobs et al. (2003) has also shown that silencing of AtGsl5 enhances the resistance of silenced lines to Sphaerotheca fusca, Golovinomyces orontii, and Blumeria graminis. In contrast to the role of callose in acting as a physical barrier to prevent pathogen invasion, the reports by Nishimura et al. (2003) and Jacobs et al. (2003) have demonstrated the resistance of Arabidopsis to pathogens in the absence of callose. These reports have proved the importance to study the molecular and physiological roles of Gsl proteins in response to biotic as well as abiotic stress, and the present investigation has identified 12 SiGsl genes which could serve as interesting candidates for functional characterization as foxtail millet is tolerant to environmental stresses.
In the case of monolignol biosynthesis, ten key enzymes namely PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, COMT, CCR, and CAD have been identified and characterized in the present study. Through systematic analysis, 10, 3, 17, 2, 2, 5, 2, 3, 26, and 13 proteins belonging to PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, COMT, CCR, and CAD families, respectively were identified (Supplementary Table S6). These numbers compared with the genes reported in Arabidopsis, poplar and eucalyptus has shown that foxtail millet has higher number of PAL genes (10) whereas other three organisms have 4, 5, and 9 genes, respectively (Raes et al., 2003; Shi et al., 2010; Carocha et al., 2015). Both foxtail millet and poplar have 2 C4H and 17 4CL genes, whereas Arabidopsis and eucalyptus have lesser number of C4H and 4CL genes. Of note, foxtail millet has a maximum of 26 CCR genes, while Arabidopsis has 7 and eucalyptus has 2 genes (Raes et al., 2003; Shi et al., 2010; Carocha et al., 2015). The identified monolignol biosynthesis genes were distributed in all the nine chromosomes of foxtail millet, of which two gene-pairs each of SiPAL (SiPAL4-SiPAL5; SiPAL8-SiPAL9) and SiCAD (SiCAD2-SiCAD3; SiCAD8-SiCAD9), and one pair each of Si4CL (Si4CL11-Si4CL12) and SiCCoAOMT (SiCCoAOMT3-SiCCoAOMT4) were identified to be tandemly duplicated (Figure 4). Phylogenetic analysis of foxtail millet monolignol biosynthesis proteins with bona fide proteins of eucalyptus, tobacco, poplar, Arabidopsis, maize, medicago and grape revealed that predominant proteins of foxtail millet are highly divergent (Figure 3).
Furthermore, promoter analysis has been performed for foxtail millet lignocellulose biosynthesis genes, which revealed the presence of diverse cis-regulatory elements that fall under the following categories; (i) cis-elements which are universally present in all the gene family members, (ii) cis-elements which are present in all the gene family members except one gene, and (iii) cis-element which is unique to any one gene of its corresponding gene family (Supplementary Tables S8, S9). These data suggest the transcriptional control of cell wall genes by the action of network of transcription factors. This would assist in understanding gene regulatory mechanism controlling the expression of lignocellulose genes and fine tuning them to achieve the optimal pattern of secondary cell-wall deposition. Since gene expression is also regulated at post-transcriptional level through miRNAs, the present study also identified foxtail millet miRNAs which target the transcripts of lignocellulose biosynthesis genes (Supplementary Table S10). Moreover, different kinds of molecular markers including SSRs, eSSRs, and ILPs present in both upstream and genic region of lignocellulose biosynthesis genes have been identified (Supplementary Table S11), which could be useful for conducting genomics-assisted breeding for biofuel traits in foxtail millet. In silico expression profiles of all the lignocellulose biosynthesis genes in four tissues as well as dehydration library revealed the differential expression of these genes in these tissues and during stress, thus signifying their putative involvement in biological functions other than cell wall structuring. This is supported by the reports on mutants of studied genes in Arabidopsis and other plants in which severe phenotypic defects have been observed.
In addition to being potential targets for biofuel traits, the lignocellulose biosynthesis genes have also been reported to play vital role in abiotic stress responses. Chen et al. (2005) have shown that Arabidopsis CesA8 mutants accumulate increased levels of ABA, proline and sugars, and express higher levels of stress-related genes, and thus possess enhanced tolerance to drought and osmotic stress. Considering this, Guerriero et al. (2014) analyzed the expression of nine putative CesA genes in response to cold, heat and salt stress in Medicago sativa and identified a salt/heat-induced and a cold/heat-repressed group of genes, which suggest the putative involvement of cellulose synthases in conferring abiotic stress tolerance. Similar to CesA genes, Csl genes have also been shown to participate in stress responsive machinery. Characterization of the salt-overly sensitive6 allele of AtCslD5 has demonstrated reactive oxygen species-based signaling mechanism in response to osmotic stress in Arabidopsis (Zhu et al., 2010). Similarly, accumulation of callose in response to environmental stimuli through overexpression of Gsl genes has been extensively studied (Nedukha, 2015). Stass and Horst (2009) have reported the production of abiotic stress-induced callose in all the plants through a highly conserved signaling pathway. Lignification has also been reported to be induced during abiotic stresses (Moura et al., 2010). In view of these, expression profiling of candidate genes in response to dehydration, salinity and cold stress as well as ABA, SA, and MeJA treatments was performed, which showed significant higher expression of SiGsl2 and SiGsl12 in all the stress conditions. Few genes including SiCAD6, SiC4H2, SiPAL2, SiF5H2, Si4CL10, SiHCT1, and SiCCR7 were evidenced to be up-regulated either at early or late or both the phases of stresses. Similarly, differential expression patterns were observed for all the genes during hormone treatments and of note, ABA treatment has no significant impact on the expression of the majority of genes.
Noteworthy, the expression profiles of candidate lignocellulose biosynthesis genes were in correlation with the cis-regulatory elements present in the promoter regions of respective genes. The genes which are up-regulated during dehydration and salinity stress including SiGsl2, SiGSl12, SiPAL2, SiC4H2, Si4Cl10, SiF5H2, SiHCT1, SiCAD1, and SiCAD6 have one or more “response to dehydration stress” cis-motifs ABRELATERD1, ACGTATERD1 and MYCATRD22 in their promoter regions (Vandepoele et al., 2009; Yan et al., 2014). Similarly, SiGsl2, SiGSl12 and Si4Cl10 that showed higher expression under cold stress have CACGT motif, which was reported to be responsible for response to cold stress (Vandepoele et al., 2009). In case of hormonal treatments, methyl jasmonate responsive cis-element BOXLCOREDCPAL (Yan et al., 2014) was found in the promoter regions of SiCesA5, SiGsl2, SiGSl12, Si4Cl10, SiPAL2, SiC4H2, and SiCCR7. These genes showed significant up-regulation at either early or late or both the phases of methyl jasmonate treatment. Similarly, ABA-responsive genes such as SiC4H2, SiCCR7, SiGsl2, and SiCesA9 have both MYCCONSENSUSAT and MYCATRD22 cis-motifs, which have been reported to be MYC recognition site in the promoter of dehydration responsive rd22 gene which in turn was ABA-dependent (Yan et al., 2014), suggesting that these genes were activated in response to ABA. Thus the present study demonstrates that the interaction of cis-elements and transcription factors has resulted in differential gene expression through activation or repression respective genes in response to various environmental stresses and hormone treatments (Lee et al., 2002; Benitez et al., 2013). The findings and potential correlation between the cis-elements to response to a specific elicitor condition are indirect. It is possible that they are linked, but such primary evidence is not provided here. It is also not known if there were any changes to cell walls in the plants used for expression analyses. Altogether, the present investigation suggests the putative involvement of these genes in strengthening the cell wall for tolerance to abiotic stresses, and they could serve as potential candidates for further functional characterization.
Conclusions
The present study has identified the genes belonging to CesA/Csl, Gsl, PAL, C4H, 4CL, HCT, C3H, CCoAOMT, F5H, COMT, CCR, and CAD superfamilies in foxtail millet and the genes were mapped onto nine chromosomes. In silico analyses of putative protein properties and gene structures revealed diverse characteristic features of these proteins and their gene duplication analysis showed that few gene family members underwent tandem duplication. Phylogenetic analysis of respective proteins demonstrated that except CesA/Csl and Gsl superfamily, the monolignol biosynthesis proteins are highly diverse. Promoter analysis showed the presence of various unique and common cis-regulatory elements in the upstream of lignocellulose biosynthesis genes and potential miRNAs of foxtail millet were identified to target few genes for post-transcriptional gene silencing. In addition, three types of molecular markers were found in lignocellulose biosynthesis genes, which could be used in genomics-assisted breeding. Comparative genome mapping of foxtail millet lignocellulose biosynthesis genes with the sequenced C4 panicoid genomes revealed higher homology with switchgrass, followed by sorghum and maize. Evolutionary analysis showed that both paralogous and homologous gene-pairs underwent intense positive purifying selection, and duplication occurred ~25 mya, whereas divergence of foxtail millet and switchgrass occurred ~4 mya. Similarly, divergence of foxtail millet from sorghum and maize was predicted to occur ~27 mya. In silico expression analysis of all the identified genes in four tissues and dehydration stress library of foxtail millet revealed their differential expression pattern, and also suggested the putative biological function of these genes in processes other than cell wall biosynthesis. Expression profiling of candidate genes in response to dehydration, salinity and cold stress along with ABA, SA and MeJA treatments supported the differential expression of these genes with significant higher expression of SiGsl12, SiHCT1, and SiCAD6 genes. The results suggested that these genes could be used as potential candidates for functional characterization for biofuel traits. Though similar studies have already been completed in switchgrass, sorghum and maize, the present study conducted in biofuel model foxtail millet would facilitate improving the crop for efficient biofuel production.
Author contributions
MP conceived and designed the experiments. MM, YK, JJ, SS, CL performed the experiments. MM, CL, MP analyzed the results. MM, MP wrote the manuscript. MP approved the final version of the manuscript.
Funding
Research on foxtail millet genomics at MP's laboratory is funded by the Core Grant of National Institute of Plant Genome Research, New Delhi, India.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MM acknowledges University Grants Commission, New Delhi, India for providing Research Fellowship. The authors thank Mr. Rohit Khandelwal for critically reading the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00965
Multiple sequence alignment of SiCesA proteins.
Multiple sequence alignment of SiCsl proteins.
Multiple sequence alignment of SiGsl proteins.
Prediction of transmembrane domains in the SiGsl proteins. Red line represents transmembrane, blue line represents inside and pink line represents outside orientation.
Multiple sequence alignment of monolignol biosynthesis proteins.
Gene structure of SiCesA genes.
Gene structure of SiCsl genes.
Gene structure of SiGsl genes.
Gene structure of monolignol biosynthesis genes.
Details of primers used in qRT-PCR analysis.
Details of SiCesA/Csl and SiGsl superfamily genes of foxtail millet.
Details of various domains present in SiCesA proteins.
Details of various domains present in SiCsl proteins.
Details of various domains present in SiGsl proteins.
Details of monolignol biosynthesis pathway genes of foxtail millet.
Details of various domains present in monolignol biosynthesis pathway proteins.
Summary of cis-regulatory elements present in SiCesA/Csl and SiGsl superfamily genes.
Summary of cis-regulatory elements present in monolignol biosynthesis pathway genes.
Details of foxtail millet miRNAs identified to target the transcripts of lignocellulose pathway genes.
Summary of molecular markers present in lignocellulose pathway genes.
The Ka/Ks ratios and estimated divergence time for homologous lignocellulose pathway proteins between Setaria italica and Panicum virgatum.
The Ka/Ks ratios and estimated divergence time for homologous lignocellulose pathway proteins between Setaria italica and Sorghum bicolor.
The Ka/Ks ratios and estimated divergence time for homologous lignocellulose pathway proteins between Setaria italica and Zea mays.
The Ka/Ks ratios and estimated divergence time for tandemly duplicated lignocellulose pathway proteins.
References
- Appenzeller L., Doblin M., Barreiro R., Wang H. Y., Niu X. M., Kollipara K., et al. (2004). Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11, 287–299. 10.1023/B:CELL.0000046417.84715.27 [DOI] [Google Scholar]
- Benitez L. C., da Maia L. C., Ribiero M. V., Pegoraro C., Peters J. A., de Oliveira A. C., et al. (2013). Salt induced change of gene expression in salt sensitive and tolerant rice species. J. Agri. Sci. 5, 251–260. 10.5539/jas.v5n10p251 [DOI] [Google Scholar]
- Bennetzen J. L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. 10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- Bernal A. J., Jensen J. K., Harholt J., Sørensen S., Moller I., Blaukopf C., et al. (2008a). Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 52, 791–802. 10.1111/j.1365-313X.2007.03281.x [DOI] [PubMed] [Google Scholar]
- Bernal A. J., Yoo C. M., Mutwil M., Jensen J. K., Hou G., Blaukopf C., et al. (2008b). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148, 1238–1253. 10.1104/pp.108.121939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D., Chapple C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. 10.1146/annurev-genet-102209-163508 [DOI] [PubMed] [Google Scholar]
- Burton R. A., Shirley N. J., King B. J., Harvey A. J., Fincher G. B. (2004). The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 134, 224–236. 10.1104/pp.103.032904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R. A., Wilson S. M., Hrmova M., Harvey A. J., Shirley N. J., Medhurst A., et al. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1, 3;1, 4)-beta-D-glucans. Science 311, 1940–1942. 10.1126/science.1122975 [DOI] [PubMed] [Google Scholar]
- Byrt C. S., Grof C. P. L., Furbank R. T. (2011). C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integ. Plant Biol. 53, 120–135. 10.1111/j.1744-7909.2010.01023.x [DOI] [PubMed] [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carocha V., Soler M., Hefer C., Cassan-Wang H., Fevereiro P., Myburg A. A., et al. (2015). Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytol. 206, 1297–1313. 10.1111/nph.13313 [DOI] [PubMed] [Google Scholar]
- Chantreau M., Chabbert B., Billiard S., Hawkins S., Neutelings G. (2015). Functional analyses of cellulose synthase genes in flax (Linum usitatissimum) by virus-induced gene silencing. Plant Biotechnol J. [Epub ahead of print]. 10.1111/pbi.12350 [DOI] [PubMed] [Google Scholar]
- Chen Z., Hong X., Zhang H., Wang Y., Li X., Zhu J. K., et al. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43, 273–283. 10.1111/j.1365-313X.2005.02452.x [DOI] [PubMed] [Google Scholar]
- Cocuron J. C., Lerouxel O., Drakakaki G., Alonso A. P., Liepman A. H., Keegstra K., et al. (2007). A gene from the cellulose synthase-like C family encodes a β-1, 4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 8550–8555. 10.1073/pnas.0703133104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. J., Shin H. S., Song C., Laosinchai W., Amano Y., Brown R. M. (2001). A putative plant homolog of the yeast β-1, 3-glucan synthase subunit FKS1 from cotton (Gossypium hirsutum L.) fibers. Planta 213, 223–230. 10.1007/s004250000496 [DOI] [PubMed] [Google Scholar]
- Dai X., Zhao P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J., Brandizzi F., Liepman A. H., Keegstra K. (2010). Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 64, 1028–1037. 10.1111/j.1365-313X.2010.04392.x [DOI] [PubMed] [Google Scholar]
- Del Bem L. E., Vincentz M. G. (2010). Evolution of xyloglucan-related genes in green plants. BMC Evol. Biol. 10:341. 10.1186/1471-2148-10-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga K. S., Barreiro R., Whitten B., Stecca K., Hazebroek J., Randhawa G. S., et al. (2004). Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 303, 363–366. 10.1126/science.1090908 [DOI] [PubMed] [Google Scholar]
- Diao X., Schnable J., Bennetzen J. L., Li J. (2014). Initiation of Setaria as a model plant. Front. Agri. Sci. Eng. 1, 16–20. 10.15302/J-FASE-2014011 [DOI] [Google Scholar]
- Djerbi S., Lindskog M., Arvestad L., Sterky F., Teeri T. T. (2005). The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221, 739–746. 10.1007/s00425-005-1498-4 [DOI] [PubMed] [Google Scholar]
- Doblin M. S., Pettolino F. A., Wilson S. M., Campbell R., Burton R. A., Fincher G. B., et al. (2009). A barley cellulose synthase-like CSLH gene mediates (1, 3;1, 4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5996–6001. 10.1073/pnas.0902019106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin M. S., Pettolino F., Bacic A. (2010). Plant cell walls: the skeleton of the plant world. Funct. Plant Biol. 37, 357–381. 10.1071/FP09279 [DOI] [Google Scholar]
- Farrokhi N., Burton R. A., Brownfield L., Hrmova M., Wilson S. M., Bacic A., et al. (2006). Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotech. J. 4, 145–167. 10.1111/j.1467-7652.2005.00169.x [DOI] [PubMed] [Google Scholar]
- Fincher G. B. (2009). Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 149, 27–37. 10.1104/pp.108.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J. C., Taylor N. G., Turner S. R. (2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15, 1740–1748. 10.1105/tpc.012815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Cheng K., Skinner M. E., Liepman A. H., Wilkerson C. G., Pauly M. (2011). Deep sequencing of voodoo lily (Amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 234, 515–526. 10.1007/s00425-011-1422-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F., Barton C. J., Mortimer J. C., Yu X., Zhang Z., Miles G. P., et al. (2009). Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60, 527–538. 10.1111/j.1365-313X.2009.03977.x [DOI] [PubMed] [Google Scholar]
- Griffiths J. S., Šola K., Kushwaha R., Lam P., Tateno M., Young R., et al. (2015). Unidirectional movement of cellulose synthase complexes in Arabidopsis seed coat epidermal cells deposit cellulose involved in mucilage extrusion, adherence, and ray formation. Plant Physiol. 168, 502–520. 10.1104/pp.15.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero G., Legay S., Hausman J.-F. (2014). Alfalfa cellulose synthase gene expression under abiotic stress: a hitchhiker's guide to RT-qPCR normalization. PLoS ONE 9:e103808. 10.1371/journal.pone.0103808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Osborne E., Youngs H., Misson J., Nussaume L., Somerville C. (2004). Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11, 279–286. 10.1023/B:CELL.0000046340.99925.57 [DOI] [Google Scholar]
- Harakava R. (2005). Genes encoding enzymes of the lignin biosynthesis pathway in Eucalyptus. Genet. Mol. Biol. 28, 601–607. 10.1590/s1415-47572005000400015 [DOI] [Google Scholar]
- Hazen S. P., Scott-Craig J. S., Walton J. D. (2002). Cellulose synthase-like genes of rice. Plant Physiol. 128, 336–340. 10.1104/pp.010875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Burton C., Hong Z., Jackson E. (2014). A mutation of the cellulose-synthase-like (CslF6) gene in barley (Hordeum vulgare L.) partially affects the β-glucan content in grains. J. Cereal Sci. 59, 189–195. 10.1016/j.jcs.2013.12.009 [DOI] [Google Scholar]
- Humphreys J. M., Hemm M. R., Chapple C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 96, 10045–10050. 10.1073/pnas.96.18.10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. K., Lipka V., Burton R. A., Panstruga R., Strizhov N., Schulze-Lefert P., et al. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2513. 10.1105/tpc.016097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Fang C., Yuan H., Wang S., Wu Y., Liu X., et al. (2015). Interaction between carbon metabolism and phosphate accumulation is revealed by a mutation of a cellulose synthase-like protein, CSLF6. J. Exp. Bot. 66, 2557–2567. 10.1093/jxb/erv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Y., Yadav A., Suresh B. V., Muthamilarasan M., Yadav C. B., Prasad M. (2014). Comprehensive genome-wide identification and expression profiling of foxtail millet [Setaria italica (L.)] miRNAs in response to abiotic stress and development of miRNA database. Plant Cell Tiss. Organ Cult. 118, 279–292. 10.1007/s11240-014-0480-x [DOI] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A., Bajaj D., Upadhyaya H. D., Das S., Ranjan R., Shree T., et al. (2015). A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Sci. Rep. 5, 11166. 10.1038/srep11166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Muthamilarasan M., Bonthala V. S., Roy R., Prasad M. (2015). Unraveling 14-3-3 proteins in C4 panicoids with emphasis on model plant Setaria italica reveals phosphorylation-dependent subcellular localization of RS splicing factor. PLoS ONE 10:e0123236. 10.1371/journal.pone.0123236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari K., Muthamilarasan M., Misra G., Gupta S., Subramanian A., Parida S. K., et al. (2013). Development of eSSR-markers in Setaria italica and their applicability in studying genetic diversity, cross-transferability and comparative mapping in millet and non-millet species. PLoS ONE 8:e67742. 10.1371/journal.pone.0067742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen K. (2004). Cloning and characterization of a ryegrass (Lolium perenne) gene encoding cinnamoyl-CoA reductase (CCR). Plant Sci. 166, 569–581. 10.1016/j.plantsci.2003.09.026 [DOI] [PubMed] [Google Scholar]
- Lata C., Gupta S., Prasad M. (2013). Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 33, 328–343. 10.3109/07388551.2012.716809 [DOI] [PubMed] [Google Scholar]
- Lata C., Mishra A. K., Muthamilarasan M., Bonthala V. S., Khan Y., Prasad M. (2014). Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 9:e113092. 10.1371/journal.pone.0113092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Tang H., Wang X., Paterson A. H. (2012). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158. 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., Gerber G. K., et al. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804. 10.1126/science.1075090 [DOI] [PubMed] [Google Scholar]
- Lerouxel O., Cavalier D. M., Liepman A. H., Keegstra K. (2006). Biosynthesis of plant cell wall polysaccharides - a complex process. Curr. Opin. Plant Biol. 9, 621–630. 10.1016/j.pbi.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Li H. J., Bacic A., Read S. M. (1999). Role of a callose synthase zymogen in regulating wall deposition in pollen tubes of Nicotiana alata Link et Otto. Planta 208, 528–538. 10.1007/s004250050590 [DOI] [Google Scholar]
- Li M., Xiong G. Y., Cui J. J., Tang D., Zhang B. C., Pauly M., et al. (2009). Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 60, 1055–1069. 10.1111/j.1365-313X.2009.04022.x [DOI] [PubMed] [Google Scholar]
- Li P., Brutnell T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J. Exp. Bot. 62, 3031–3037. 10.1093/jxb/err096 [DOI] [PubMed] [Google Scholar]
- Liepman A. H., Cavalier D. M. (2012). The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front. Plant Sci. 3:109. 10.3389/fpls.2012.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A. H., Nairn C. J., Willats W. G., Sørensen I., Roberts A. W., Keegstra K. (2007). Functional genomic analysis supports conservation of function among CELLULOSE SYNTHASE-LIKE A gene family members and suggests diverse roles of mannans in plants. Plant Physiol. 143, 1881–1893. 10.1104/pp.106.093989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A. H., Wilkerson C. G., Keegstra K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U.S.A. 102, 2221–2226. 10.1073/pnas.0409179102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H. E., Döring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. 10.1146/annurev-arplant-050213-040240 [DOI] [PubMed] [Google Scholar]
- Mishra A. K., Muthamilarasan M., Khan Y., Parida S. K., Prasad M. (2013). Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS ONE 9:e86852. 10.1371/journal.pone.0086852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura J. C. M. S., Bonine C. A. V., De Oliveira Fernandes Viana J., Dornelas M. C., Mazzafera P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. 10.1111/j.1744-7909.2010.00892.x [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Bonthala V. S., Mishra A. K., Khandelwal R., Khan Y., Roy R., et al. (2014b). C2H2-type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct. Integr. Genomics 14, 531–543. 10.1007/s10142-014-0383-2 [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Dhaka A., Yadav R., Prasad M. (2015). Exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci. 10.1016/j.plantsci.2015.08.023. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Khandelwal R., Yadav C. B., Bonthala V. S., Khan Y., Prasad M. (2014a). Identification and molecular characterization of MYB Transcription Factor Superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS ONE 9:e109920. 10.1371/journal.pone.0109920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamilarasan M., Prasad M. (2013). Plant innate immunity: an updated insight into defense mechanism. J. Biosci. 38, 433–449. 10.1007/s12038-013-9302-2 [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Prasad M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. 10.1007/s00122-014-2399-3 [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Venkata Suresh B., Pandey G., Kumari K., Parida S. K., Prasad M. (2014c). Development of 5123 intron-length polymorphic markers for large-scale genotyping applications in foxtail millet. DNA Res. 21, 41–52. 10.1093/dnares/dst039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedukha O. M. (2015). Callose: localization, functions, and synthesis in plant cells. Cytol. Genet. 49, 49–57. 10.3103/S0095452715010090 [DOI] [Google Scholar]
- Nishimura M. T., Stein M., Hou B. H., Vogel J. P., Edwards H., Somerville S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. 10.1126/science.1086716 [DOI] [PubMed] [Google Scholar]
- Osakabe K., Tsao C. C., Li L., Popko J. L., Umezawa T., Carraway D. T., et al. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 96, 8955–8960. 10.1073/pnas.96.16.8955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G., Misra G., Kumari K., Gupta S., Parida S. K., Chattopadhyay D., et al. (2013). Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. 20, 197–207. 10.1093/dnares/dst002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K., Chen F., Guo D., Blount J. W., Dixon R. A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25, 193–202. 10.1046/j.1365-313x.2001.00956.x [DOI] [PubMed] [Google Scholar]
- Pear J. R., Kawagoe Y., Schreckengost W. E., Delmer D. P., Stalker D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 93, 12637–12642. 10.1073/pnas.93.22.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571. 10.1073/pnas.0706592104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C., Shearer A., Tateno M., Ruwaya M., Nokes S., Brutnell T., et al. (2013). Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and panicoid crop species. Front. Plant Sci. 4:181. 10.3389/fpls.2013.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon M., Courbou I., Beckert M., Boudet A. M., Grima-Pettenati J. (1998). Cloning and characterization of two maize cDNAs encoding Cinnamoyl-CoA Reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 38, 671–676. 10.1023/A:1006060101866 [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Mandal S. N., Venkata Suresh B., Parida S. K., Prasad M. (2013). Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 8:e64594. 10.1371/journal.pone.0064594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Xie S., Liu Y., Yi F., Yu J. (2013). Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 83, 459–473. 10.1007/s11103-013-0104-6 [DOI] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J. H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. 10.1104/pp.103.026484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. A., Somerville C. R. (2000). The cellulose synthase superfamily. Plant Physiol. 124, 495–498. 10.1104/pp.124.2.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saathoff A. J., Hargrove M. S., Haas E. J., Tobias C. M., Twigg P., Sattler S., et al. (2012). Switchgrass PviCAD1: understanding Residues Important for Substrate Preferences and Activity. Appl. Biochem. Biotechnol. 168, 1086–1100. 10.1007/s12010-012-9843-0 [DOI] [PubMed] [Google Scholar]
- Saathoff A. J., Sarath G., Chow E. K., Dien B. S., Tobias C. M. (2011b). Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 6:el6416. 10.1371/journal.pone.0016416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saathoff A. J., Tobias C. M., Sattler S. E., Haas E. E., Twigg P., Sarath G. (2011a). Switchgrass contains two cinnamyl alcohol dehydrogenase involved in lignin formation. Bioener. Res. 4, 120–133. 10.1007/s12155-010-9106-2 [DOI] [Google Scholar]
- Saballos A., Ejeta G., Sanchez E., Kang C., Vermerris W. (2009). A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181, 783–795. 10.1534/genetics.108.098996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Sattler S., Palmer N., Saballos A., Greene A., Xin Z., Sarath G., et al. (2012). Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenergy Res. 5, 855–865. 10.1007/s12155-012-9197-z [DOI] [Google Scholar]
- Saxena I. M., Brown R. M. (2000). Cellulose synthases and related enzymes. Curr. Opin. Plant Biol. 3, 523–531. 10.1016/S1369-5266(00)00125-4 [DOI] [PubMed] [Google Scholar]
- Schmer M. R., Vogel K. P., Mitchel R. B., Perrin R. K. (2008). Net energy of cellulosic ethanol from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 105, 464–469. 10.1073/pnas.0704767105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Mazarei M., Hisano H., Escamilla-Trevino L., Fu C., Pu Y., et al. (2013). A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell 25, 4342–4361. 10.1105/tpc.113.118828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Sun Y. H., Li Q., Heber S., Sederoff R., Chiang V. L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51, 144–163. 10.1093/pcp/pcp175 [DOI] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., et al. (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211. 10.1126/science.1102765 [DOI] [PubMed] [Google Scholar]
- Stass A., Horst W. J. (2009). Callose in abiotic stress, in Chemistry, Biochemistry and Biology of (1 → 3)-β-Glucans and Related Polysaccharides, eds Bacic A., Fincher G. B., Stone B. A., (New York, NY: Academic; ), 499–524. [Google Scholar]
- Stone B. A., Clarke A. E. (1992). Chemistry and Biology of (1, 3)-β-Glucans. Victoria: La Trobe University Press. [Google Scholar]
- Suresh B. V., Muthamilarasan M., Misra G., Prasad M. (2013). FmMDb: a versatile database of foxtail millet markers for millets and bioenergy grasses research. PLoS One 8:e71418. 10.1371/journal.pone.0071418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Li L., Sun Y. H., Chiang V. L. (2006). The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142, 1233–1245. 10.1104/pp.106.086678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. G., Howells R. M., Huttly A. K., Vickers K., Turner S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455. 10.1073/pnas.0337628100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucco G. M., Matos D. A., Lee S. J., Saathoff A. J., Priest H. D., Mockler T. C., et al. (2013). Functional characterization of cinnamyl alcohol dehydrogenase and caffeic acid O-methyltransferase in Brachypodium distachyon. BMC Biotechnol. 13:61. 10.1186/1472-6750-13-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Quimbaya M., Casneuf T., Veylder D. L., de Peer Y. V. (2009). Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 150, 535–546. 10.1104/pp.109.136028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weijde T., Alvim Kamei C. L., Torres A. F., Vermerris W., Dolstra O., Visser R. G., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:107. 10.3389/fpls.2013.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J. H., Goeminne G., et al. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24, 3506–3529. 10.1105/tpc.112.102574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Parijs F. R., Ruttink T., Boerjan W., Haesaert G., Byrne S. L., Asp T., et al. (2015). Clade classification of monolignol biosynthesis gene family members reveals target genes to decrease lignin in Lolium perenne. Plant Biol. 17, 877–892. 10.1111/plb.12316 [DOI] [PubMed] [Google Scholar]
- Voorrips R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Wang L., Guo K., Li Y., Tu Y., Hu H., Wang B., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10:282. 10.1186/1471-2229-10-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang L., Chen C., Xiong G., Tan X. Y., Yang K. Z., et al. (2011). Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 62, 5161–5177. 10.1093/jxb/err221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chantreau M., Sibout R., Hawkins S. (2013). Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 4:220. 10.3389/fpls.2013.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnasooriya S. N., Brutnell T. P. (2014). Enhancing the productivity of grasses under high-density planting by engineering light responses: from model systems to feedstocks. J. Exp. Bot. 65, 2825–2834. 10.1093/jxb/eru221 [DOI] [PubMed] [Google Scholar]
- Wong S. C., Shirley N. J., Little A., Khoo K. H., Schwerdt J., Fincher G. B., et al. (2015). Differential expression of the HvCslF6 gene late in grain development may explain quantitative differences in (1, 3;1, 4)-β-glucan concentration in barley. Mol. Breed. 35, 20. 10.1007/s11032-015-0208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav C. B., Muthamilarasan M., Pandey G., Prasad M. (2015). Identification, characterization and expression profiling of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in foxtail millet. Plant Mol. Biol. Rep. 33, 43–55. 10.1007/s11105-014-0736-y [DOI] [Google Scholar]
- Yan J., Wang B., Jiang Y., Cheng L., Wu T. (2014). GmFNSII-controlled soybean flavone metabolism responds to abiotic stresses and regulates plant salt tolerance. Plant Cell Physiol. 55, 74–86. 10.1093/pcp/pct159 [DOI] [PubMed] [Google Scholar]
- Yin Y., Huang J., Xu Y. (2009). The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9:99. 10.1186/1471-2229-9-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Johns M. A., Cao H., Rupani M. (2014). A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genomics 15:260. 10.1186/1471-2164-15-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn B., Camacho R., Moinuddin S. G. A., Lee C., Davin L. B., Lewis N. G., et al. (2006). Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org. Biomol. Chem. 4, 1687–1697. 10.1039/b601672c [DOI] [PubMed] [Google Scholar]
- Youngs H. L., Hamann T., Osborne E., Somerville C. R. (2007). The cellulose synthase superfamily, in Cellulose: Molecular and Structural Biology, eds Brown M., Saxena I. M. (Berlin: Springer; ), 35–49. [Google Scholar]
- Zhang G., Liu X., Quan Z., Cheng S., Xu X., Pan S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. 10.1038/nbt.2195 [DOI] [PubMed] [Google Scholar]
- Zhong R., Ye Z. H. (2015). Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56, 195–214. 10.1093/pcp/pcu140 [DOI] [PubMed] [Google Scholar]
- Zhu J., Lee B. H., Dellinger M., Cui X., Zhang C., Wu S., et al. (2010). A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 63, 128–140. 10.1111/j.1365-313x.2010.04227.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of SiCesA proteins.
Multiple sequence alignment of SiCsl proteins.
Multiple sequence alignment of SiGsl proteins.
Prediction of transmembrane domains in the SiGsl proteins. Red line represents transmembrane, blue line represents inside and pink line represents outside orientation.
Multiple sequence alignment of monolignol biosynthesis proteins.
Gene structure of SiCesA genes.
Gene structure of SiCsl genes.
Gene structure of SiGsl genes.
Gene structure of monolignol biosynthesis genes.
Details of primers used in qRT-PCR analysis.
Details of SiCesA/Csl and SiGsl superfamily genes of foxtail millet.
Details of various domains present in SiCesA proteins.
Details of various domains present in SiCsl proteins.
Details of various domains present in SiGsl proteins.
Details of monolignol biosynthesis pathway genes of foxtail millet.
Details of various domains present in monolignol biosynthesis pathway proteins.
Summary of cis-regulatory elements present in SiCesA/Csl and SiGsl superfamily genes.
Summary of cis-regulatory elements present in monolignol biosynthesis pathway genes.
Details of foxtail millet miRNAs identified to target the transcripts of lignocellulose pathway genes.
Summary of molecular markers present in lignocellulose pathway genes.
The Ka/Ks ratios and estimated divergence time for homologous lignocellulose pathway proteins between Setaria italica and Panicum virgatum.
The Ka/Ks ratios and estimated divergence time for homologous lignocellulose pathway proteins between Setaria italica and Sorghum bicolor.
The Ka/Ks ratios and estimated divergence time for homologous lignocellulose pathway proteins between Setaria italica and Zea mays.
The Ka/Ks ratios and estimated divergence time for tandemly duplicated lignocellulose pathway proteins.